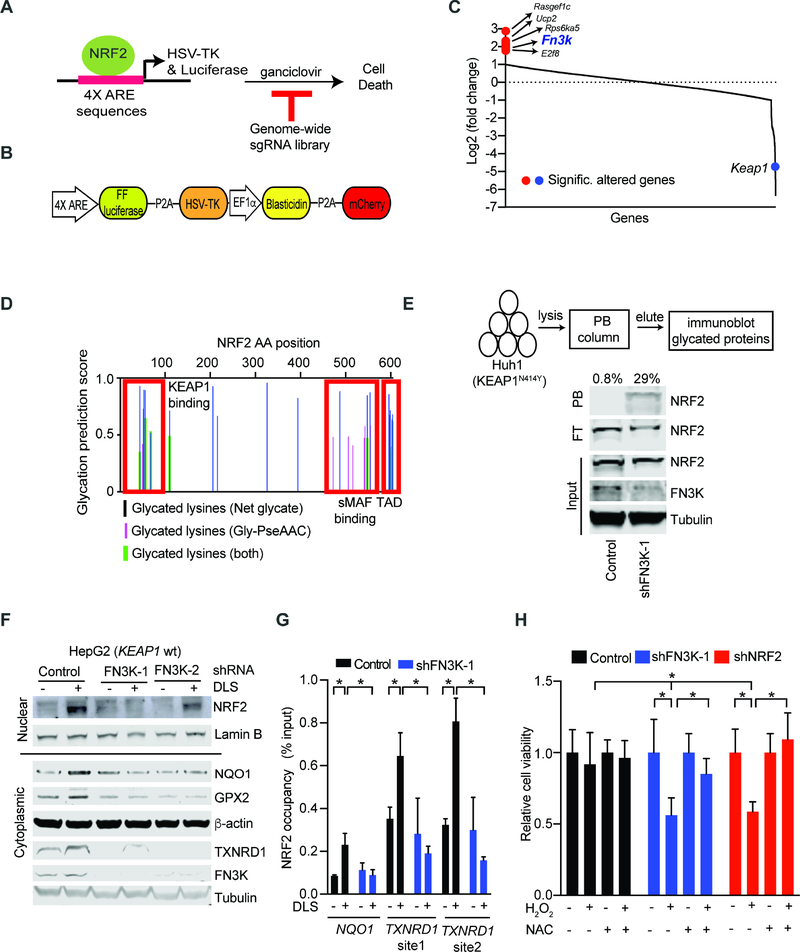
Figure 3: Genome-wide CRISPR screen identifies the de-glycating kinase FN3K as a requirement for NRF2 function.
A) Diagram of our strategy for a genome-wide screen against NRF2 driven expression of the HSV-TK suicide gene; B) Map of the lentiviral vector directing ARE-controlled HSV-TK and luciferase expression; C) Change in sgRNA library representation comparing untreated cells and cells treated with the NRF2 inducer tBHQ and ganciclovir; D) Predicted sites of NRF2 protein glycation using indicated algorithms; TAD: Transactivation domain; E) Phenyl borate affinity purification and immunoblotting reveals NRF2 glycation upon FN3K knockdown in KEAP1 mutant Huh1 cells; values on top refers to % of glycated NRF2 represented by the ratio of NRF2 signal intensity in PB-bound (PB) to the sum PB-bound and flow through (FT); F) Immunoblot for nuclear (upper panel) and cytoplasmic (lower panel) levels of the indicated proteins in KEAP1 wild type HepG2 cells transduced and treated as indicated; G) Chromatin immunoprecipitation (ChIP) on indicated HepG2 nuclear lysates with anti-NRF2 antibody followed by amplification of indicated promoters; shown as % of input DNA and error bar is SD of 4 replicates; H) Viability of HepG2 cells untreated or treated with H2O2 (400 μM, 24 hours) with and with without pre-incubation with NAC (10 mM, 3 hours); mean of 9 replicates ± SD. (* indicates p-value < 0.05 by two-tailed student t test). See also Figure S3 and Table S3.