Summary
Allergic rhinitis is thought to be an allergic disease associated with immunoglobulin (Ig)E‐mediated immune response, characterized by increased T helper type 2 (Th2) cytokine production, elevated eosinophil levels in the nasal mucosa and induced nasal secretions. MicroRNA (miRNA) microarray data revealed that the expression level of miR‐466a‐3p was significantly decreased. Notably, GATA binding protein (GATA‐3) was identified as one of its target genes through miRNA target prediction web tools. The expression levels of miR‐466a‐3p were altered by mimics and lentivirus both in vivo and in vitro, similar to those of GATA‐3. Furthermore, the symptoms and histology of allergic rhinitis as well as the levels of serum IgE and interleukin (IL)‐4 were examined in different groups of mice. Interestingly, the results for lentiviral miR‐466a‐3p‐treated allergic rhinitis mice were relatively similar to normal mice, compared to allergic rhinitis mice without treatment. Also, miR‐466a‐3p negatively regulated GATA‐3 expression in allergic rhinitis mice, indicating the participant of Th2‐cell responses in allergic rhinitis. Taken together, our findings highlight a new perspective on the role of miR‐466a‐3p in allergic rhinitis. In addition, this study provides a theoretical framework and experimental reference for future research targeting microRNAs as therapeutic targets and diagnostic biomarkers of allergic rhinitis.
Keywords: allergic rhinitis, GATA‐3, microRNA, miR‐466a‐3p, Th2 cell
Introduction
Due to changing environmental and social factors, the incidence of allergic rhinitis is rising 1, 2. According to a national telephone survey, the prevalence of allergic rhinitis ranges from 8·5 to 21·3% among 11 cities in China 3. Allergic rhinitis has a great influence on daily life, especially sleep, memory, mood and work performance. It is noteworthy that the risk of developing asthma in patients with allergic rhinitis is three times greater than in normal individuals. Moreover, allergic rhinitis may serve as a stimulating factor for asthma. Therefore, allergic rhinitis has been considered a disease, rather than a condition, which deserves more attention.
The pathogenesis of allergic rhinitis is complex, which involves the production of multiple types of inflammatory cells and cytokines during the course of its development. Treatment for single types of inflammatory cells and cytokines can only alleviate the symptoms of allergic rhinitis patients to a certain extent. Therefore, changing the immune state of imbalance may overcome allergic rhinitis 4, 5, 6. A previous study has reported that the molecular mechanism underlying the allergic rhinitis immune response involves the interaction of a complex cascade of cellular events 7. The abnormal differentiation of CD4+ helper T lymphocytes [T helper (Th) cells] and the expression patterns of related cytokines are the developmental basis of allergic rhinitis 8. CD4+ Th cells secrete the corresponding cytokines after being activated, and the cytokines can be divided into two subsets, Th1 and Th2, depending on their specific functions. In the normal state, both Th1 and Th2 cells maintain the immune homeostasis. During the disease state (i.e. allergic rhinitis and asthma), Th1 cells appear as immune response suppression, whereas Th2 cells are responsible for hyperfunction. The differentiation of Th cells depends mainly on the cytokines in the local environment and the modulation of key transcription factors 9. Hence, transcription factors play an important role in the regulation of Th cell differentiation.
MicroRNA (miRNA) is a type of small molecule non‐coding RNA, which plays an important role in post‐transcriptional gene regulation 10, 11. MiRNA acts through the inhibition of target gene translation and/or degradation of target mRNAs. Recent studies have found that miRNAs are involved in the generation and differentiation of immune cells, regulate both innate and adaptive immune responses and are closely related to immune system disorders 12, 13, 14.
In the present study, ovalbumin (OVA) was used to construct a mouse model of allergic rhinitis. Differential miRNA expression profiling in the nasal mucosa of allergic rhinitis and normal mice was detected using miRNA array technology. The relationship between the identified miRNA and its target genes was predicted by correlation software. In the subsequent analyses, the role of miRNA in murine allergic rhinitis was explored.
Materials and methods
Mice
Five‐week‐old male BALB/c mice, raised and maintained under specific pathogen‐free conditions, were obtained from the Center for Disease Control and Prevention in Hubei province, China. BALB/c mice were sensitized by intraperitoneal injection with 500 μl phosphate‐buffered saline (PBS) containing 10 μg of ovalbumin (OVA; grade V; Sigma, St Louis, MO, USA) and 1 mg of aluminum hydroxide on days 0, 7 and 14. Allergic rhinitis was elicited by intranasal challenge with 20 μL PBS containing 500 μg of OVA for 7 days, from days 21 to 27. The mice in the normal group were injected with PBS alone, and PBS was administered intranasally using the same schedule. Along with sensitization and challenge, parts of allergic rhinitis mice were given lentiviral miR‐466a‐3p or empty lentiviral vector (GeneChem, Shanghai, China) by tail‐vein injection before intranasal OVA challenge on days 22, 24 and 26. Each group comprised eight to 10 mice. This study was approved by the Animal Ethics Committee of Renmin Hospital of Wuhan University (Fig. 1).
Figure 1.
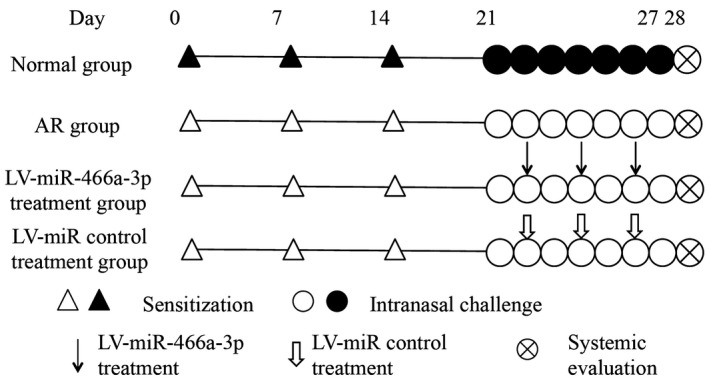
The experimental protocol of the present study. Mice were sensitized with ovalbumin (OVA) and aluminum hydroxide on days 0, 7 and 14, while phosphate‐buffered saline (PBS) was used for the mice in normal group. All groups received intranasal OVA between days 21 and 27, except for the normal group. Selected groups of mice were treated with lentiviral (LV)‐miR‐466a‐3p or lentivirus (LV)‐miR control via tail‐vein injection 2 h before intranasal OVA challenge on days 22, 24 and 26.
Measurement of nasal symptoms and tissue preparation
Twenty min after final OVA/PBS challenge on day 27, blinded observers recorded the frequencies of nasal rubbing and sneezing in each group. Twenty‐four h after the last challenge with OVA/PBS, the nasal mucosa was taken out meticulously using a small curette under a microscope and was immersed immediately in liquid nitrogen and stored until use for microRNA array detection, real‐time reverse transcription–polymerase chain reaction (RT–PCR) and Western blotting.
MicroRNA array detection
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) from nasal mucosa in allergic rhinitis and control mice, respectively, according to the manufacturer’s instructions. We then submitted the samples to KangChen‐Biotech (Shanghai, China) for array hybridization on a miRCURY LNA microRNA array (version 16.0; Exiqon, Vedbaek, Denmark). Each microarray chip was hybridized with a single sample labeled with either cyanin 3 (Cy3) or Cy5. Background subtraction and normalization were performed.
Real‐time quantitative RT–PCR (RT–qPCR)
Total RNA from the nasal mucosa was extracted using TRIzol reagent. The RNA (2 μg) was then reverse‐transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instruction. The expression of miR‐466a‐3p and GATA binding protein (GATA‐3) was quantified by real‐time RT–qPCR. We utilized specific target gene primers for miR‐466a‐3p, U6, GATA‐3 and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) in conjunction with SYBR‐Green PCR Master Mix (TaKaRa, Tokyo, Japan), according to the manufacturer’s protocol. qPCR was performed by an ABI PRISM7500 system (Applied Biosystems, Foster City, CA, USA). Data were analyzed using the 2−ΔΔCT method, as previously described 15.
Luciferase activity assay
For 3′‐untranslated region (3′‐UTR) assays, human embryonic kidney cell line 293T cells were co‐transfected with plasmid containing miR‐control or miR‐466a‐3p and firefly luciferase reporter constructs containing wild‐type (WT) or mutated (MU) 3′‐UTR of GATA‐3 using lipofectamine 2000 reagent (Invitrogen) and a renilla luciferase reporter vector to normalize the transfection efficiency. Renilla and firefly luciferase activities were measured 24 h post‐transfection using the dual‐luciferase reporter assay system (Promega, Madison, WI, USA). The luciferase activities represent the firefly/renilla luciferase ratios, and transfection efficiency was normalized to the control luciferase.
Histopathological evaluation of nasal cavity
Nasal tissues were removed 24 h after the last challenge with OVA/PBS, and part of them were fixed in 10% neutral buffered formalin and decalcified. Coronal nasal section (5 μm) was stained with hematoxylin and eosin (H&E), and the number of eosinophils was counted under a microscope at four random fields of submucosal region of nasal cavity with ×400 magnification.
Enzyme‐linked immunosorbent assay (ELISA)
Serum levels of anti‐OVA specific immunoglobulin (Ig)E (R&D Systems, Minneapolis, MN, USA) and interleukin (IL)‐4 (e‐Bioscience, San Diego, CA, USA) in animals were measured using commercially available ELISA, according to the manufacturer’s recommendations. The concentrations of anti‐OVA specific IgE and IL‐4 were calculated from the equations obtained from standard curve plots for the standard solutions in the kits.
Western blot
Proteins were obtained from the nasal mucosa of each group using lysing buffer (0·5% Triton X‐100, 150 mM NaCl, 15 mM Tris (pH 7.4) 1 mM CaCl2, 1 mM MgCl2). Protein concentrations were determined using BCA protein assay reagent (Thermo Fisher Scientific). Samples were separated on 12% sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gels and transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Billerica, MA, USA) and immunoblotted with primary antibodies against GATA‐3 and GAPDH (Cell Signaling Technology, Danvers, MA, USA). After immunoblotting with secondary antibodies, proteins were detected with enhanced chemiluminescence (ECL) reagent.
Splenocyte culture
Spleens from euthanized mice were collected aseptically. Single‐cell suspensions were prepared in complete RPMI‐1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin and were stimulated with 5 μg/ml concanavalin A (Sigma) for 48 h. The cells were cultured at 37°C in a CO2 incubator. Splenocyte were transfected with 50 nM miR‐466a‐3p mimics and mimics control, according to the manufacturer’s instructions. The plates were centrifuged at 1000 g, cells collected and used for analysis 12.
MicroRNA transfection
Scramble miRNA mimics (miR‐control), miR‐466a‐3p mimics were purchased from Ribobio (Guangzhou, China). Before transfection, splenocyte cells were seeded in six‐well plates (80 000 cells per well) and cultured in RPMI‐1640 medium containing 10% FBS for 12 h. For transient over‐expression of miR‐466a‐3p, cells were transfected with 50 nM miR‐466a‐3p or miR‐control using lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Samples were collected after 24 h for quantication of miRNA or protein expression.
Statistical analysis
All data are presented as the mean ± standard deviation (s.d.) or mean ± standard error of the mean (s.e.m.). Differences between groups were compared using the non‐parametric Kruskal–Wallis test, followed by the Mann–Whitney U‐test. For all other analyses, two‐tailed t‐test or analysis of variance (ANOVA) with Bonferroni adjustment for multiple comparisons were used. Statistical calculations were performed using IBM SPSS Statistics version 19.0 software.
Results
Differential miRNA expression in the nasal mucosa of allergic rhinitis mice
To investigate the differences in microRNA expression between the allergic rhinitis and control groups, miRNA array was used to profile global microRNA expression in the nasal mucosa of allergic rhinitis and control mice. It was found that the microRNAs were differentially expressed in mice with allergic rhinitis, including 25 down‐regulated and 57 up‐regulated miRNAs (Supporting information, Table S1). The heat‐map of differentially expressed miRNAs obtained from the miRNA array is presented in Fig. 2.
Figure 2.
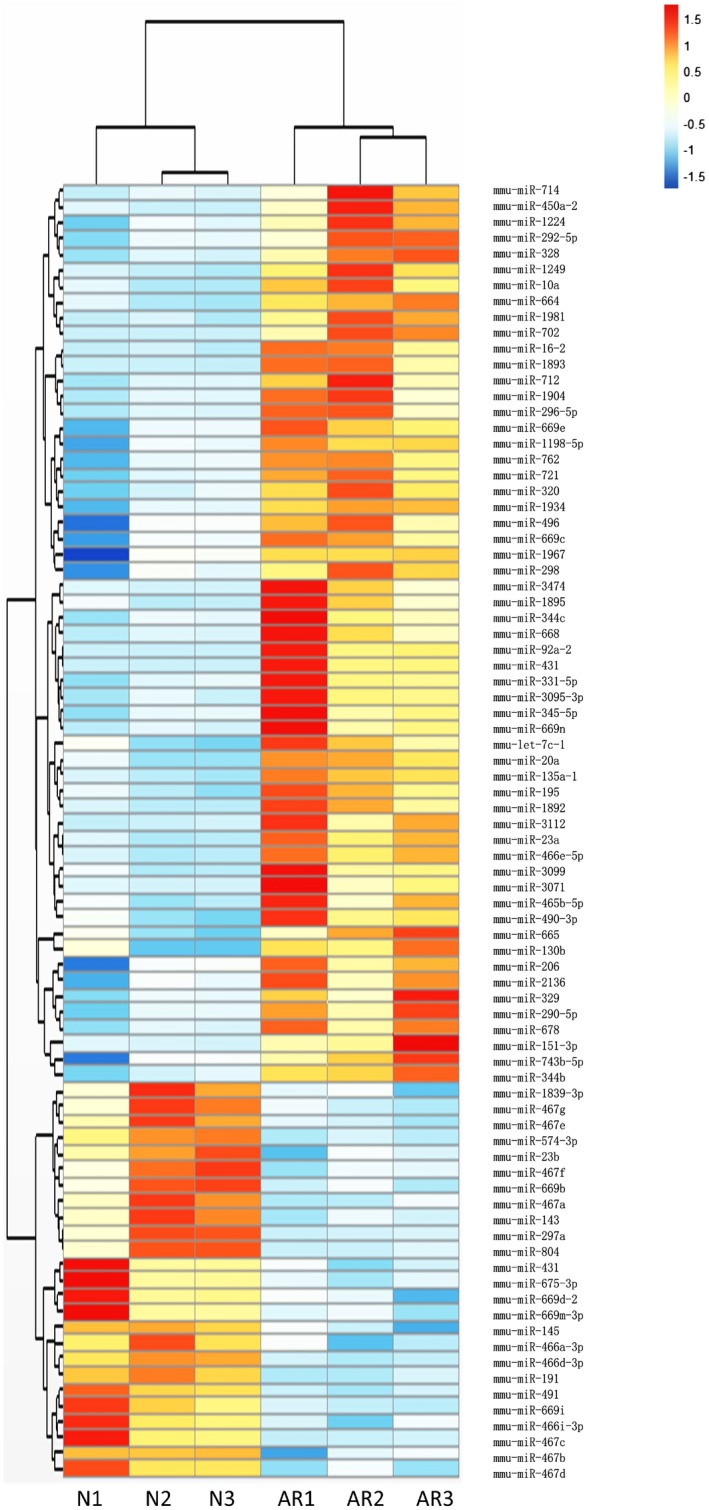
Heat‐map of the differentially expressed MicroRNAs (miRNAs) between the normal group and the allergic rhinitis group.
miR‐466a‐3p is down‐regulated in the nasal mucosa of allergic rhinitis mice
We focus on each of the 15 most fold‐changed miRNAs in up‐ and down‐regulated groups. Three miRNA target gene prediction web tools, such as Miranda, miRBase and targetscan, were used to predict the target gene of those miRNAs. The identical target genes predicted by the three algorithms were recognized as the candidate gene targets for differentially expressed miRNAs. Coincidently, GATA‐3 is one target gene of miR‐466a‐3p (Fig. 3a). As Th2 cells play important roles in allergic rhinitis and GATA‐3 is the defining transcription factor of Th2 cells, miR‐466a‐3p was investigated in the present study. The array data revealed that the expression level of miR‐466a‐3p was significantly decreased in nasal mucosa of mice with allergic rhinitis compared to control mice. The expression pattern of the down‐regulated miR‐466a‐3p was further verified by qPCR using the same set of nasal mucosa samples.
Figure 3.
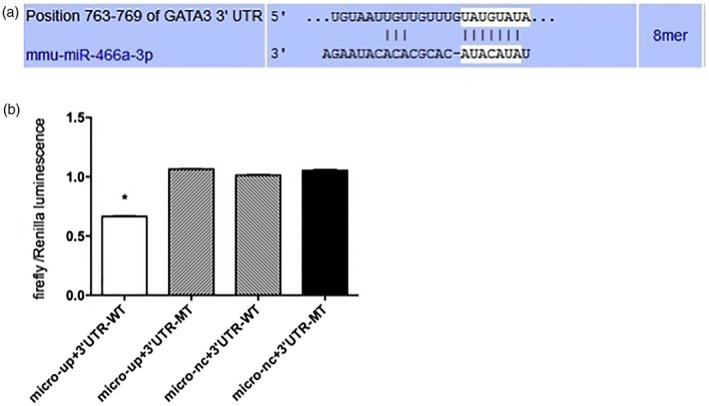
GATA binding protein (GATA‐3) is a direct target of miR‐466a‐3p. (a) The putative miR‐466a‐3p‐specific miRNA recognition elements on mouse GATA‐3 3′‐untranslated region (UTR) predicted by TargetScan (http://www.targetscan.org). (b) MiR‐466a‐3p decreased the luciferase activity after co‐transfected with the vector harboring wild‐type GATA‐3 3′‐UTR, but exerted no effect on the luciferase activity after co‐transfection with the vector harboring mutant GATA‐3 3′‐UTR. The data are presented as mean ± standard error of the mean (s.e.m.) (n = 3). *P < 0·05 versus miR control.
The 3′‐UTR region of GATA‐3 is the direct target of miR‐466a‐3p
To verify that GATA‐3 is a target of miR‐466a‐3p, the dual‐luciferase method was used. We examined whether miR‐466a‐3p can directly target the 3′‐UTR of GATA‐3 gene by co‐transfecting miR‐466a‐3p mimic or negative control with a luciferase vector harboring WT or mutant GATA‐3 3′‐UTR. The results showed that miR‐466a‐3p decreased the luciferase activity after co‐transfection with the vector harboring WT GATA‐3 3′‐UTR, but exerted no effect on the luciferase activity after co‐transfection with the vector harboring mutant GATA‐3 3′‐UTR (Fig. 3b). Overall, these data indicate that miR‐466a‐3p directly targets the 3′‐UTR of GATA‐3.
miR‐466a‐3p suppressed the protein expression of GATA‐3 in splenocytes
To explore the relationship between GATA‐3 and miR‐466a‐3p in vitro, the protein expression levels of GATA‐3 were detected after transfecting miR‐466a‐3p mimics or miR control into splenocytes. The expression level of miR‐466a‐3p was significantly increased following transfection with miR‐466a‐3p mimic. It was found that the protein level of GATA‐3 was decreased in splenocytes transfected with miR‐466a‐3p mimic, but did not change in those transfected with miR control. These results demonstrate that miR‐466a‐3p can regulate the expression of GATA‐3 at protein level (Fig. 4).
Figure 4.
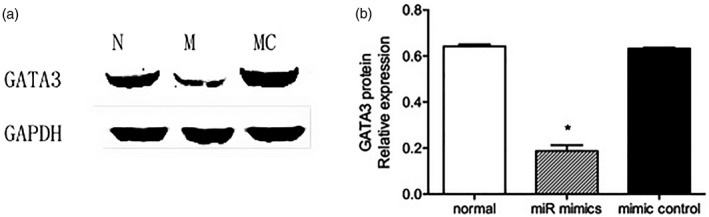
Transfection with mimics‐miR‐466a‐3p inhibited the protein expression of GATA binding protein (GATA‐3). (a) Transfection with mimics‐miR‐466a‐3p significantly reduced the protein expression level of GATA‐3 compared to the normal group. (b) Quantitative data of GATA‐3 protein levels in splenocytes transfected with mimics‐miR‐466a‐3p compared to those transfected with *P < 0·05 versus the normal group.
miR‐466a‐3p suppressed GATA‐3 protein expression in the nasal mucosa of mice
It was found that miR‐466a‐3p specifically based‐paired to the 3′‐UTR of GATA‐3 mRNA, and its expression was down‐regulated in the allergic rhinitis mouse model. To further elucidate the role of miR‐466a‐3p in allergic rhinitis, miR‐466a‐3p was over‐expressed in allergic rhinitis mice via tail‐vein injection of lentiviral miR‐466a‐3p vector. Following lentiviral miR‐466a‐3p transduction, the expression level of miR‐466a‐3p was significantly increased in nasal mucosa, indicating a successful lentiviral–host gene transduction in allergic rhinitis mice. In addition, the protein expression of GATA‐3 was determined in nasal mucosa. The results demonstrated that the expression level of GATA‐3 was decreased in allergic rhinitis mice transfected with lentiviral miR‐466a‐3p compared to allergic rhinitis mice without lentiviral miR‐466a‐3p transfection. Also, the protein level of GATA‐3 did not change after transfection with empty lentiviral vector (Fig. 5). Collectively, these data suggest that GATA‐3 is a target of miR‐466a‐3p.
Figure 5.
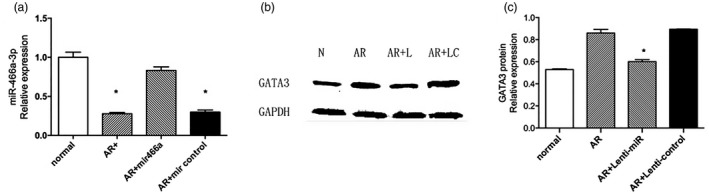
Treatment with allergic rhinitis (AR) + lenti‐miR‐466a‐3p significantly decreased the protein expression of GATA binding protein (GATA‐3). (a) After transfection with lenti‐miR‐466a‐3p, the expression level of miR‐466a‐3p was increased in the mucosa of allergic rhinitis mice. Its expression level did not change in allergic rhinitis mice receiving empty lentiviral vector transfection. (b) Transfection of lenti‐miR‐466a‐3p via tail‐vein injection reduced the protein expression level of GATA‐3 compared to the AR group with or without lenti‐miR control. (c) Quantitative data of GATA‐3 protein levels in the mouse nasal mucosa. *P < 0·05 versus the AR group.
miR‐466a‐3p alleviated symptoms and histology of allergic rhinitis
The numbers of nose‐scratching and sneezing were decreased in allergic rhinitis mice treated with lentiviral miR‐466a‐3p compared to those without treatment. Meanwhile, the mice that received empty lentiviral vector transfection exhibited no changes in allergic rhinitis symptoms (Fig. 6). Increased infiltration of eosinophils is a primary index that used for allergic rhinitis diagnosis. Thus, to assess the effects of miR‐466a‐3p on the histological profile of OVA‐induced allergic rhinitis mouse model, H&E staining was performed on the nasal mucosa of mice. As shown in Fig. 7, the cytoplasm of eosinophils was stained red. Notably, there was a significant influx of eosinophils in the nasal mucosa of allergic rhinitis mice compared to control mice, indicating the occurrence of nasal sensitization. Furthermore, the number of eosinophils was significantly reduced in the nasal mucosa of allergic rhinitis mice with lentiviral miR‐466a‐3p transfection compared to non‐transfected allergic rhinitis mice (Fig. 7).
Figure 6.
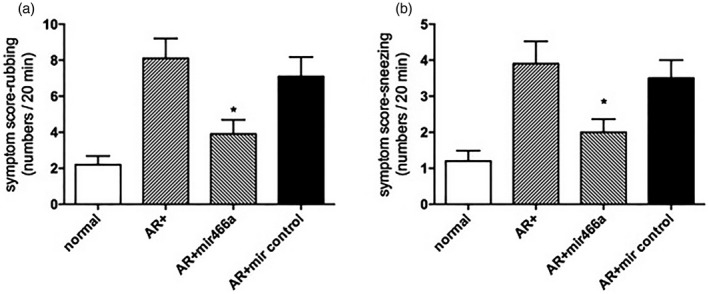
Transfection with lenti‐miR suppressed allergic symptoms. (a) Rubbing symptom score. (b) Sneezing symptom score. *P < 0·05 versus the allergic rhinitis (AR) group.
Figure 7.
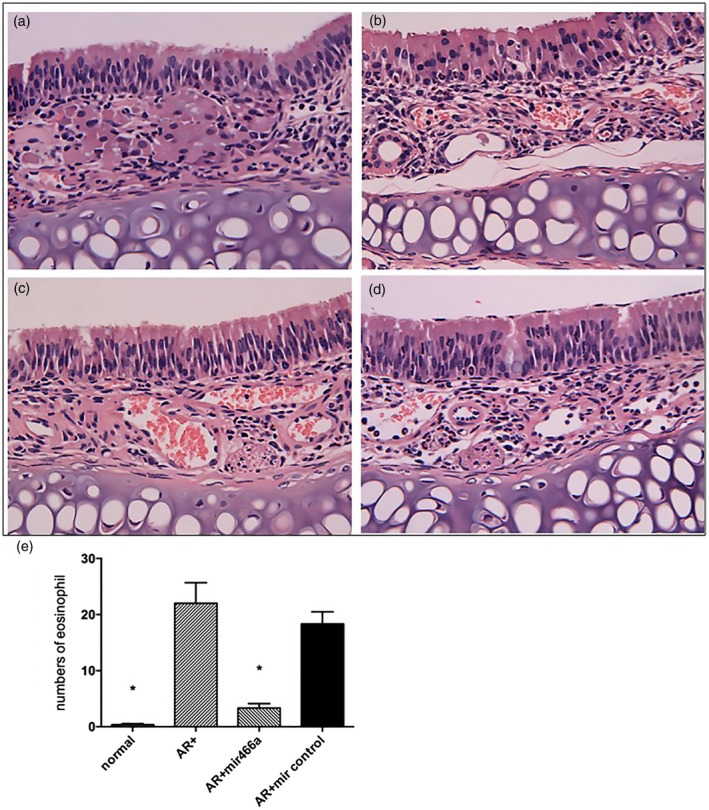
Histological analysis of nasal mucosa in the (a) normal group, (b) allergic rhinitis (AR) group, (c) AR + lenti‐miR‐466a‐3p group and (d) AR + lenti‐miR control group (magnification ×400). The normal group demonstrated no inflammatory changes, while the nasal mucosa was infiltrated with eosinophils in the AR group and the AR + lenti‐miR control group (white arrow). Eosinophilic infiltration was markedly reduced in the AR + lenti‐miR‐466a‐3p group compared to the AR group. (e) The number of eosinophils in the four groups (per ×400 microscopic field).
miR‐466a‐3p reduced the levels of serum IgE and IL‐4 in allergic rhinitis mice
To determine the effect of miR‐466a‐3p in mice with allergic rhinitis, lentiviral vector with miR‐466a‐3p or miR control was transfected into the allergic rhinitis mice via tail‐vein injection. Subsequently, the levels of serum IgE and IL‐4 were measured. The level of serum total IgE was significantly elevated in allergic rhinitis mice compared to normal mice. However, allergic rhinitis mice treated with or without lentiviral miR‐466a‐3p exhibited similar IgE levels. No significant change in IgE level was observed in mice injected with empty lentiviral vector compared to non‐treated allergic rhinitis mice. Meanwhile, the level of IL‐4, an essential Th2 cytokines involved in allergic rhinitis pathogenesis, was detected. The obtained result was the same as the result of serum IgE (Fig. 8). In addition, the data on GATA‐3 expression decreased by lentiviral miR‐466a‐3p vector further supported that miR‐466a‐3p inhibited or attenuated the proinflammatory and immune responses in allergic rhinitis mice, especially in the Th2‐type immune response, by targeting GATA‐3 down‐regulation.
Figure 8.
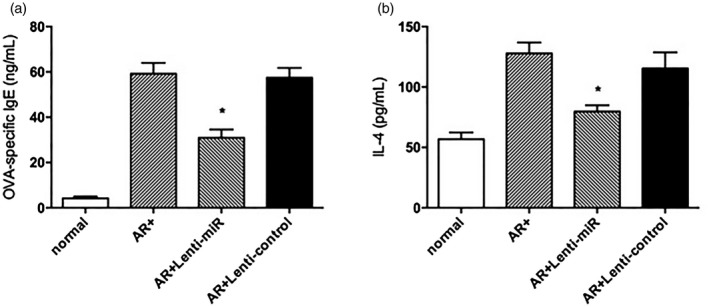
The levels of (a) ovalbumin (OVA)‐specific immunoglobulin (Ig)E and (b) interleukin (IL)‐4 in the four groups, as determined by enzyme‐linked immunosorbent assay (ELISA). Treatment with lenti‐miR‐466a‐3p significantly decreased the levels of OVA‐specific IgE and T helper type 1 (Th2) cytokine IL‐4 in the AR group. *P < 0·05 versus the AR group.
Discussion
There is no doubt that miRNAs act as important gene regulators in controlling almost every aspect of cellular functions. The role of miRNAs in the regulation of allergic rhinitis is emerging rapidly, but there is a lack of studies that investigate the association between miR‐466a‐3p and allergic disease. Recent studies have indicated that some miRNAs may influence the pathogenesis of allergic rhinitis. For instance, mir‐146a induces the levels of transforming growth factor (TGF)‐β and IL‐10 in dendritic cells and monocytes, respectively, and exerts an immune suppressor effect on allergic rhinitis mice 16, 17. Also, intranasal administration of lentiviral miR‐135a into the OVA‐sensitized allergic rhinitis mice reduces the levels of serum IgE and GATA‐3 expression, as well as submucosal mast cell and eosinophilic infiltration, which favor the Th2 response 18. MiR‐106b negatively regulates the pro‐allergic properties of bone marrow‐derived dendritic cells and subsequent Th2 polarization upon OVA stimulation, and thus may represent a promising therapeutic target for allergic inflammation 19. Furthermore, miR‐143 regulates IL‐13‐induced inflammatory cytokine and mucus production in the nasal epithelial cells derived from allergic rhinitis patients 20. In this study, a well‐characterized model of OVA‐induced allergic rhinitis was constructed to reveal the transient down‐regulation of miR‐466a‐3p in nasal mucosa after allergen exposure, and its important role in the pathogenesis of allergic rhinitis. The findings provide evidence for a novel cross‐talk between miR‐466a‐3p and GATA‐3 upon allergen challenge.
Several miRNAs have been demonstrated to influence the development, phenotypical stability and functions of Th2 cells. For example, miR‐138 and miR‐135a regulate the balance of Th1/Th2 in psoriasis by inhibiting runt‐related transcription factor 3 (RUNX3) expression 21, 22. MiR‐155 deficiency alleviates allergic airway inflammation in mice by diminishing Th2 priming capacity and ATP‐/P2R‐induced activation of dendritic cells 23. The up‐regulation of miR‐19a in asthma may be an indicator and a cause of increased TH2 cytokine production in the airways 24. MiR‐21 expression is increased in Th2‐type inflammation by targeting Bcl6 25. Selective blockade of miR‐126 suppresses the asthmatic phenotype, resulting in diminished Th2 responses, inflammation, airways hyperresponsiveness, eosinophil recruitment and mucus hypersecretion, as well as altering Th2 cell function via negative regulation of GATA‐3 expression 26. GATA‐3 has been identified as an important transcription factor for Th2 cells, the key effector cells that are involved in the pathogenesis of allergic rhinitis via the release of inflammatory mediators and cytokines upon allergen exposure. In this study, it was found that lentiviral miR‐466a‐3p directly suppressed the expression of GATA‐3 and reduced the secretion of Th2 cytokine IL‐4 and of OVA‐specific IgE, which are regarded as the characteristic alteration in allergic disease. Due to these changes, lentiviral miR‐466a‐3p‐treated allergic rhinitis mice exhibited lower numbers of nose‐scratching and sneezing. In addition, miR‐466a‐3p treatment decreased the recruitment of eosinophils in allergic rhinitis mice. These findings provide evidence that miR‐466a‐3p contributes to the regulation of Th2 responses in allergic nasal inflammation by directly modulating GATA‐3 expression.
Only miR‐466a‐3p and GATA‐3 were investigated in the present study because of certain factors. There are many other miRNAs accounting for allergic rhinitis. Further research focused on these miRNAs is required, and will be achieved in our future study.
Taken altogether, the present study indicates that miR‐466a‐3p contributes to the pathogenesis of allergic rhinitis in vivo. Although there is not enough evidence to support that miR‐466a‐3p can affect T cells in allergic rhinitis mechanistically, we show that miR‐466a‐3p inhibited Th2 cells priming by directly repressing GATA‐3 transcription. Given that lentiviral miR‐466a‐3p alleviated the symptoms of allergic rhinitis in mice and suppressed the recruitment of eosinophils, targeted miR‐466a‐3p delivery may be a promising option for treating allergic rhinitis and other Th2‐cell‐mediated diseases.
Supporting information
Table S1. Down‐regulated and up‐regulated miRNAs in nasal mucosa of allergic rhinitis mice
Acknowledgement
This study was supported by a grant from the Natural Science Foundation of China (no. 81070766).
Contributor Information
Z. Chen, Email: wb002987@whu.edu.cn.
Z. Tao, Email: taozezhang@hotmail.com
References
- 1. Codispoti CD, Levin L, LeMasters GK et al Breast‐feeding, aeroallergen sensitization, and environmental exposures during infancy are determinants of childhood allergic rhinitis. J Allergy Clin Immunol 2010; 125:1054–60 e1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunlop J, Matsui E, Sharma HP. Allergic rhinitis: environmental determinants. Immunol Allergy Clin North Am 2016; 36:367–77. [DOI] [PubMed] [Google Scholar]
- 3. Han DM, Zhang L, Huang D et al Self‐reported prevalence of allergic rhinitis in eleven cities in China. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2007; 42:378–84. [PubMed] [Google Scholar]
- 4. Cohen SG, Richard E III. Allergen immunotherapy in historical perspective. Clin Allergy Immunol 2008; 21:1–29. [PubMed] [Google Scholar]
- 5. Nathan RA. Management of patients with allergic rhinitis and asthma: literature review. South Med J 2009; 102:935–41. [DOI] [PubMed] [Google Scholar]
- 6. Ciprandi G, Comite P, Ferrero F, Fontana V, Bruzzone M, Mussap M. Serum allergen‐specific IgE, allergic rhinitis severity, and age. Rhinology 2016; 54:231–8. [DOI] [PubMed] [Google Scholar]
- 7. Shamji MH, Bellido V, Scadding GW et al Effector cell signature in peripheral blood following nasal allergen challenge in grass pollen allergic individuals. Allergy 2015; 70:171–9. [DOI] [PubMed] [Google Scholar]
- 8. Schmitt N, Ueno H. Regulation of human helper T cell subset differentiation by cytokines. Curr Opin Immunol 2015; 34:130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaminuma O. Selective inhibitors of nuclear factor of activated T cells: potential therapeutic drugs for the treatment of immunological and inflammatory diseases. Inflamm Allergy Drug Targets 2008; 7:35–40. [DOI] [PubMed] [Google Scholar]
- 10. Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 2011; 3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 11. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 2010; 10:111–22. [DOI] [PubMed] [Google Scholar]
- 13. Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol 2013; 132:3–13; quiz 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Z, Zhang XH, Callejas‐Diaz B, Mullol J. MicroRNA in united airway diseases. Int J Mol Sci 2016;17:716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 2001; 25:402–8. [DOI] [PubMed] [Google Scholar]
- 16. Luo X, Hong H, Tang J et al Increased expression of miR‐146a in children with allergic rhinitis after allergen‐specific immunotherapy. Allergy Asthma Immunol Res 2016; 8:132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu HJ, Zhang AF, Zhao N, Li XZ. Role of miR‐146a in enforcing effect of specific immunotherapy on allergic rhinitis. Immunol Invest 2016; 45:1–10. [DOI] [PubMed] [Google Scholar]
- 18. Deng YQ, Yang YQ, Wang SB et al Intranasal administration of lentiviral miR‐135a regulates mast cell and allergen‐induced inflammation by targeting GATA‐3. PLOS ONE 2015; 10:e0139322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang H, Jiang H, Zheng J et al MicroRNA‐106b regulates pro‐allergic properties of dendritic cells and Th2 polarisation by targeting early growth response‐2 in vitro . Int Immunopharmacol 2015; 28:866–74. [DOI] [PubMed] [Google Scholar]
- 20. Teng Y, Zhang R, Liu C et al miR‐143 inhibits interleukin‐13‐induced inflammatory cytokine and mucus production in nasal epithelial cells from allergic rhinitis patients by targeting IL13Ralpha1. Biochem Biophys Res Commun 2015; 457:58–64. [DOI] [PubMed] [Google Scholar]
- 21. Fu D, Yu W, Li M et al MicroRNA‐138 regulates the balance of Th1/Th2 via targeting RUNX3 in psoriasis. Immunol Lett 2015; 166:55–62. [DOI] [PubMed] [Google Scholar]
- 22. Luo Y, Deng Y, Tao Z et al Regulatory effect of microRNA‐135a on the Th1/Th2 imbalance in a murine model of allergic rhinitis. Exp Ther Med 2014; 8:1105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zech A, Ayata CK, Pankratz F et al MicroRNA‐155 modulates P2R signaling and Th2 priming of dendritic cells during allergic airway inflammation in mice. Allergy 2015; 70:1121–9. [DOI] [PubMed] [Google Scholar]
- 24. Simpson LJ, Patel S, Bhakta NR et al A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol 2014; 15:1162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA‐21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol 2013; 54:435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA‐126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA 2009; 106:18704–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Down‐regulated and up‐regulated miRNAs in nasal mucosa of allergic rhinitis mice


