ABSTRACT
DEAD-box RNA-helicases catalyze the reorganization of structured RNAs and the formation of RNP complexes. The cyanobacterium Synechocystis sp. PCC 6803 encodes a single DEAD-box RNA helicase, CrhR (Slr0083), whose expression is regulated by abiotic stresses that alter the redox potential of the photosynthetic electron transport chain, including temperature downshift. Despite its proposed effect on RNA metabolism and its known relevance in cold-stress adaptation, the reported impact of a CrhR knockout on the cold adaption of the transcriptome only identified eight affected genes. Here, we utilized a custom designed microarray to assess the impact of the absence of CrhR RNA helicase activity on the transcriptome, independent of cold stress. CrhR truncation impacts an RNA subset comprising ~10% of the ncRNA and also ~10% of the mRNA transcripts. While equal numbers of mRNAs showed increased as well as decreased abundance, more than 90% of the ncRNAs showed enhanced expression in the absence of CrhR, indicative of a negative effect on ncRNA transcription or stability. We further tested the effect of CrhR on the stability of strongly responding RNAs that identify examples of post-transcriptional and transcriptional regulation. The data suggest that CrhR impacts multiple aspects of RNA metabolism in Synechocystis.
KEYWORDS: Small regulatory RNA, cyanobacteria, gene expression regulation, CrhR RNA helicase, RNA:RNA interaction
Introduction
Regulatory non-coding RNAs (ncRNAs) are important players in the regulation of gene expression in all domains of life [1,2]. Functional ncRNAs may originate from intergenic regions (sRNAs), from the strand opposite to coding regions (antisense RNAs, asRNAs), from 5ʹ UTRs (5ʹ UTR-ncRNAs) and internal mRNA fragments in the sense orientation (iRNAs) that originate from either their own transcription start sites (TSSs) or processing or degradation products [3–5]. In bacteria, although some ncRNAs sequester proteins, the majority target other RNA molecules. The regulatory effect depends upon base-pairing interactions between the ncRNA and target mRNA, the resulting intermolecular heteroduplex RNA alters gene expression, the duplex is either rapidly degraded or enhances translation initiation [6,7]. In the case of trans-acting regulatory sRNAs, the complementary regions are often short and imperfect while cis-encoded asRNAs have perfect target sequence complementarity [4,8]. Interaction of the ncRNA and target mRNAs require locally accessible single strands in order to form the intermolecular heteroduplex, a process that competes with the secondary structures of the individual RNAs [9]. These RNA-RNA interactions therefore frequently require the presence of RNA chaperones in vivo such as Hfq, an Sm-like protein widely distributed in the bacterial kingdom and extensively studied in enterobacteria [10,11], or the more recently described ProQ [12]. However, RNA:RNA interactions and folding can also be facilitated passively by cold shock domain proteins, bacterial histone-like proteins and ribosomal proteins or actively by RNA helicases [11,13,14].
RNA helicases belong to helicase superfamily 2 (SF2), the largest family within SF2 being the DEAD-box family [15]. RNA helicase activity is associated with all aspects of RNA metabolism and thus are important players in the regulation of gene expression, including involvement in housekeeping functions including transcription, translation initiation, RNA decay and processing and also developmental pathways [16]. In addition, RNA helicases are often expressed in response to abiotic stress, presumably to facilitate acclimatization to the altered environmental conditions [17,18]. Biochemically, DEAD-box RNA-helicases rearrange RNA secondary structure, potentially catalyzing duplex RNA unwinding, annealing of complementary ssRNAs, RNA strand exchange and remodeling of RNA-protein (RNP) complexes in vitro [14,16,19,20]. These biochemical attributes indicate that these enzymes have the capacity to perform an important role in ncRNA-mRNA target regulation by facilitating proper interaction or folding of the ncRNA regulator with its mRNA target and/or the half-lives of the interacting RNAs, as observed in higher plant systems [21]. Evidence for RNA helicase mediated ncRNA-mRNA interaction in bacteria has been provided by the Escherichia coli DEAD-box helicase CsdA which is necessary for the translational activation of rpoS by the regulatory sRNA, DsrA, at low temperature [22].
Cyanobacteria are the only bacteria performing oxygenic photosynthesis. A single DEAD box RNA helicase, CrhR (Slr0083), is encoded in the model cyanobacterium Synechocystis sp. PCC 6803 (from here on Synechocystis). This is in striking contrast to the five different DEAD-box RNA helicases in E. coli [23] and raises the question as to the role(s) CrhR performs in RNA metabolism.
Expression of crhR is regulated by light-driven reduction of the electron transport chain [24], levels which are augmented by conditions that increase reduction of the chain, including salt and cold stress [25,26]. Temperature-downshift induction of crhR expression is a complex, auto-regulatory process involving both CrhR-dependent and -independent mechanisms [26], while conditional proteolysis contributes to the temperature upshift repression [27]. Induction of RNA helicases upon cold stress has been observed in several bacteria, conditions under which helicase activity has been proposed to be required to overcome the thermodynamically enhanced stability of RNA secondary structures at temperatures below the growth optimum [18]. Lack of CrhR RNA helicase activity has drastic effects on the physiology of Synechocystis in response to cold stress but also at the standard growth temperature of 30°C [28–30]. In a partial crhR deletion mutant, crhRTR, a pleiotropic range of effects were observed after a temperature downshift from 30 to 20°C including a rapid cessation of photosynthesis, impaired cell growth, decreased viability, cell size and DNA content, as well as structural abnormalities associated with a reduction in photosynthetic carbon fixation [30].
The molecular effects of crhR deletion have been studied at both the transcriptome [28] and proteome level [29]. Microarray analysis revealed no significant changes between wild type (WT) and a complete crhR mutant at 34°C [28]. At 24°C, genes encoding the chaperones and the fatty acid desaturase Sll1611 showed a reduced cold induction, whereas slr0082, pyrB, gifA and gifB mRNAs were upregulated [28]. In contrast, differential 2D gel electrophoresis of soluble proteins identified 16 proteins differentially expressed in the complete crhR mutant at 34°C and 25 proteins at 24°C, including the GroES, GroEL-1 and GroEL-protein chaperones [29]. The mechanism(s) generating these altered expression profiles are not known but may partially result from CrhR-regulated post-transcriptional changes in gene expression. The impact of CrhR on transcript classes other than mRNAs were not addressed in these previous studies.
Bioinformatic, tiling microarray and differential RNAseq analysis of the Synechocystis transcriptome has identified numerous ncRNAs [31–34], some of which (PsrR1, IsaR1 and NsiR4) perform crucial regulatory functions [35–37]. In this report, we exploited a customized Agilent microarray to investigate the transcriptomic consequences created by the absence of CrhR RNA helicase activity in Synechocystis. The data revealed that a CrhR-dependent mechanism influences a specific subset of the total RNA repertoire in Synechocystis.
Results
Transcriptome – wide effects of partial crhR deletion
To identify transcripts whose expression was altered in response to crhR mutation, total RNA extracted from the partial crhR deletion mutant, crhRTR, and wild type. The crhRTR mutant was generated by insertion of a spectinomycin-streptomycin resistance cassette into the PmlI site between the codons encoding RNA helicase motifs III (SAT) and IV (FVRTK), thereby removing the second RecA domain and C-terminal extension [30]. The strains were cultivated at standard temperature (30°C) or exposed to cold stress (20°C) for 3 h. The RNA was directly labeled and hybridized to Agilent microarrays (GEO accession number GSE58544). In total, 252 transcripts responded to crhR truncation representing ~10% of all annotated mRNAs and ncRNAs (Figure 1, Table 1, Table S1). The transcript levels of 90% (108/115) of the affected ncRNAs increased in the crhRTR strain (Figure 1, left panel), while this strong asymmetric effect was not observed for mRNAs (Figure 1, right panel). This observation suggested that CrhR has a direct or indirect negative effect on the accumulation of a subset of ncRNAs. In addition, the crhRTR mutation affected transcript abundance at both 20°C and 30°C. As depicted in the scatter plots and the Venn diagram in Figure 1, 80 RNAs were differentially expressed only at 20°C (blue), 43 only at 30°C (orange) and 30 only if the temperature response from crhRTR and WT was compared (FC20°C – FC30°C, green). RNAs that were equally strongly affected at 20°C and 30°C are depicted in black. In general, at low temperature roughly twice as many RNAs responded to the crhRTR mutation than at 30°C, which is consistent with the low temperature specific phenotype of CrhR.
Figure 1.
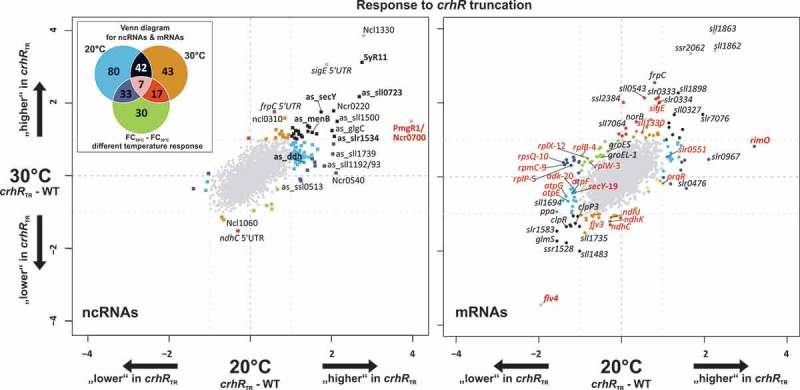
Microarray results showing the transcriptome response to crhR inactivation. The left scatter plot displays the log2 fold changes of the comparison crhRTR versus wild type (WT) at 20°C (x-axis) and 30°C (y-axis) for sRNAs and asRNAs, while the right plot shows the FCs for mRNAs. Significantly differentially expressed genes (absolute log2 FC ≥ 1, adjusted p-value ≤ 0.05) are color-coded, matching the colors in the Venn diagram in the upper left corner. RNAs that are only differentially expressed at 20°C, 30°C or in the comparison of the temperature responses (FC20°C – FC30°C) are colored blue, orange or green, respectively. The colors of RNAs that are significantly and differentially expressed at more than one condition can be taken from the overlapping areas in the Venn diagram. The numbers in the fields of the Venn diagram are the total numbers of differential expressed genes at the specific comparison including sRNAs, asRNAs and mRNAs. Selected members of the ribosomal operon sll1799-ssl3441 have red labels, with a number following the gene name to indicate the position in the operon. Transcripts specifically mentioned in the discussion are also labeled in red. The expression profiles of RNAs shown in bold face were verified by northern blot.
Table 1.
Differentially expressed transcript types. Absolute and relative numbers of transcripts differentially expressed in the crhRTR mutant compared to wild type in response to growth at 20 and 30°C (log2FC ≥1, adj. p -value ≤0.05).
|
crhRTR/WT (20°C) |
crhRTR/WT (30°C) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Transcript type |
Increase | Decrease | Increase | Decrease | ||||
| mRNA | 34 | 40% | 50 | 60% | 38 | 46.3% | 44 | 53.7% |
| 5ʹ UTR-ncRNA | 23 | 92% | 2 | 8% | 17 | 77.3% | 5 | 22.7% |
| iRNA | 26 | 86.7% | 4 | 13.3% | 16 | 69.6% | 7 | 30.4% |
| asRNA | 48 | 96% | 2 | 4% | 22 | 95.7% | 1 | 4.3% |
| sRNA | 31 | 94% | 2 | 6% | 19 | 90.5% | 2 | 9.5% |
| Total | 162 | 73% | 60 | 27% | 112 | 65.5% | 59 | 34.5% |
In contrast to previous studies, we detected additional mRNAs that responded significantly to CrhR inactivation. Expression levels of the hypothetical proteins sll1862, sll1863 and ssr2062 were significantly enhanced in the crhRTR mutant, while levels of flv4 (sll0217) were strongly reduced (Table S1). Similar to groES/EL, the 21 gene ribosomal protein operon sll1799-ssl3441 showed a reduced cold induction in crhRTR. While the first two genes in the operon showed no differential expression, the differential expression between crhRTR and wild type at 20°C increased with increasing operon length. Transcript levels of rimO (slr0082) and crhR (slr0083), which are encoded as an operon (Figure 2A), were higher in crhRTR than in the wild type at 20°C, consistent with previous studies [26,28]. Furthermore, we also detected a reduced cold induction of both groES and groEL in the crhRTR mutant strain.
Figure 2.
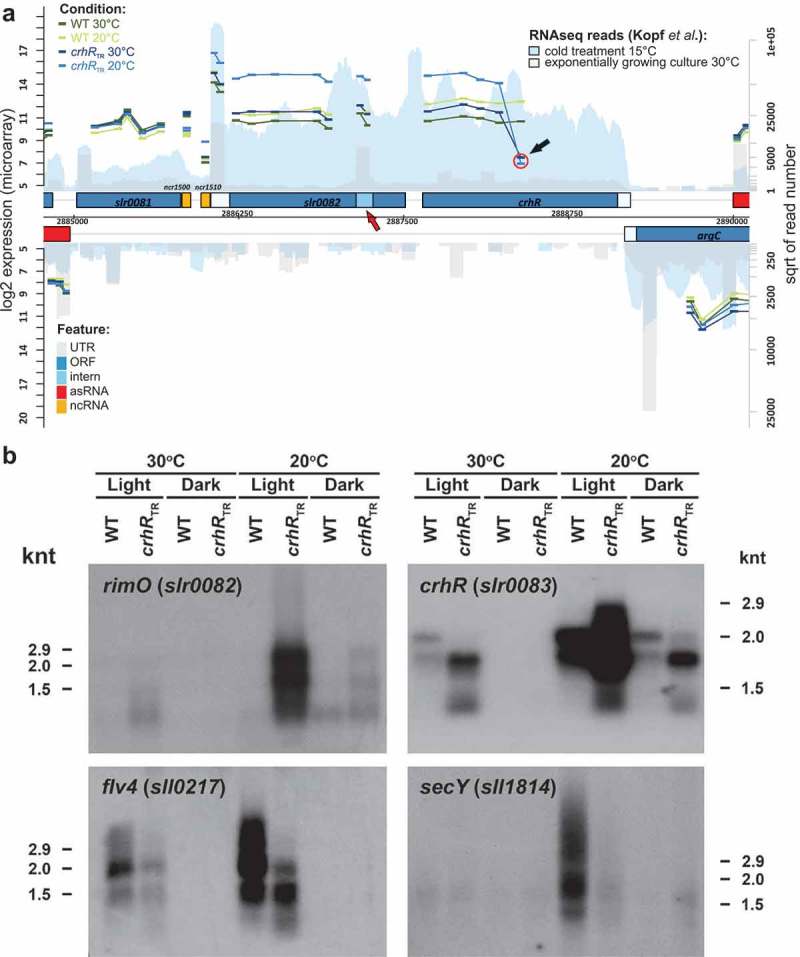
Verification of mRNA abundance changes. (A) Transcript accumulation in the genomic region containing the rimO(miaB)-crhR operon. The results of this study (microarray experiments) are combined with the coverage from a previous RNAseq analysis [34]. Both DNA strands are shown. Genes are drawn as blue boxes, UTRs as white-blue boxes, internal RNAs as light blue boxes, asRNAs as red boxes and intergenic sRNAs as yellow boxes. The numbers of RNAseq reads from an exponentially growing culture at 30°C are plotted in light grey and after transfer to 15°C for 30 min in blue. See also the legend given in the upper right corner. The read numbers are given in a log2 scale represented by the right y-axis. The normalized log2 expression values of the microarray experiments are plotted for each probe as short vertical tabs, which span the corresponding hybridization region. The scale for the microarray data is given at the left y-axis (normalized fluorescence values). The values for the wild type (WT) at 30°C and at 20°C are pictured in dark and light green, for crhRTR at 30°C and at 20°C in dark and light blue. All probes (thicker bars) corresponding to a single RNA feature are connected by lines. The red circle and the black arrow indicate the position of the first probe following the antibiotic cassette insertion used to generate the truncation. The red arrow points to a potential transcription start site within the rimO(miaB) slr0082 gene. (B) Northern analysis of microarray-predicted mRNA abundance changes in response to crhR mutation. Gene-specific probing was performed on total RNA (5 µg) isolated from wild type or crhRTR cells grown at 30°C, cold stressed at 20°C or incubated in the dark for three h, as indicated.
Altered expression profiles of selected mRNAs and ncRNAs were verified by northern analysis (Figures 2B and 3). These analyses confirmed the microarray data indicating CrhR-dependent alteration of mRNA transcript abundance with respect to both direction and magnitude of change. We additionally included dark treatment in the analysis of selected mRNAs to determine possible effects on the transcripts selected for validation (Figure 2B). Specifically, we confirmed the upregulation of slr0082 in the crhRTR mutant at 20°C, the enhanced cold induction of crhR (slr0083) in both strains but even more pronounced in crhRTR. The hybridizations also confirmed the cold induction of flv4 (sll0217) in both strains but more pronounced in wild type and the cold induction of secY (sll1814) transcript in the wild type but not in crhRTR. Dark treatment significantly decreased expression of all tested mRNAs in both wild type and crhRTR at both temperatures, implying that a light signal is crucial for the accumulation of these transcripts (Figure 2B).
Figure 3.
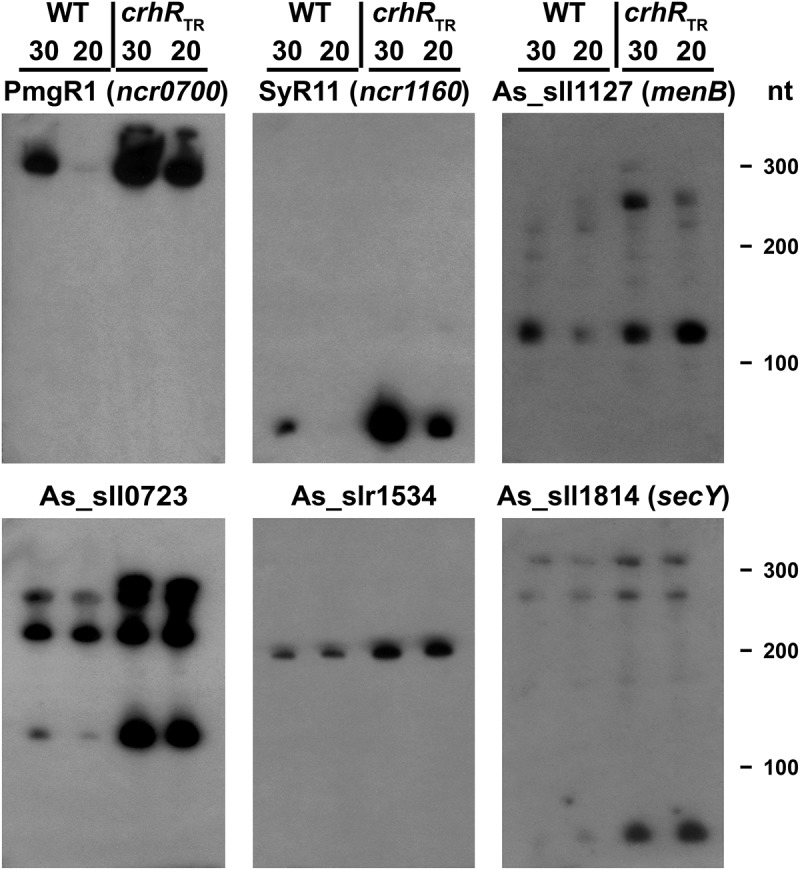
Northern analysis of microarray-predicted ncRNA abundance changes in response to crhR mutation. Transcript abundances of the indicated ncRNAs was determined using 5 µg of total RNA isolated from wild type or crhRTR cells grown at 30°C or cold stressed at 20°C for three h.
The microarray data indicating CrhR-dependent alteration of ncRNA abundance was validated with respect to both direction and magnitude of change for four asRNAs and two sRNAs (Figure 3). This experiment independently confirmed not only the strong induction of PmgR1 in crhRTR but showed that indeed all of the tested ncRNAs increased in abundance in crhRTR, an effect observed at both temperatures. This implies that CrhR performs a role associated with expression of a subset of the Synechocystis non-coding transcriptome, a function that is temperature dependent, functioning primarily at low temperature.
Half-life estimation of selected transcripts
The asymmetric effect on the accumulation of asRNAs and sRNAs in the crhRTR strain raised the question if CrhR might negatively affect the stability of these transcripts. In order to distinguish between transcriptional and post-transcriptional effects of the crhR mutation, we estimated the half-life of four significantly altered transcripts, PmgR1, SyR11, Ncl1330 and crhR mRNA (Figure 4). The data show that for all four candidate targets, the half-life normally increases in wild type cells at low temperature, an effect that is significantly enhanced in the crhRTR strain (Table 2). In the case of the crhR mRNA, the change in the RNA stability largely explained the changes in transcript accumulation at 20°C. Based on the northern blot data (Figure 2B), the transcript accumulation of crhR at 20°C was 3.42 ± 0.23 higher in the crhRTR mutant and the calculated degradation constants could explain a 2.17 times difference suggesting that an alteration in transcription rate accounted for the remaining 1.6-fold increase.
Figure 4.
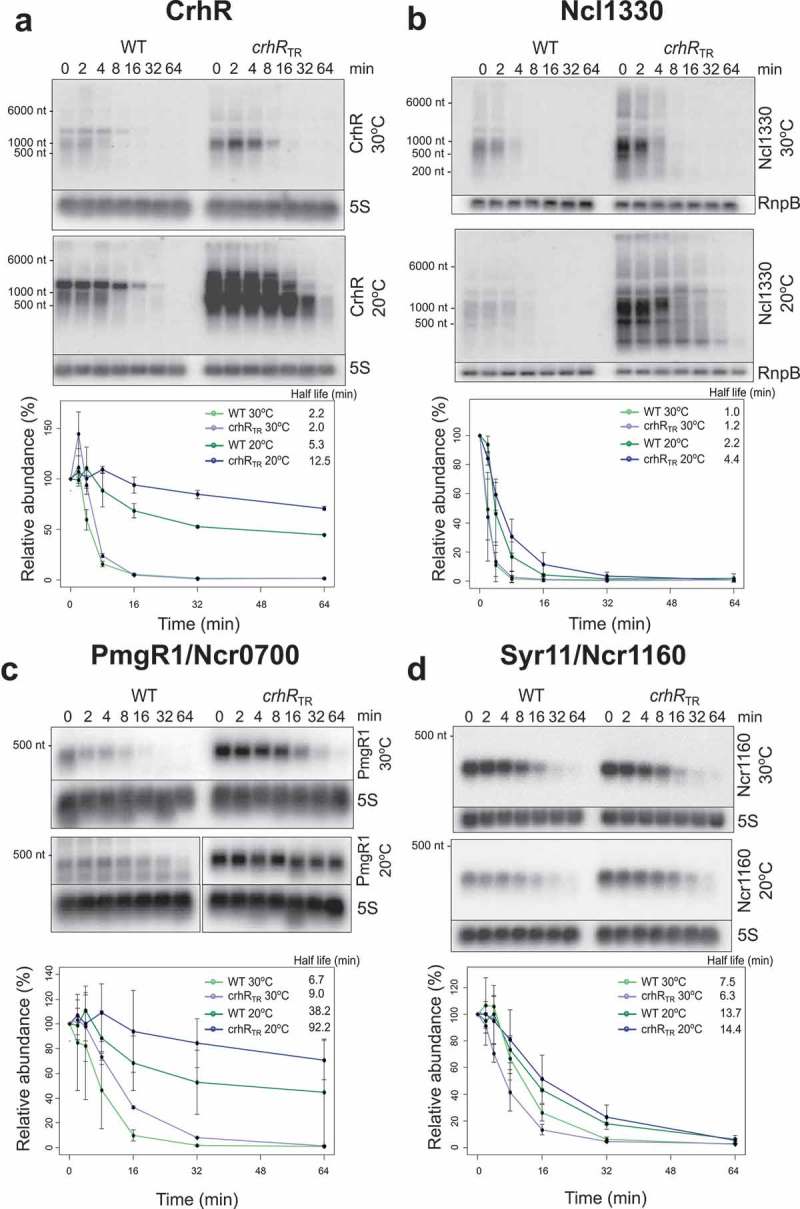
RNA half-life determination. Northern analysis was performed in order to determine the effect of crhR mutation on the stability of selected RNA transcripts whose abundance dramatically changed in the microarray analysis. RNA half-lives were determined from total RNA (3 µg) isolated from wild type and crhRTR cells for: (A) the mRNA crhR, (B) the sRNA Ncl1330, (C) the sRNA PmgR1 (Ncr0700) and (D) the sRNA SyR11/Ncr1160. 5S rRNA or rnpB abundance was utilized as a loading control for normalization. All experiments were performed in biological duplicates.
Table 2.
Expression fold changes, stability and synthesis parameters based on the rifampicin time series northern blots (Figure 4). FC, Fold change crhRTR/WT; λ, degradation constant [1/min]; αcrhRTR/αWT, ratio of transcription rates. Data are based on two biological replicates.
| Degradation constant |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Norm. intensity crhRTR/WT |
λ [1/min] = ln(2)/t1/2 |
20°C |
30°C |
20°C |
30°C |
|||||
| FC 20°C | FC 30°C | WT 20°C | WT 30°C | crhRTR 20°C | crhRTR 30°C | λWT/λcrhR | λWT/λcrhR | αcrhR/αWT | αcrhR/αWT | |
| PmgR1 | 186.3 ± 84.8 | 5.2 ± 1.7 | 0.018 | 0.103 | 0.0075 | 0.077 | 2.4 | 1.33 | 77.6 | 3.9 |
| SyR11 | 2.1 ± 0.1 | 1.2 ± 0.1 | 0.05 | 0.09 | 0.048 | 0.11 | 1.04 | 0.82 | 2.01 | 1.5 |
| Ncl1330 | 12.9 ± 1.4 | 6.2 ± 4.4 | 0.31 | 0.69 | 0.17 | 0.58 | 1.8 | 1.2 | 7.2 | 5.2 |
| crhR | 3.42 ± 0.23 | 1.6 ± 0.26 | 0.13 | 0.31 | 0.06 | 0.33 | 2.17 | 0.94 | 1.6 | 1.7 |
In contrast, the observed changes in half-life did not completely explain the fold increases in the abundance of other transcripts. In case of PmgR1, SyR11 and Ncl1330, the expression fold-changes at 20°C were 186.3 ± 84.8, 2.1 ± 0.1 and 12.9 ± 1.4, respectively, while the changes of the degradation constants could explain only a fold-change of 2.4, 1.04 and 1.8, respectively (Table 2). This analysis indicates that altered expression of PmgR, SyR11 and Ncl1330 predominantly resulted from a ~ 78-, ~2- and ~7-fold increase in the transcription rate in the crhRTR mutant.
Discussion
The ability of bacteria to acclimate to rapidly changing environmental conditions is frequently associated with the sRNA-mediated regulation of gene expression [1,35–37]. In many bacterial taxa, RNA chaperones, such as Hfq or ProQ/FinO-domain proteins, mediate the underlying sRNA-mRNA interactions [1,38]. Cyanobacteria, including Synechocystis, express a diverse range of ncRNAs [35–37]. However, the Hfq homolog from Synechocystis does not exhibit obvious RNA binding or RNA chaperone activity [39,40] and searches we performed for homologues of ProQ in the genome did not identify any possible candidates. The mechanism by which ncRNA-mRNA interaction is regulated in cyanobacteria is not known, thus it was investigated if CrhR RNA helicase activity is relevant in the context of ncRNA metabolism in Synechocystis. We demonstrate that CrhR RNA helicase activity is associated with alteration of a specific subset of the Synechocystis transcriptome, comprised of both mRNAs and sRNAs.
Comparison with previous studies
In agreement with previous studies, we detected changes in rimO (slr0082) and groES/groEL (slr2075-slr2076) transcript abundances, while other reported changes were not observed. Our crhRTR transcriptome results differed in several other aspects from previous reports [28,41]. Intriguingly, although CrhR abundance is maximal at low temperature [26], our transcriptome data indicate that while more effects were observed in response to cold stress, significant alterations in gene expression also existed at the standard growth temperature of 30°C [28]. The experiments differed in the previous use of arrays targeting only coding sequences, from which changes in ncRNA abundance could not be extracted and the use of a complete crhR deletion mutant [28] versus the crhRTR mutant used here. Importantly, the growth conditions differed, 34°C and 24°C [28], or 34°C and 22°C for 20 min [41] versus 30°C and 20°C for 3 h (this study). These differences would be expected to differentially affect the transcriptome and proteome, as a reduced level of crhR cold induction was observed at 25°C [26].
Differential expression of the crhR operon
The data indicate that rimO (slr0082) and crhR (slr0083) form a dicistronic operon, portions of which are differentially expressed. slr0082 and crhR transcript abundance is enhanced in response to cold stress in wild type cells, a level that is enhanced further by crhR mutation. Factors, potentially contributing to these changes include a putative additional transcription start site within the slr0082 ORF at position 2,887,140 [34], which may give rise to an alternative 5ʹ UTR. At the transcript level, the expression differences between crhRTR and wild type at 20°C can be largely explained by a post-transcriptional stabilization of the crhR transcript in the absence of functional RNA helicase (Figure 4). Together, the data support a model in which the strong induction observed at 20°C in the crhRTR strain would be explained by the lack of a CrhR-dependent auto-regulatory, negative feedback loop, in which CrhR binds and decreases stability of its own transcript.
Connection between CrhR and the ribosome maturation system
The genomic organization of the rimO (slr0082) and crhR (slr0083) as dicistronic operon is widely conserved in cyanobacteria (Figure S1). This synteny combined with the co-regulation of expression, implies the two genes function in a common pathway. slr0082 encodes a methyltransferase targeting either ribosomal protein S12 (RimO) or tRNAs (MiaB), respectively [42], and thus potentially affects the biogenesis or activity of ribosomes at low temperature. Concurrently, the ribosomal protein operon sll1799-ssl3441 also showed a reduced cold induction in crhRTR. CrhR association with translation and ribosome biogenesis is not unexpected, as prokaryotic DEAD-box RNA helicases are frequently associated with these processes, for example three of five members are associated with ribosome biogenesis in E. coli [23].
Altered mRNA expression correlates with altered cellular morphology and physiology
Our data frequently revealed directional alteration of expression for genes encoded in operons including decreases in the ribosomal and ATP synthase operons and an increased abundance of rimO (slr0082) and crhR (slr0083) operon at 20°C. Similarly, expression of the ndhCKJ operon was reduced at 30°C, while expression of the operon sll0217-sll0219 encoding flavoproteins Flv2 and Flv4, and in addition sll0550 encoding Flv3, and genes encoding components of the CLP protease were reduced at both temperatures. The genes with the strongest positive response, sll1862 and sll1863, have no annotated function but are strongly induced at iron starvation [34].
Altered levels of the sigma factor sigE (sll1689), the sugar catabolism regulator Rre37 (sll1330), the negative regulator of glucose metabolism and oxidative stress acclimation PrqR (slr0895) and also PmgR1 (ncr0700) suggest an interesting link between CrhR and sugar/energy/heterotrophic metabolism, which is the major functional target category of these regulators [43–47]. PmgR1 (ncr0700), the most strongly induced sRNA in crhRTR, is involved in the switch to photomixotrophic growth and the regulation of glycogen metabolism [48]. CrhR association with glucose metabolism is consistent with the previous report that the Dbp2 DEAD-box RNA helicase regulates this process in yeast [49]. Overall, the observed transcriptomic changes are reflected physiologically by the progressive degeneration of thylakoid membrane structure at 20°C, reduced electron transport and signs of oxidative stress and photoinhibition exhibited by the crhRTR mutant [30]. These morphological effects are anticipated to reduce energy production, thereby affecting photosynthetic rate and cell growth, as observed in the crhRTR mutant.
The non-coding share of the transcriptome is strongly affected by crhR mutation
In contrast to previous studies, we also investigated the non-coding transcriptome of Synechocystis. The CrhR truncation only affected a small proportion of the Synechocystis ncRNA pool, altering roughly 10% of the annotated sRNAs and asRNAs. The non-global nature of these effects are illustrated by the strong effect on PmgR1 (ncr0700) while only minor effects were observed for other sRNAs, such as the well-studied IsaR1 [36] and PsrR1 [35]. The altered ncRNA abundance exhibited temperature dependence, with stronger responses occurring at 20°C. Surprisingly, >90% of the affected ncRNAs increased in abundance in the absence of CrhR RNA helicase activity, which may indicate a common mechanism. It is tempting to speculate that CrhR reduces the stability of these transcripts by unwinding stabilizing secondary structures at the post-transcriptional level. However, at least in the case of the investigated sRNAs, only a minor fraction of the fold-change in abundance resulted from altered transcript stability. Thus, CrhR alteration of ncRNA levels may not occur solely at the post-transcriptional level or may partially be indirect. While the mechanism by which CrhR could alter transcription is not known, similar data has been provided in E. coli where Hfq affects the abundance of specific mRNAs through a transcriptional mechanism, independently of degradation [50]. The authors proposed that Hfq affects an early step in elongation, preventing premature transcript release [50]. In addition, CrhR interaction with RNA polymerase subunits or the Rho transcription termination factor could potentially be associated with such transcriptional regulation, similar to Hfq [51,52]. The indirect effects of CrhR truncation on RNA turnover could be associated with the observed up-regulation of RNase J (slr0551). Although not studied in cyanobacteria, RNase J is known to perform functions in all aspects of RNA metabolism in a range of organisms [53–55]. Moreover, yeast two-hybrid screens indicated that the DEAD-box RNA helicase HP4 interacted with the RNase J homolog in C. reinhardtii [53]. Thus, the CrhR-induced alteration in RNase J mRNA abundance could be associated with the observed changes in mRNA and ncRNA accumulation, possibly similar in relevance to RNase E in cyanobacteria [56].
Overall, the data suggest that CrhR seemingly performs roles that affect a limited set of the mRNA and ncRNA repertoire in Synechocystis. These roles presumably include combinations of the duplex RNA unwinding and annealing activities CrhR catalyzes [20] that likely involve temperature-dependent mechanisms. Deciphering which of the observed effects is a direct or indirect consequence of CrhR RNA helicase activity will be a major goal of future research.
Materials and methods
Bacterial strains and culture conditions
The Synechocystis strains used in this study were the non-motile wild type and a strain in which crhR was partially inactivated, crhRTR [30]. The crhRTR mutant was generated by insertion of a spectinomycin-streptomycin resistance cassette into the PmlI site between RNA helicase motifs III (SAT) and IV (FVRTK), thereby removing the second RecA domain and C-terminal extension from crhR [30]. The partial crhR deletion also removed the 3ʹ end of the downstream gene, argC which is transcribed in the opposite direction [30]; however, the resulting mutant cells are not auxotrophs [30]. The completely segregated crhRTR strain constitutively produces a 27 kDa truncated CrhR polypeptide, CrhRTR, the abundance of which is not repressed at 30°C [26,27] and which is inactive at both gene expression level [26,27] and at physiological and morphological level [30]. Microarray analysis was performed using the CrhRTR partial deletion mutant since this strain was utilized in previous investigations and provided crucial insights into the regulation of crhR expression that would not have been obtained with a complete deletion strain. Cells were cultured in BG-11 liquid medium at 30°C under continuous illumination (50 µmol photons m−2s−1) with continuous bubbling with sterile, humidified air essentially as described previously [57]. Media supporting crhRTR mutant growth was supplemented with sodium thiosulphate (0.3%) and buffered with tricine (10 mM, pH 8.0) plus spectinomycin (50 µg/mL each) for crhRTR. Cells were cold-stressed by incubation at 20°C for three hours [57] and dark stress by wrapping culture flasks in aluminum foil in a dark walk-in incubator.
RNA isolation, analysis and half-life estimation
RNA isolation and northern analysis were performed as described previously [26,57,58]. Briefly, mid-log phase Synechocystis cells were inactivated at the stated growth temperature by addition of a 1:1 (vol:vol) dilution of the culture with ice-cold 5% phenol in ethanol. Total RNA was subsequently extracted by glass bead lysis in the presence of phenol, followed by extensive phenol-chloroform extraction and lithium chloride precipitation [57]. RNA quantity and quality was determined using an Agilent Bioanalyzer, via fluorometry and RIN number analysis, respectively. mRNA and ncRNA transcripts were detected in total RNA (3 or 5 μg), separated on 1% agarose and 8 M urea-8%-PAA gels, respectively. Transcripts were detected on Northern blots containing total RNA using 32P-labelled riboprobes specific for each gene, generated by in vitro transcription from PCR-generated templates as described previously [26,58] and using the oligonucleotide primers listed in Table S2. The blots were exposed to X-ray films (Figures 2 and 3) or detected with a storage phosphor screen (Kodak) and a GE Healthcare Typhoon FLA 9500 imaging system (Figure 4). Transcript size was estimated using Fermentas RiboRulerTM RNA markers. When indicated, transcript half-life was measured in the presence of rifampicin (300 µg/mL) in total RNA extracted from duplicate samples taken over the indicated time courses. The half-life (was calculated by fitting the data to an exponential decay curve with the R nls function. The degradation constant is .
At steady state expression of a given gene the transcript levels are defined by the ratio of the synthesis rate/transcription rate ( and the degradation constant (): and the fold-change of expression levels at two different conditions is
With knowledge about the fold-change and the half-life/degradation constant the impact of the transcription rate can be calculated.
Synteny analysis
Homologs of CrhR were detected by BLASTp and all proteins encoded by genes in a window 3000 nt upstream and downstream of the start codon of crhR were analyzed. Homolog classification was done on amino acid level with Cd-hit [59] using a word size of 2, minimal overlap of 60% and an identity threshold of 40%.
Microarray analysis
The microarray design, hybridization procedure and data analysis have been described previously [60]. The Agilent microarrays contain oligonucleotide probes representing all annotated mRNAs plus the majority of other expressed transcripts, allowing precise determination of transcripts with respect to both DNA strand and genomic location. Total RNA (5 µg) extracted from the wild type and crhRTR mutant strains grown at standard temperature (30°C) and after 3 h of cold stress (20°C) was directly labeled without cDNA synthesis using Kreatech’s ULS labeling kit for Agilent gene expression arrays with Cy5 according to the manufacturer’s protocol (Kreatech Diagnostics, B.V., Netherlands). RNA fragmentation and hybridization followed the manufacturer’s instructions for Agilent one color microarrays with 3 to 5 µg of labeled RNA. Array analysis was performed using two biological replicates and in addition, each array had an internal technical replicate. Raw data were normexp background corrected [61] and quantile normalized using the limma R package [62]. The transcriptome differences between wild type and crhRTR mutant were determined for the two different temperatures. A transcript was taken as differentially expressed, when it the significance criteria log2FC ≥│1│, adj. p -value ≤ 0.05). P-values were adjusted for multiple testing with the Benjamini-Hochberg method. The array data have been deposited in the GEO database under the accession number GSE58544.
Abbreviations
| asRNAs | cis-encoded antisense RNAs |
| CrhR | cyanobacterial RNA helicase Redox |
| ncRNAs | non-coding regulatory RNAs |
| sRNA | small regulatory RNA |
Funding Statement
This work was supported by a grant from the Baden-Wuerttemberg Foundation BWST_NCRNA_008 to WRH and by the Natural Sciences and Engineering Research Council of Canada (NSERC), grant number 171319, to GWO.
Acknowledgments
We thank Loubna Youssar for support in the calculation of RNA half lives.
Author contributions
GWO, WRH and JG designed the study; JG, ARRR, DC, AM performed experiments; AM, GWO, JG, WRH analyzed data; JG, GWO and WRH wrote the paper with contributions from all authors.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here
References
- [1].Wagner EGH, Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208. [DOI] [PubMed] [Google Scholar]
- [2].Carrier M-C, Lalaouna D, Massé E. Broadening the definition of bacterial small RNAs: characteristics and mechanisms of action. Annu Rev Microbiol. 2018;72:141–161. [DOI] [PubMed] [Google Scholar]
- [3].Chao Y, Papenfort K, Reinhardt R, et al. An atlas of Hfq-bound transcripts reveals 3ʹ UTRs as a genomic reservoir of regulatory small RNAs. Embo J. 2012;31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Georg J, Hess WR. Widespread antisense transcription in prokaryotes. Microbiol Spectr. 2018;6(4). DOI: 10.1128/microbiolspec.RWR-0029-2018 [DOI] [PubMed] [Google Scholar]
- [5].Dar D, Sorek R. Bacterial noncoding RNAs excised from within protein-coding transcripts. MBio. 2018;9:e01730–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. [DOI] [PubMed] [Google Scholar]
- [8].Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Malmgren C, Wagner EG, Ehresmann C, et al. Antisense RNA control of plasmid R1 replication. The dominant product of the antisense RNA-mRNA binding is not a full RNA duplex. J Biol Chem. 1997;272:12508–12512. [DOI] [PubMed] [Google Scholar]
- [10].Woodson SA, Panja S, Santiago-Frangos A.Proteins that chaperone RNA regulation. Microbiol Spectr. 2018;6:(4).10.1128/microbiolspec.RWR-0026-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Santiago-Frangos A, Woodson SA. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA. 2018;9:e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smirnov A, Förstner KU, Holmqvist E, et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci USA. 2016;113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Khemici V, Linder P. RNA helicases in bacteria. Curr Opin Microbiol. 2016;30:58–66. [DOI] [PubMed] [Google Scholar]
- [14].Rajkowitsch L, Chen D, Stampfl S, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. [DOI] [PubMed] [Google Scholar]
- [15].Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. [DOI] [PubMed] [Google Scholar]
- [16].Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. [DOI] [PubMed] [Google Scholar]
- [17].Owttrim GW. RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol. 2013;10:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34:3220–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fairman ME, Maroney PA, Wang W, et al. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. [DOI] [PubMed] [Google Scholar]
- [20].Chamot D, Colvin KR, Kujat-Choy SL, et al. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J Biol Chem. 2005;280:2036–2044. [DOI] [PubMed] [Google Scholar]
- [21].Linder P, Owttrim GW. Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci. 2009;14:344–352. [DOI] [PubMed] [Google Scholar]
- [22].Resch A, Većerek B, Palavra K, et al. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol. 2010;7:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kujat SL, Owttrim GW. Redox-regulated RNA helicase expression. Plant Physiol. 2000;124:703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vinnemeier J, Hagemann M. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch Microbiol. 1999;172:377–386. [DOI] [PubMed] [Google Scholar]
- [26].Rosana ARR, Chamot D, Owttrim GW. Autoregulation of RNA helicase expression in response to temperature stress in Synechocystis sp. PCC 6803. PLoS ONE. 2012;7:e48683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tarassova OS, Chamot D, Owttrim GW. Conditional, temperature-induced proteolytic regulation of cyanobacterial RNA helicase expression. J Bacteriol. 2014;196:1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Prakash JSS, Krishna PS, Sirisha K, et al. An RNA helicase, CrhR, regulates the low-temperature-inducible expression of heat-shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiol. 2010;156:442–451. [DOI] [PubMed] [Google Scholar]
- [29].Rowland JG, Simon WJ, Prakash JSS, et al. Proteomics reveals a role for the RNA helicase crhR in the modulation of multiple metabolic pathways during cold acclimation of Synechocystis sp. PCC6803. J Proteome Res. 2011;10:3674–3689. [DOI] [PubMed] [Google Scholar]
- [30].Rosana ARR, Ventakesh M, Chamot D, et al. Inactivation of a low temperature-induced RNA helicase in synechocystis sp. PCC 6803: physiological and morphological consequences. Plant Cell Physiol. 2012;53:646–658. [DOI] [PubMed] [Google Scholar]
- [31].Voss B, Georg J, Schön V, et al. Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics. 2009;10:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Georg J, Voss B, Scholz I, et al. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mitschke J, Georg J, Scholz I, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kopf M, Klähn S, Scholz I, et al. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014;21:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Georg J, Dienst D, Schürgers N, et al. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell. 2014;26:3661–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Georg J, Kostova G, Vuorijoki L, et al. Acclimation of oxygenic photosynthesis to iron starvation is controlled by the sRNA IsaR1. Curr Biol. 2017;27:1425–1436, e7 [DOI] [PubMed] [Google Scholar]
- [37].Klähn S, Schaal C, Georg J, et al. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA. 2015;112:E6243–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Olejniczak M, Storz G. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol. 2017;104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dienst D, Dühring U, Mollenkopf H-J, et al. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiol. 2008;154:3134–3143. [DOI] [PubMed] [Google Scholar]
- [40].Schuergers N, Ruppert U, Watanabe S, et al. Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol Microbiol. 2014;92:840–852. [DOI] [PubMed] [Google Scholar]
- [41].Suzuki I, Kanesaki Y, Mikami K, et al. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol. 2001;40:235–244. [DOI] [PubMed] [Google Scholar]
- [42].Anton BP, Saleh L, Benner JS, et al. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:1826–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Azuma M, Osanai T, Hirai MY, et al. A response regulator Rre37 and an RNA polymerase sigma factor SigE represent two parallel pathways to activate sugar catabolism in a cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011;52:404–412. [DOI] [PubMed] [Google Scholar]
- [44].Joseph A, Aikawa S, Sasaki K, et al. Rre37 stimulates accumulation of 2-oxoglutarate and glycogen under nitrogen starvation in Synechocystis sp. PCC 6803. FEBS Lett. 2014;588:466–471. [DOI] [PubMed] [Google Scholar]
- [45].Osanai T, Kuwahara A, Iijima H, et al. Pleiotropic effect of sigE over-expression on cell morphology, photosynthesis and hydrogen production in Synechocystis sp. PCC 6803. Plant J. 2013;76:456–465. [DOI] [PubMed] [Google Scholar]
- [46].Nakaya Y, Iijima H, Takanobu J, et al. One day of nitrogen starvation reveals the effect of sigE and rre37 overexpression on the expression of genes related to carbon and nitrogen metabolism in Synechocystis sp. PCC 6803. J Biosci Bioeng. 2015;120:128–134. [DOI] [PubMed] [Google Scholar]
- [47].Khan RI, Wang Y, Afrin S, et al. Transcriptional regulator PrqR plays a negative role in glucose metabolism and oxidative stress acclimation in Synechocystis sp. PCC 6803. Sci Rep. 2016;6:32507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].de Porcellinis AJ, Klähn S, Rosgaard L, et al. The non-coding RNA Ncr0700/PmgR1 is required for photomixotrophic growth and the regulation of glycogen accumulation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016;57:2091–2103. [DOI] [PubMed] [Google Scholar]
- [49].Beck ZT, Cloutier SC, Schipma MJ, et al. Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics. 2014;198:1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Le Derout J, Boni IV, Régnier P, et al. Hfq affects mRNA levels independently of degradation. BMC Mol Biol. 2010;11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. [DOI] [PubMed] [Google Scholar]
- [52].Rabhi M, Espéli O, Schwartz A, et al. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011;30:2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liponska A, Jamalli A, Kuras R, et al. Tracking the elusive 5ʹ exonuclease activity of Chlamydomonas reinhardtii RNase J. Plant Mol Biol. 2018;96:641–653. [DOI] [PubMed] [Google Scholar]
- [54].Linder P, Lemeille S, Redder P. Transcriptome-wide analyses of 5ʹ-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 2014;10:e1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rische-Grahl T, Weber L, Remes B, et al. RNase J is required for processing of a small number of RNAs in Rhodobacter sphaeroides. RNA Biol. 2014;11:855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cameron JC, Gordon GC, Pfleger BF. Genetic and genomic analysis of RNases in model cyanobacteria. Photosyn Res. 2015;126:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chamot D, Owttrim GW. Regulation of cold shock-induced RNA helicase gene expression in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2000;182:1251–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Steglich C, Futschik ME, Lindell D, et al. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. [DOI] [PubMed] [Google Scholar]
- [60].Voß B, Hess WR. The identification of bacterial non-coding RNAs through complementary approaches. In: Hartmann RK, Bindereif A, Schön A, Westhof E, editors. Handbook of RNA biochemistry. 2nd ed. Weinheim: Wiley‐VCH Verlag; 2014. p. 787–800. [Google Scholar]
- [61].Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. [DOI] [PubMed] [Google Scholar]
- [62].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


