Advanced detection technologies, such as next‐generation sequencing, make possible the identification of rare variants of EGFR. This case report describes a patient with lung adenocarcinoma harboring a rare EGFR‐RAD51 fusion, with results that might help establish personalized treatment approaches for patients with rare EGFR fusions.
Abstract
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (EGFR‐TKIs) have become the first choice for patients with sensitive mutations and have significantly improved prognosis. EGFR exon 19 deletions and L858R mutation in exon 21 are the most common sensitive mutations in lung adenocarcinoma. With advances in detection technology, some rare variants of EGFR have been detected, including EGFR kinase domain duplications and EGFR fusions. Only a few reports have revealed the effectiveness of EGFR‐TKIs in patients with these rare variants. In this study, we report a case of EGFR‐RAD51 fusion in lung adenocarcinoma that showed a response to icotinib; these findings provide additional support for the use of EGFR‐TKIs for patients with these atypical variants.
Key Points.
A young patient with lung adenocarcinoma harboring a rare EGFR‐RAD51 fusion who responded to icotinib with a PFS of longer than 15 months.
All reported EGFR‐RAD51 fusions have the same breakpoints and show responses to EGFR‐TKIs including icotinib, except for one patient who responded to chemotherapy.
Although EGFR fusion is a rare EGFR variant type, the efficacy of EGFR‐TKIs suggests the necessity for new detection technology, such as NGS, for patients with lung adenocarcinoma.
The clinical usage of NGS could maximize the benefits of precision medicine in patients with cancer.
The current case provides new evidence for the efficacy of icotinib in patients with the rare EGFR‐RAD51 fusion and EGFR‐activating mutations.
Introduction
Lung cancer is the leading cause of cancer‐related deaths. The main type of lung cancer is non‐small cell lung cancer (NSCLC), which includes two major histological subtypes, adenocarcinoma and squamous cell carcinoma [1]. Genetic analyses have revealed driver genes in lung adenocarcinoma and have changed the treatment paradigm [2]. Epidermal growth factor receptor (EGFR) mutations are commonly associated with adenocarcinoma. The frequency of EGFR mutations differs between Western and Asia‐Pacific regions (12% vs. 47%) [3], indicating that more patients with lung adenocarcinoma would benefit from EGFR‐tyrosine kinase inhibitors (TKIs) in Asia than in other regions. Exon 19 deletions and L858R in EGFR are the most common variants with sensitivity to EGFR‐TKIs [4]. Resistance mutations have also been identified, such as T790M and exon 20 insertions. Furthermore, rare genomic events can activate the kinase domain of EGFR, such as EGFR kinase domain duplications (EGFR‐KDD) and EGFR rearrangements [5], [6]. Advanced detection technologies, such as next‐generation sequencing (NGS), have facilitated the identification of rare variants. We present a case report of a patient with lung adenocarcinoma harboring a rare EGFR‐RAD51 fusion; treatment with icotinib resulted in a progression‐free survival (PFS) of longer than 15 months. These results might help establish personalized treatment approaches for patients with rare EGFR fusions.
Patient Story
In a 26‐year‐old male patient with no smoking history or symptoms, the right lower lung exhibited a patchy appearance with an area of about 2.0 × 2.3 × 3.2 cm during a routine chest x‐ray included in medical examination on October 24, 2016. An enhanced scan revealed that the density was not uniform, and the right oblique fissure and horizontal fissure exhibited pleural thickening. On November 25, 2016, the patient underwent thoracoscopic resection of the tumor in the right lower lobe and thoracoscopic electrocautery of pleural nodules. The postoperative pathology showed that the tumor in the right lower lobe was a 2 × 1.5 × 1.3 cm alveolar infiltrative adenocarcinoma involving the visceral pleura, invading the cartilage of the bronchial wall, invading the nerve and vessel wall, and without clear intravascular thrombosis. The chest wall nodules showed infiltrating adenocarcinoma components in fibers and adipose tissues. Surface nodules on the right middle lobe contained a few invasive adenocarcinoma components. The diagnosis was right lung adenocarcinoma (pT4NxM1, stage IV). The patient underwent thoracic perfusion with cisplatin and pemetrexed + cisplatin chemotherapy until March 2017. However, control of the pleural effusion and chest pain was poor.
Molecular Tumor Board
A surgical tissue sample was obtained for NGS using a panel of 47 cancer‐related genes (OrigiMed, Shanghai, China) on January 11, 2017. TP53 mutation (p.G244D) and EGFR‐RAD51 fusion were detected.
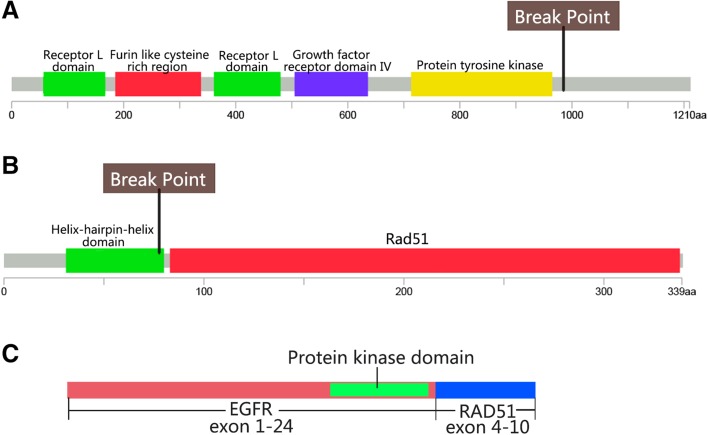
EGFR‐RAD51 was a fusion of exons 1–24 of EGFR with exons 4–10 of RAD51 (Fig. 1), resulting in the deletion of the EGFR C‐terminal CBL binding domain, which is related to EGFR degradation. This fusion retains the complete kinase domain of EGFR and could activate downstream signaling pathways via MAPK and PI3K/Akt. EGFR‐RAD51 fusion cells are sensitive to the EGFR inhibitor erlotinib, afatinib, and the EGFR monoclonal antibody cetuximab [6].
Figure 1.
Schematic diagram of gene structure of EGFR and RAD51. Break points of EGFR and RAD51 were shown in (A) and (B). Schematic fusion protein was shown in (C).
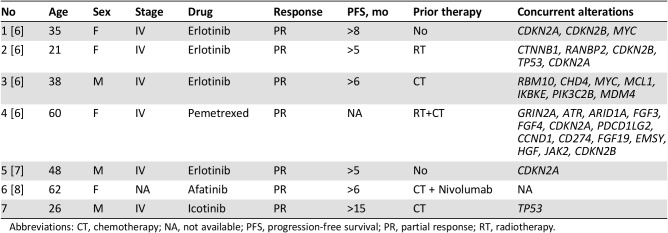
In addition to this case, seven clinically treated EGFR‐RAD51 fusion cases have been reported (Table 1) [6], [7], [8]. All patients were diagnosed at stage IV, except for one patient with an unknown stage and wild‐type EGFR at initial diagnosis but an EGFR‐RAD51 fusion during metastasis. All reported breakpoints of the EGFR‐RAD51 fusion are in intron 24 of EGFR and intron 3 of RAD51, resulting in the fusion of EGFR exons 1–24 and RAD51 exons 4–10. One patient showed a continuous response to chemotherapy. The other five patients showed partial responses to EGFR‐TKIs, including erlotinib (4 patients) and afatinib (1 patient).
Table 1. Clinical characteristics of seven treated patients with EGFR‐RAD51 fusion lung adenocarcinoma.
Abbreviations: CT, chemotherapy; NA, not available; PFS, progression‐free survival; PR, partial response; RT, radiotherapy.
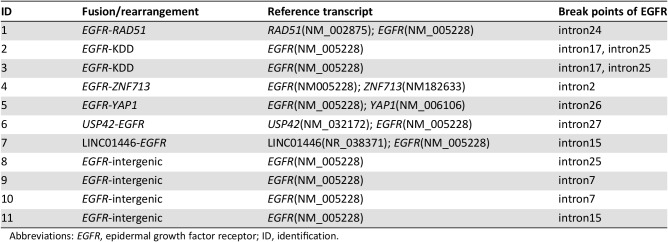
Fusions and rearrangements are rare genomic events in EGFR. They are relatively common (7.6%, 11/185) in glioblastoma, including EGFR‐SEPT14 and EGFR‐PSPH fusions [9]. EGFRvIII (n = 65) generated by an intragenic rearrangement resulting in the deletion of exons 2–7 is a common rearrangement [10]. Raez summarized EGFR fusion based on publicly available genomic data and found a frequency of 0.05% in Foundation Medicine NSCLC data and 0.13% in MSK‐IMPACT NSCLC data [8]. Using data from OrigiMed, we found an EGFR fusion/rearrangement frequency of 1.1% (11/989) in Chinese patients with lung adenocarcinoma. We further analyzed the break points of all EGFR fusion/rearrangements in OrigiMed (Table 2). Along with the reported EGFR‐RAD51 fusion and EGFR‐KDD (patients 1–3 in Table 2), we found eight new EGFR rearrangements. In addition to the previously established breakpoints in EGFR exon 23 through intron 25 [6], we found three EGFR rearrangements with new partners and EGFR breakpoints in introns 2, 26, and 27 (patients 4–6 in Table 2). We also found one EGFR rearrangement with a lncRNA and four EGFR rearrangements involving intergenic regions, with breakpoints in EGFR introns 7, 15, and 25 (patients 7–11 in Table 2). These results emphasize the advantage of NGS for finding new variants, but their oncogenic potential and clinical significance need further exploration.
Table 2. Break points of EGFR fusion/rearrangement detected in the Chinese patients with lung adenocarcinoma.
Abbreviations: EGFR, epidermal growth factor receptor; ID, identification.
EGFR fusions are rare in NSCLC. Gene fusion detection techniques vary with respect to sensitivity and specificity [11], [12]. Immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and quantitative polymerase chain reaction (qPCR) have various limitations for the accurate discovery and detection of rare fusions. IHC and FISH yield false‐positive and false‐negative results. Break‐apart FISH cannot assess the fusion partner of the target gene. Additionally, FISH is unrealistic for screening rare gene fusions, such as EGFR fusions, in clinical practice. qPCR can only detect known fusion types; its high specificity and low sensitivity limit its use for the discovery of new fusions and the screening of rare fusions. Targeted panel sequencing covering common fusion breakpoints located in intronic regions could provide information about known fusion types and could be helpful for finding new fusion types. Considered the efficiency of targeted therapies for some rare gene fusions and the importance of screening multiple gene variants simultaneously in clinical practice, NGS might be the optimal choice in certain cases (e.g., for patients with lung adenocarcinoma).
Icotinib (Conmana, Betta Pharmaceuticals Co., Ltd., Hangzhou, China) is a self‐developed first‐generation EGFR‐TKI and has been widely used in clinics in China [13]. A phase III ICOGEN trial proved its noninferiority to gefitinib in terms of PFS, and it was approved for patients with NSCLC by the China Food and Drug Administration [14]. The result of a network meta‐analysis showed that icotinib had similar effectiveness to geftinib and erlotinib for the treatment of patients with EGFR‐mutated NSCLC [15].
Patient Update
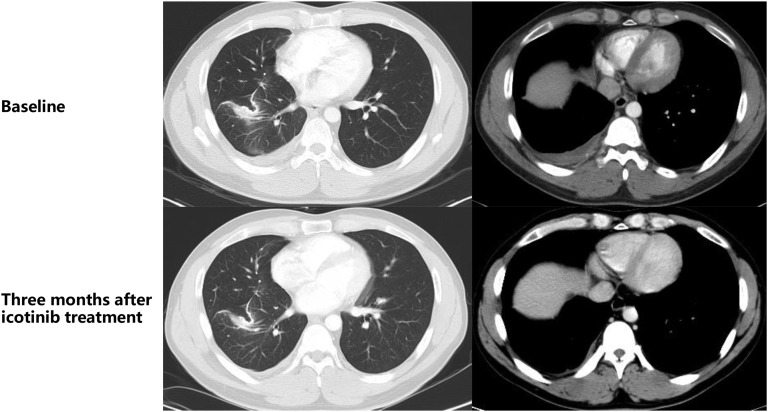
The imaging review on March 27, 2017, revealed pleural effusion and right interlobular pleural nodules. On March 28, 2017, oral icotinib was started. One week later, the chest pain was released. Three months later, chest CT showed a reduction in right interlobular pleural nodules and no pleural effusion, and the symptom of chest pain was significantly relieved (Fig. 2). The patient continued to use icotinib (as of June 13, 2018) with good disease control and no pleural effusion. The PFS was longer than 15 months.
Figure 2.
Imaging evaluation of the therapeutic effects of targeted therapy.
Glossary of Genomic Terms and Nomenclature
Fusion/Rearrangement: Recombination of two unlinked segments of human DNA, exhibited as sequencing reads uniquely aligned to two different genes or tow apart DNA segments.
Acknowledgments
The authors thank Dr. Hui Chen, Dr. Jinwei Hu, and Dr. Kai Wang for their technological consultation for the manuscript.
Author Contributions
Conception/design: Yan Guan, Zhanshuai Song, Yan Li
Provision of study material or patients: Yan Guan
Collection and/or assembly of data: Xuemei Zhang
Data analysis and interpretation: Honglin Guo, Junping Shi
Manuscript writing: Honglin Guo, Junping Shi
Final approval of manuscript: Yan Guan, Ming Yao
Disclosures
Honglin Guo: OrigiMed (E); Junping Shi: OrigiMed (E); Xuemei Zhang: OrigiMed (E); Ming Yao: OrigiMed (E). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Ferlay J, Soerjomataram I, Dikshit R et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int J Cancer 2015;136:E359–386. [DOI] [PubMed] [Google Scholar]
- 2.Li T, Kung HJ, Mack PC et al. Genotyping and genomic profiling of non‐small‐cell lung cancer: Implications for current and future therapies. J Clin Oncol 2013;31:1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non‐small‐cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 4.Murray S, Dahabreh IJ, Linardou H et al. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non‐small cell lung cancer: An analytical database. J Thorac Oncol 2008;3:832–839. [DOI] [PubMed] [Google Scholar]
- 5.Gallant JN, Sheehan JH, Shaver TM et al. EGFR kinase domain duplication (EGFR‐KDD) is a novel oncogenic driver in lung cancer that is clinically responsive to afatinib. Cancer Discov 2015;5:1155–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konduri K, Gallant JN, Chae YK et al. EGFR fusions as novel therapeutic targets in lung cancer. Cancer Discov 2016;6:601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu YC, Wang WX, Xu CW et al. EGFR‐RAD51 fusion variant in lung adenocarcinoma and response to erlotinib: A case report. Lung cancer 2018;115:131–134. [DOI] [PubMed] [Google Scholar]
- 8.Raez LE, Pinto JA, Schrock AB et al. EGFR‐RAD51 fusion: A targetable partnership originated from the tumor evolution? J Thorac Oncol 2018;13:e33–e34. [DOI] [PubMed] [Google Scholar]
- 9.Frattini V, Trifonov V, Chan JM et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 2013;45:1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zehir A, Benayed R, Shah RH et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letovanec I, Finn S, Zygoura P et al. Evaluation of NGS and RT‐PCR methods for ALK rearrangement in european NSCLC patients: Results from the European Thoracic Oncology Platform Lungscape project. J Thorac Oncol 2018;13:413–425. [DOI] [PubMed] [Google Scholar]
- 12.Blackhall FH, Peters S, Bubendorf L et al. Prevalence and clinical outcomes for patients with alk‐positive resected stage I to III adenocarcinoma: Results from the european thoracic oncology platform lungscape project. J Clin Oncol 2014;32:2780–2787. [DOI] [PubMed] [Google Scholar]
- 13.Liang JL, Ren XC, Lin Q. Treating advanced non‐small‐cell lung cancer in chinese patients: Focus on icotinib. Onco Targets Ther 2014;7:761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Zhang L, Liu X et al. Icotinib versus gefitinib in previously treated advanced non‐small‐cell lung cancer (ICOGEN): A randomised, double‐blind phase 3 non‐inferiority trial. Lancet Oncol 2013;14:953–961. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Zhang Y, Feng G et al. Comparison of effectiveness and adverse effects of gefitinib, erlotinib and icotinib among patients with non‐small cell lung cancer: A network meta‐analysis. Exp Ther Med 2017;14:4017–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]