Abstract
For years, antibodies (Abs) have been used as a paradigm for understanding how protein structure contributes to molecular recognition. However, with the ability to evolve Abs that recognize specific chromophores, they also have great potential as models for how protein dynamics contribute to molecular recognition. We previously raised murine Abs to different chromophores, and with the use of 3-pulse photon echo peak shift spectroscopy, we demonstrated that the immune system is capable of producing Abs with widely varying flexibility. We now report the characterization of the complexes formed between two Abs, 5D11 and 10A6, and the chromophoric ligand they were evolved to recognize, 8-methoxypyrene-1,3,6-trisulfonic acid (MPTS). The sequences of the Ab genes indicate that they evolved from a common precursor. We also used a variety of spectroscopic methods to probe the photophysics and dynamics of the Ab-MPTS complexes, and found that they are similar to each other but distinct from previously characterized anti-MPTS Abs. Structure studies revealed that this likely results from a unique mode of binding wherein MPTS is sandwiched between the side chain of PheH98, which interacts with the chromophore via T-stacking, and the side chain of TrpL91, which interacts with the chromophore via parallel stacking. The T-stacking interaction appears to mediate relaxation on the picosecond timescale, while the parallel stacking appears to mediate relaxation on an ultrafast, femtosecond timescale, which dominates the response. The anti-MPTS Abs thus not only demonstrate the simultaneous use of the two limiting modes of stacking for molecular recognition, but also provide a unique opportunity to characterize how dynamics might contribute to molecular recognition. Both types of stacking are common in proteins and protein complexes where they may similarly contribute to dynamics and molecular recognition.
Graphical Abstract
INTRODUCTION
Molecular recognition underlies virtually all of the activities a protein can possess. Perhaps the most remarkable example of molecular recognition, and its evolution, is the adaptive immune system, in which antibodies (Abs) specific for virtually any foreign molecule (i.e. antigen, Ag) can be produced via somatic mutation from a germline precursor (affinity maturation) within weeks of their first encounter. Since the earliest days of structural biology, Abs have served as a paradigm for understanding how structure contributes to molecular recognition.1 However, dynamics must also contribute, because the limiting models of induced fit, conformational selection, and lock-and-key recognition are differentiated by the level of flexibility attributed to the protein.
In recent years, increasing attention has been paid to the study of conformational dynamics and its link to Ab specificity.2,3 Structural studies have provided evidence that affinity maturation can pre-order the Ab combining site for specific Ag recognition,2,4–11 while other studies have demonstrated that mature Abs retain the ability to adopt multiple conformations that facilitate the recognition of different Ags.12,13 The effects of affinity maturation on the entropy of binding have also provided evidence of conformational restriction.14,15 Finally, computational studies have provided evidence that Ab rigidity is linked to specificity.16–19 However, our understanding of Ab dynamics, how it contributes to Ag recognition, and if it is tailored during affinity maturation remains incomplete, at least in part due to the experimental challenges associated with the direct characterization of protein dynamics.
In addition to their unprecedented level of molecular recognition, the use of Ab-Ag complexes to study protein dynamics is attractive because Abs can be evolved to bind specific chromophores that facilitate characterization of dynamics, for example through the use of ultrafast nonlinear optical methods such as 3-pulse photon echo peak shift (3PEPS) spectroscopy.20–22 Unlike the conjugation of chromophores to a protein of interest, which is likely to be perturbative if the chromophore is buried within the protein or insensitive to the protein environment if surface exposed, the use of a chromophoric Ag, to which the Ab evolved to bind, is by definition non-perturbative and likely to be sensitive to biologically relevant dynamics.
The response of the environment to the force exerted by a photoinduced change in a chromophore’s electronic structure is typically discussed in terms of impulsive and diffusive motions, in analogy to solvation dynamics.22 However, inspired by the field of materials science, we have adopted the language of elasticity, anelasticity, and plasticity.23–25 Anelasticity corresponds to motions associated with barrier crossings that occur during the timescale of an experiment, while plasticity corresponds to the population of multiple deep minima separated by barriers that are sufficiently large to prevent interconversion on the timescale of the experiment and thus result in static inhomogeneity. In contrast, elasticity corresponds to motion within a single potential energy minimum.
During our efforts to develop Abs as models for the study of protein dynamics, we have used 3PEPS spectroscopy to characterize how different Abs evolved to bind the chromophoric Ag fluorescein23,26–29 or 8-methoxypyrene-1,3,6-trisulfonic acid (MPTS).24,25,30 Generally, a broad range of anelasticity and plasticity was observed, with ultrafast elastic motions contributing up to 85% of the total response to chromophore excitation. While we have demonstrated that a wide range of dynamics is available to Abs, and even that the dynamics can be tailored by evolution,27,28 structural data have only been available with two systems,24,26–28 and even then, the structural origins of the observed dynamics were not readily apparent, limiting our ability to understand the relationship between structure and dynamics.
Here, we characterize the sequence, affinity, photophysics, and dynamics of two anti-MPTS Abs, 10A6 and 5D11, which interestingly are siblings that arose via divergent evolution from a common precursor germline Ab. Despite having acquired different mutations during divergence, the affinities for MPTS, as well as the photophysics and dynamics of the resulting complexes are virtually identical. However, the observed photophysics and dynamics of the siblings are very different than those of the previously characterized Ab-MPTS complexes. Structural studies of the 10A6 complex reveal that MPTS is sandwiched by T-stacking and parallel stacking interactions with Ab side chains, which represents a binding motif that is distinct from those observed in previously characterized Abs, and likely the source of the unique dynamics. T-stacking and parallel stacking represent the two limiting modes of stacking, and the simultaneous use of both appears to provide the Abs with unique dynamics and the ability to respond to forces on different timescales, and also makes these Abs excellent models with which to characterize the dynamics associated with stacking interactions.
MATERIALS AND METHODS
Generation and Sequencing of Anti-MPTS Abs.
Anti-MPTS Abs were raised in Swiss Webster mice immunized with MPTS conjugated to keyhole limpet hemocyanin. Monoclonal IgG Abs were produced and purified from hybridoma culture supernatants using standard methods.31 To sequence the heavy-chain variable (VH) and light-chain variable (VL) gene regions, mRNA was isolated from freshly grown hybridoma cells (Qiagen RNeasy Kit) and used with an oligonucleotide (dT)18 primer for cDNA synthesis (Superscript III Kit, Invitrogen). The VH and VL gene regions were PCR-amplified from this cDNA using a specifically designed primer set for cloning mouse immunoglobulin regions and then sequenced.
Isothermal Titration Calorimetry (ITC).
ITC of each Ab with MPTS in 1× PBS (phosphate buffered saline, pH 7.4) was carried out at 25 °C using an MCS-ITC or a VP-ITC calorimeter (GE Healthcare). For each titration, an 8 μM solution of antibody was titrated with 19 consecutive injections of MPTS at 240 s intervals to a final concentration of 150 μM MPTS. The heat dilution of the titrant was corrected by performing a titration of MPTS into the buffer alone. ITC data is shown in Figure S1.
3PEPS and Transient Grating Spectroscopy.
The 3PEPS setup32 has been described in detail previously.24,30 Nonlinear response function theory was used to recover the two-point correlation function CB(t) from the experimental 3PEPS decay via least-square fitting and to deconvolute the contributions of elasticity, anelasticity, and plasticity into Brownian oscillator terms.22,33 In brief, elastic motions are described as underdamped Brownian oscillators, yielding the amplitude (λBO) and timescale (τBO) of the motion. Anelastic motions are described as strongly overdamped Brownian oscillations (the so-called Kubo term), again yielding amplitudes (λK) and timescales (τK). Plastic motions are described as a strongly overdamped Brownian oscillation on an ‘infinite’ timescale (i.e. a timescale longer than the experimental time window), yielding the amplitude (λinh) of this contribution. The overall two-point correlation function used to deconvolute the experimental 3PEPS decay is thus:
| (1) |
for one or more Kubo contributions to describe multiple anelastic components.
Transient Absorption Spectroscopy.
The transient absorption apparatus includes a Ti:sapphire regenerative amplifier (Spectra-Physics Spitfire XP-Pro) which produces ~100-fs pulses with an energy of ~3 mJ at 800 nm and 1 kHz repetition rate. A 10/90 beam splitter was used to split the output of the regenerative amplifier: the larger fraction of the 800-nm pulse was frequency doubled in a 1-mm type I BBO crystal to produce the second harmonic (400 nm) pump pulses, and the smaller fraction of the 800-nm probe was used to generate a white light continuum probe pulse in a 2-mm thick sapphire plate mounted on a constantly moving translation stage (Zaber Technologies, T-NA08A). The pump beam was delayed relative to the probe beam with an optical delay line (Newport IMS600CCHA). The pump beam was mechanically chopped at half the repetition rate of the amplifier by an optical chopper (Thorlabs, Inc., MC200B). The pump and probe beams were focused into a 2-mm cuvette containing the sample and a micro-stir bar for sample mixing. The beam diameter at the focal point was ~200 μm with the pump pulse diameter slightly larger than the probe pulse diameter. The probe beam was directed into a spectrograph (Horiba Jobin-Yvon iHR-320 with a 300 groove/mm grating) coupled to a thermoelectrically cooled CCD detector (Andor Newton DU920N-BN). The CCD detection was synchronized to the amplifier repetition rate, and each pulse of the probe beam was recorded. LabView software was used to control the delay line and readout of the CCD. The instrument response (FWHM ~200 fs) was estimated from the coherent artifact produced by the solvent.
X-ray Crystallography.
Crystals of the Fab:MPTS complex were grown and X-ray diffraction data collected at APS 23-ID-D using an ADXV m300 CCD detector and processed with HKL-2000.34 Scaling of the data indicated an apparent space group of either P3112 or P3212, and molecular replacement (MR) was initially carried out in space groups P3112/P3212 using the individual variable and constant domains from Fab 29G12 (PDB entry 1MEX) as models, with Phaser V 2.1.4.35 However, refinement with Phenix36 stalled with Rfree values of about 33%. We then carried out MR in space group P1 and subsequently tested different space group choices with Zanuda.37 The results from Zanuda confirmed that the correct space group was indeed P32, but revealed that, in addition to the correct crystallographic origin, there were two pseudo-origins for this crystal form. Choosing the correct origin finally led to better refinement results, and twin refinement further improved R-values by about 4%. Thus, our final model is refined as a twinned crystal, with twin fraction of 0.49. Final Rwork and Rfree values were 22.1% and 25.2%. Final data collection and refinement statistics are listed in Table S1. Coordinates and data have been deposited in the Protein Data Bank as entry 6M87. Molecular graphics images were produced using PyMOL38 and the UCSF Chimera package.39
RESULTS
Characterization of Ab Sequence, Evolution, and MPTS Recognition.
Abs 5D11 and 10A6 were identified from a panel of Abs raised against MPTS in Swiss Webster mice. The Abs were sequenced from hybridoma mRNA and were found to be nearly identical (Figure 1). Their virtually identical VH recombination junctions indicate that they arose from a common germline precursor.32 The eight amino-acid differences between the Abs thus must have arisen during affinity maturation. Three of the mutations are located in the light chain (VL) and five are in the heavy chain (VH). To identify which amino acid was encoded in the progenitor germline Ab, we determined the likely germline sequence by homology search using the IMGT V-Quest database.41 The identified germline variable (V) and joining (J) genes for the light chain (IGKV4–59*01 and IGKJ5*01) and for the heavy chain (IGHV1–39*01 and IGHJ2*01) suggest that Ab 10A6 acquired three somatic mutations (MetL33Val, TyrH54Ser, GlyH65Asp) while Ab 5D11 acquired five (LysL53Thr, TyrL86His, AsnH41Ser, ValH50Leu, SerH76Asn). All three somatic mutations of 10A6 are located in complementarity-determining regions (CDRs, as defined by Kabat40), while three of the five somatic mutations of 5D11 are located in framework regions. In addition, comparison with the putative germline genes identified SerH58Asn, GlnL89His, SerL92Arg, and SerL93Thr as potential somatic mutations that were acquired prior to divergence; however, it is not possible to exclude allelic variation as a source of these differences. ITC revealed that both Abs have similar, low nanomolar affinities (KD) for MPTS achieved by a different combination of entropic and enthalpic contributions (Table 1).
Figure 1.
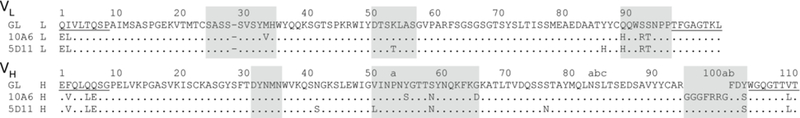
Amino-acid sequences of Abs 10A6 and 5D11, and their predicted germline precursor (GL; only sequence corresponding to the identified V and J gene segments is shown), with differences indicated. CDRs as defined by Kabat40 are highlighted. Regions installed by cloning primers (and thus where differences do not correspond to somatic mutations) are underlined. The dash in the VL sequence corresponds to a deletion according to Kabat numbering (Kabat numbering shown above40).
Table 1.
Thermodynamics Parameters at 25 °C Determined by ITC
| KD (nM) | ∆G° (kcal/mol) | ∆H° (kcal/mol) | T∆S° (kcal/mol) | |
|---|---|---|---|---|
| 5D11 | 6 ± 2 | −11.3 ± 0.2 | −15.2 ± 0.2 | −4.0 ± 0.1 |
| 10A6 | 8 ± 1 | −11.0 ± 0.1 | −10.4 ± 0.2 | 0.6 ± 0.1 |
Photophysical Characterization of the Ab-MPTS Complexes.
The UV/vis absorption spectrum of MPTS is similar when bound to Ab 10A6 or 5D11 but significantly different from that of the free chromophore (Figure 2). This is in stark contrast to the absorption spectra of all previously characterized Ab-MPTS complexes which, with the exception of small red-shifts, closely resemble that of the free chromophore.24,25,30 When bound to Ab 10A6 or 5D11, the absorption of the lowest-energy transition of MPTS (~410 nm) is significantly broadened and appears to result from the superposition of at least two vibronic transitions, while the vibronic band at ~375 nm remains mostly unchanged. Also in contrast to previously characterized MPTS-Ab complexes, the fluorescence of MPTS is strongly quenched when bound to Abs 10A6 or 5D11 (data not shown). While fluorescence quenching and the observed changes in the UV/vis absorption spectrum are consistent with the formation of H-type dimers or higher aggregates of MPTS, MPTS titrations revealed that at the concentrations employed, MPTS and Ab form a one-to-one complex,32 which is also supported by structural data (see below). Therefore the observed spectroscopic changes must originate from an interaction between MPTS and the Ab binding site.
Figure 2.
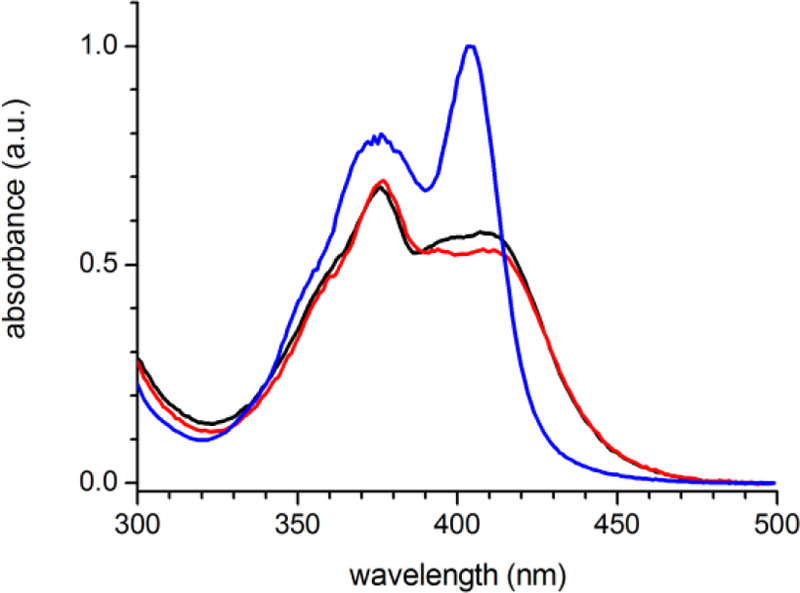
Absorption spectra of MPTS in buffer (blue line) and bound to Abs 5D11 (black line) or 10A6 (red line).
To determine the origin of the fluorescence quenching, we performed transient grating (TG) experiments with each complex, and somewhat surprisingly found long-lived TG signals with decay times of ~700 ps that are not significantly decreased relative to MPTS free in solution (Table 2 and Figure S2). Hence, the observed fluorescence quenching is not due to a fast radiationless decay of the fluorescent S1 state of MPTS back to the ground state. In addition, the lack of spectral changes in the transient absorption spectra of the complex (Figure 3)32 show that there is no excited-state population transfer. We therefore conclude that the observed spectral changes in absorption and the fluorescence quenching is due to the direct excitation of a long-lived, non-fluorescent excited state.
Table 2.
Fit Parameters for Multi-exponential TG Decays of MPTS
| buffer | 5D11 | 10Α6 | |
|---|---|---|---|
| Α1 | 1 | 1 | 1 |
| τ1 (ps) | 800 | 700 | 660 |
| Α2 | 0.053 | 0.024 | |
| τ2 (ps) | 3.6 | 3.6 | |
| Α3 | −0.11 | −0.12 | |
| τ3 (ps) | 0.079 | 0.090 |
Figure 3.
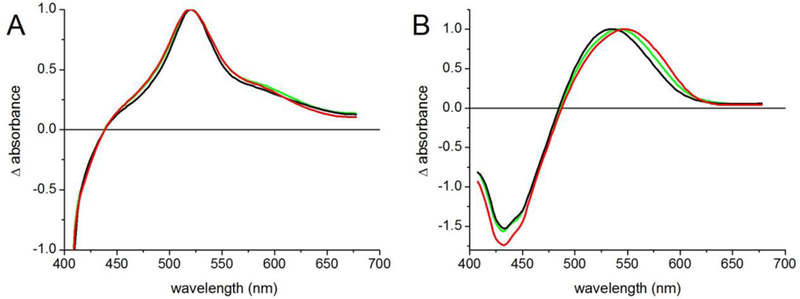
Normalized transient absorption spectra after 0.2 ps (red), 1 ps (green) and 50 ps (black) delay time for (A) MPTS bound to Ab 5D11, (B) MPTS in buffer.
Characterization of Ab-MPTS Complex Dynamics.
We employed 3PEPS spectroscopy to characterize the dynamics of the Ab complexes. The 3PEPS decay of MPTS bound to each Ab is virtually identical (Figure 4). Surprisingly, and in contrast with all previously characterized anti-MPTS23,26–29 or anti-fluorescein Abs,24,25,30 the peak shift decay is almost complete within the first 100 fs of population time. The remaining 3PEPS amplitude decays within a few picoseconds to virtually zero.
Figure 4.
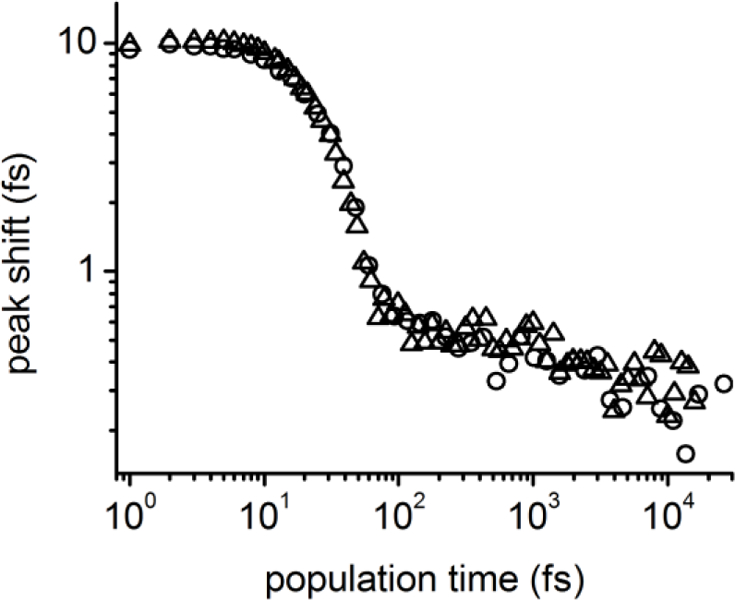
3PEPS decay for Ab 5D11 (circles) and 10A6 (triangles).
We used nonlinear response function theory to deconvolute the 3PEPS decay into contributions from elasticity, anelasticity, and plasticity. Specifically, a Brownian oscillator (BO) term was used to describe the sub-100 fs elastic motions, a Kubo term was used to describe the anelastic protein motions on the picosecond time scale, and static inhomogeneity was included to describe the plastic motions that occur on time scales longer than the experimental time window.
The spectral changes observed in the Ab-MPTS complexes (UV/vis absorbance, radiationless excited state relaxation) clearly demonstrate that the nature of the excited state is significantly altered compared to MPTS in solution. While the origins of the complexity, which could include stabilization of the mostly forbidden 1Lb state such that it is the lowest-energy state of the Ab-bound chromophore42 or formation of a non-fluorescent ground-state charge-transfer complex,43,44 remain to be elucidated, there is no evidence of excited state population transfer. We therefore modeled the MPTS-Ab complexes as two-level systems (ground state and a single excited state).22,33 Note that it is inconsequential for the fit results whether the system is modeled with a single excited state or several excited states, at least at room temperature where the imaginary part of the response functions of ground and excited state pathways can be neglected, as long as the quenching of the initially excited state does not preserve coherence.45
Simultaneous least-square fitting of the 3PEPS decays and absorption spectra of the MPTS-Ab complexes (Table 3 and Figure S3) reveals, as expected, that the Brownian oscillator term has by far the largest amplitude (λBO), while the Kubo term has a relatively low amplitude (λK) and there is virtually no static inhomogeneity (λinh).
Table 3.
3PEPS Fit Parameters
| λBO (cm−1) | ωBOa (cm−1) | ΓBOa (cm−1) | λK (cm−1) | τK (ps) | Δinh (cm−1) | |
|---|---|---|---|---|---|---|
| 10A6 | 730 ± 100 | 550 | 320 | 28 ± 3 | 3.9 ± 0.5 | 11 ± 4 |
| 5D11 | 680 ± 40 | 550 | 320 | 35 ± 8 | 3.5 ± 0.8 | 8.5 ± 5 |
Parameter fixed during fit.
Characterization of Ab Structure.
The crystal structure of the 10A6-MPTS complex was determined by molecular replacement to 2.6 Å resolution. Of the four likely pre-divergence mutations (SerH58Asn, GlnL89His, SerL92Arg, and SerL93Thr), the structure reveals that only AsnH58 appears to interact directly with MPTS and does so via H-bonding to one of the sulfonate groups. None of the post-divergence mutations acquired by Ab 10A6 (MetL33Val, TyrH54Ser, GlyH65Asp) make direct contact with MPTS, and based on the 10A6 structure, none of the post-divergence mutations acquired by 5D11 (LysL53Thr, TyrL86His, AsnH41Ser, ValH50Leu, and SerH76Asn in 5D11) are predicted to directly contact MPTS.
The structure reveals that MPTS binds at an approximate 50° angle relative to the pseudo 2-fold axis relating VL and VH domains in a shallow cleft that resembles a cupped hand (Figure 5A and 5B). Within this cleft, MPTS engages in eleven hydrogen bonds, one with a buried water molecule and ten with the Ab, six of which involve Ab side chains and four of which involve main-chain amides (Figure 5C). Two of the three sulfonate groups are buried and oriented into a pocket formed by a kink in the CDR H3 loop, which joins the palm to the thumb of the cupped hand. The two buried sulfonates engage in multiple H-bonds with the Ab: AsnH33 (ND2), AsnH35 (ND2), and GlyH97 (NH) form H-bonds with one sulfonate, which is also held in place via a water-mediated H-bonding network with the backbone of AsnH33, while TyrL32 (OH), HisL34 (NE2), ArgH99 (NH), ArgH100 (NH), and GlyH100a (NH) form H-bonds with the other. The remaining sulfonate is positioned near the outer edge of the palm, where it forms an H-bond with the side chains of AsnL94 and AsnH58. The palm itself is formed by the indole side chain of TrpL91, which engages in a parallel-stacking interaction with the MPTS. Finally, the tip of CDR H3 curves over the Ag and forms the thumb of the cupped hand. Interestingly, the different molecules in the crystal asymmetric unit present two different rotamers for PheH98 at the tip of the loop (Figures 5A and 5B). In one, the side chain is swung out and away from the center of MPTS, while in the other it is swung in and positioned directly over MPTS in a T-stacking interaction. However, even in the swung-out conformation, electron density is observed at the swung-in position, likely reflecting side-chain dynamics (Figure S10), although we cannot exclude the possibility that the density results from the presence of an unidentified solvent molecule.
Figure 5.
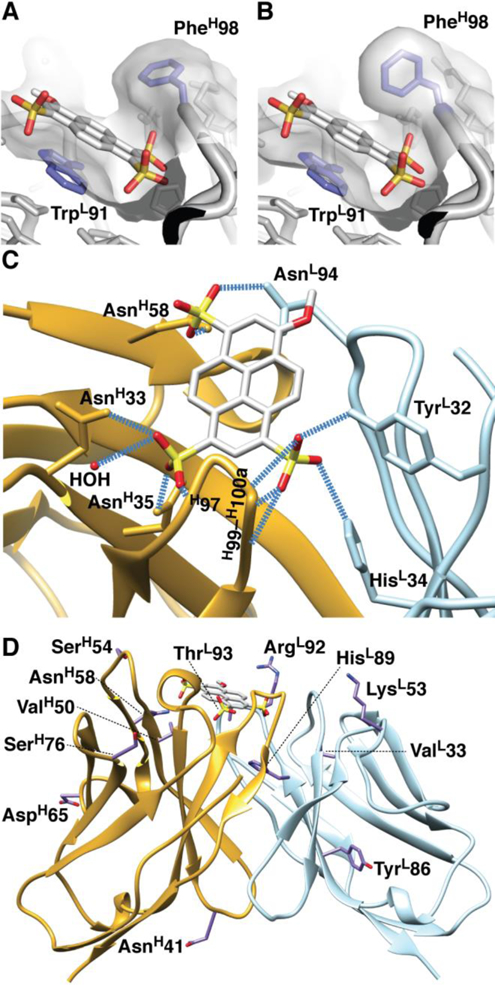
Crystal structure of 10A6-MPTS complex. (A) Swung-out and (B) swung-in conformations of PheH98 over MPTS. (C) H-bonds formed between Ab and Ag. (D) Sites of assigned or potential somatic mutations in 10A6 and/or 5D11 indicated on the 10A6 structure.
DISCUSSION
Abs have served as the paradigm for characterizing the structural-based aspects of molecular recognition. However, dynamics also contributes to molecular recognition, and with the ability to evolve Abs to specific chromophores that facilitate the characterization of binding site dynamics, Abs can also serve as the paradigm for characterizing the contribution of dynamics,23–26,29,30 and even how it is evolved.27,28 Towards this goal, we have raised Abs to several chromophores, including fluorescein23,26–29 and MPTS.24–28,30 Here, we characterized the anti-MPTS Abs 5D11 and 10A6, which based on analysis of their rearranged gene sequences, evolved from a common germline Ab precursor. Sequence analysis also suggests that four mutations were acquired before divergence: SerH58Asn, GlnL89His, SerL92Arg, and SerL93Thr (note that it is not possible to definitively identify these as somatic mutations as opposed to allelic variation). By comparison of the Ab gene sequences themselves, it is possible to unambiguously conclude that three and five mutations were acquired by 10A6 and 5D11, respectively, after divergence, MetL33Val, TyrH54Ser, and GlyH65Asp in 10A6, and LysL53Thr, TyrL86His, AsnH41Ser, ValH50Leu, and SerH76Asn in 5D11.
The ITC data reveals that the binding of MPTS by Ab 5D11 is more enthalpically favorable than binding by 10A6, while binding by 10A6 is more entropically favorable. Although it is not possible to interpret this data mechanistically in the absence of unbound Ab structures, especially considering the possible contribution of solvation,46–49 the data reveal that the mutations acquired by the two Abs have different effects on MPTS recognition. The similar and low nM affinity achieved by both pathways of affinity maturation likely reflects the threshold affinity required for Ab function during the immune response. The absorption spectra of MPTS bound to Abs 5D11 or 10A6 are very similar, but relative to either MPTS free in solution or bound to the previously characterized Abs, they are unusually broad and MPTS fluorescence is strongly quenched. While such spectral changes are reminiscent of H-aggregates, titration and structural data show that MPTS and the Ab form a one-to-one complex. Therefore the spectral changes must originate from an interaction between MPTS and the protein.
Regardless of the complex photophysics, there is no evidence of excited-state population transfer on timescales longer than 50 fs and even if it occurs, the 3PEPS data can be interpreted in terms of protein dynamics as long as it does not preserve coherence.45 Within this model, the dynamics observed with Ab 10A6 and 5D11 are virtually identical, and remarkably, the 3PEPS decays are nearly complete by 100 fs. Sub-100 fs components often contribute to 3PEPS decays and are attributed to a combination of intramolecular vibrational relaxation and elastic (impulsive) motions of solvent molecules and/or protein side chains in the vicinity of the chromophore. However, after the initial decay, a significant peak shift amplitude usually remains, due to persisting inhomogeneity that decays on longer time scales and that reflects the presence of anelasticity and plasticity.24,25 When fit using nonlinear response function theory, we found that elastic motions mediate 95% of the response of Abs 10A6 and 5D11 to MPTS excitation. In contrast, for the eleven previously characterized anti-MPTS Abs, significant contributions of anelasticity and plasticity limited the contribution of elasticity to 67% to 85% of the total response, with an average of 77% (Table 4). To our knowledge, the only other protein in which such a dominance of elastic (impulsive) motion has been observed is bacteriorhodopsin, where it is attributed to small-amplitude motions of many residues dispersed around the chromophore.50
Table 4.
Relative Amplitudes of Elastic (λBO), Anelastic (λK) and Plastic (λinh) Motions
The structure of the 10A6-MPTS complex (and presumably the 5D11-MPTS complex) reveals that MPTS binds in a cleft that resembles a cupped hand (Figures 5A and 5B). One sulfonate forms an H-bond with the Ab residue at the open side of the hand, while the other two are buried and form H-bonds with residues at the connection of the palm and thumb. The planar core of MPTS is sandwiched between the side chains of TrpL91, which forms the palm of the cupped-like hand, and PheH98, which is centrally located in the CDR H3 loop that forms the thumb. While a majority of the somatic mutations are located in CDR loops, the structure also reveals that none actually directly contact MPTS, with the single exception of AsnH58 of Ab 10A6, which participates in an H-bond with MPTS. Thus, the majority of the somatic mutations must exert any effect they have on binding indirectly. For example, in 10A6, the MetL33Val mutation in CDR L1 may optimize MPTS recognition, as this residue’s side chain is anchored to the core of the Ab via packing interactions with the side chains of TyrL71 and ValL30, and its backbone carbonyl links the CDR L1 and L2 loops via an H-bond with the backbone NH of ThrL51. In 5D11, ThrL53 is located near the tip of CDR L2 with its side chain predicted to be directed toward solvent; however its methyl group is predicted to be positioned ~4–5 Å from the methylene group of AspL50, which in turn is positioned ~5 Å from the methylene of TyrL32, thereby potentially providing some stabilization between CDR L1 and CDR L2. AsnH76 is located in a framework region with its side chain predicted to be packed against PheH29, which is at the N-terminal base of CDR H1. LeuH50 is positioned at the floor of the Ag binding site where the additional methylene may contribute to binding via packing interactions with AsnH35 and AsnH58 (the latter is a likely pre-divergence mutation and forms an H-bond with a sulfonate of MPTS).
While the sequence and ITC data reveal that the two Abs evolved from their common germline precursor via unique pathways, the observed dynamics of the complexes are virtually identical. This likely results from the relaxation of both complexes after MPTS excitation being dominated by interactions that are present in both of the 10A6 and 5D11 complexes, but absent in previously characterized complexes.24 The differences in entropic and enthalpic contributions to Ag-Ab binding revealed by the ITC data may reflect differences in the dynamics that are masked by the common interactions that dominate the response in the complexes, and/or differences in the motions or solvation of the free Abs.
When considering the possible origin of the observed motion in the two complexes, it is interesting to note that MPTS bound to 10A6 (and presumably 5D11) is significantly more buried than in previously reported complexes. Consequently, it is significantly more sequestered from water and its sulfonates form more H-bonds with the Ab. While this suggests that the increase in elasticity observed with 10A6 and 5D11 relative to the previously characterized Abs does not result from increased solvent exposure, the H-bonding interactions are not expected to be qualitatively different. The most obvious qualitative difference between the 10A6 and 5D11 complexes and those previously characterized is the sandwiching of MPTS between the side chains of TrpL91 and PheH98. It is likely that the relaxation of both complexes after MPTS excitation is dominated by these interactions. Both the unusual photophysical properties and the ultrafast, elastic motion likely result from the parallel stacking interaction between MPTS and the indole side chain of TrpL91. Indeed, similar stacking interactions in other proteins are known to result in the formation of ground state charge transfer complexes,51–55 which is consistent with the observed fluorescence quenching. Moreover, it is likely that the indole moiety and MPTS are strongly electronically coupled,56–58 such that even small, elastic motions of the side chain significantly broaden the absorption spectrum and provide an efficient channel for vibrational relaxation of the excited complex. Nonetheless, it is perhaps surprising that the induced motions appear to be exclusively elastic. This appears to result from the indole side chain being rigidly held in place by intra-protein interactions, apparently within a single potential energy minimum. In particular, the indole side chain appears to be rigidly held against MPTS through tight packing with the side chains of ValH50, AsnH35, PheH100b, HisL89, ProL96, and TrpH47, and also by AsnL94, which H-bonds to both the indole nitrogen of TrpL91 and MPTS (Figure S11). Hence, the relative motion between MPTS and the “palm” of the binding site appears to be limited to elastic motion, resulting in the unusually large amplitude of the sub-100 fs 3PEPS decay.
The structure of the 10A6 (and presumably the 5D11) complex also reveals that the side chain of PheH98, which is part of the “thumb” of the binding site, directly interacts with MPTS, and therefore its motion is also expected to make significant contributions to the 3PEPS decay. Interestingly, two different rotamers in the different molecules in the asymmetric unit of the crystal structure were observed for the PheH98 side chain, one centered above and engaged in a T-stacking interaction with the bound MPTS, and a second flipped away from the center of the MPTS via a 120° rotation about its Cα-Cβ bond. No static inhomogeneity was detected in the 3PEPS decay, which would be expected if the PheH98 side chain, which is also likely electrostatically coupled to MPTS, interconverted between the observed states on a timescale longer than a few hundred picoseconds. This, along with the previously characterized picosecond timescale observed for the motion of Phe side chains in other proteins,59–65 suggests that interconversion between the crystallographically observed conformations of PheH98 give rise to the picosecond-timescale anelastic dynamics observed in both Ab-MPTS complexes.
CONCLUSION
The dynamics of the 10A6 and 5D11 Abs are distinct from previously characterized Abs, and the differences appear to result from the motions associated with the T-stacking and parallel stacking interactions employed by both Abs to recognize MPTS. Despite the absence of these motifs in the previously characterized Ab structures, they are ubiquitous in proteins and protein-ligand complexes.52–55,66–68 Our data suggest that the two motifs contribute to dynamics on distinct timescales, and it is possible that such dynamics are functionally relevant. For example, it seems unlikely that MPTS could dissociate from the Ab with PheH98 in the T-stacked conformation, and thus the picosecond timescale motion assigned to this residue could have important implications for gated ligand recognition.69,70 The ultrafast motion assigned to TrpL91 suggests that similar parallel stacking interactions might contribute to the accommodation of structural and electronic changes in other proteins and protein complexes and provide efficient channels for energy dissipation.
We have shown previously that the immune system is capable of generating Abs with either rigid or flexible binding sites, and the current work extends these results and demonstrates that the immune system is capable of using different stacking interactions to generate dynamically diverse binding sites. This may contribute to the immune system’s ability to evolve Abs to virtually any foreign molecule, and possibly, as we have previously speculated,27,28,71 to its ability to tailor dynamics such that mature Abs have a sufficient level of specificity to be produced at the high levels required to eradicate an infection without problematic self-binding.27,28,71
Supplementary Material
ACKNOWLEDGMENT
We thank Julie Vanhasy and Kendra Pivaroff-Ward for Fab preparation and purification. We gratefully acknowledge Prof. Michael Tauber (UCSD) and Joel Rivera (UCSD) for assistance with the transient absorption spectroscopy.
Funding Sources
This work was supported by the National Institute of Allergy and Infectious Diseases (AI079319 to F.E.R.) and R56 AI127371 (to I.A.W.). Crystallographic data were collected at GM/CA@APS, which has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357.
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.
Supplementary methods and discussion, Ab gene sequences, Supplementary Table S1 and Figures S1–S11 (PDF)
REFERENCES
- (1).Davies DR, Padlan EA, and Sheriff S (1990) Antibody-antigen complexes, Annu. Rev. Biochem 59, 439–473. [DOI] [PubMed] [Google Scholar]
- (2).Wedemayer GJ, Patten PA, Wang LH, Schultz PG, and Stevens RC (1997) Structural insights into the evolution of an antibody combining site, Science 276, 1665–1669. [DOI] [PubMed] [Google Scholar]
- (3).Ma B, Kumar S, Tsai CJ, and Nussinov R (1999) Folding funnels and binding mechanisms, Protein Eng 12, 713–720. [DOI] [PubMed] [Google Scholar]
- (4).Patten PA, Gray NS, Yang PL, Marks CB, Wedemayer GJ, Boniface JJ, Stevens RC, and Schultz PG (1996) The immunological evolution of catalysis, Science 271, 1086–1091. [DOI] [PubMed] [Google Scholar]
- (5).Yin J, Andryski SE, Beuscher A. E. t., Stevens RC, and Schultz PG (2003) Structural evidence for substrate strain in antibody catalysis, Proc. Natl. Acad. Sci. USA 100, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Li Y, Li H, Smith-Gill SJ, and Mariuzza RA (2000) Three-dimensional structures of the free and antigen-bound Fab from monoclonal antilysozyme antibody HyHEL-63, Biochemistry 39, 6296–6309. [DOI] [PubMed] [Google Scholar]
- (7).Kondo H, Shiroishi M, Matsushima M, Tsumoto K, and Kumagai I (1999) Crystal structure of anti-hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex - Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain-heavy chain interfaces, J. Biol. Chem 274, 27623–27631. [DOI] [PubMed] [Google Scholar]
- (8).Mohan S, Sinha N, and Smith-Gill SJ (2003) Modeling the binding sites of anti-hen egg white lysozyme antibodies HyHEL-8 and HyHEL-26: an insight into the molecular basis of antibody cross-reactivity and specificity, Biophys J 85, 3221–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Li YL, Li HM, Yang F, Smith-Gill SJ, and Mariuzza RA (2003) X-ray snapshots of the maturation of an antibody response to a protein antigen, Nat. Struct. Biol 10, 482–488. [DOI] [PubMed] [Google Scholar]
- (10).Cauerhff A, Goldbaum FA, and Braden BC (2004) Structural mechanism for affinity maturation of an anti-lysozyme antibody, Proc. Natl. Acad. Sci. USA 101, 3539–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Khan T, and Salunke DM (2012) Structural elucidation of the mechanistic basis of degeneracy in the primary humoral response, J. Immunol 188, 1819–1827. [DOI] [PubMed] [Google Scholar]
- (12).Wang W, Ye W, Yu Q, Jiang C, Zhang J, Luo R, and Chen HF (2013) Conformational selection and induced fit in specific antibody and antigen recognition: SPE7 as a case study, J. Phys. Chem. B 117, 4912–4923. [DOI] [PubMed] [Google Scholar]
- (13).James LC, Roversi P, and Tawfik DS (2003) Antibody multispecificity mediated by conformational diversity, Science 299, 1362–1367. [DOI] [PubMed] [Google Scholar]
- (14).Manivel V, Sahoo NC, Salunke DM, and Rao KVS (2000) Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site, Immunity 13, 611–620. [DOI] [PubMed] [Google Scholar]
- (15).Sagawa T, Oda M, Ishimura M, Furukawa K, and Azuma T (2003) Thermodynamic and kinetic aspects of antibody evolution during the immune response to hapten, Mol. Immunol 39, 801–808. [DOI] [PubMed] [Google Scholar]
- (16).Wong SE, Sellers BD, and Jacobson MP (2011) Effects of somatic mutations on CDR loop flexibility during affinity maturation, Proteins 79, 821–829. [DOI] [PubMed] [Google Scholar]
- (17).Li T, Tracka MB, Uddin S, Casas-Finet J, Jacobs DJ, and Livesay DR (2015) Rigidity emerges during antibody evolution in three distinct antibody systems: evidence from QSFR analysis of Fab fragments, PLoS Comput. Biol 11, e1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Babor M, and Kortemme T (2009) Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility, Proteins 75, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Fernández-Quintero ML, Loeffler JR, Kraml J, Kahler U, Kamenik AS, and Liedl KR (2019) Characterizing the diversity of the CDR-H3 loop conformational ensembles in relationship to antibody binding properties, Front Immunol 9, 3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cho M, Yu J-Y, Joo T, Nagasawa Y, Passino SA, and Fleming GR (1996) The integrated photon echo and solvation dynamics, J. Phys. Chem 100, 11944–11953. [Google Scholar]
- (21).de Boeij WP, Pshenichnikov MS, and Wiersma DA (1996) System-bath correlation function probed by conventional and time-grated stimulated photon echo, J. Phys. Chem 100, 11806–11823. [Google Scholar]
- (22).Fleming GR, and Cho MH (1996) Chromophore-solvent dynamics, Annu. Rev. Phys. Chem 47, 109–134. [Google Scholar]
- (23).Thielges MC, Zimmermann J, Yu W, Oda M, and Romesberg FE (2008) Exploring the energy landscape of antibody-antigen complexes: protein dynamics, flexibility, and molecular recognition, Biochemistry 47, 7237–7247. [DOI] [PubMed] [Google Scholar]
- (24).Adhikary R, Yu W, Oda M, Walker RC, Chen T, Stanfield RL, Wilson IA, Zimmermann J, and Romesberg FE (2015) Adaptive mutations alter antibody structure and dynamics during affinity maturation, Biochemistry 54, 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Adhikary R, Yu W, Oda M, Zimmermann J, and Romesberg FE (2012) Protein dynamics and the diversity of an antibody response, J. Biol. Chem 287, 27139–27147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jimenez R, Salazar G, Baldridge KK, and Romesberg FE (2003) Flexibility and molecular recognition in the immune system, Proc. Natl. Acad. Sci. USA 100, 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jimenez R, Salazar G, Yin J, Joo T, and Romesberg FE (2004) Protein dynamics and the immunological evolution of molecular recognition, Proc. Natl. Acad. Sci. USA 101, 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zimmermann J, Oakman EL, Thorpe IF, Shi XH, Abbyad P, Brooks CL, Boxer SG, and Romesberg FE (2006) Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics, Proc. Natl. Acad. Sci. USA 103, 13722–13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zimmermann J, Romesberg FE, Brooks CL 3rd, and Thorpe IF (2010) Molecular description of flexibility in an antibody combining site, J. Phys. Chem. B 114, 7359–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Jimenez R, Case DA, and Romesberg FE (2002) Flexibility of an antibody binding site measured with photon echo spectroscopy, J. Phys. Chem. B 106, 1090–1103. [Google Scholar]
- (31).Harlow E, and Lane D (1988) Antibodies: a Laboratory Manual, pp 371–396, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- (32). See the Supporting Information for complete details.
- (33).Mukamel S (1995) Principles of Nonlinear Optical Spectroscopy, Oxford University Press, New York. [Google Scholar]
- (34).Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode, Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- (35).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software, J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine, Acta Cryst. Sect. D, Biol. Cryst 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lebedev AA, and Isupov MN (2014) Space-group and origin ambiguity in macromolecular structures with pseudo-symmetry and its treatment with the program Zanuda, Acta Cryst. Sect. D, Biol. Cryst 70, 2430–2443. [DOI] [PubMed] [Google Scholar]
- (38).The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC. [Google Scholar]
- (39).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis, J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (40).Kabat EA, and Wu TT (1991) Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites, J. Immunol 147, 1709–1719. [PubMed] [Google Scholar]
- (41).Brochet X, Lefranc MP, and Giudicelli V (2008) IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis, Nucleic Acids Res 36, W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Spry DB, Goun A, Bell CB 3rd, and Fayer MD (2006) Identification and properties of the 1La and 1Lb states of pyranine, J. Chem. Phys 125, 144514. [DOI] [PubMed] [Google Scholar]
- (43).Fornasiero D, and Grieser F (1990) Study of the absorption spectra of pyrene complexed to paraquat in pentanol-sodium docecylsulphate micelles, J. Chem. Soc. Faraday Trans 86, 2955–2960. [Google Scholar]
- (44).Gehlen M, and De Schryver F (1993) Fluorescence quenching in micelles in the presence of a probe-quencher ground-state charge-transfer complex, J. Phys. Chem 97, 11242–11248. [Google Scholar]
- (45).Yang M, Ohta K, and Fleming GR (1999) Three-pulse photon echoes for model reactive systems, J. Chem. Phys 110, 10243–10252. [Google Scholar]
- (46).Bhat TN, Bentley GA, Boulot G, Greene MI, Tello D, Dall’Acqua W, Souchon H, Schwarz FP, Mariuzza RA, and Poljak RJ (1994) Bound water molecules and conformational stabilization help mediate an antigen-antibody association, Proc. Natl. Acad. Sci. USA 91, 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Guinto ER, and Di Cera E (1996) Large heat capacity change in a protein-monovalent cation interaction, Biochemistry 35, 8800–8804. [DOI] [PubMed] [Google Scholar]
- (48).Yokota A, Tsumoto K, Shiroishi M, Kondo H, and Kumagai I (2003) The role of hydrogen bonding via interfacial water molecules in antigen-antibody complexation. The HyHEL-10-HEL interaction, J. Biol. Chem 278, 5410–5418. [DOI] [PubMed] [Google Scholar]
- (49).Stites WE (1997) Protein-protein interactions: Interface structure, binding thermodynamics, and mutational analysis, Chem. Rev 97, 1233–1250. [DOI] [PubMed] [Google Scholar]
- (50).Kennis JTM, Larsen DS, Ohta K, Facciotti MT, Glaeser RM, and Fleming GR (2002) Ultrafast protein dynamics of bacteriorhodopsin probed by photon echo and transient absorption spectroscopy, J. Phys. Chem. B 106, 6067–6080. [Google Scholar]
- (51).Watt RM, and Voss EW Jr. (1977) Mechanism of quenching of fluorescein by anti-fluorescein IgG antibodies, Immunochemistry 14, 533–551. [DOI] [PubMed] [Google Scholar]
- (52).Jackson WR, and Dwek RA (1981) On the contribution of tryptophan to the affinity and specificity of anti-dinitrophenyl antibodies, Mol. Immunol 18, 499–506. [DOI] [PubMed] [Google Scholar]
- (53).Gudgin Templeton EF, and Ware WR (1985) Charge transfer between fluorescein and tryptophan as a possible interaction in the binding of fluorescein to anti-fluorescein antibody, Mol. Immunol 22, 45–55. [DOI] [PubMed] [Google Scholar]
- (54).Droupadi PR, Meyers EA, and Linthicum DS (1994) Spectroscopic evidence for charge-transfer complexation in monoclonal antibodies that bind opiates, J. Protein Chem 13, 297–306. [DOI] [PubMed] [Google Scholar]
- (55).Little JR, and Eisen HN (1967) Evidence for tryptophan in the active sites of antibodies to polynitrobenzenes, Biochemistry 6, 3119–3125. [DOI] [PubMed] [Google Scholar]
- (56).Xu Q-H, Scholes GD, Yang M, and Fleming GR (1999) Probing solvation and reaction coordinates of ultrafast photoinduced electron-transfer reactions using nonlinear spectroscopies: rhodamine 6G in electron-donating solvents, J. Phys. Chem. A 103, 10348–10358. [Google Scholar]
- (57).Bruckner RC, Zhao G, Ferreira P, and Jorns MS (2007) A mobile tryptophan is the intrinsic charge transfer donor in a flavoenzyme essential for nikkomycin antibiotic biosynthesis, Biochemistry 46, 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Markel F, Ferris NS, Gould IR, and Myers AB (1992) Mode-specific vibrational reorganization energies accompanying photoinduced electron transfer in the hexamethylbenzene/tetracyanoethylene charge-transfer complex, J. Am. Chem. Soc 114, 6208–6219. [Google Scholar]
- (59).Frey MH, DiVerdi JA, and Opella SJ (1985) Dynamics of phenylalanine in the solid state by NMR, J. Am. Chem. Soc 107, 7311–7315. [Google Scholar]
- (60).Gall CM, DiVerdi JA, and Opella SJ (1981) Phenylalanine ring dynamics by solid-state deuterium NMR, J. Am. Chem. Soc 103, 5039–5043. [Google Scholar]
- (61).Wagner G, DeMarco A, and Wuthrich K (1976) Dynamics of the aromatic amino acid residues in the globular conformation of the basic pancreatic trypsin inhibitor (BPTI). I. 1H NMR studies, Biophys. Struct. Mech 2, 139–158. [DOI] [PubMed] [Google Scholar]
- (62).Gall CM, Cross TA, DiVerdi JA, and Opella SJ (1982) Protein dynamics by solid-state NMR: aromatic rings of the coat protein in fd bacteriophage, Proc. Natl. Acad. Sci. USA 79, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kasinath V, Valentine KG, and Wand AJ (2013) A 13C labeling strategy reveals a range of aromatic side chain motion in calmodulin, J. Am. Chem. Soc 135, 9560–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Kasinath V, Fu Y, Sharp KA, and Wand AJ (2015) A sharp thermal transition of fast aromatic-ring dynamics in ubiquitin, Angew. Chem. Int. Ed 54, 102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Levy RM, and Szabo A (1982) Initial fluorescence depolarization of tyrosines in proteins, J. Am. Chem. Soc 104, 2073–2075. [Google Scholar]
- (66).McGaughey GB, Gagne M, and Rappe AK (1998) pi-Stacking interactions. Alive and well in proteins, J. Biol. Chem 273, 15458–15463. [DOI] [PubMed] [Google Scholar]
- (67).Stornaiuolo M, De Kloe GE, Rucktooa P, Fish A, van Elk R, Edink ES, Bertrand D, Smit AB, de Esch IJ, and Sixma TK (2013) Assembly of a pi-pi stack of ligands in the binding site of an acetylcholine-binding protein, Nat. Commun 4, 1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).An Y, Bloom JWG, and Wheeler SE (2015) Quantifying the π-Stacking Interactions in Nitroarene Binding Sites of Proteins, J. Phys. Chem. B 119, 14441–14450. [DOI] [PubMed] [Google Scholar]
- (69).Karplus M, and McCammon JA (2002) Molecular dynamics simulations of biomolecules, Nat. Struct. Biol 9, 646–652. [DOI] [PubMed] [Google Scholar]
- (70).Zhou HX, and McCammon JA (2010) The gates of ion channels and enzymes, Trends Biochem. Sci 35, 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Goodman MF, Scharff MD, and Romesberg FE (2007) AID-initiated purposeful mutations in immunoglobulin genes, Adv. Immunol 94, 127–155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


