Abstract
Solid organ transplantation can treat end-stage organ failure, but the half-life of transplanted organs colonized with commensals is much shorter than that of sterile organs. Whether organ colonization plays a role in this shorter half-life is not known. We have previously shown that an intact whole-body microbiota can accelerate the kinetics of solid organ allograft rejection in untreated colonized mice, when compared with germ-free (GF) or with antibiotic-pretreated colonized mice, by enhancing the capacity of antigen-presenting cells (APCs) to activate graft-reactive T cells. However, the contribution of intestinal versus skin microbiota to these effects was unknown. Here, we demonstrate that colonizing the skin of GF mice with a single commensal, Staphylococcus epidermidis, while preventing intestinal colonization with oral vancomycin, was sufficient to accelerate skin graft rejection. Notably, unlike the mechanism by which whole-body microbiota accelerates skin graft rejection, cutaneous S. epidermidis did not enhance the priming of alloreactive T cells in the skin-draining lymph nodes. Rather, cutaneous S. epidermidis augmented the ability of skin APCs to drive the differentiation of alloreactive T cells. This study reveals that the extraintestinal donor microbiota can affect transplant outcome and may contribute to the shorter half-life of colonized organs.
Keywords: Immunology, Transplantation
Keywords: Adaptive immunity, Mouse models, T cells
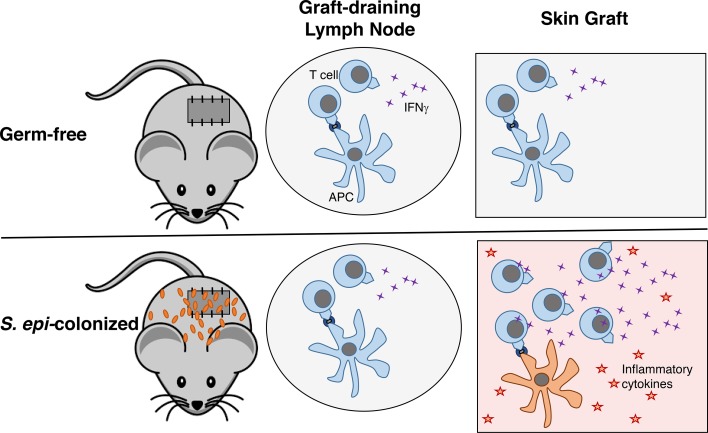
The cutaneous microbiota drives accelerated skin graft rejection following transplantation when compared with uncolonized skin.
Introduction
Solid organ transplantation is an accepted therapy for end-stage organ failure. Depending on the type and location of the transplanted organ, the level of commensal colonization varies. Sterile organs, such as the heart and kidneys, have a longer half-life following transplantation than barrier organs that are colonized with commensal bacteria, such as intestines and lungs (Organ Procurement and Transplantation Network, https://optn.transplant.hrsa.gov/data/; Scientific Registry of Transplant Recipients, http://www.srtr.org/, December 2012). This clinical observation prompted us to investigate whether the microbiota in barrier organs could promote graft rejection.
Numerous studies have established that the gut microbiota plays a critical role in local immune development and function (1), for instance, inducing lamina propria regulatory T cells (2) and Th17 cells (3). The gut microbiota can also affect immune responses at distal locations (4–6), such that, in theory, the gut microbiota might affect alloimmunity to any transplanted organ, whether sterile or colonized. Indeed, we previously showed that oral administration of broad-spectrum antibiotics that reduced the diversity of gut microbiota resulted in prolonged survival of both skin and heart transplants (7). However, whether the microbiota within the barrier organ itself can affect alloimmunity or graft outcome after it is transplanted remains to be determined. In fact, our mice gavaged with broad-spectrum antibiotics displayed reduced microbial diversity not only of the intestine but also of the skin (7), making it critical to understand whether graft-restricted microbiota can affect the outcome of that transplanted organ.
Staphylococcus epidermidis, a skin commensal, has been shown to calibrate cutaneous T cell responses and local inflammation, improving local immunity to Leishmania major and Candidia albicans (8, 9). This tissue-specific barrier immunity was independent of the effect of the gut microbiota and attributed to interactions between S. epidermidis and dermal CD103+ and CD11b+ dendritic cells (DCs) (9). Given the influence of a skin commensal to a local infectious immune response, we investigated the impact of skin S. epidermidis on skin allograft outcome.
Results
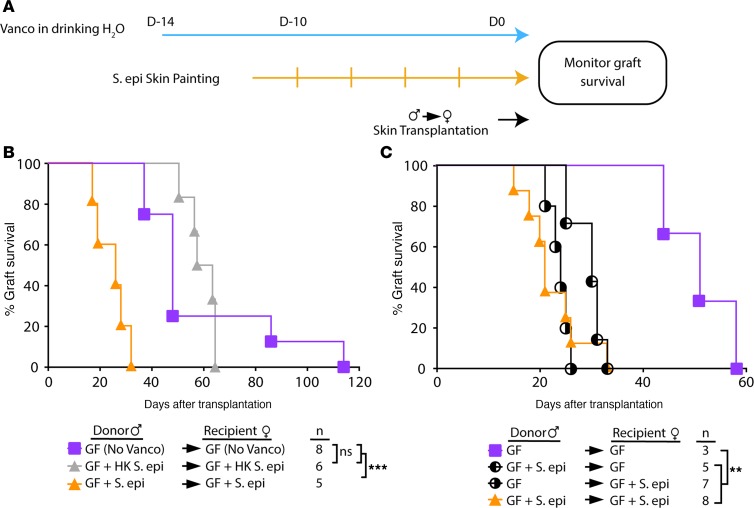
To investigate the impact on skin graft rejection of a commensal present exclusively on the skin, S. epidermidis was painted on the skin of male and female C57BL/6 germ-free (GF) mice, before transplantation of painted or unpainted male skin onto painted or unpainted female recipients (Figure 1A). To prevent gut colonization from self-grooming, the nonabsorbable antibiotic vancomycin was supplemented in the drinking water, starting before S. epidermidis painting and continuing until animal sacrifice. Cultures of skin swabs and fecal samples before transplantation confirmed skin but not gut colonization of the S. epidermidis–painted gnotobiotic mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127569DS1).
Figure 1. Cutaneous S. epidermidis colonization is sufficient to accelerate skin allograft rejection.
(A) Schematics of the transplant logistics in the gnotobiotic facility. (B) Both germ-free (GF) male donors and female recipients were colonized with live or heat-killed (HK) S. epidermidis for 10 days before male-to-female skin transplantation. (C) GF male donors and/or female recipients were left uncolonized or colonized with S. epidermidis for 10 days before skin transplantation. (B and C) All mice received vancomycin-supplemented water except the group labeled “no Vanco.” Statistical analysis by log-rank test. **P < 0.01; ***P < 0.001.
Transplantation of S. epidermidis–colonized male skin onto S. epidermidis–colonized female gnotobiotic mice resulted in marked acceleration of skin graft rejection over that of GF male skin onto female GF recipients (Figure 1B). Skin colonization did not drive local infection because grafts appeared healed and healthy at the time of bandage removal. Importantly, S. epidermidis needed to be alive to promote rejection because colonization with heat-killed S. epidermidis did not accelerate rejection, suggesting that pattern recognition alone of molecular components in S. epidermidis was not sufficient to promote rejection.
To determine whether accelerated rejection was driven by donor or recipient skin microbiota, only male donors or female recipients were painted with S. epidermidis before transplantation. Either donor or recipient skin colonization was sufficient to accelerate rejection (Figure 1C). We also investigated the impact of donor skin S. epidermidis on the outcome of major mismatched skin grafts. Although rejection of fully mismatched skin grafts is extremely rapid, rejection of S. epidermidis–painted monocolonized FVB (H-2q) skin grafts by B6 (H-2b) hosts trended toward even faster rejection than that of GF FVB grafts (Supplemental Figure 2).
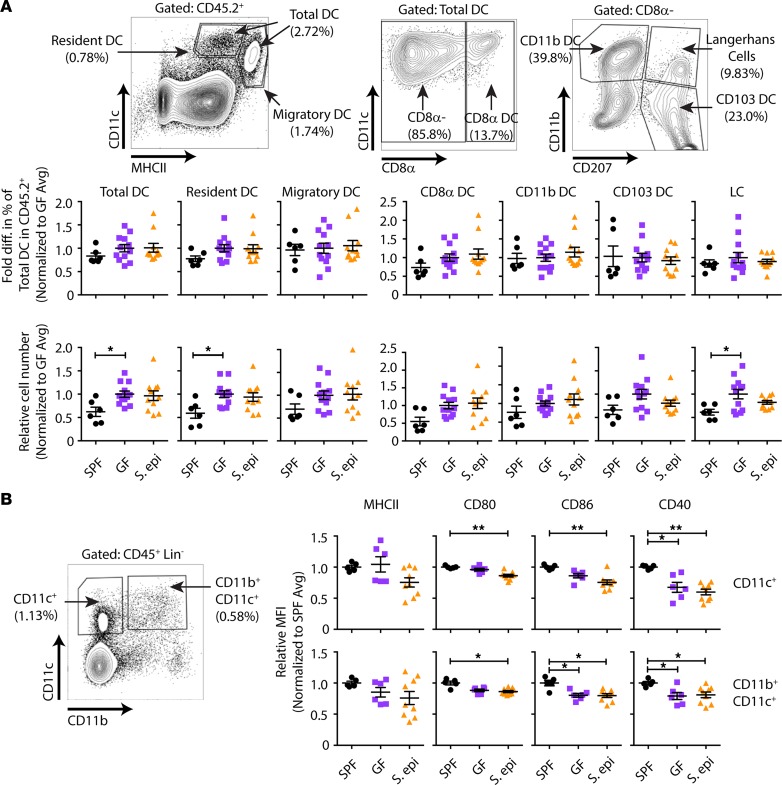
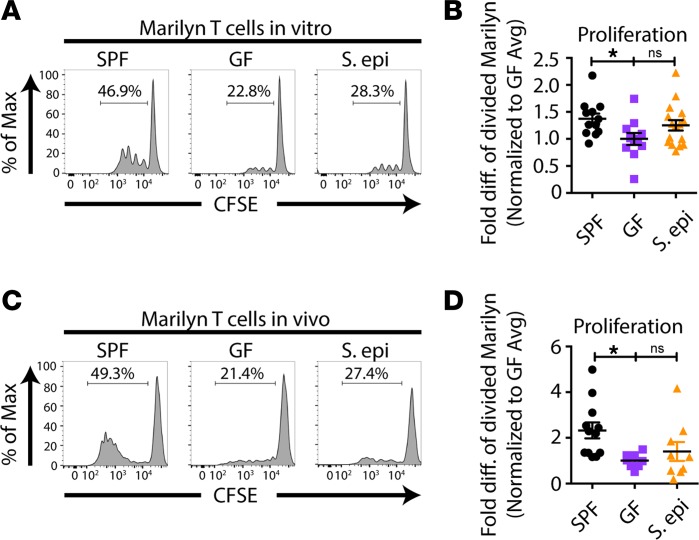
Skin colonization–dependent acceleration of skin graft rejection might be due to increased host alloreactivity at the priming phase of the anti–skin graft response. Previously, we found that normal global microbiota enhances the ability of antigen-presenting cells (APCs) in the skin-draining lymph nodes (LNs) to prime alloreactive T cells when compared with APCs from antibiotic-pretreated or GF mice, without changing the composition or the costimulatory profile of APCs (7). In keeping with these data, S. epidermidis skin colonization did not change either the composition of LN CD8α+CD11b+CD103+ DCs and CD207+ Langerhans cells (Figure 2A) or their expression of MHC class II, CD80, CD86, or CD40 when compared with GF DCs (Figure 2B). To determine whether LN APCs from S. epidermidis–skin-colonized mice acquire better T cell priming capacity, we cultured CFSE-labeled, male-specific, CD4+ TCR-transgenic T cells (Marilyn T cells) with LN APCs from S. epidermidis–painted GF male mice in vitro. Unlike LN APCs from globally colonized SPF mice, which induced greater Marilyn T cell proliferation than APCs from GF mice, LN APCs from S. epidermidis–painted monocolonized mice failed to significantly enhance Marilyn T cell division relative to that induced by APCs from GF mice (Figure 3, A and B). This was likely not due to a fixed immaturity of the innate immune system in GF mice because we have previously reported that fecal transfer from colonized mice rapidly restores, in less than a week, the ability of peripheral LN APCs from ex-GF mice to activate Marilyn T cells in vitro and in vivo (7); in contrast, skin S. epidermidis monocolonization did not increase the stimulatory capacity of LN APCs. To investigate the impact of skin S. epidermidis painting on the priming of T cells in vivo, CFSE-labeled Marilyn T cells were transferred into uncolonized or S. epidermidis–colonized female GF mice on the day of transplantation with skin from uncolonized or S. epidermidis–colonized male GF mice, and T cells were harvested from skin-draining LNs on day 4 or 6 after transplantation. Surprisingly, but consistent with the in vitro priming data, Marilyn T cell proliferation was not enhanced in S. epidermidis–colonized mice, suggesting that S. epidermidis skin colonization does not accelerate rejection by increasing the priming phase of the alloresponse (Figure 3, C and D).
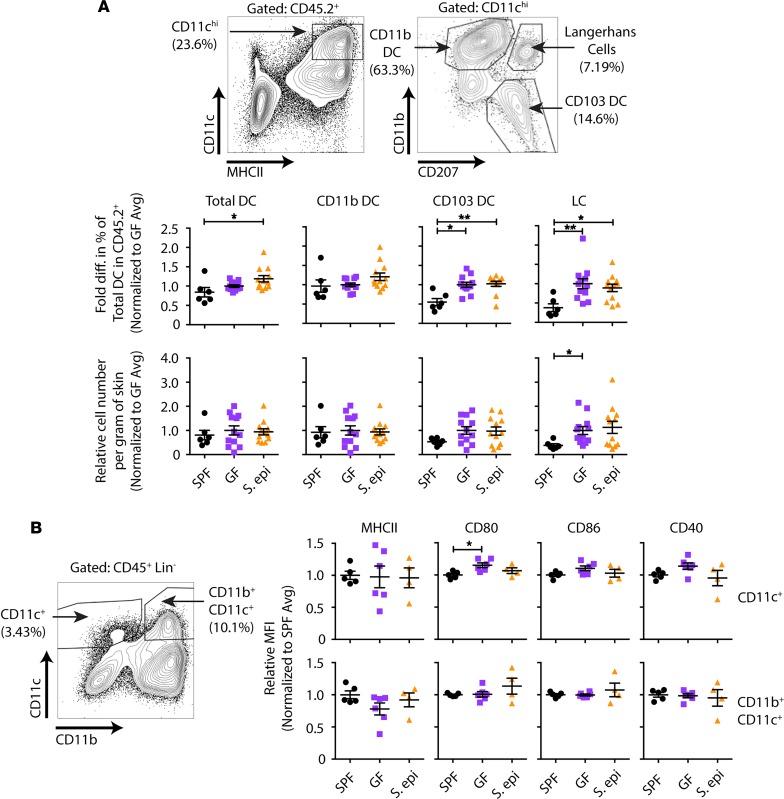
Figure 2. S. epidermidis skin colonization does not change the composition or costimulatory molecule expression of LN APCs relative to GF APCs.
APCs from the peripheral LNs of specific pathogen–free (SPF), GF, and 10-day-painted S. epidermidis–monocolonized GF mice were analyzed by flow cytometry. (A) Gating strategy for different dendritic cell (DC) populations and fold change relative to the average in the percentage of DC subsets observed in GF controls within each experiment. LC, Langerhans cell. (B) Fold change in mean fluorescence intensity (MFI) relative to that in GF mice for costimulatory molecules on CD11c+-gated cells. Data were normalized from 3 individual experiments with n = 2–5 per group. Data represent the mean ± SEM. Statistical analyses were performed using the Kruskal-Wallis test for multiple comparisons. *P < 0.05; **P < 0.01.
Figure 3. Skin S. epidermidis colonization does not enhance the proliferation of alloreactive T cells in the skin-draining LNs.
(A and B) APCs from skin-draining LNs were isolated from male SPF, GF, or S. epidermidis–painted GF mice 10 days after initial painting and cultured with CFSE-labeled T cells from Marilyn females for 3 days, followed by flow cytometric analysis. (C and D) Congenic Marilyn T cells were labeled with CFSE and transferred into GF and S. epidermidis–painted GF recipients on the day of transplantation with male GF or GF plus S. epidermidis skin grafts. Mice were sacrificed 4 or 6 days after transplantation, and cells were isolated from the graft draining LNs for analysis of CFSE dilution. Representative plots (A and C) and quantitation of CFSE dilution (B and D) of divided Marilyn T cells. (B and D) Quantitation represents normalized data with normalization to the average of percentage of divided Marilyn T cells in GF mice within each experiment from 3–5 experiments with n = 2–4 mice per group. Data represent the mean ± SEM. Statistical analysis was done using the Kruskal-Wallis test for multiple comparisons. *P < 0.05.
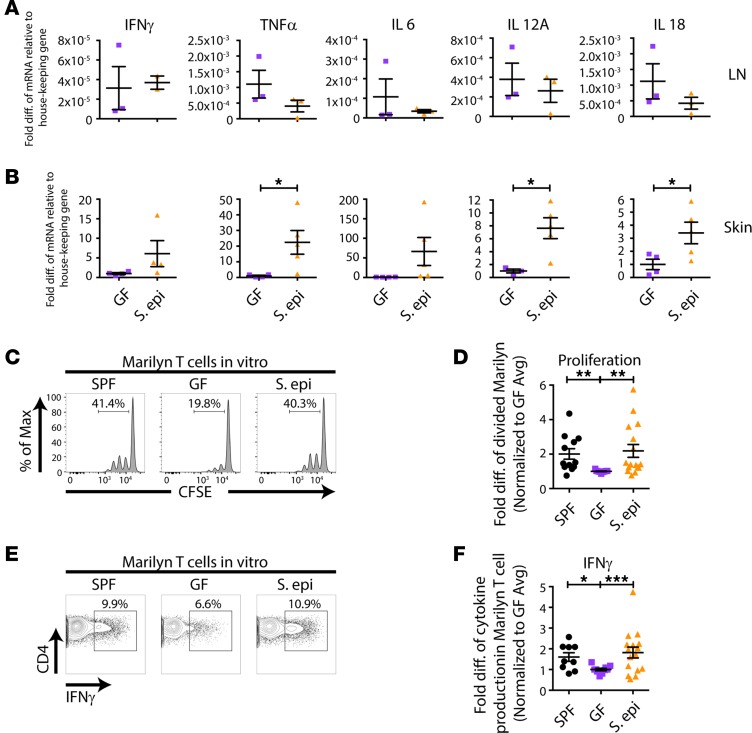
S. epidermidis colonization can modulate local cutaneous immune responses (8, 9). Although the proportion of skin DCs and their expression of costimulatory molecules were unchanged in S. epidermidis–colonized mice compared with uncolonized GF mice (Figure 4), it was possible that skin S. epidermidis affected their function. To determine whether skin S. epidermidis accelerates skin graft rejection by driving a stronger effector rather than priming phase of the alloresponse, we harvested skin-draining LNs and skin grafts from GF or S. epidermidis–colonized animals on day 10 after transplantation, a time point before any evidence of skin graft rejection. cDNA was prepared from the homogenized LNs and skin grafts, and real-time quantitative PCR for inflammatory cytokines was performed. Whereas cytokine expression was similar in the LNs of recipients of GF and S. epidermidis–monocolonized skin grafts (Figure 5A), TNF-α, IL-12A, and IL-18 were significantly elevated in the colonized versus GF skin grafts (Figure 5B), suggesting a local skin environment favorable to Th1 differentiation, which often correlates with graft rejection (10). To investigate the impact of this microenvironment on the ability of local APCs to activate graft-reactive T cells, we isolated APCs from the skin of GF and S. epidermidis–painted monocolonized male mice. Upon culture with CFSE-labeled Marilyn T cells, skin APCs from S. epidermidis–colonized mice induced T cell proliferation (Figure 5, C and D) and IFN-γ production upon PMA and ionomycin restimulation (Figure 5, E and F) that was as robust as skin APCs from SPF mice and greater than skin APCs from GF mice. Moreover, supporting a functional role for the inflammatory cytokines found in the day 10 S. epidermidis–colonized skin graft, exogenous addition of IL-6 and IL-12 enhanced the ability of both skin and LN APCs to induce IFN-γ production by Marilyn T cells (Supplemental Figure 3). In vivo, approximately 7-fold more Marilyn T cells were retrieved from S. epidermidis–colonized than GF skin grafts on day 10 after transplantation (Supplemental Figure 4). Together, these results suggest that the skin graft S. epidermidis acts locally in the skin, at the target site rather than at the priming site of the alloimmune response, by augmenting the effector function of the alloreactive T cells.
Figure 4. S. epidermidis skin colonization does not change the composition or costimulatory molecule expression of skin APCs relative to GF APCs.
APCs from the flank skin of SPF, GF, and 10 day–painted S. epidermidis–monocolonized GF mice were analyzed by flow cytometry. (A) Gating strategy for different DC populations and fold change relative to the average percentage of DC subsets within GF controls in each experiment. (B) Fold change in MFI relative to that in GF mice for costimulatory molecules on CD11c+-gated cells. Data were normalized from 3 individual experiments with n = 2–5 per group. Data represent the mean ± SEM. Statistical analyses were performed using the Kruskal-Wallis test for multiple comparisons. *P < 0.05; **P < 0.01.
Figure 5. Skin S.
epidermidiscolonization enhances skin APC activation of alloreactive T cells. (A and B) GF or S. epidermidis–colonized skin-draining LNs (A) and skin grafts (B) were isolated on day 10 after transplantation of male skin onto female recipients, cDNA was prepared, and qPCR for cytokines was performed. Pooled results from 2 independent experiments are shown for the skin graft, normalized to GF average values. (C–F) Skin APCs were isolated from male SPF, GF, or S. epidermidis–painted GF mice and cultured with CFSE-labeled T cells from Marilyn females, followed by flow cytometric analysis. (E and F) For IFN-γ detection, cells were stimulated with PMA and ionomycin before staining. Representative plots (C and E) and quantitation of CFSE dilution of Marilyn T cells (D) and IFN-γ production by Marilyn T cells (F). Quantitation represents data normalized to the average of the percentage of divided Marilyn T cells (D) or IFN-γ production (F) in GF mice within each experiment from 2–3 experiments with n = 2–4 mice per group. Data represent the mean ± SEM. Statistical analyses were done using the unpaired t test (A and B) or Kruskal-Wallis test for multiple comparisons (D and F). *P < 0.05; **P < 0.01; and ***P < 0.001.
Discussion
Our results demonstrate that monocolonization with S. epidermidis, restricted to the donor skin, causes accelerated rejection following transplantation of this skin onto GF recipients. This supports the sufficiency of the transplanted organ’s microbiota for the tuning up of host alloreactivity. Moreover, our data support the conclusion that cutaneous S. epidermidis modulates alloimmunity in the skin graft itself, at the effector, rather than during the priming phase of the alloresponse, and therefore by a different mechanism than the enhanced priming observed in mice also harboring intestinal colonization (7). These results provide a possible explanation for why colonized organs have a shorter half-life than sterile organs following transplantation in the clinic.
The kinetics of skin graft rejection in skin only–colonized GF mice were similar to those of globally colonized SPF mice or of GF mice that were conventionalized and became globally colonized, in the gut and skin, following SPF fecal transfer by gavage (7). However, it is important to note that not all skin microbiota accelerates skin graft rejection. Indeed, we previously showed that fecal transfer from antibiotic-pretreated mice into GF mice by oral gavage, which resulted in both gut and skin colonization, did not hasten skin graft rejection (7), suggesting that different skin microbiota communities have different consequences on alloimmunity. Indeed, association of mice with a defined clade of S. epidermidis, but not S. xylosus or S. aureus, enhanced IFN-γ and IL-17A production by dermal CD8+ T cells (9, 11), indicating that even single commensals of the same genus can have different immune impacts. Whether skin commensals other than S. epidermidis can accelerate skin allograft rejection, and if so whether their mechanism of action is the same, remains to be investigated.
The fact that donor microbiota is sufficient to accelerate skin graft rejection is consistent with our previous findings (7) that global colonization of either the donor or the recipient was sufficient to drive fast skin graft rejection, although those experiments did not distinguish which microbial compartment (intestine or skin) affected graft outcome. Our current results demonstrate that skin microbiota in either the donor or the recipient is sufficient to accelerate skin graft rejection. We acknowledge that a GF graft will be rapidly colonized when transplanted onto an S. epidermidis–colonized host, and vice versa, that a host GF skin will be rapidly colonized following transplantation of an S. epidermidis–painted skin graft, but such cross-colonization will occur after transplantation. Thus, if donor colonization requires the donor commensals to colonize the host skin to accelerate graft rejection, our results still suggest an extremely rapid impact of the donor microbiota on the host alloimmune response.
The exact mechanism by which skin S. epidermidis accelerates skin graft rejection is not fully elucidated. The observation that painting with heat-killed S. epidermidis did not result in accelerated graft rejection suggests that pattern recognition receptor (PRR) signaling is not sufficient. In addition to engaging PRRs, live S. epidermidis may produce metabolites that enable local cutaneous but not LN APCs to better reactivate alloreactive T cells when they reach the skin, either because of closer proximity of the skin APCs to the cutaneous S. epidermidis or because the APC subset composition in the skin is more susceptible to modulation by S. epidermidis metabolites. This is consistent with our data that skin but not LN APCs from S. epidermidis–colonized mice enhanced activation of donor-reactive T cells. S. epidermidis has been shown to elicit local commensal-specific immune responses, such as the accumulation of Tregs in the skin (12) and the activation of cutaneous nonclassical MHC class I–restricted S. epidermidis–specific CD8+ T cells (11). The impact of these local changes in cutaneous immune homeostasis on skin graft rejection remains to be elucidated. Skin S. epidermidis has also been linked to faster skin wound closure (11), which might allow faster infiltration of alloreactive T cells following skin transplantation.
Cutaneous S. epidermidis resulted in higher mRNA expression levels of inflammatory cytokines in the skin graft 10 days after transplantation, before signs of rejection were visible, but not in the graft draining LNs. Whether cytokines are also elevated in the skin graft at the protein level remains to be confirmed, but some of the cytokines identified, such as IL-6, are known to enhance T cell proliferation (ref. 13 and our in vitro results), suggesting that they may enhance the effector function of alloreactive T cells when they enter the skin graft. Alternatively, the lymphatic drainage of the skin to the LNs or lymphatic repair after surgical transplantation may be impaired in GF mice such that S. epidermidis–induced cytokines in the skin or S. epidermidis–modulated skin APCs may not be able to drain normally to the LNs. Indeed, the blunt-end lacteal lymphatics located in each intestinal villus and helping lipid absorption were recently reported to be reduced in antibiotic-treated SPF and GF mice, though rapidly restored by conventionalization (14). Whether this also occurs with skin lymphatics remains to be investigated and would be relevant to cutaneous immune responses.
Although our data demonstrate that a skin commensal can modulate alloresponses locally, it remains possible that the gut microbiota can affect alloreactivity distally, such that both commensal body locations, skin and intestine, may independently modulate alloreactivity to skin grafts, albeit by different mechanisms. Future studies will need to investigate whether colonization of only the intestine also affects skin graft outcomes and whether it does so by influencing a different stage of the immune response than the effector stage modulated by skin S. epidermidis. This will require a commensal that colonizes the gut but not the anaerobic hair follicle despite skin contact with fecal material in the cage.
The discovery that an extraintestinal, graft-restricted commensal can act locally to enhance the effector phase of alloimmunity in the graft microenvironment provides a potential mechanistic explanation for the clinical observation that colonized organs have a shorter half-life than sterile organs and suggests that modulation of the graft microbiota before transplantation may improve graft outcomes.
Methods
Mice.
SPF C57BL/6NHsd male and female mice were purchased from Envigo. CD45.1+ RAG2-knockout TCR-transgenic C57BL/6 mice whose CD4+ T cells are specific for an H-Y antigen (15) (Marilyn mice) were obtained from Charles Mainhart via Taconic Biosciences (Rensselaer, New York, USA) and bred in-house under SPF conditions. GF FVB mice (H-2q) were obtained from Tatyana Golovkina (University of Chicago gnotobiotic facility, Chicago, Illinois, USA). GF mice were kept in sterile isolators at the University of Chicago’s Gnotobiotic Research Animal Facility and were fed autoclaved food and water. Fecal aerobic and anaerobic cultures were carried out weekly to verify the status of microbial colonization. All animals were used in agreement with the University of Chicago’s Institutional Animal Care and Use Committee, according to the NIH guidelines for animal use.
S. epidermidis painting.
S. epidermidis bacterial stock (strain NIHLM087) was cultured as described previously (8). Briefly, S. epidermidis was inoculated and cultured in tryptic soy broth for 18 hours. The resulting culture was painted onto the skin of GF mice using cotton swabs every 2 days for 10 days until sacrifice or transplantation. Vancomycin (Hospira) was supplemented to the drinking water (500 mg/L) starting 1 week before skin painting and continued until sacrifice to prevent intestinal colonization. Skin swabs and fecal samples were cultured before skin transplantation to confirm colonization status.
Transplantation.
Aseptic surgery for SPF mice and sterile surgery for gnotobiotic mice were performed as previously described (16). Tail skin from male C57BL/6 or FVB donor mice was transplanted onto the flank of female C57BL/6 recipients. Bandages were removed after 7 days. Graft survival was then monitored every 2 days for minor mismatched and daily for major mismatched skin grafts. The day when less than 20% viable skin graft tissue remained was called the day of rejection.
Leukocyte isolation.
Skin-draining LN (inguinal, axillary, and brachial) cells were isolated at the indicated times (10-day painted, 4 or 6 days after transplantation, and 10 days after transplantation), resuspended in complete DMEM (Corning, with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% l-glutamine, 1% nonessential amino acids, 1% HEPES, and 0.028 mM β-mercaptoethanol), and counted using a Countess cell counter. For skin leukocyte isolation, skin grafts or shaved flanks were harvested. The skin was cut into small pieces, resuspended in Roswell Park Memorial Institute (RPMI, Corning) medium supplemented with Liberase (0.4 mg/mL, Roche) and DNAse (0.01%, MP Biomedicals), and incubated for 2 hours at 37°C. The skin was then homogenized using a 70-μm cell strainer, a petri dish, and a syringe plunger. Single-cell suspensions were centrifuged at 375 g and pellets resuspended in complete RPMI.
CFSE labeling.
Marilyn T cells were collected from the peripheral LNs and ACK-lysed spleens of Marilyn mice. Cells were resuspended at a concentration of 20 × 106 cells/mL in serum-free DMEM. CFSE (C34554, Life Technologies) was diluted to 5 μΜ and incubated with cells for 10 minutes at 37°C. Labeling was stopped with an equal volume of fetal bovine serum. Cells were centrifuged at 375 g and resuspended in complete DMEM. For adoptive transfer, the cells were filtered, washed, and resuspended in 1× phosphate-buffered saline. CFSE-labeled Marilyn T cells (1 × 106) were transferred i.v. day 1, day 0, or day +4 relative to skin transplantation. At day 4 or 6, or day 10 after transplantation, skin-draining LN leukocytes were analyzed by flow cytometry to assess T cell proliferation. On day 10 after transplantation, graft-infiltrating Marilyn T (TCRβ+CD45.1+) cells were enumerated.
Ex vivo stimulation of Marilyn T cells.
APCs were isolated from the skin-draining LNs of male or female mice following magnetic negative enrichment using biotinylated anti-Thy1.2 (53-2.1, eBioscience) and anti-NK1.1 (mAb facility, University of Chicago) antibodies. Marilyn T cells were magnetically enriched from the peripheral LNs and spleens of Marilyn mice by negative selection using streptavidin magnetic beads (88817, Pierce), anti–Ter119-biotin (TER-119, eBioscience), anti–CD11b-biotin (M1/70, eBioscience), and anti–NK1.1-biotin (PK136, Frank Fitch Monoclonal Antibody Facility at the University of Chicago) before CFSE labeling. APCs (1 × 105 to 2 × 105/well) were cultured in duplicate with Marilyn T cells (1 × 105/well) in complete DMEM for 3 days in the presence or absence of IL-6 (50 ng/mL) and IL-12 (5 ng/mL). For detection of IFN-γ production, T cells were restimulated with PMA (50 ng/mL) and ionomycin (0.5 μg/mL) in the presence of brefeldin A (5 μg/mL) for 46 hours at 37°C and analyzed by flow cytometry.
Quantitative real-time PCR for skin and LN cytokine signatures.
Skin grafts and graft-draining LNs were isolated and homogenized on ice with TissueRuptor II handheld homogenizer (9002755, Qiagen) in RLT buffer at the volume suggested by the manufacturer’s protocol for RNeasy Micro Kit (74004, Qiagen). The supernatant of homogenized skin was then processed with RNeasy Micro Kit for RNA isolation. The resulting RNA was used for cDNA amplification according to the manufacturer’s protocol for iScript cDNA Synthesis Kit (1708891, Bio-Rad). The expression level of cytokines was determined by quantitative real-time PCR with primers specific for IFN-γ, TNF-α, IL-6, IL-12A, and IL-18 genes (Supplemental Table 1) and normalized for expression of β-actin or 18S, using iQ SYBR Green Supermix (Bio-Rad) and 7300 Real Time PCR system (Applied Biosystems).
Flow cytometry.
All cells were stained with 1:1000 Live Aqua Live/Dead stain (Life Technologies) before phenotypic staining. For cell population analyses, cells were stained with antibodies against CD45.2-APC (104, eBioscience), TCRβ-BV421 (H57597, BioLegend), CD19-PacBlue (eBio1D3, eBioscience), CD8α-BV711 (53-6.7, BioLegend), NK1.1-BV421 (PK136, BioLegend), CD11c-PECy7 (N418, eBioscience), CD11b-BV605 (M1/70, BD Horizon), I-A/I-E (M5/114, BD Horizon), SiglecH-PerCP-Cy5.5 (551, BioLegend), CD207-PE (eBioL31, eBioscience), and CD103-FITC (2E7, eBioscience). For adoptive transfer and in vitro stimulation analyses, cultured cells were stained with anti–CD45.1-APC-eF780 (A20, eBioscience), anti–CD45.2-APC (104, eBioscience), anti–TCRβ-PerCP-Cy5.5 (H57597, eBioscience), anti–CD4-BV605 (RM4-5, BioLegend), and anti–IFN-γ-PE/PE-Cy7 (XMG1.2, eBioscience) mAbs. For intracellular cytokine staining, cells were fixed, permeabilized, and stained with BD Fixation/Permeabilization Solution Kit (554714, BD Biosciences), as described in the manufacturer’s protocol. All samples were analyzed with the LSR Fortessa (BD Biosciences).
Statistics.
Graft survival curves were analyzed using Kaplan-Meier plots and log-rank post hoc tests. Comparisons of APC populations, costimulatory expression, Marilyn T cell division, and cytokine production between groups were analyzed with unpaired t test or Kruskal-Wallis test for multiple comparisons as appropriate. P values of less than 0.05 were considered statistically significant. Statistical analyses were performed using GraphPad Prism 6: ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001; and ****P < 0.0001.
Study approval.
All mouse studies were approved by the Institutional Animal Care and Use Committee of the University of Chicago (protocol 71095), Chicago, Illinois, USA.
Author contributions
YML, LC, ASC, and MLA designed the study. BT, LC, and YW developed the gnotobiotic surgical techniques. YML, IP, MS, and LC conducted experiments and analyzed data. LC and YW performed microsurgery and monitored graft survival. YB provided S. epidermidis bacterial stock and experimental advice. YML and MLA wrote the manuscript and all authors edited it.
Supplementary Material
Acknowledgments
This work was supported by grants R01 AI071080 to MLA, 1ZIA A1001115 to YB, and ROTRF 979162997 to YML. The authors wish to acknowledge the help of Nicole Martinec in the Gnotobiotic Research Animal Facility of the University of Chicago, as well as the gift of GF C57BL/6 mice by Vanessa Leone and of GF FVB mice by Tatyana Golovkina, both at the University of Chicago.
Version 1. 07/16/2019
In-Press Preview
Version 2. 08/08/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(15):e127569.https://doi.org/10.1172/jci.insight.127569.
Contributor Information
Yuk Man Lei, Email: leik@uchicago.edu.
Martin Sepulveda, Email: msepulveda@uchicago.edu.
Luqiu Chen, Email: lchen@medicine.bsd.uchicago.edu.
Ying Wang, Email: yingwang@medicine.bsd.uchicago.edu.
Isabella Pirozzolo, Email: ipirozzolo@gmail.com.
Betty Theriault, Email: btheriault@bsd.uchicago.edu.
Anita S. Chong, Email: achong@surgery.bsd.uchicago.edu.
Yasmine Belkaid, Email: ybelkaid@niaid.nih.gov.
Maria-Luisa Alegre, Email: malegre@midway.uchicago.edu.
References
- 1.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilmore JR, et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe. 2018;23(3):302–311.e3. doi: 10.1016/j.chom.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teng F, et al. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity. 2016;44(4):875–888. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei YM, et al. The composition of the microbiota modulates allograft rejection. J Clin Invest. 2016;126(7):2736–2744. doi: 10.1172/JCI85295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naik S, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520(7545):104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atalar K, Afzali B, Lord G, Lombardi G. Relative roles of Th1 and Th17 effector cells in allograft rejection. Curr Opin Organ Transplant. 2009;14(1):23–29. doi: 10.1097/MOT.0b013e32831b70c2. [DOI] [PubMed] [Google Scholar]
- 11.Linehan JL, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. 2018;172(4):784–796.e18. doi: 10.1016/j.cell.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharschmidt TC, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43(5):1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299(5609):1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 14.Suh SH, et al. Gut microbiota regulates lacteal integrity by inducing VEGF-C in intestinal villus macrophages. EMBO Rep. 2019;20(4):e46927. doi: 10.15252/embr.201846927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Diez A, et al. CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood. 2007;109(12):5346–5354. doi: 10.1182/blood-2006-10-051318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theriault B, Wang Y, Chen L, Vest A, Bartman C, Alegre ML. Long-term maintenance of sterility following skin transplantation in germ-free mice. Transplant Direct. 2015;1(8):e28. doi: 10.1097/TXD.0000000000000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.