Abstract
Background
The cells that form the arterial wall contribute to multiple vascular diseases. The extent of cellular heterogeneity within these populations has not been fully characterized. Recent advances in single cell RNA-sequencing makes it possible to identify and characterize cellular subpopulations.
Methods
We validate a method for generating a droplet-based single cell atlas of gene expression in a normal blood vessel. Enzymatic dissociation of four whole mouse aortas was followed by single cell sequencing of over 10,000 cells.
Results
Clustering analysis of gene expression from aortic cells identified 10 populations of cells representing each of the main arterial cell types—fibroblasts, vascular smooth muscle cells (VSMCs), endothelial cells (ECs), and immune cells including monocytes, macrophages, and lymphocytes. The most significant cellular heterogeneity was seen in the 3 distinct EC populations. Gene set enrichment analysis of these EC subpopulations identified a lymphatic EC cluster and two other populations more specialized in lipoprotein handling, angiogenesis, and extracellular matrix production. These subpopulations persist and exhibit similar changes in gene expression in response to a Western diet. Immunofluorescence for Vcam1 and Cd36 demonstrates regional heterogeneity in EC populations throughout the aorta.
Conclusions
We present a comprehensive single cell atlas of all cells in the aorta. By integrating expression from over 1,900 genes per cell we are better able to characterize cellular heterogeneity compared with conventional approaches. Gene expression signatures identify cell subpopulations with vascular disease-relevant functions.
Keywords: Vascular biology, endothelial cell, transcriptome, endothelial shear stress, aortic disease, single cell RNA-sequencing
Introduction
Blood vessels and the cells of the arterial wall contribute to risk for multiple diseases1,2. Vascular tissue is present in all organs of the body and is involved in the full spectrum of physiologic activities – from tissue development and remodeling to metabolism and inflammation3. Though the major arterial cell types are known, the molecular profile and heterogeneity of individual cells is poorly understood. Current characterization of cell types using a small number of marker genes does not capture the spectrum of functional states and gene expression programs in the vasculature.
Single cell RNA-sequencing technology now allows for the analysis of large numbers of individual cells from vascular tissue. Here we report the transcriptional profiling of individual cells from the mouse aorta. A cellular atlas of the entire aorta is possible with recent advances in droplet-based, massively-parallel single cell sequencing and computational analysis4,5. We identify 10 distinct clusters of cells, their gene signatures, and the relationship of vessel wall cell types to vascular disease-causing genes. This unbiased approach allows us to characterize multiple cells in a complex arterial tissue without the need for a priori sorting based on predefined markers.
Recent studies have used single-cell RNA-seq methods to survey leukocyte populations within the aorta6,7. These approaches separated CD45+ cells by flow cytometry, and using droplet-based single cell analysis identified the transcriptional profile for inflammatory cells in murine atherosclerosis. Non-flow sorting-based methods can improve single cell RNA-seq analysis of cellular heterogeneity by including all the cells of the blood vessel wall such as endothelial cells, vascular smooth muscle cells, or arterial fibroblasts. Recently, a single cell atlas of cell types in multiple mouse organs was published using unbiased, droplet-based RNA-sequencing8. However, a more specific profile of heterogeneity within vascular cell types and the implications of cellular subpopulations for vascular function has yet to be reported.
By surveying all the cells in the aorta we identify markers of cellular heterogeneity and distinct cellular subpopulations with disease relevant functions. We also examine the changes in these cellular subpopulations in response to a Western Diet. The recent advances in single cell RNA-sequencing allow for this type of large-scale analysis to comprehensively profile all arterial cell types.
Methods
Mice
Whole mouse aortas were harvested from 12-week old female C57/BL6 mice on either chow diet or 8 weeks of Western Diet (Research Diets). Four mice were included in each group, with 2 dissociated aortas from each condition sequenced at low-depth (17,000 reads/cell) and 2 samples sequenced at high-depth (145,000 reads/cell). The high-depth samples from the chow diet (n=2) and Western diet (n=2) were used for subsequent analyses. All mouse protocols were approved by the Broad Institute IACUC and all protocols were in accordance with institutional guidelines. The aorta was dissected from the root (distal to the aortic valve) to the femoral artery bifurcation. The isolated aorta included aortic arch, ascending, descending, thoracic, and abdominal portions. Perivascular fat was dissected from the vascular tissue prior to dissociation and single cell analysis.
Aortic dissociation
Preparation of a single cell suspension of aortic cells was performed using a previously described enzymatic digestion protocol9. Briefly, the isolated whole aorta was finely cut and incubated in 1X Aortic Dissociation Enzyme Solution (125 U/mL collagenase type XI, 60 U/mL hyaluronidase type 1-s, 60 U/mL DNase I, and 450 U/mL collagenase type I) for 1 hour at 37°C. The cell suspension was strained through a 30 μm filter, treated with ACK lysis buffer for 5 minutes at room temperature, and washed twice with PBS. The cells were resuspended in 0.4% BSA-PBS at a final concentration of 8×105 cells/mL. To determine if the dissociation protocol resulted in under-representation of certain cell types, a second aortic dissociation protocol10 using elastase (0.5mg/mL) and collagenase A (2mg/mL) for 30 minutes at 37ºC was analyzed by flow cytometry and droplet-based single cell RNA-seq. Both dissociation protocols had a similar yield of endothelial cells and identified the six major vascular cell types (Supplemental Figure 1).
Droplet-based scRNA-sequencing
Single cells were processed through the GemCode Single Cell Platform using GemCode Gel Bead, Chip and Library Kits (10X Genomics) as per the manufacturer’s protocol. In brief, single cells were sorted into 0.4% BSA–PBS solution. 9,000 cells were added to each channel. The cells were then partitioned into Gel Beads in emulsion in the GemCode instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina NextSeq 500.
Single-cell data analysis
Dimensional reduction, clustering, and analysis of single-cell RNA sequencing data were performed using the R package Seurat (Version 2.3.1)11. Cells with expression of fewer than 200 or more than 4000 genes and cells with greater than 25% expression of mitochondrial genes were filtered out of the analysis. Normalized expression values Ei,j for gene i in cell j were calculated by dividing unique molecular identifier (UMI) counts for gene i by the sum of the UMI counts in cell j, to normalize for differences in coverage, multiplying by 10,000 to create transcript per million (TPM)-like values, and finally computing log2(TPM + 1). Variable genes were identified using the Seurat FindVariableGenes method with the LogVMR dispersion function parameter; genes with log-normalized expression values between 0.125 and 4 and with a dispersion of at least 0.5 were considered variable. The Seurat ScaleData function was used to scale and center expression values in the dataset for dimensional reduction.
Principal component analysis (PCA) for dimensional reduction was performed using Seurat functions based on the variable genes previously identified. PCs 1–12 were selected for further study based on manual examination of the contribution of each PC to overall variability and based on the genes contributing to each PC. t-SNE was performed using Seurat functions based on PCs 1–12, and clustering to define cell identity was performed using the Seurat FindClusters function with resolution=0.5. Marker genes for each cluster were determined using the Wilcoxon rank-sum test via the FindAllMarkers function in Seurat.
Data Sharing
The raw counts table and the normalized expression table for the high-depth normal mouse aorta transcriptional profile are publicly available via the Broad Institute Single Cell Portal: https://portals.broadinstitute.org/single_cell/study/SCP289/single-cell-analysis-of-the-normal-mouse-aorta-reveals-functionally-distinct-endothelial-cell-populations.
Endothelial cluster gene set signatures
Gene Ontology (GO) biological process-associated genes were identified using the R package biomaRt12,13 and gene set scores for each set were defined as described previously. The FindAllMarkers Seurat function was utilized to find positive markers of each EC subpopulation using the Wilcoxon rank-sum test. Pathway enrichment analysis was performed using ReactomePA14 to identify gene sets from the Reactome database15 with false discovery rate (FDR)<0.05 enrichment in marker sets. Functional gene signatures for each endothelial cell subpopulation were defined as the intersection between the enriched Reactome pathway and the subpopulation markers. The Seurat function AddModuleScore was used to define a score for each of the gene signatures defined this way as previously described.
Partitioning cell type contribution to aortopathy-related gene expression
A list of all genes linked to Mendelian forms of inherited aortic dissection syndromes was compiled from the Online Mendelian Inheritance in Man (OMIM) database. The average expression of each aortopathy gene in each stromal cell type (EC, Fibroblast, and VSMC) was computed using the AverageExpression function in Seurat. Average expression values for each gene were normalized to determine proportion of stromal expression in each cell type for ternary plotting using the R package ggtern16.
Statistical analysis
Marker genes for transcriptional subpopulations in scRNA-seq profiles were identified using the FindAllMarkers Seurat function with a minimum log-fold change threshold of 0.25 and with p-values computed using a Wilcoxon rank-sum test. Pathway enrichment analysis was implemented using the enrichPathway ReactomePA function; p-values were computed using a hypergeometric test and adjusted for multiple hypothesis correction with a Benjamini–Hochberg procedure. Gene set scores and imaging characteristics were compared between cell populations using the Mann-Whitney U test.
Additional methods included in Supplemental Methods
Results
Single cell profile of the normal aorta
We transcriptionally profiled four wild-type C57/BL6 mouse aortas using droplet-based massively parallel scRNA-sequencing. Two aortas were sequenced at low-sequencing depth (17,000 reads/cell), and two aortas were sequenced at high-sequencing depth (145,000 reads/cell). There was no difference in the number of clustered cell populations based on sequencing depth. The high-sequencing depth data was used for all subsequent analyses, and yielded approximately 6,200 cells and 1,900 genes/ cell (Figure 1). Cells were enzymatically dissociated over 1 hour using either collagenase/hyaluronidase or collagenase/elastase protocols, with no difference in the number of isolated cell types (Supplemental Figure 1). Single cells were then individually bar-coded and sequenced using massively parallel droplet-based sequencing (Figure 1A). The individual samples were independently analyzed to confirm correlation between replicates, and then normalized and aggregated for joint analysis. The samples yielded a mean of 145,000 post-normalization reads per cell, which corresponds to a median of 1,900 genes per cell. We eliminated cells that expressed gene counts greater than 4,000 or less than 200 prior to analysis. The reproducibility between samples was validated by ensuring that all biological replicates were represented in all cell clusters. (Supplemental Figure 2).
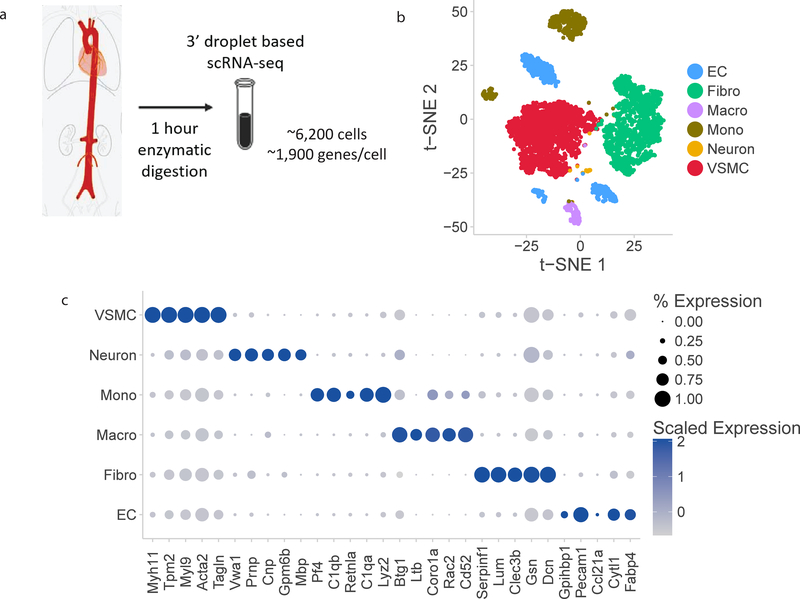
Figure 1:
Single cell RNA-sequencing atlas of aortic cells types. a) Schematic overview of experimental approach. Four mouse aortas were dissected from aortic root to femoral take-off, enzymatically dissociated for 1 hour at 37ºC, and then the single cell suspension was sequenced at a depth of 1,900 median genes per cell using droplet-based RNA-sequencing methods. b) t-SNE representation of single cell gene expression shows the 7 identified major aortic cell types. c) Dotplot demonstrates the top markers of each aortic cell type. Dot size corresponds to proportion of cells within the group expressing each transcript and dot color corresponds to expression level. d) Heatmap identifying all genes with log fold change>2 for each aortic cell type relative to all other cells. EC indicates endothelial cells; Fibro, fibroblasts; Macro, macrophages; Mono, monocytes; VSMC, vascular smooth muscle cells.
Unsupervised graph clustering partitioned the cells into groups, which we visualized using t-distributed stochastic neighbor embedding (t-SNE, Figure 1B). Individual clusters were labelled for cell type using known marker genes (Supplemental Table 1). The full set of transcriptional data was used to generate the clusters. Here we present clustering analysis using 1,100 variable genes per cell to identify cells with similar profiles. Vascular cell types represented in the distinct clusters included ECs, VSMCs, arterial fibroblasts, and immune cells, as well as a small neuronal cluster likely arising from adjacent tissue. A small population of red blood cells (RBCs) was also present following whole-aorta preparation; these cells were excluded from further analysis.
scRNA-sequencing identifies novel cell type-specific gene expression markers
The full set of differentially expressed markers for each cell population were identified from the RNA-sequencing data. Cell type specific markers for a cluster were defined as the five genes with the highest differential expression relative to all other cells (Figure 1C). For example, the top 5 VSMC specific markers are Myh11, Tpm2, Myl9, Tagln and Acta2. These genes include canonical VSMC genes, and therefore confirm the assignment of the cell clusters as VSMCs. We also identified all genes with log fold enrichment greater than 2 for each cell type relative to all other cells (Figure 1D). The heatmap of the gene expression shows the number of genes with significantly different expression (p-value<0.01 by Wilcoxon rank-sum test and log fold enrichment > 2) for each cell type. Monocytes have the largest number of unique transcripts as defined by this specificity standard compared with other cell clusters.
With expression data for ~1,900 genes per cell, we were able to identify different categories of cell type-specific markers. The first set of markers are widely expressed in all cells of that type, and not expressed in other clusters. In Figure 1C the size of each dot corresponds to the percentage of cells within a cluster expressing the gene, and the color represents the level of expression. Several markers are expressed in over 75% of the cells and with an average normalized expression of >0.75. Examples of these markers are Myh11 (VSMCs), C1qa (Monocytes), and Pecam1 (ECs). These markers represent the most specific and sensitive markers for the entire population of the given cell type. This is in contrast to markers that are highly expressed in a subset of a cell type. These genes remain specific markers, as they are not expressed in other cell types, but are not sensitive to capture all the cells of a pre-specified group. Examples include Gpihbp1 (ECs) and Cd52/Rac2 (monocytes). A third marker type we identified were genes with high expression in one cell type, but also significant expression in other cell clusters. While these genes are not classically considered cell type specific markers, they did contribute to identification of cell identity in our clustering algorithm. Using all genes to cluster the cells allows for higher resolution detection of cell subpopulations and cellular heterogeneity compared with using only a small number of canonical cell-type specific markers.
Whole transcriptomic data identifies cellular subpopulations within aortic cell types
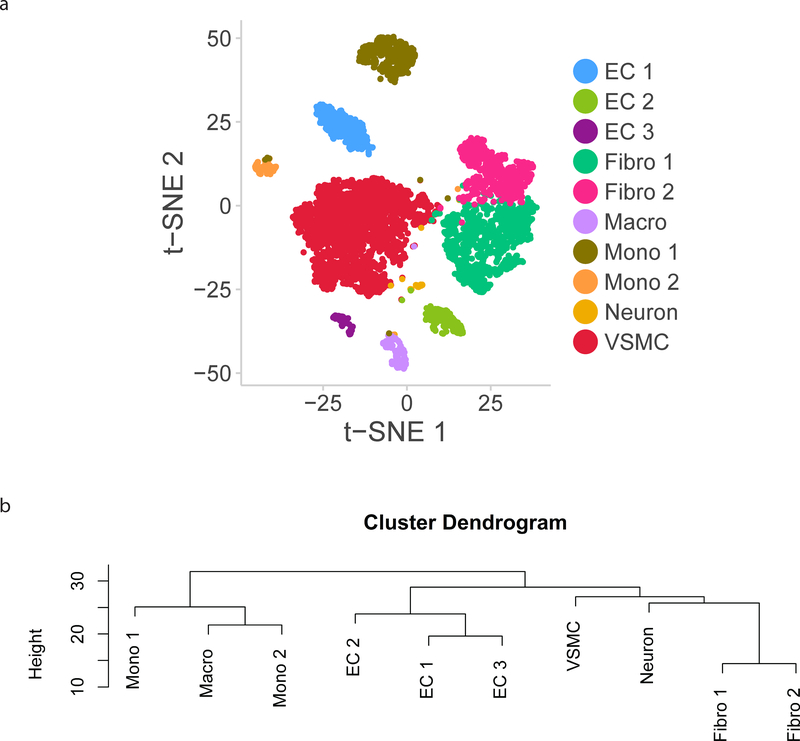
Following the analysis of cell types, we examined the 10 individual groups defined by clustering to define subpopulations within each aortic cell type (Figure 2A). Fibroblasts, monocytes/macrophages, and ECs all clustered into multiple subpopulations which were more similar to each other than to other aortic cell types as described by a cluster dendrogram (Figure 2B).
Figure 2:
Subpopulations within cell types identified from single cell RNA-sequencing. a) t-SNE demonstrating 12 clusters identified as comprising the aorta. b) Dendrogram summarizing similarity between aortic cell subpopulations. c) Heatmap identifying markers of each cellular subpopulation. EC indicates endothelial cells; Fibro, fibroblasts; Macro, macrophages; Mono, monocytes; VSMC, vascular smooth muscle cells.
VSMCs comprise the largest population of cells in our analysis, accounting for 39% of all cells. VSMCs clustered into one subpopulation which expresses the canonical VSMC markers Myh11 and Cnn1.
The second largest population of cells were fibroblasts, which account for 33% of all cells. These cells are defined by higher expression of Pdgfra and collagens/collagen-binding proteins (Col1a1, Col1a2, Dcn, Lum) along with reduced expression of VSMC-associated contractile proteins (Myh11, Cnn1) and were split into 2 subgroups by clustering. One canonical marker of VSMC identity, alpha smooth muscle actin (Acta2), was highly expressed in VSMCs, but also present in a large proportion of fibroblasts, though at lower levels (Supplemental Figure 3).
Endothelial cells cluster into three distinct groups, defined by their common expression of canonical marker Cdh5. Certain canonical EC-specific genes such as Von Willebrand Factor (Vwf) and VEGF-receptors (Flt1, Kdr) showed heterogeneous expression by cluster, and therefore do not serve as effective markers for identifying all ECs in the aorta.
Immune cells clustered into 3 broadly defined groups – macrophages (H2-Ab1) and two subpopulations of monocytes (both expressing Lyz2). There is overlap in gene expression in the monocyte and macrophage/DC clusters, and multiple markers were necessary to discriminate the two clusters (Supplemental Table 2). Finally, a small population of cells expressed markers of neuronal or nerve-associated identity including Mbp and Cnp. These likely represent a small amount of contamination from neighboring neuronal tissue during dissection of the aorta.
In order to determine whether separate clusters identified within each cell type represent discrete subpopulations or a continuous phenotypic gradient, the markers for each of the 10 cellular subpopulations were plotted via heatmap (Figure 2C). Each of the three EC clusters express distinct and non-overlapping markers, suggesting the presence of discrete subpopulations. Conversely, the two fibroblast clusters expressed overlapping markers, suggesting the presence of a continuous phenotypic gradient rather than true subpopulations.
Previous work on small vessels in the brain17 has suggested that immediate early genes (IEG) including Fos, Fosb, Jun, and Junb may be enriched in cells isolated from solid organs or vascular tissue for single-cell profiling. This IEG signature likely represents an artifact of single cell dissociation, as there is no evidence these genes are expressed in situ. The IEG score for each of the cellular subpopulations shows similar levels of expression in each cluster (Supplemental Figure 4), which suggests that IEG activation is not an explanation for the cellular heterogeneity in ECs and fibroblasts.
In order to explore the minimum number of sequencing reads per cell to achieve effective partitioning of aortic cell types using single-cell RNA-sequencing, we also performed clustering and identification of cell types in a single cell RNA-seq library of much lower depth (~17,000 reads/cell). The clustering of cells was essentially identical to data obtained from higher depth RNA sequencing. The same major vascular wall cell types and immune populations were identified with lower depth RNA sequencing (Supplemental Figure 5). The clustering of ECs into three discrete subpopulations is present at low sequencing depth (~17,000 reads/cell) and high sequencing depth (~145,000 reads/cell).
Three endothelial cell subpopulations have distinct gene expression profiles
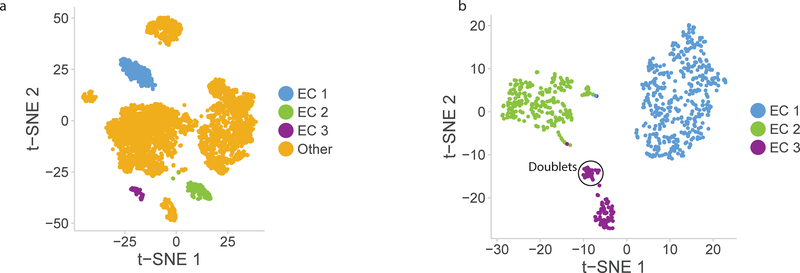
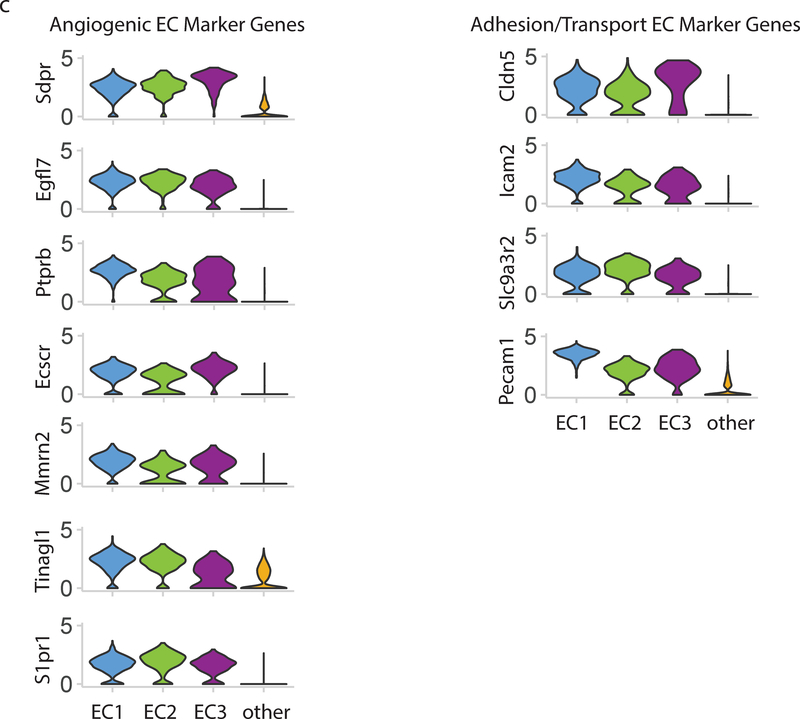
Clustering analysis of all cells in the wild-type mouse aorta identified three distinct subpopulations of ECs as determined using the canonical marker Cdh5, which has previously been identified as one of the few markers constitutively expressed throughout the vascular tree3 (Figure 3A–B). Analysis of these clusters relative to other vascular cell populations using a Wilcoxon rank sum test reveals novel transcriptional markers of all aortic ECs (Figure 3C). These include known endothelial-specific participants in angiogenesis and blood vessel formation including Sdpr18, Egfl7 19, Ptprb 20, and Ecscr21 as well as several adhesion and transport molecules that have previously been reported in ECs and other cell types including Cldn522, Icam2 23, and Slc9a3r224.
Figure 3:
Endothelial cells cluster into 3 distinct populations. a) t-SNE plot of EC subpopulations highlighted relative to other aortic cell types. The three EC subpopulations have distinct transcriptional profiles compared with each other and with all other aortic cells. b) t-SNE plot of all VE-Cadherin positive cells in mouse aorta. EC subpopulations were extracted from whole aorta data, and separately analyzed. In this t-SNE plot the 3 subpopulations are re-identified along with a small (5%) population of doublet cells which are an artifact of droplet-based single cell RNA-sequencing. c) Multiple genes show EC-specific expression and are expressed in all 3 EC subpopulations. Genetic markers expressed in all EC clusters compared with all other cells are divided into angiogenic EC marker genes and adhesion/transport EC marker genes. Violin plot y-axis demonstrates normalized transcript expression values.
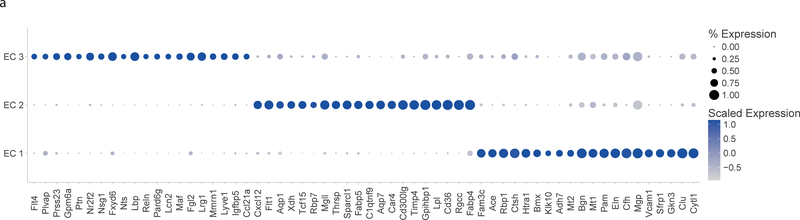
In order to characterize these subpopulations, we identified the transcriptional markers that differentiate each cluster. Unique markers were defined by highest differential expression using the Wilcoxon rank sum test with Bonferroni correction (Figure 4A). The largest population (EC 1) is defined by higher expression of many “canonical” EC markers (Vcam1) as well as other genes with known functions in ECs such as Clu, Gkn3, and Eln. The second EC population (EC 2) expresses genes involved in lipid transport (Cd36, Fabp4, Lpl, and Gpihbp1) and angiogenesis markers (Flt1). The EC 3 population expresses markers characteristic of lymphatic endothelium, including Lyve125. In order to elucidate the distinction between the two distinct subpopulations of blood endothelial cells, the lymphatic ECs in EC 3 were filtered out from downstream analysis.
Figure 4:
Gene expression signatures of EC subpopulations. a) Top 20 genes with specific expression for each EC subpopulation. Dot plot shows the percentage of cells expressing each gene (dot size) and the expression level (dot color). b) Differential expression of transcription factors by EC subpopulation. c) Expression of PPAR-G targets in EC 1 and EC 2 subpopulations. *** indicates Mann-Whitney U-test p-value < 0.001.
Each of the subpopulations is characterized by a unique transcription factor profile (Figure 4B). In order to characterize the contribution of these transcription factor-based signaling networks to subpopulation identity, the known targets activated by each transcription factor were identified using the TRRUST database26. These targets were used to define a transcription factor network signature that was computed for all endothelial cells and compared between the two major subpopulations EC 1 and EC 2. EC 2 showed significantly greater expression of Pparg-activated genes (Figure 4C, p-value < 2.2 × 10−16 by Mann-Whitney U test), consistent with the greater expression of Pparg and other lipid-handling genes in that subpopulation. The differential expression of multiple transcription factors and their targets in EC subpopulations suggests that they have broadly different functional properties.
Gene set enrichment profiles identify distinct functions for the EC subpopulations
To systematically identify cellular functions that differ between EC subpopulations, we investigated the expression profiles enriched for each cell subpopulation. In order to confirm the functional identity of identified EC populations, two endothelial cell-relevant biological process gene sets for EC development and EC differentiation were located in the Gene Ontology (GO) database. These annotated gene sets were used to create EC gene set scores (Supplemental Table 3). Generating expression scores for all cells in the dataset demonstrated that these well-characterized EC signatures did differentiate all the endothelial subpopulations from other aortic cells, but were not sufficient to resolve the differences between the subpopulations (Figure 5A).
Figure 5:
Pathway analysis of EC subpopulations. a) GO-annotated EC development and differentiation gene set scores in endothelial subpopulations and non-endothelial aortic cells (Other). b) Functional gene sets identified from Reactome pathway enrichment and subpopulation markers differentiate the two major EC populations EC 1 and EC 2. *** indicates Mann-Whitney U-test p-value < 0.001. c) Angiogenic tip cell gene set score differentiates EC 1 and EC 2. *** indicates Mann-Whitney U-test p-value < 0.001.
In order to identify the EC functions that showed the greatest heterogeneity between the major subpopulations EC 1 and EC 2, pathway enrichment was performed on the top 50 markers of each subpopulation against the Reactome pathway database15 using the R package ReactomePA14 (Supplemental Figure 6). Extracellular matrix organization and integrin cell surface interaction pathways showed selective enrichment in the markers of EC 1, while the plasma lipoprotein assembly, remodeling, and clearance pathway demonstrated selective enrichment in EC 2. Functional pathway scores based on these signatures (Supplemental Table 4) were significantly different (p-value < 2.2 × 10−16 by Mann-Whitney U test) between the two EC subpopulations and provided a functional biological hypothesis for the observed separation in transcriptional space (Figure 5B).
Because the subpopulation markers specific to EC 2 included multiple angiogenic factors, we also investigated the ability of angiogenic gene sets to differentiate the two subpopulations. Angiogenic heterogeneity in ECs is often summarized in the distinction between “tip” and “stalk” cells27. We used a previously identified gene signature for tip cells28 (Supplemental Table 5) to generate a tip cell score for all observed endothelial cells in our data set. This score significantly differentiated EC 1 and EC 2, with increased expression in EC 2 (Figure 5C, p-value < 2.2 × 10−16 by Mann-Whitney U test).
Single-cell RNA-seq in aortas from Western diet-fed mice reveal conserved and diet-dependent markers of endothelial subpopulations
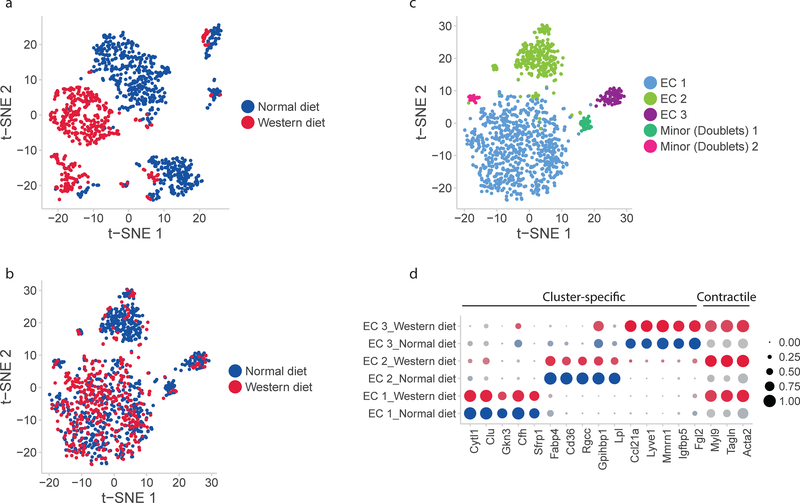
In order to determine the effect of diet on the endothelial subpopulation markers, we analyzed aortic scRNA-seq profiles from mice fed normal and Western diet. Differentiating conserved subpopulation markers from markers altered by diet requires two distinct methods for dataset alignment, as previously described5. PCA-based alignment (Figure 6A) highlights variation between the datasets. CCA-based alignment (Figure 6B) accentuates the similarities between the two datasets. A clustering algorithm applied to the datasets aligned for similarity (Figure 6C) showed three major subpopulations as previously identified in normal aortic data. These subpopulations are distinguished by the same marker profiles as previously determined. EC 1 specifically expresses Cytl1, Clu, and Gkn3; EC 2 specifically expresses lipid-handling genes including Fabp4, Gpihbp1, and Lpl; and EC 3 expresses markers of lymphatic identity including Lyve1. All EC subpopulations expressed the EC-specific markers differentiating them from other aortic cell types under both dietary conditions. Two minor populations identified via clustering consisted of far fewer cells than the major populations and expressed markers from multiple major clusters, possibly signifying the presence of cell doublets.
Figure 6:
Conserved and diet-dependent markers of EC subpopulations. a) Unaligned normal- and Western diet EC scRNAseq profiles represented in a PCA-based space with t-SNE. b) Aligned normal- and Western diet scRNAseq profiles represented with CCA and t-SNE. c) Conserved subpopulations of ECs identified in both datasets. d) Conserved diet-independent markers of EC subpopulation and pan-subpopulation markers of diet-induced change. Diet-independent subpopulation markers are the same as those identified previously while Western diet induces contractile proteins Myl9, Tagln, and Acta2 in all EC subpopulations.
This analysis also identified markers of the response to Western diet which are common to all EC subpopulations (Figure 6D). Markers of contractile function including myosin light-chain 9 (Myl9), transgelin (Tagln), and alpha smooth muscle actin (Acta2) were increased in all subgroups with exposure to high-fat diet. Comparison of the two scRNA-seq datasets using CCA-based alignment demonstrates that the presence of distinct EC subpopulations persists after exposure to Western Diet, and they share a common response feature of upregulation in contractile gene expression.
Identification of EC heterogeneity in situ with subpopulation-derived markers
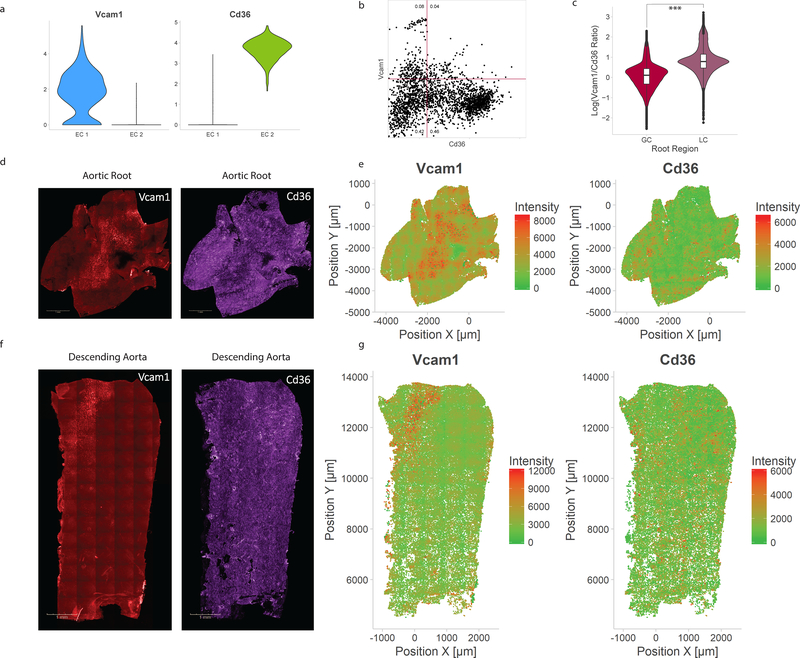
Representative markers for EC 1 and EC 2 were chosen to identify the spatial location of the EC subpopulations in situ. From the scRNA-seq data, Vcam1 and Cd36 are markers of EC 1 and EC 2, respectively (Figure 7A). These single markers exhibit heterogeneity within the subpopulations which were defined using the combined expression of 1,100 variable genes.
Figure 7:
Histological correlates of identified EC subpopulations. a) Violin plot comparison of Vcam1, an EC 1-associated marker, and Cd36, an EC 2-associated marker. b) Flow cytometry of mouse aortic endothelial cells demonstrating heterogeneity corresponding to subpopulation markers, with both Vcam1+/Cd36− and Vcam1−/Cd36+ populations. Numbers indicate fraction of cells in each quadrant. c) Quantification of log-transformed Vcam1/Cd36 ratio in greater (GC) and lesser (LC) curvatures of aortic root. *** indicates Mann-Whitney U-test p-value < 0.001. d) Representative immunofluorescence microscopy images for Vcam1 and Cd36 in the aortic root. e) Quantification of Vcam1 fluorescence intensity and Cd36 fluorescence intensity in the aortic root. f) Representative immunofluorescence microscopy images for Vcam1 and Cd36 in the descending aorta. g) Quantification of Vcam1 fluorescence intensity and Cd36 fluorescence intensity in the descending aorta.
The presence of Vcam1+/Cd36− and Vcam1−/Cd36+ EC populations in situ was confirmed by flow cytometry (Figure 7B, Supplemental Figure 7). Immunofluorescence of the aortic root and descending thoracic aorta (Supplemental Figure 8) shows distinct regions of Vcam1 and Cd36 expression. The average of the log-transformed Vcam1/Cd36 ratio in three aortic root specimens was significantly higher in the lesser curvature compared with the greater curvature (Figure 7C, p <2.2 × 10−16 by Mann-Whitney U test). The ECs in the greater curvature of the aortic root demonstrate low Vcam1 expression and high Cd36 expression with an elongated morphology from flow alignment (Figure 7D–E, Supplemental Figure 9A). The ECs in lesser curvature of the aortic root show a cuboidal morphology along with high Vcam1 expression and lower Cd36 expression (Figure 7D–E, Supplemental Figure 9B). The proximal portion of the descending thoracic aorta also has a region of high Vcam1 and low Cd36 corresponding to the lesser curvature (Figure 7F–G). A defined boundary exists between high Cd36 and high Vcam1 regions in the transition zone between the lesser curvature and greater curvature in the aortic root (Supplemental Figure 10).
Expression of Mendelian aortopathy genes identifies relevant cells and biologic pathways
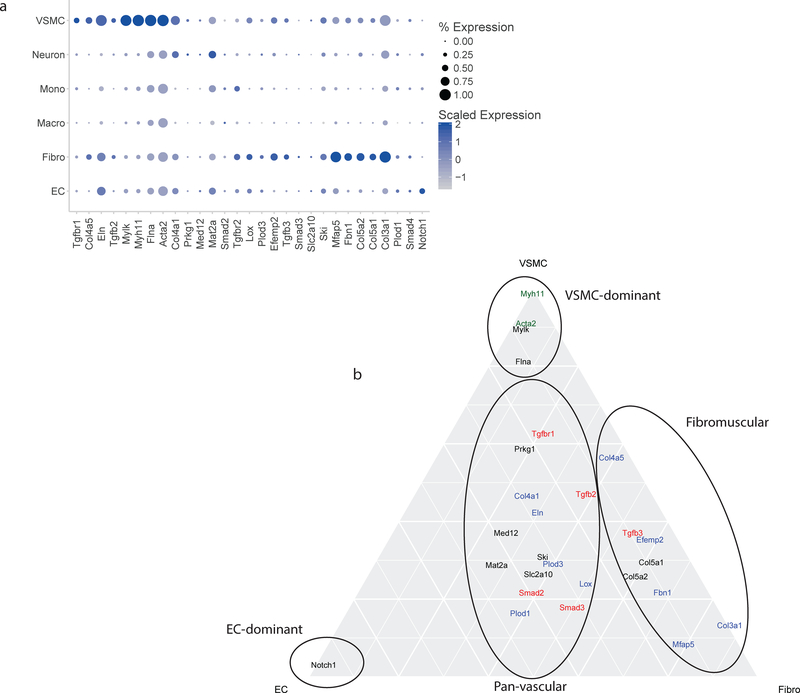
We next applied this aortic single cell gene expression data to determine the role different cell types play in the pathogenesis of aortic diseases. We determined the cell-specific expression of 30 genes with known coding mutations associated with disease29. The plurality of the genes show highest expression in VSMCs, though several display significant expression in other cell types (Figure 8A). As expected, genes known to affect muscle contraction (e.g. Myhlk, Myh11) are predominantly expressed in VSMCs. Genes with extracellular matrix functions are expressed most highly in fibroblasts (e.g. Col5a2, Col5a1, Col3a1). Several genes associated with thoracic aortic aneurysm and dissection (TAAD) syndromes are expressed in multiple cell types, though the major cell types are VSMCs, fibroblasts, and ECs. When cell type specific expression is limited to these 3 relevant cell types, the TAAD-causing mutations separate into distinct patterns of cellular expression (Figure 8B). First, four VSMC-specific genes—Myhlk, Myh11, Acta2, and Flna. Several ECM-related genes lie along the VSMC-Fibroblast axis, and have minimal EC expression. These genes include Col4a5, Col5a1, Col5a2, Fbn1, Mfap5, and Col3a1. Only Notch1 has exclusive expression in ECs. There are, however, several genes with significant expression in all three cell types. These include the TGFb pathway genes (Tgfb2, Smad2, Smad3, Smad4, Tgfbr1) and two ECM-related genes (Lox, Eln).
Figure 8:
Expression of aortopathy genes by cell identity. a) Expression of thirty genes associated with Mendelian aortopathy by cell type. The majority of these genes have highest expression in VSMCs. Several show highest expression in fibroblasts. Two genes (Mat2a and Notch1) have highest expression in ECs. b) Ternary plot to show expression of aortopathy genes in ECs, VSMCs, and fibroblasts. Several genes are exclusively expressed in VSMCs, while the majority show expression predominantly in VSMCs and fibroblasts. Genes with contractile function are in green, TFGb response genes are in red, ECM-related genes are in blue, and unassigned genes are in black. EC indicates endothelial cells; Fibro, fibroblasts; VSMC, vascular smooth muscle cells.
Discussion
Vascular tissue is composed of many multifunctional cell populations, each of which contributes to cardiovascular disease. Heterogeneity in gene expression differentiates major cell types and defines multiple subpopulations with distinct functions. Advances in single cell RNA-sequencing provide the resolution to identify these heterogeneous subpopulations and to profile the pan-cellular landscape of the vasculature. Here we use the latest droplet-based single cell RNA-sequencing methods to characterize the genes and functional pathways expressed in 6,200 cells from the whole murine aorta. We identify 10 clusters of cells, which form the main cell types in this large artery—fibroblasts, VSMCs, ECs, and immune cell populations. Our approach also identified three distinct subpopulations of ECs within the murine aorta. The gene expression profiles for these EC subpopulations reveals their distinct functional capabilities. Immunofluorescence confirms the presence of these subpopulations, with different relative abundance of the two major EC populations in the greater and lesser curvatures of the aortic root. To demonstrate a clinical application of these cell-type specific transcriptional profiles, we partition the contribution of different cell types to the genes associated with Mendelian aortopathy syndromes.
Our approach to transcriptional profiling of the mouse aorta is unique for its incorporation of all vascular cell types into a single gene expression dataset. Recent papers have identified markers of cellular heterogeneity in subsets of vascular cells, such as CD45+ leukocytes6,7 and neurovascular cells17. In these studies, subsets of cells were presorted using flow cytometry prior to single cell RNA-sequencing. In contrast, our study generated droplet-based single cell RNA-seq profiles for all aortic cells without presorting. Our approach is unbiased and utilizes the full transcriptome to cluster similar cells, allowing for analysis of populations previously lost to sorting and for computational identification of cluster markers without reliance on prior knowledge. The fact that many previously described “pan-EC” markers, such as Vwf, are only expressed in a subpopulation of ECs demonstrates the importance of this unbiased approach. The presence of EC heterogeneity in the form of 3 discrete subpopulations may have been missed if we had used a marker gene to sort for ECs before single cell RNA-sequencing. By sequencing all cells from the dissociated aorta we were able to determine the new pan-EC markers and transcripts specific to EC subpopulations.
To ensure that our methods provide an accurate representation of all aortic cell types, we compare two different dissociation protocols and sequence cells at high and low read number per cell. Comparison of the collagenase- and elastase-based enzymatic dissociation methods demonstrates that the overall cell population clustering achieved by these methods is similar with both approaches. Certain methods, however, may be better suited to particular cell types—such as elastase to improve VSMC yield. Comparison between high-depth and low-depth sequencing-based profiles demonstrates that identification of cell populations in vascular tissue may be achieved with relatively shallow sequencing—as low as 17,000 reads/cell in the case of our data.
The identification of three distinct EC subpopulations in the aorta contributes to the growing importance of EC heterogeneity in vascular function. EC heterogeneity is recognized in different organs, at different levels of the vascular tree, and more recently within a single vessel30–32. Previous work in defining EC heterogeneity has focused on variation in individual markers, such as vWF32. We demonstrate that single cell RNA-sequencing in the aorta can identify the full set of transcriptional profiles that are dynamically regulated in ECs. We identify 3 distinct profiles of ECs in the aorta, which share some markers, but differ in expression of multiple genes. These differences suggest functional specialization of EC subpopulations in either ECM production, lipid handling and angiogenesis, or lymphatic function.
The spatial distribution of the two major EC populations further demonstrates that transcriptional heterogeneity can identify functionally distinct subpopulations of cells. We observed unique distribution patterns for EC 1 (Vcam1) and EC 2 (Cd36) in the lesser and greater curvatures of the aorta. This pattern may be the result of differences in blood flow and shear stress in these regions of the aorta. Shear stress is a known driver of arterial EC heterogeneity, and has previously been shown to induce Vcam1 expression and reduce p65 expression in atherosclerosis-prone ECs33–35. With single cell RNA-sequencing we now identify the full transcriptional profile of these ECs. The high-dimensional transcriptional signatures we have generated enable identification of EC subpopulations in a way that cannot be accomplished with any individual histological marker. Using transcriptomics alone, we cannot say whether these subpopulations arise solely from environmental exposures such as shear stress, or are the result of developmentally distinct precursors. The phenotypic drift of cultured ECs suggests that the microenvironment is responsible for heterogeneous gene expression36. However, other studies suggest the epigenetic signature of EC subpopulations are fixed, and gene expression remains site-specific after multiple ex vivo passages37,38. Further studies are needed to determine the developmental origin and functional consequences of EC heterogeneity, and our comprehensive gene expression signatures of each subpopulation in the normal aorta will serve as a starting point for this analysis.
Additionally, our analysis suggests next steps for examining how these EC subpopulations respond to pathological stressors. The upregulation of contractile gene expression in ECs after exposure to Western diet is particularly interesting given that endothelial-mesenchymal transition (EndMT) is a known “final common pathway” In EC dysfunction. EndMT in atherosclerosis has previously been linked to an increase in smooth muscle cell markers39, including Acta2. Our data suggest that in the setting of a Western diet, this alteration is part of a set of increased contractile transcripts that can be detected in all three EC subpopulations.
Profiling the complete vascular milieu using single-cell RNA-seq also provides key insights for disease pathogenesis. By simultaneously analyzing the transcriptional signature of all vascular cell types, we partition the relative contribution of the three major vascular wall cell types to Mendelian aortopathies. Our analysis reveals that aortopathies derived from contraction-related genes may be mediated by VSMCs, while those arising from alterations in extracellular matrix metabolism are likely the consequence of VSMC and fibroblast biology with a minimal contribution from ECs. Aortopathies arising from alterations in TGF-beta signaling may have pathology arising from all three cell types. These findings can guide future efforts to understand genotype-phenotype correlations in genetic aortopathies and develop targeted therapies for these diseases.
Our demonstration that it is feasible to sequence and analyze whole transcriptome data on a single cell level for 6,200 cells representing all cell types in the aorta will be a resource for the extension of this method to other experiments in vascular biology. As the technology continues to improve, greater sequencing depth will enable a more refined pathway for genes with lower expression (i.e. enzymes, transcription factors). This will allow augmentation of our aortic transcriptional profile to better appreciate the transcriptional networks characterizing major cell types and cellular subpopulations.
Transcriptional profiling with droplet-based single cell RNA-sequencing is proving to be the standard by which to define cellular identity. We use this method to show that the integration of data from the 1,900 profiled genes per cell identifies novel cell subpopulations unidentified by previous antibody-based detection methods. Several recent papers have also demonstrated the higher resolution of transcriptional profiling in gut epithelium40, dendritic cells41, retina42, and the kidney43. With the international efforts of the Human Cell Atlas consortium, the methods for single cell transcriptomics are rapidly improving44. Our data from the murine aorta contributes to these efforts and will serve as a reference for full characterization of vascular cells in healthy and diseased tissue.
Supplementary Material
Clinical Perspective.
1. What is new?
A single-cell transcriptional profile of the aorta identifies 3 distinct endothelial cell subpopulations with differences driven by major functional gene programs including adhesion and lipid handling.
Comparison of aortic single-cell RNA-seq datasets from normal and Western diet-fed mice suggests that these subpopulations exist under both dietary conditions and have some unified responses to diet alteration.
Immunofluorescence using single marker genes to identify endothelial cell subpopulations shows that the Vcam1+ population is spatially located in regions of disturbed blood flow like the lesser curvature of the aorta.
2. What are the clinical implications?
Characterizing functional subpopulations may serve as a novel method for understanding endothelial health in patients with vascular disease.
Although aortic endothelial cell subpopulations demonstrate some unified responses to vascular disease-relevant stimuli like a Western diet, functionally different subpopulations may contribute differentially to vascular diseases enabling subpopulation-targeted therapies.
Single-cell RNA-sequencing can help determine the relative contribution of different vascular cells (i.e. ECs, VSMCs, and fibroblasts) in the pathogenesis of Mendelian aortopathies.
Acknowledgements
The authors thank Yevgenia Tesmenitsky, Eric Engelbrect, Kevin Croce, Timothy Hla and Thomas Michel for technical assistance and helpful discussions.
Funding Sources
The single cell sequencing was funded through a research grant from Bayer to the Broad Institute. Addition funding from NIH NHLBI K08HL128810 (to R.M.G.); Donovan Family Foundation, R01 HL127564 (to S.K.); NIH NIGMS T32-GM008313, NIH NIGMS T32-GM007753, and the American Heart Association Predoctoral Fellowship (to A.S.K.); NIH NIGMS 5R01GM049039-23 (to E.R.E.); NIH 1R01HL092577, R01HL128914, K24HL105780 and Fondation Leducq (14CVD01) (to P.T.E).
Disclosures
A.S.K. has served as a consultant to and holds equity in nference, inc. S.K. has received a research grant from Bayer Healthcare; and consulting fees from Merck, Novartis, Sanofi, AstraZeneca, Alnylam Pharmaceuticals, Leerink Partners, Noble Insights, MedGenome, Aegerion Pharmaceuticals, Regeneron Pharmaceuticals, Quest Diagnostics, Genomics PLC, Third Rock Ventures, and Eli Lilly and Company; and holds equity in San Therapeutics, and Catabasis Pharmaceuticals. P.T.E. has served as a consultant to Bayer AG, Novartis and Quest Diagnostics.
References
- 1.Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, Pober JS, Wick TM, Konkle BA, Schwartz BS, Barnathan ES, McCrae KR, Hug BA, Schmidt AM, Stern DM. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. [DOI] [PubMed] [Google Scholar]
- 3.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotech. 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh H, Kobiyama K, Hamers A, Cochain C, Vafadarnejad E, Saliba A-E, Zernecke A, Pramod A, Ghosh A, Anto Michel N, Hoppe N, Hilgendorf I, Zirlik A, Hedrick C, Ley K, Wolf D. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ Res. 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochain C, Vafadarnejad E, Arampatzi P, Jaroslav P, Winkels H, Ley K, Wolf D, Saliba A-E, Zernecke A. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res. 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 8.Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;18:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butcher MJ, Herre M, Ley K, Galkina E. Flow cytometry analysis of immune cells within murine aortas. J Vis Exp. 2011;53:2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Li Q, Jiao Y, Qin L, Ali R, Zhou J, Ferruzzi J, Kim RW, Geirsson A, Dietz HC, Offermanns S, Humphrey JD, Tellides G. Tgfbr2 disruption in postnatal smooth muscle impairs aortic wall homeostasis. J Clin Invest. 2014;124:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. [DOI] [PubMed] [Google Scholar]
- 13.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protocol. 2009;4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G, He Q-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–479. [DOI] [PubMed] [Google Scholar]
- 15.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton N, Ferry M. ggtern: Ternary Diagrams Using ggplot2. Journal of Statistical Software, Code Snippets. 2018;87:1–17. [Google Scholar]
- 17.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, Sun Y, Raschperger E, Räsänen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. [DOI] [PubMed] [Google Scholar]
- 18.Boopathy GTK, Kulkarni M, Ho SY, Boey A, Chua EWM, Barathi VA, Carney TJ, Wang X, Hong W. Cavin-2 regulates the activity and stability of endothelial nitric-oxide synthase (eNOS) in angiogenesis. J Biologic Chem. 2017;292:17760–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong G, Kuek V, Shi J, Zhou L, Han X, He W, Tickner J, Qiu H, Wei Q, Xu J. EGFL7: Master regulator of cancer pathogenesis, angiogenesis and an emerging mediator of bone homeostasis. J Cell Physiol. 2018;35:6223. [DOI] [PubMed] [Google Scholar]
- 20.Mori M, Murata Y, Kotani T, Kusakari S, Ohnishi H, Saito Y, Okazawa H, Ishizuka T, Mori M, Matozaki T. Promotion of cell spreading and migration by vascular endothelial-protein tyrosine phosphatase (VE-PTP) in cooperation with integrins. J Cell Physiol. 2010;224:195–204. [DOI] [PubMed] [Google Scholar]
- 21.Xiang H, Ma J, Shen P, Wang Y, Huang H, Shi C. ECSM2, an endothelial specific VE-cadherin binding protein, has a tyrosine phosphorylation site essential to cell migration. Gene. 2018;662:131–138. [DOI] [PubMed] [Google Scholar]
- 22.Miettinen M, Sarlomo-Rikala M, Wang Z-F. Claudin-5 as an immunohistochemical marker for angiosarcoma and hemangioendotheliomas. Am J Surg Pathol. 2011;35:1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyck R, Enzmann G. The physiological roles of ICAM-1 and ICAM-2 in neutrophil migration into tissues. Curr Opin Hematol. 2015;22:53–59. [DOI] [PubMed] [Google Scholar]
- 24.Wallgard E, Larsson E, He L, Hellström M, Armulik A, Nisancioglu MH, Genove G, Lindahl P, Betsholtz C. Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler Thromb Vasc Biol. 2008;28:1469–1476. [DOI] [PubMed] [Google Scholar]
- 25.Shinoda K, Hirahara K, Iinuma T, Ichikawa T, Suzuki AS, Sugaya K, Tumes DJ, Yamamoto H, Hara T, Tani-Ichi S, Ikuta K, Okamoto Y, Nakayama T. Thy1+IL-7+ lymphatic endothelial cells in iBALT provide a survival niche for memory T-helper cells in allergic airway inflammation. Proc Natl Acad Sci USA. 2016;113:E2842–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han H, Cho J-W, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon H-N, Jung H, Nam S, Chung M, Kim J-H, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tung JJ, Tattersall IW, Kitajewski J. Tips, stalks, tubes: notch-mediated cell fate determination and mechanisms of tubulogenesis during angiogenesis. Cold Spring Harb Perspect Med. 2012;2:a006601–a006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siemerink MJ, Klaassen I, Vogels IMC, Griffioen AW, Van Noorden CJF, Schlingemann RO. CD34 marks angiogenic tip cells in human vascular endothelial cell cultures. Angiogenesis. 2012;15:151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isselbacher EM, Lino Cardenas CL, Lindsay ME. Hereditary Influence in Thoracic Aortic Aneurysm and Dissection. Circulation. 2016;133:2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augustin HG, Koh GY. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science. 2017;357:eaal2379. [DOI] [PubMed] [Google Scholar]
- 31.Potente M, Mäkinen T. Vascular heterogeneity and specialization in development and disease. Nat Rev Mol Cell Biol. 2017;18:477–494. [DOI] [PubMed] [Google Scholar]
- 32.Yuan L, Chan GC, Beeler D, Janes L, Spokes KC, Dharaneeswaran H, Mojiri A, Adams WJ, Sciuto T, Garcia-Cardeña G, Molema G, Kang PM, Jahroudi N, Marsden PA, Dvorak A, Regan ER, Aird WC. A role of stochastic phenotype switching in generating mosaic endothelial cell heterogeneity. Nat Commun. 2016;7:10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu J-J, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol Rev. 2011;91:327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iiyama K, Hajra L, Li H, DiChiara M, Medoff BD, Cybulsky MI. Patterns of vascular cell adhesion molecule-1 and intercellular adhesion molecule-1 expression in rabbit and mouse atherosclerotic lesions and at sites predisposed to lesion formation. Circ Res. 1999;85:199–207. [DOI] [PubMed] [Google Scholar]
- 35.Won D, Zhu S-N, Chen M, Teichert A-M, Fish JE, Matouk CC, Bonert M, Ojha M, Marsden PA, Cybulsky MI. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am J Pathol. 2007;171:1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burridge KA, Friedman MH. Environment and vascular bed origin influence differences in endothelial transcriptional profiles of coronary and iliac arteries. Am J Physiol Heart Circ Physiol. 2010;299:H837–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chi J-T, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, van de Rijn M, Botstein D, Brown PO. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci USA. 2003;100:10623–10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacorre D-A, Baekkevold ES, Garrido I, Brandtzaeg P, Haraldsen G, Amalric F, Girard J-P. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood. 2004;103:4164–4172. [DOI] [PubMed] [Google Scholar]
- 39.Chen P-Y, Qin L, Baeyens N, Li G, Afolabi T, Budatha M, Tellides G, Schwartz MA, Simons M. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest. 2015;125:4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sivakamasundari V, Bolisetty M, Sivajothi S, Bessonett S, Ruan D, Robson P. Comprehensive Cell Type Specific Transcriptomics of the Human Kidney. bioRxiv. 2017; 238063:1–36. Doi: 10.1101/238063. [DOI] [Google Scholar]
- 44.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, Clevers H, Deplancke B, Dunham I, Eberwine J, Eils R, Enard W, Farmer A, Fugger L, Göttgens B, Hacohen N, Haniffa M, Hemberg M, Kim S, Klenerman P, Kriegstein A, Lein E, Linnarsson S, Lundberg E, Lundeberg J, Majumder P, Marioni JC, Merad M, Mhlanga M, Nawijn M, Netea M, Nolan G, Pe’er D, Phillipakis A, Ponting CP, Quake S, Reik W, Rozenblatt-Rosen O, Sanes J, Satija R, Schumacher TN, Shalek A, Shapiro E, Sharma P, Shin JW, Stegle O, Stratton M, Stubbington MJT, Theis FJ, Uhlen M, van Oudenaarden A, Wagner A, Watt F, Weissman J, Wold B, Xavier R, Yosef N, Human Cell Atlas Meeting Participants. The Human Cell Atlas. Elife. 2017;6:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.