ABSTRACT
Circular RNAs (circRNAs) are a recently identified class of non-coding RNAs that participate in multiple biological processes and tumour progression. However, circRNA expression pattern in parathyroid neoplasms remains unknown. The circRNA profile of 6 parathyroid carcinomas (PCs), 6 parathyroid adenomas (PAs) and 4 normal parathyroid tissues was assessed by a microarray. Bioinformatic analyses were performed to investigate potential core circRNAs via co-expression network. CircRNA and corresponding mRNA expression were validated in a cohort of parathyroid neoplasms by RT-qPCR and fluorescence in situ hybridization (FISH). Compared to normal parathyroid, 5310 and 1055 circRNAs were differentially expressed in PC and PA tissues, respectively. The differential expression of 4 circRNAs (hsa_circRNA_0035563 (p = 0.006), hsa_circRNA_0017545 (p = 0.009), hsa_circRNA_0001687 (p = 0.005) and hsa_circRNA_0075005 (p = 0.001)) and 4 mRNAs (MYC, FSCN1, ANXA2 and AKR1C3) between PC and PA tissues were confirmed by RT-qPCR. In addition, high expression of hsa_circ_0035563 was related to CDC73 mutations (p = 0.022) and recurrence in PC patients (p = 0.042). Furthermore, hsa_circ_0075005 helped distinguish PCs from benign lesions using FISH, and the area under the curve was 0.779 (p = 0.013). Our findings describe the circRNA profile of PC for the first time and suggest that circRNAs and mRNAs interact in parathyroid tumourigenesis. This study demonstrates that hsa_circ_0075005 and MYC mRNA may be used for the differential diagnosis of PC and PA. The expression levels of hsa_circ_0035563 are related to CDC73 mutations and recurrence in malignancy, highlighting the significance of this parameter in prognosis of PC patients.
KEYWORDS: Parathyroid carcinoma, circular RNA, mRNA, co-expression network, CDC73
Introduction
Parathyroid carcinoma (PC) is a rare malignancy with a poor prognosis. PC accounts for less than 1% of cases of primary hyperparathyroidism in Western countries, while the incidence of PC is greater than 5% in China and other Eastern countries [1]. Compared with benign parathyroid adenomas (PAs), PC is characterized by more severe clinical symptoms and more remarkable biochemical parameters, which may indicate the risk of malignancy in patients with hyperparathyroidism. However, diagnosis remains challenging because of the overlap in the clinical presentation and histopathological findings between PC and PA unless recurrence or metastasis has occurred. The 5-year and 10-year overall survival rates were reported to be 82.3% and 66%, respectively [2]. The main reason for mortality due to PC is uncontrollable hypercalcaemia during the late stage rather than tumour invasion. Radical resection in the initial operation is suggested to decrease the risk of recurrence and metastasis, but more than 60% of patients suffer from local relapse or distant metastasis. Furthermore, thus far, PC has responded poorly to chemotherapy and radiotherapy [3]. Early diagnosis is critical for radical resection and close follow-up, which may be the only way to cure this malignancy. A set of molecular markers is desperately needed to screen patients for radical surgery before the initial operation or timely remedial operation after the initial operation. Unfortunately, the exact molecular mechanisms involved in the tumourigenesis, recurrence, metastasis and chemoradiotherapy resistance of PC are still poorly understood [4]. Although 50–70% of patients with sporadic PC have been found to carry CDC73 inactivating mutations, gene sequencing and immunohistochemical staining still cannot identify any mutation in CDC73 in a variable percentage of sporadic PC cases [5]. In addition, some PA tissues can also carry CDC73 mutations [6,7]. Therefore, other diagnostic markers and molecular therapeutic targets are needed to identify these patients.
Circular RNAs (circRNAs) belong to a novel class of non-coding RNAs with regulatory potency in biological processes. In contrast to linear RNAs with terminal 5ʹ caps and 3ʹ tails, the 3ʹ and 5ʹ ends of circRNAs are covalently linked to form a closed circular structure, rendering them more stable than their linear counterparts [8]. Initially, circRNAs were mistaken as outcomes of mis-splicing. However, since 2012, their biological functions have been recognized due to technological breakthroughs in high-throughput deep RNA sequencing (RNA-seq) [9,10]. With the development of RNA-seq technology and bioinformatics, more than 200,000 circRNAs have been identified in tumor tissue [11]. Accumulating evidence has revealed that circRNAs play important roles in multiple biological and pathological processes [12]. CircRNAs play different roles in cancer development via multiple mechanisms, such as acting as microRNA (miRNA) sponges, regulators of RNA binding proteins, and modulators of the Wnt signalling pathway [13,14]. Notably, ‘miRNA sponge’ represents the most prominent and definite function of circRNAs [15]. In a type of post-transcriptional regulation, some circRNAs compete with miRNAs for the same binding sites in corresponding mRNAs [16]. Recently, some endogenous circRNAs have been reported to be translatable in living human cells [17,18]. This finding suggests that circRNAs may also play an important role in transcription levels, even though the extent of their participation is not currently known.
Studies have increasingly shown that the aberrant expression of circRNAs is related to a variety of tumours, including laryngeal cancer, colorectal cancer, ovarian cancer, gastric cancer, bladder cancer and oesophageal cancer [19–21]. In oesophageal squamous cell carcinoma and colorectal carcinoma, circular RNA ITCH has been found to exert its inhibitory effect through the Wnt/β-catenin pathway [22]. In oral cancer, circRNA_100290 has been found to regulate CDK6 expression by sponging miR-29b family members [23]. Because circRNAs are resistant to ribonucleases due to their unique circular structure, circRNA molecules are speculated to be more stable in plasma than linear RNAs [24,25]. The levels of hundreds of circRNAs in the blood have been shown to be higher than those of their homologous linear transcripts [26]. These features suggest that circRNAs may serve as novel diagnostic markers and therapeutic targets for the treatment of malignancy. The hsa_circ_0001017 and hsa_circ_0061276 circRNAs in plasma were identified as new potential biomarkers of gastric cancer [27]. Furthermore, more than 400 circRNAs have been identified in human cell-free saliva [28]. However, the significance of circRNAs in parathyroid neoplasms remains unknown.
So far, whether PA intermediates the process of PC development remains unclear and meanwhile PA is responsible for the majority of primary hyperparathyroidism. Lack of effective diagnostic markers to identify the PC from PA may lead to inaccurate diagnosis and interfere the optimal treatment strategy. In this study, we investigated the global circRNA expression profile with a circRNA microarray in a Chinese cohort of patients. We focused on the most differentially expressed circRNAs in PC that are potentially associated with known pathogenic genes. The results of the mRNA expression profiles were also used to construct a circRNA-mRNA co-expression network. A group of differentially expressed circRNAs and mRNAs was validated with quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR) and fluorescence in situ hybridization (FISH). The significance of these circRNAs as diagnostic markers and relevant mechanisms of cancer progression was tentatively explored in a series of parathyroid tissues.
Results
Identification of differentially expressed circRNAs among PC, PA and normal parathyroid tissues
An SBC Human CircRNA expression microarray (Shanghai Biotechnology Corporation, Shanghai, China) containing 88371 probes for circRNAs was used in our study. CircRNAs were analysed by the high-throughput human microarray of 6 PC, 6 PA and 4 normal parathyroid (PaN) tissues. To obtain an overview of the circRNAs in the parathyroid disease, PaN were deemed as the control. In sum, 2668 upregulated and 2642 downregulated circRNAs were identified in PC, with a fold change ≥ 2.0 or ≤ 0.5 and a p-value < 0.05. In the PA tissues, there were 420 upregulated and 635 downregulated differentially expressed circRNAs, which was much lower than that the number found in the PC tissues (Supplementary Figure 1). Figure 1 shows the distribution of these differentially expressed circRNAs on human chromosomes. To further explore the distinct circRNAs for potential markers, PC was compared to PA. Consequently, 2521 upregulated and 2232 downregulated circRNAs were identified in PC. The volcano plot (Figure 2(a)) and heat map (Figure 2(b)) were constructed to visualize the differential circRNA expression between PC and PA tissues. These data reveal distinct circRNA expression in PC and PA.
Figure 1.
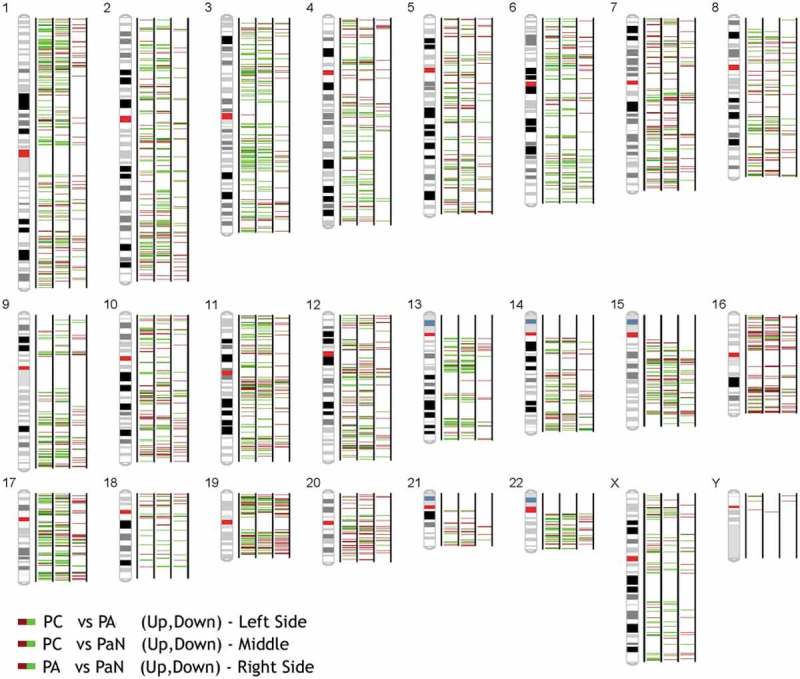
The distribution of the differentially expressed circRNAs on human chromosomes among parathyroid carcinomas (PCs), parathyroid adenomas (PAs) and normal parathyroid tissues (PaN). The brown and green bars represent the up- and downregulated circRNAs between the tumour and normal samples, respectively.
Figure 2.
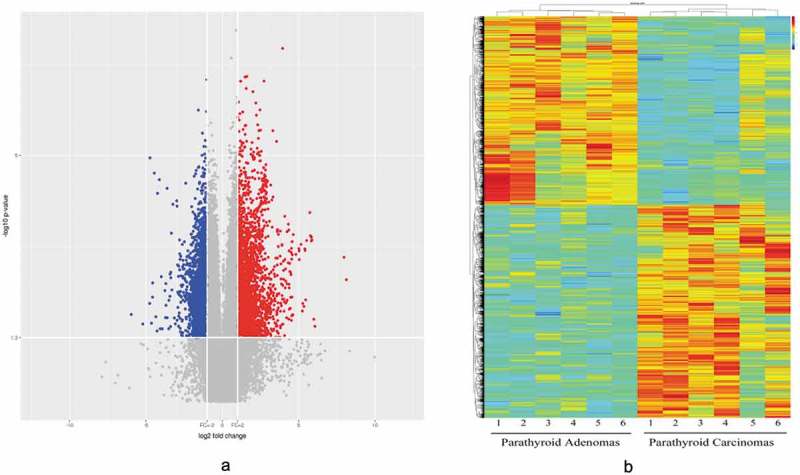
A volcano plot (2A) and heat map (2B) were used to identify the difference in circRNA expression between the PC and PA samples. In the volcano plot, the log2 fold change is plotted on the x-axis, and the negative log10 p-value is plotted on the y-axis. The heat map visualizes the differential circRNA expression (p < 0.05 and fold change ≥ 2.0 or ≤ 0.5) between the PC and PA samples. Each column represents one sample, and each row indicates a circRNA transcript. The colour scale ranges from blue (low intensity) and yellow (medium intensity) to red (strong intensity). PC, parathyroid carcinoma; PA, parathyroid adenoma.
Co-expression network analysis of circRNAs and mRNAs
To identify the critical circRNA in PCs, a circRNA-mRNA co-expression network was constructed based on the degree of the correlation between PC and PA (Supplementary Fig. 2A and B). Briefly, the Pearson correlation of each RNA pair was calculated. The higher the degree of the correlation, the more important is the RNA in the network. Based on the microarray data and analysis, the candidates for validation were selected based on the following criteria: (1) fold changes in circRNA expression ≥ 2.0 or ≤ 0.5; (2) p-value < 0.01; (3) raw intensity > 200; (4) host genes known to be related to cancer; and 5) absolute network difference > 0.1. In total, 6 circRNAs and 4 mRNAs from the co-expression network were selected as candidates for further validation. The 4 mRNAs (FSCN1, ANXA2, AKR1C3, and MYC) were the corresponding linear transcripts of hsa_circ_0079278, hsa_circ_0035563, hsa_circ_0017545 and hsa_circ_0085534. Hsa_circ_0001687 and hsa_circ_0075005 were also selected. CDC73 is also believed to be a critical gene in PC. However, in the present study, the circRNAs related to CDC73 were not aberrantly expressed in PC.
Validation of dysregulated circRNAs and the corresponding mRNAs in parathyroid neoplasm tissues
The expression of the selected circRNAs and mRNAs was evaluated by an RT-qPCR analysis of 20 PC and 41 PA tissues. Based on Mann-Whitney U test or t-test analysis, PC samples were compared with the PA samples. The expression of hsa_circRNA_0035563 (p = 0.006), hsa_circRNA_0017545 (p = 0.009), hsa_circRNA_0001687 (p = 0.005) and hsa_circRNA_0075005 (p = 0.001) were significantly upregulated in the PC samples. The detected circRNAs, i.e., hsa_circRNA_0079278 and hsa_circRNA_0085534, did not significantly differ between the PC and PA samples (Figure 3(a-e, j)).
Figure 3.
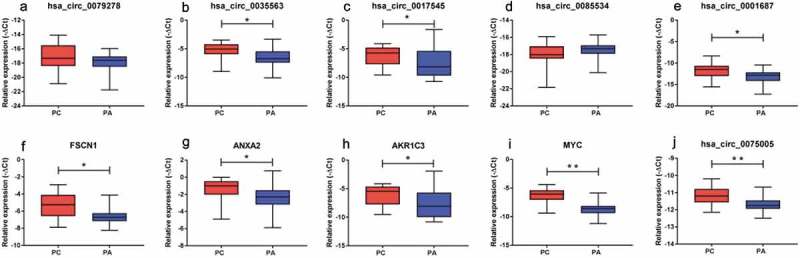
Validation of the differentially expressed circRNAs (a-e, j) and mRNAs (f-i) between the PC and PA tissues. FSCN1, ANXA2, AKR1C3 and MYC represent the host genes of hsa_circ_0079278, hsa_circ_0035563, hsa_circ_0017545 and hsa_circ_0085534. hsa_circ_0035563, hsa_circ_0017545, hsa_circ_0001687 and hsa_circ_0075005 were upregulated in the PC tissues. All transcripts were verified in a cohort of PC (n = 20) and PA (n = 41) tissues. * p < 0.05, ** p < 0.01.
We also detected the linear transcripts (mRNAs) of the circRNA host genes in the 20 PC and 41 PA samples. The mRNA expression of MYC, FSCN1, ANXA2 and AKR1C3 was significantly upregulated in the PC specimens compared with that in the PA tissues (Figure 3(f-i)). A significant positive correlation was observed between circ_0079278 and FSCN1 mRNA, between hsa_circ_0035563 and ANXA2 mRNA, and between hsa_circ_0017545 and AKR1C3 mRNA (Spearman’s correlation, p < 0.001, Figures 4 and 5). No correlation was identified between hsa_circ_0085534 and MYC mRNA (Spearman’s correlation, p = 0.117). In each sample, the expression levels of hsa_circ_0079278, hsa_circ_35563 and hsa_circ_0085534 were much lower than those of their linear counterparts FSCN1, ANXA2 and MYC (Figure 4).
Figure 4.
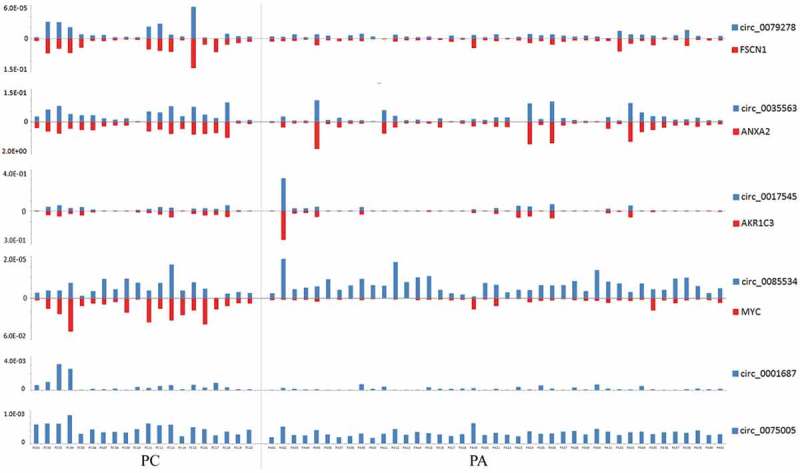
The relative expression levels of circRNAs and mRNAs (2−ΔCt) in each PC and PA tissue. Each column represents one sample, and each row indicates the expression level of an RNA transcript. For the convenience, the ordinate scale differs for each RNA in this diagram. The expression levels of hsa_circ_79278 and hsa_circ_85534 were much lower than those of their linear counterparts FSCN1 and MYC. The expression levels of hsa_circ_0079278 and FSCN1 mRNA, hsa_circ_0035563 and ANXA2 mRNA, and hsa_circ_0017545 and AKR1C3 mRNA were found to be correlated in each parathyroid neoplasm tissue.
Figure 5.
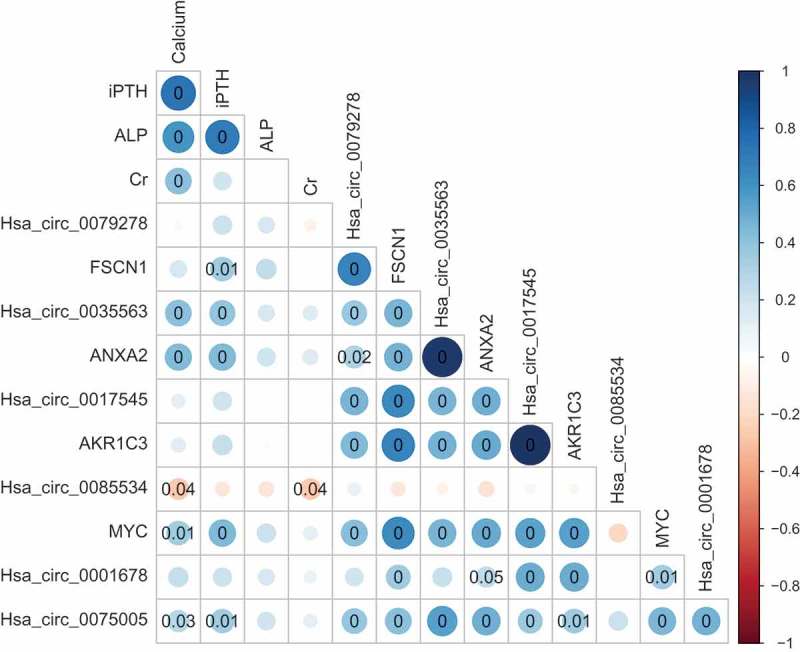
The Spearman’s correlation matrix between the clinical parameters and detected circRNAs and mRNAs. The size and colour intensity of the circle denote the correlation coefficient value. Positive correlations are visualized in blue and negative correlations in red scale. P-value is marked in the matrix if P-value is below 0.05, which represents statistical significance. iPTH, intact parathyroid hormone; ALP, alkaline phosphatase.
Correlations between circRNA and mRNA expression and clinicopathological characteristics
Binary logistic regression analysis indicated that hsa_circ_0075005 was the most promising diagnostic marker for distinguishing PC from PA. The receiver operating characteristic (ROC) curves are created to select possibly the optimal models and the area under the curve (AUC) varying between 0 and 1 was calculated to demonstrate the performance of a diagnostic marker. Here, the ROC analysis revealed that the AUC of hsa_circ_0075005 was 0.770 (95% confidence interval (CI), 0.633–0.906; p = 0.001). Regarding the mRNAs, MYC showed a better distinguishing potential with an AUC of 0.909 (95% CI, 0.823–0.994; p < 0.001, Supplementary Fig. 3).
The Mann-Whitney U test showed that hsa_circ_0035563 (p = 0.002), hsa_circ_0017545 (p = 0.012), AKR1C3 (p = 0.003), hsa_circ_001687 (p = 0.047), FSCN1 (p = 0.012), ANXA2 (p < 0.001), and MYC mRNA (p = 0.020) were dysregulated between PC patients with and without CDC73 inactivating mutations. The expression of hsa_circ_0035563 (p = 0.002), hsa_circ_001687 (p = 0.002), and ANXA2 (p = 0.002) mRNA differed between PC patients with and without recurrence. Furthermore, the binary logistic regression analysis indicated that high expression of hsa_circ_0035563 was related to CDC73 mutations (p = 0.022) and recurrence in the patients with PC (p = 0.042).
Correlations between circRNA or mRNA expression levels and clinical characteristics were further analysed (Spearman’s correlation, Figure 5). The preoperative serum calcium levels of the patients with parathyroid neoplasms were correlated with the levels of hsa_circ_0035563 (rs = 0.413, p = 0.001), hsa_circ_0085534 (rs = −0.269, p = 0.036), hsa_circ_0075005 (rs = 0.283, p = 0.027), MYC mRNA (rs = 0.347, p = 0.006) and ANXA2 mRNA (rs = 0.432, p = 0.001). The serum intact parathyroid hormone (iPTH) levels were also correlated with hsa_circ_0035563 (rs = 0.397, p = 0.002), hsa_circ_0075005 (rs = 0.342, p = 0.007), FSCN1 mRNA (rs = 0.349, p = 0.006), ANXA2 mRNA (rs = 0.438, p < 0.001), and MYC mRNA (rs = 0.440, p < 0.001). No circRNA or mRNA expression levels were correlated with alkaline phosphatase (ALP) levels in the patients. No other RNA expression levels were correlated with the serum creatinine level in these patients, except for hsa_circ_0085534 (rs = −0.266, p = 0.039).
Validation of circRNA expression using FISH
Two circRNAs, hsa_circ_0035563 and hsa_circ_0075005, were selected to be validated in formalin-fixed paraffin-embedded (FFPE) samples of PC and PA using FISH. The expression levels of hsa_circ_0035563 and hsa_circ_0075005 in FFPE samples of 12 PC and 16 parathyroid benign neoplasms were compared (Figure 6). The expression levels of both circRNAs in the PC samples were higher than those in the benign lesions. The AUC of hsa_circ_0075005 was 0.779 (95% CI, 0.599–0.959; p = 0.013).
Figure 6.
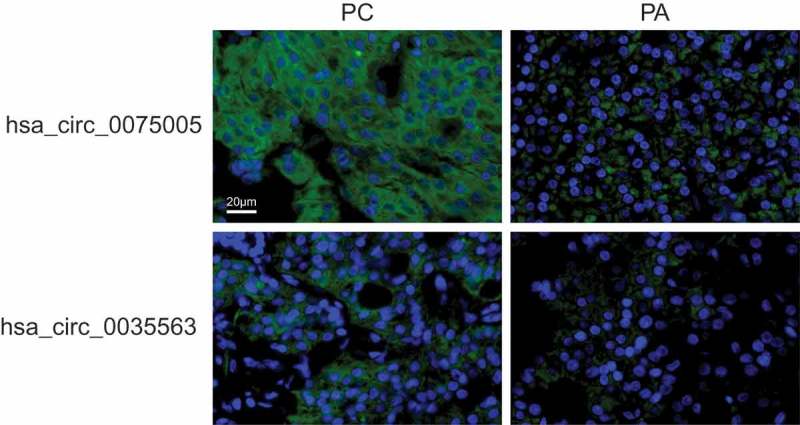
The expression of hsa_circ_0075005 and hsa_circ_0035563 was assessed by fluorescence in situ hybridization of differentiating malignant and benign parathyroid lesions. The upper images show the typical positive and negative staining of hsa_circ_0075005 in the PC and PA tissues, respectively. The lower images show the typical positive and negative staining of hsa_circ_0035563 in the PC and PA tissues, respectively. The blue fluorescence shows the position of the nucleus, while the circRNA is indicated by green fluorescence.
Discussion
Recent accumulating evidence has shown that regulation of the transcriptome is an important mechanism responsible for oncogenesis and the development of parathyroid neoplasms [4,29]. Increasing evidence indicates that circRNAs play an important role in both transcriptional and post-transcriptional regulation during the initiation and progression of a wide range of cancers [30]. As new members of the RNA regulation network, circRNAs have drawn extensive attention as diagnostic markers with substantial diversity, high molecular stability and tissue specificity. This discovery extended our understanding of the complex network of non-coding RNAs. To the best of our knowledge, this study is the first attempt to explore the significance of circRNAs in parathyroid carcinoma. A set of dysregulated circRNAs was identified, and their significance was verified.
Compared with PaN, 5310 dysregulated circRNAs were identified using a circRNA microarray of PC, which was much higher than the 1055 dysregulated circRNAs identified in PA. Therefore, more complex regulatory processes may be involved in malignancy. Furthermore, the host genes of these dysregulated circRNAs were distributed across all chromosomes, suggesting that circRNAs are widely involved in the regulation of many genes.
A prominent function of circRNAs as a part of the competing endogenous RNA network is their potency in regulating mRNA expression at the transcriptional and post-transcriptional levels. We believe that an integrated analysis of both circRNAs and mRNAs may be useful as a preliminary exploration of the effects of circRNAs in PC. The mRNA data originate from the same parathyroid tissue investigated in our previous study [31]. From this network, 6 circRNAs and 4 corresponding mRNAs were selected as candidates for further verification. Using RT-qPCR, 4 circRNAs were validated to be differentially expressed between PCs and PAs. Furthermore, high expression of hsa_circ_0035563 was found to be related to CDC73 mutations and recurrence in patients with PC. The expression of hsa_circ_0075005 and hsa_circ_0035563 was correlated with the serum levels of calcium and iPTH. The host gene of hsa_circ_0035563 was ANXA2 encoding Annexin A2, whose overexpression is related to advanced stage, reduced survival and resistance to treatment regimens in many kinds of malignancy [32,33]. In the present study, the ANXA2 mRNA was also found to be highly expressed in PC. The NPM1 gene, which is the host gene of hsa_circ_0075005, has also been reported to be linked to high-grade tumours and poor prognosis in leukaemia and some solid cancers [34]. Hence, these two circRNAs deserve further investigation in the diagnosis and treatment of PC. To explore the significance of these circRNAs in differentiating benign and malignant parathyroid neoplasms, FISH was performed, and the AUC of hsa_circ_0075005 was 0.779 (p = 0.013).
Regarding mRNA expression, four candidate mRNAs were found to be differentially expressed between PC and PA. Among these mRNAs, the MYC mRNA was identified as the best diagnostic marker of PC and PA with an AUC of 0.909. MYC gene activation is believed to be a hallmark of the initiation and maintenance of many different types of cancers [35,36]. As a target gene of β-catenin in the Wnt signalling pathway, MYC is also a critical component in parathyroid neoplasm [37]. Compared with PaN, MYC mRNA has been reported to be overexpressed in some benign parathyroid tumours [38]. In the present study, MYC mRNA expression in PC tissues was significantly higher than that in PA tissues. Furthermore, we found that MYC mRNA expression in PC tissues with the CDC73 mutation was higher than that in tissues without the CDC73 mutation. Parafibromin, which is the coding protein of CDC73, could inhibit the expression of MYC by occupying the c-myc promoter [39]. The inactivating CDC73 mutation could result in a loss of parafibromin, which may weaken this inhibitory effect on cell proliferation. In the present study, the expression of hsa_circ_0088534 was not correlated with that of the linear RNA transcript of MYC, indicating that circRNAs may not be the main factor regulating the expression of MYC. The expression of three other pairs of circRNAs and their corresponding linear transcripts (hsa_circ_0079278 and FSCN1 mRNA, hsa_circ_0035563 and ANXA2 mRNA, and hsa_circ_0017545 and AKR1C3 mRNA) was found to be correlated in each parathyroid neoplasm tissue. These genes were all previously reported to be tumour related.
Several limitations of this study should be mentioned. The main shortcoming was the limited cohort size of PC patients due to its rarity, although the present cohort may be the largest in recent years. The validation of these markers in more patients from multiple centres is necessary to explore their clinical value. Second, the PC diagnosis of some patients in our study was based on a histopathological evaluation without evidence of metastases. Long-term follow-up is needed to validate these results. Third, differential expressed circRNAs from microarray were screened without FDR correction, suggesting a potentially higher false positive rate in statistics. Moreover, due to the lack of a PC cell line, intervention experiments that could further confirm the regulatory effects of these circRNAs on their host genes were not completed.
Patients and methods
Patients and specimens
In total, 18 patients with PC and 41 patients with PA who underwent operations at the Peking Union Medical College Hospital were included in this study. During the surgery, the excised specimens were immediately immersed in RNAlater RNA Stabilization Reagent (Ambion Inc., USA) at 4 °C and preserved at −80 °C until use. In two patients, two carcinoma tissues from independent recurrences were included in the study. All samples were confirmed by a pathological examination in accordance with the WHO criteria [40]. The diagnosis of PC was based on histopathological criteria, including invasion into adjacent structures and/or extracapsular blood vessels or clinical evidence of metastasis. Six PC samples, six PA samples and four normal parathyroid gland samples were used for the microarray chip detection. The tiny normal parathyroid tissues were incidentally obtained during surgery in patients with thyroid diseases and normal parathyroid function. Some samples were collected from patients who were also recruited in our previous studies investigating long non-coding RNAs [31] and miRNAs [41]. The clinical characteristics of these PC patients are listed in Supplementary Table 1. Another 14 PC samples and 35 PA samples were used for RT-qPCR validation. FFPE samples from 12 patients with PC and 16 patients with benign parathyroid lesions (12 PA and 4 parathyroid hyperplasia) were randomly selected for the FISH analysis of the circRNAs.
This study was approved by the Institutional Ethics Review Board of Peking Union Medical College Hospital (China) (approval number: S-K470). Written informed consent was obtained from all participants in this study. All individuals’ data have been deidentified.
RNA isolation and quality control
Total RNA was extracted from the samples and purified using a mirVana™ miRNA Isolation Kit (Cat# AM1561, Ambion, Austin, TX, US) following the manufacturer’s instructions. The concentration of RNA was determined using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, US). The RNA integrity was detected by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, US).
Microarray and data analysis
The probes on the microarray were designed according to the databases from circBase (http://www.circbase.org/). In general, total RNA was amplified and labelled with a Low Input Quick Amp WT Labelling Kit (Cat. #5190–2305, Agilent Technologies) following the manufacturer’s protocol. Labelled cRNA was purified by the RNeasy Mini Kit (Cat.# 74106, QIAGEN, GmBH, Germany). Each slide was hybridized with 1.65 μg Cy3-labelled cRNA using a Gene Expression Hybridization Kit (Cat. #5188–5242, Agilent technologies, Santa Clara, CA, US) according to the manufacturer’s instructions. After 17 hours of hybridization, the slides were washed in staining dishes (Cat.# 121, Thermo Shandon, Waltham, MA, US) with a Gene Expression Wash Buffer Kit (Cat.# 5188–5327, Agilent Technologies, Santa Clara, CA, US) according to the manufacturer’s instructions. The processed slides were finally scanned by an Agilent Microarray Scanner (Cat. #G2565CA, Agilent Technologies, Santa Clara, CA, US) with the default settings (dye channel: green, scan resolution = 3 μm, PMT 100%, 20 bit). The data were extracted with Feature Extraction software 10.7 (Agilent Technologies, Santa Clara, CA, US). Raw data were normalized by the Quantile algorithm/limma package in R. The microarray analysis was conducted by Shanghai Biotechnology Corporation (Shanghai, China). Differentially expressed circRNAs were identified using Student’s t-test, with expression fold changes ≥ 2.0 or ≤ 0.5 and p-values < 0.05. A volcano plot and hierarchical clustering were used to show the different circRNA expression patterns in the PA and PC samples.
Construction of the circRNA-mRNA co-expression network
To explore the potential functions of the circRNAs, we constructed a circRNA-mRNA co-expression network based on the normalized mRNA signal intensities of samples from our previous study [31]. Co-expression networks were constructed to identify the interactions between the differentially expressed circRNAs and mRNAs in the PC and PA samples (fold changes ≥ 2.0 or ≤ 0.5 and p-values < 0.05). For each pair of analysed RNAs, we calculated the Pearson correlation and chose pairs with significant correlations to construct the network [42]. If the expression levels of RNA pairs were similar and above a preselected threshold in the Pearson analysis, the pairs were considered to exhibit a co-expression relationship and could be connected. The networks of both the PC group and PA group were constructed using Cytoscape (http://www.cytoscape.org). Each circRNA or mRNA corresponded to a node, and 2 RNAs were connected by a string, indicating a close correlation. The degree of the correlation reflected the RNA’s importance in the network. Then, the degree value of each mRNA or circRNA was normalized by dividing by the largest degree value in each network. An absolute network difference degree value was obtained for each mRNA or circRNA by comparing the normalized degree value between the PC and PA networks.
Validation with RT-qPCR
RT-qPCR was carried out to further confirm and extend the results from microarray to a larger disease cohort, which included 20 PC samples and 41 PA samples. The convergent primers for the circRNAs were designed, and GAPDH was used as an internal reference. The mRNA expression of four corresponding host genes, i.e., MYC, FSCN1, AKR1C3 and ANXA2, was also included in the validation. The primers were designed with Primer Express 3.0.1 (Supplementary Table 2). Primer location and annotated result of the circRNAs was visualized using CircPrimer 1.2 (Supplementary Fig. 4) [43].
RT-qPCR was conducted in triplicate on a 7900 HT Sequence Detection System (ABI, USA) with SYBR® Green Master Mix (ABI, 4368708). Briefly, the circRNA or mRNA from each sample was reverse transcribed with a ReverTra Ace qPCR Kit (TOYOBO, FSQ-101). Then, 20 μL of qPCR mixture containing 2× SYBR Green PCR buffer, forward primer, reverse primer, synthesized cDNA template and H2O was incubated in a 384-well plate. The PCR cycling programme was 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The cycle threshold (Ct) represented the quantitative endpoint. The relative expression level of each circRNA or mRNA was calculated using the 2−ΔCt formula, where ΔCt = Ct (RNA) – Ct (GAPDH).
FISH of RNA
FISH of the circRNAs was performed in FFPE tissues of malignant and benign parathyroid neoplasms. The digoxigenin (DIG)-labelled primers used for the circRNAs were as follows: hsa_circ_0035563, 5ʹ-DIG-GGGGTGTAGAGTGCTTTGGTCTTGATGGC-3ʹ; and hsa_circ_0075005, 5ʹ-DIG-CAGCCCCTAAACTGACTTTCTTCACTGGCG-3ʹ. The probe (8 ng/µL) was incubated at 37°C overnight. After removing the hybrids by washing and blocking with BSA, the probe was visualized with DyLight 488-conjugated IgG Fraction Monoclonal Mouse Anti-Digoxin Antibody (Jackson). Then, DAPI (Servicebio) was used to stain the cell nuclei (blue). A fluorescence microscope with an imaging system (NIKON ECLIPSE CI and NIKON DS-U3) was used to record the image of FISH. An assessment of the FISH signals was performed independently by two doctors who were blinded to the clinical features of the slides.
Statistical analysis
Continuous variables are described as the mean ± standard deviation (SD), and discrete data are reported as numbers or corresponding percentages. The differences in the expression of the circRNAs and mRNAs between the groups were evaluated by the Mann-Whitney U test or t-test. Categorical data were compared using Fisher exact tests. Binary logistic regression analysis was used for the multivariate analysis. Spearman’s rank test was conducted to assess the correlation between the circRNAs and clinicopathologic features of the patients. ROC curves were constructed, and the AUC was calculated to evaluate the diagnostic value of the selected RNAs. A two-sided p-value < 0.05 was considered statistically significant. GraphPad Prism 5 (GraphPad Software, CA, USA) and SPSS version 16.0 for Windows were used for all statistical analyses.
Conclusions
In summary, this study is the first to reveal the expression profile of circRNAs in PC. A set of circRNAs potentially involved in the tumourigenesis of parathyroid tumours was identified. hsa_circ_0075005 and MYC mRNA may be used for the differential diagnosis of PC. A high expression level of hsa_circ_0035563 was found to be related to CDC73 mutations and recurrence in patients with PC.
Funding Statement
This work was supported by the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine (CAMS-I2M) (2017-I2M-1-001), the Peking Union Medical College Innovative Team Development Program, and the 2016 Peking Union Medical College Hospital Science Foundation for Junior Faculty (pumch-2016.2.7).
Acknowledgments
None.
Clinical trial registration
None
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The datasets generated and/or analysed in the present study are available from the corresponding authors upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Wang O, Wang C, Nie M, et al. Novel HRPT2/CDC73 gene mutations and loss of expression of parafibromin in Chinese patients with clinically sporadic parathyroid carcinomas. PLoS One. 2012;7(9):e45567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Asare EA, Sturgeon C, Winchester DJ, et al. Parathyroid carcinoma: an update on treatment outcomes and prognostic factors from the National Cancer Data Base (NCDB). Ann Surg Oncol. 2015;22(12):3990–3995. [DOI] [PubMed] [Google Scholar]
- [3].Salcuni AS, Cetani F, Guarnieri V, et al. Parathyroid carcinoma. Best Pract Res Clin Endocrinol Metab. 2018;32(6):877–889. [DOI] [PubMed] [Google Scholar]
- [4].Verdelli C, Corbetta S.. Epigenetic alterations in parathyroid cancers. Int J Mol Sci. 2017;18(2):310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cetani F, Pardi E, Marcocci C. Update on parathyroid carcinoma. J Endocrinol Invest. 2016;39(6):595–606. [DOI] [PubMed] [Google Scholar]
- [6].Cetani F, Banti C, Pardi E, et al. CDC73 mutational status and loss of parafibromin in the outcome of parathyroid cancer. Endocr Connect. 2013;2(4):186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guarnieri V, Battista C, Muscarella LA, et al. CDC73 mutations and parafibromin immunohistochemistry in parathyroid tumors: clinical correlations in a single-centre patient cohort. Cell Oncol (Dordr). 2012;35(6):411–422. [DOI] [PubMed] [Google Scholar]
- [8].Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Danan M, Schwartz S, Edelheit S, et al. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40(7):3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- [11].Xia S, Feng J, Chen K, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bi W, Huang J, Nie C, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys. 2019;661:196–202. [DOI] [PubMed] [Google Scholar]
- [15].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- [16].Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859(10):1245–1251. [DOI] [PubMed] [Google Scholar]
- [18].Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lai Z, Yang Y, Yan Y, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16(23):2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang X, Yuan W, Tao J, et al. Identification of circular RNA signature in bladder cancer. J Cancer. 2017;8(17):3456–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [24].Huang M, He YR, Liang LC, et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ouyang Q, Huang Q, Jiang Z, et al. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 2018;101:531–538. [DOI] [PubMed] [Google Scholar]
- [26].Memczak S, Papavasileiou P, Peters O, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96(1):85–96. [DOI] [PubMed] [Google Scholar]
- [28].Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Silva-Figueroa AM, Perrier ND. Epigenetic processes in sporadic parathyroid neoplasms. Mol Cell Endocrinol. 2018;469:54–59. [DOI] [PubMed] [Google Scholar]
- [30].Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365. [DOI] [PubMed] [Google Scholar]
- [31].Zhang X, Hu Y, Wang M, et al. Profiling analysis of long non-coding RNA and mRNA in parathyroid carcinoma. Endocr Relat Cancer. 2018;163–176. DOI: 10.1530/ERC-18-0480 [DOI] [PubMed] [Google Scholar]
- [32].Christensen MV, Hogdall CK, Jochumsen KM, et al. Annexin A2 and cancer: a systematic review. Int J Oncol. 2018;52(1):5–18. [DOI] [PubMed] [Google Scholar]
- [33].Liu X, Ma D, Jing X, et al. Overexpression of ANXA2 predicts adverse outcomes of patients with malignant tumors: a systematic review and meta-analysis. Med Oncol. 2015;32(1):392. [DOI] [PubMed] [Google Scholar]
- [34].Box JK, Paquet N, Adams MN, et al. Nucleophosmin: from structure and function to disease development. BMC Mol Biol. 2016;17(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lin CY, Lovén J, Rahl PB, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6):a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Westin G. Molecular genetics and epigenetics of nonfamilial (sporadic) parathyroid tumours. J Intern Med. 2016;280(6):551–558. [DOI] [PubMed] [Google Scholar]
- [38].Bjorklund P, Akerstrom G, Westin G. Accumulation of nonphosphorylated beta-catenin and c-myc in primary and uremic secondary hyperparathyroid tumors. J Clin Endocrinol Metab. 2007;92(1):338–344. [DOI] [PubMed] [Google Scholar]
- [39].Lin L, Zhang JH, Panicker LM, et al. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc Natl Acad Sci U S A. 2008;105(45):17420–17425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lloyd RV, Osamura RY, Klöppel G, et al. WHO classification of tumours of endocrine organs. 4th ed. Lyon: IARC; 2017. [Google Scholar]
- [41].Hu Y, Zhang X, Cui M, et al. Verification of candidate microRNA markers for parathyroid carcinoma. Endocrine. 2018;60(2):246–254. [DOI] [PubMed] [Google Scholar]
- [42].Prieto C, Risueno A, Fontanillo C, et al. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3(12):e3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhong S, Wang J, Zhang Q, et al. CircPrimer: a software for annotating circRNAs and determining the specificity of circRNA primers. BMC Bioinformatics. 2018;19(1):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed in the present study are available from the corresponding authors upon reasonable request.


