Abstract
Context
There is substantial heterogeneity in insulin sensitivity, and genetics may suggest possible mechanisms by which common variants influence this trait.
Objectives
We aimed to evaluate an 11-variant polygenic lipodystrophy genetic risk score (GRS) for association with anthropometric, glycemic and metabolic traits in the Diabetes Prevention Program (DPP). In secondary analyses, we tested the association of the GRS with cardiovascular risk factors in the DPP.
Design
In 2713 DPP participants, we evaluated a validated GRS of 11 common variants associated with fasting insulin-based measures of insulin sensitivity discovered through genome-wide association studies that cluster with a metabolic profile of lipodystrophy, conferring high metabolic risk despite low body mass index (BMI).
Results
At baseline, a higher polygenic lipodystrophy GRS was associated with lower weight, BMI, and waist circumference measurements, but with worse insulin sensitivity index (ISI) values. Despite starting at a lower weight and BMI, a higher GRS was associated with less weight and BMI reduction at one year and less improvement in ISI after adjusting for baseline values but was not associated with diabetes incidence. A higher GRS was also associated with more atherogenic low-density lipoprotein peak-particle-density at baseline but was not associated with coronary artery calcium scores in the Diabetes Prevention Program Outcomes Study.
Conclusions
In the DPP, a higher polygenic lipodystrophy GRS for insulin resistance with lower BMI was associated with diminished improvement in insulin sensitivity and potential higher cardiovascular disease risk. This GRS helps characterize insulin resistance in a cohort of individuals at high risk for diabetes, independent of adiposity.
Keywords: insulin resistance, obesity, metformin, lifestyle intervention, type 2 diabetes
The underlying pathogenesis of type 2 diabetes involves relative insufficiency in insulin secretion through β cell dysfunction in the context of increased secretory demand because of insulin resistance [1]. Genome-wide association studies (GWAS) have established that common genetic variation influences insulin resistance by identifying 19 single nucleotide polymorphisms (SNPs) that have reached genome-wide associations with fasting insulin-based measures of insulin resistance [2–4]. However, despite the importance of insulin resistance as a risk factor for type 2 diabetes, there is variation in the pathological pathways leading to diabetes, and there is considerable heterogeneity in the genetic architecture of insulin resistance. Studies have also shown that insulin resistance is an independent predictor of cardiovascular disease risk [5]. Although increased body mass index (BMI) remains an overwhelming risk factor for insulin resistance and the development of type 2 diabetes, it is now established that for a given BMI, the pattern of fat storage is associated with modification of this risk [6]. However, the role of insulin resistance independent of BMI in the pathogenesis of type 2 diabetes and cardiovascular disease risk has been difficult to comprehend, largely because of the strong correlation between obesity and insulin resistance.
Rare monogenic disorders may shed some light on the relationship between insulin resistance and diabetes in the absence of the confounding effects of BMI. For example, in primary lipodystrophy, severe insulin resistance develops in lean individuals in association with generalized or regional lack of adipose tissue. Patients with lipodystrophy exhibit severe insulin resistance, metabolic dyslipidemia, and diabetes resulting from impaired adipose tissue function [7]. To define the polygenic correlate of lipodystrophy, Yaghootkar et al. [8] selected 19 common genetic variants associated with fasting insulin-based measures of insulin resistance and used hierarchical clustering and results from GWAS of eight nondisease outcomes of monogenic insulin resistance to group these variants. A cluster of 11 common genetic variants associated with fasting insulin-based measures of insulin resistance in GWAS was associated with chronic disease outcomes, including type 2 diabetes and coronary artery disease, in directions consistent with a lipodystrophy phenotype: high metabolic disease risk despite low BMI [8].
Although this provides evidence for a genetic signature for metabolic disease, it is unknown whether these variants predict metabolic disease features in a multiethnic cohort of subjects at high risk for the development of diabetes, and whether they predict change in glycemic and metabolic parameters over time in the context of diabetes interventions designed to ameliorate insulin resistance and reduce the risk of type 2 diabetes. The Diabetes Prevention Program (DPP), a randomized controlled trial of metformin and lifestyle modification vs placebo, can be analyzed to address these questions. In the current study, we aimed to evaluate the same 11 variant polygenic lipodystrophy genetic risk score (GRS) [8] for association with relevant anthropometric, glycemic and metabolic traits, and incident diabetes in the DPP’s multiethnic cohort of individuals who are at high metabolic risk at baseline. Additionally, we aimed to evaluate the association of the GRS with progression to diabetes and response to metformin and intensive lifestyle interventions. In secondary analyses, we also tested the association of the GRS with cardiovascular disease risk factors that are intermediate measurable phenotypes that are associated with cardiovascular disease [9].
1. Materials and Methods
A. Description of DPP Study Design and Participants
The study design of the DPP (ClinicalTrials.gov Identifier: NCT00004992) and characteristics of the participants at baseline have been described previously [10, 11]. Briefly, the DPP was a multicenter trial in the United States that assessed whether intensive lifestyle intervention or metformin therapy prevented or delayed the onset of diabetes in individuals who were at high risk of developing diabetes. The DPP enrolled 3234 overweight or obese individuals without diabetes but with impaired glucose tolerance and elevated fasting glucose, and randomly assigned them to placebo, metformin (850 mg twice daily), or an intensive lifestyle intervention. The DPP showed that after a mean follow-up of 2.8 years, metformin and lifestyle interventions reduced the incidence of diabetes by 31% (95% CI 17 to 43) and 58% (95% CI 48 to 66) respectively, vs placebo [12].
All 3150 surviving DPP participants who had not withdrawn consent were eligible for a follow-up study, the Diabetes Prevention Program Outcomes Study (DPPOS), a study initiated to establish the longer term effects of the DPP interventions on the development of diabetes [13]. At the end of the DPP, after a brief metformin and placebo washout study [13], the participants in the placebo and metformin groups were subsequently unmasked to their treatment assignment and placebo was stopped. In view of the clear evidence of benefit of the lifestyle intervention, all participants were offered the lifestyle intervention in a group format during a one-year bridge period between DPP and DPPOS [14]. During DPPOS, as in DPP, metformin was provided to the group originally assigned to it, however, metformin was now unmasked. In this study, except for coronary artery calcification (CAC) scores, which were obtained at year 14 of follow-up in DPPOS, all measurements were obtained during the original DPP trial. Institutional review board approval was obtained by each participating center and all subjects included in this study provided written informed consent for the main studies and for subsequent genetic investigations.
B. Measurements
The methods for measuring glucose, insulin, total cholesterol, and triglyceride levels have been described previously [11]. Participants were excluded from the DPP if they had markedly elevated alanine aminotransferase (ALT) or aspartate aminotransferase (AST) concentrations at baseline as defined by age and sex: for age <47 years, ALT >46 U/L for women and >118 U/L for men; for age ≥47 years, ALT >58 U/L for both men and women; for AST ≥66 U/L per criteria established by the DPP central laboratory. After baseline measurements, follow-up ALT and AST levels were not measured in the intensive lifestyle group. Mean AST concentrations increased in 1999 by ∼4 U/L regardless of study visit (baseline, 3, 6 months, etc.) consistent with assay drift. A similar change in ALT concentrations was not found [15]. For these reasons, this analysis was limited to ALT.
The insulin sensitivity index (ISI) was calculated as the reciprocal of homeostasis model assessment of insulin resistance using the equation [(FI (mU/L) × fasting glucose (mmol/L)/22.5] [16] based on glucose and insulin levels during the oral glucose tolerance test at baseline and one-year follow-up. Participants were asked not to take metformin or placebo on the morning of the oral glucose tolerance test. For longitudinal analyses, the one-year end point was chosen because the sample size was largest at that time point (95% of participants completed the one-year follow-up visit) and weight loss was the most pronounced at one year in the intervention arms.
DPP participants from 18 of the 27 sites (n = 1106) volunteered for measurement of adipose tissue by CT at baseline and after one year of the study. The instruments used included the GE High Speed Advantage (General Electric, Milwaukee, WI), at five centers, the Picker PQ 5000 (Picker, Groton, CT) at five centers, the Siemens and Siemens Somatom Plus (Siemens and Siemens, New York, NY) at two centers, the GE 9800 (General Electric) at three centers, and the GE Highlite (General Electric) at two centers. Two 10-mm thick axial images were obtained at the L4-5 spaces. The data obtained were submitted to a central reading facility at the University of Colorado in Denver. The reading center calculated the total visceral adipose area on each scan, delineated visceral fat from subcutaneous fat by circumscribing the transversalis fascia, and calculated subcutaneous adipose tissue by subtracting the visceral adipose tissue from the total cross-sectional area for fat [17]. We report results using measurement of both visceral and subcutaneous fat in 618 participants.
Total circulating adiponectin was measured using a latex particle-enhanced turbidimetric assay (Otsuka Pharmaceutical, Tokyo, Japan) [18]. In participants with triglyceride >4.5 mmol/L, lipoprotein fractions were separated using preparative ultracentrifugation of plasma by β quantification. C-reactive protein and fibrinogen levels in plasma were immunochemically measured using the Behring Nephelometer auto-analyzer (Dade Behring; Marburg, Germany). Tissue plasminogen activator levels were measured in citrated plasma using an ELISA (Asserachrom tPA; Diagnostica Stago, Parsippany, NJ [19]), which measures total tissue plasminogen activator antigen [20].
Given the multiethnic demographics of the study population, cardiovascular disease risk was computed using the American Heart Association/American College of Cardiology 10-year risk score [21]. Subclinical atherosclerosis by coronary artery calcification (CAC) was measured in year 14 of DPPOS in 2029 subjects using a multidetector CT according to methods that have been published previously [22].
C. Genotyping
DNA was extracted from peripheral blood leukocytes and genotyping was performed on the customized Metabochip (Illumina, San Diego, CA). The Metabochip contains ∼200,000 SNPs chosen based on previous GWAS meta-analyses of 23 metabolic traits related to T2D, obesity, and cardiovascular diseases. Study participants with sex discrepancy (4 subjects) or familial relatedness (6 subjects) were excluded. SNPs were excluded if the call rate was less than 95% or if they failed Hardy-Weinberg equilibrium testing (P < 1.0 × 10−7) within each ethnic group. The overall genotyping success rate was excellent at >99.85%.
D. SNP Selection and Construction of Genetic Risk Score
Eleven common variants associated with fasting insulin-based measures of insulin resistance were selected for the GRS based on their association with features of metabolic syndrome including increased risk of type 2 diabetes, coronary artery disease, and higher systolic and diastolic blood pressures despite low BMI, similar to a lipodystrophic profile [8]. All these variants have been associated with fasting insulin at the accepted level of genome-wide significance (P < 5 × 10−8) in GWAS previously published by the MAGIC investigators [2–4]. The GRS was computed based on the assumption of an additive genetic effect. The GRS was calculated by accounting for the number of risk alleles present per SNP and summing the results over the 11 SNPs. Participants with more than three missing SNPs were excluded (n = 281). For participants with 1, 2, or 3 missing SNPs (total 120 individuals), the GRS was calculated by multiplying the GRS from the available SNPs by 22 and dividing by twice the number of successfully genotyped SNPs. We used an unweighted score in which equal weight was given to all the risk alleles because of the minimal differences in the published effect sizes for fasting insulin published by MAGIC (Table 1). We divided the subjects into four groups of genetic risk only for descriptive purposes in the baseline table (Table 2) and in the plots for illustration purposes (Fig. 1). Based on the distribution of the GRS, a GRS of <12 defined the first group, a GRS of ≥12 but <14 defined the second group, a GRS of ≥14 but <15 defined the third group, and a GRS of ≥15 defined the fourth group. There were numerous tied values at the boundaries of the groups, leading to uneven sample sizes in the four groups. However, this did not affect statistical analyses because the analyses were done considering the GRS as a continuous variable.
Table 1.
Effect Sizes for Fasting Insulin Along With Frequencies of Genetic Variants in Published Reports from MAGIC and in the DPP Participants
| Gene | SNP | β Estimates for ln FI (μU/mL) in MAGIC | Risk Allele Frequency in MAGIC (%) | Frequency of High-Risk Alleles in DPP (%) | |||||
| Overall | Caucasian | Black | Hispanic | Asian/ Pacific Islanders | American Indian | ||||
| N = 2713 | N = 1503 | N = 554 | N = 458 | N = 120 | N = 78 | ||||
| IRS1 | rs2943634 | 0.027 | 66 | 67 | 68 | 45 | 77 | 92 | 92 |
| COBLL1/GRB14 | rs7607980 | 0.027 | 88 | 88 | 87 | 85 | 90 | 98 | 97 |
| ARL15 | rs4865796 | 0.011 | 67 | 74 | 70 | 75 | 82 | 81 | 94 |
| FAM13A | rs3822072 | 0.013 | 48 | 49 | 49 | 53 | 42 | 54 | 40 |
| LYPLAL1 | rs2785980 | 0.015 | 67 | 70 | 67 | 84 | 53 | 71 | 44 |
| PEPD | rs731839 | 0.014 | 34 | 34 | 34 | 39 | 40 | 58 | 47 |
| PDGFC | rs4691380 | 0.017 | 67 | 59 | 66 | 29 | 61 | 64 | 71 |
| RSPO3 | rs2745353 | 0.011 | 39 | 35 | 50 | 63 | 56 | 60 | 66 |
| PPARG | rs18001282 | 0.022 | 87 | 91 | 90 | 98 | 91 | 90 | 81 |
| TET2 | rs9884482 | 0.012 | 39 | 35 | 38 | 11 | 45 | 58 | 40 |
| ANKRD55 | rs459193 | 0.013 | 73 | 70 | 75 | 58 | 73 | 53 | 74 |
Abbreviation: FI, fasting insulin.
Table 2.
Characteristics of DPP Participants at Baseline by Quartile of Polygenic Lipodystrophy GRS
| Group of GRSa,b | |||||
| Baseline Variable | 1st | 2nd | 3rd | 4th | P c |
| Nd | 394 | 773 | 476 | 1070 | |
| Sex, n, % | |||||
| Women | 286 (16.4) | 504 (28.8) | 325 (18.6) | 712 (40.7) | 0.072 |
| Men | 108 (12.2) | 269 (30.4) | 151 (17.0) | 358 (40.4) | |
| Self-reported race/ethnicity, n, % | |||||
| Caucasian | 195 (13.0) | 420 (27.9) | 276 (18.4) | 612 (40.7) | 0.115 |
| Black | 141 (25.5) | 207 (37.7) | 93 (16.8) | 113 (20.4) | |
| Hispanic | 51 (11.1) | 116 (25.3) | 75 (16.4) | 216 (47.2) | |
| American Indian | 46 (59.0) | ||||
| Asian American | 16 (13.3) | 19 (15.8) | 83 (69.2) | ||
| Age at randomization, y | 50.9 (49.9-52.0) | 51.1 (50.4-51.9) | 50.6 (49.7-51.6) | 50.3 (49.7-51.0) | 0.423 |
Data are reported as mean (95% CI) for continuous variables and as frequency (%) for categorical variables.
Empty cells indicate sample sizes ≤15 that are not reported per DPP policy for concern of compromise of confidentiality and limitations with interpretation of findings.
P values from logistic regression for categorical variables and general linear models for continuous variables.
The sample sizes in each group are uneven because of the many tied values at the boundaries of the groups.
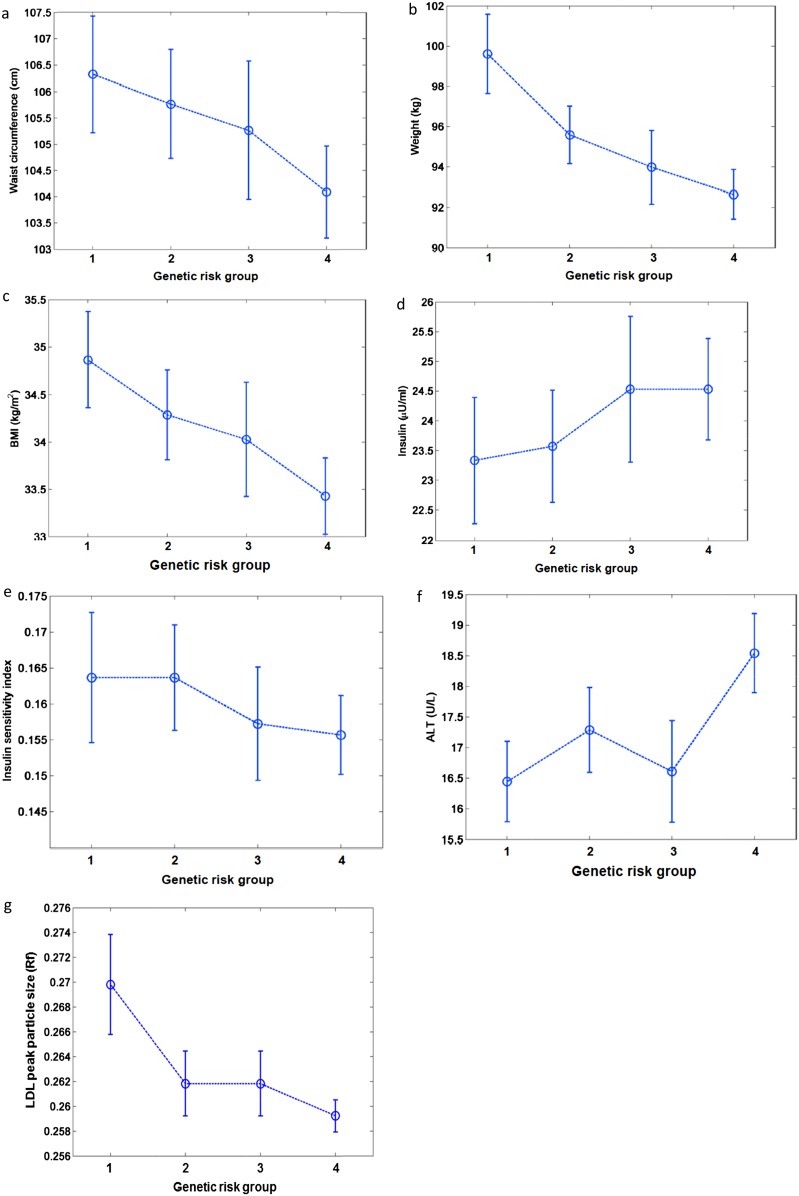
Figure 1.
Relation of baseline variables (a) weight, (b) BMI, (c) waist circumference, (d) fasting insulin, (e) ISI, (f) ALT, and (g) LDL peak particle size with the GRS across four groups (G1, G2, G3, G4) of genetic risk is shown. The circles represent the means and the heights of the bars represent the 95% CI. Log transformed variables have been reverse transformed for descriptive purposes. All measures were adjusted for age, sex, and race/ethnicity. Fasting insulin, ISI, ALT, and LDL peak particle density measurements were additionally adjusted for waist circumference.
E. Statistical Analyses
The GRS was analyzed in general linear models predicting baseline and one-year change from baseline for measured variables. Variables with a non-normal distribution were log transformed. The one-year analysis included a test for treatment × GRS interaction. The effect of the GRS on diabetes incidence over the course of the main trial (mean follow-up time of 3.2 years) was tested using a Cox proportional hazards model with genotype, treatment arm, and a genotype-treatment arm interaction test as the independent variables predicting time to diabetes. The association between quantitative traits and genotype under an additive genetic model was tested in each treatment arm using analysis of covariance (ANCOVA). CAC severity was analyzed by Tobit regression [23] of the CAC score using the QLIM procedure in SAS to account for the skewness resulting from the relatively large number of individuals with a CAC score of 0. The Tobit regression coefficient represents the log ratio of the geometric mean CAC score per unit increase in the covariate, assuming some measurable calcification for all subjects, including subjects with undetectable levels. All tests performed were two sided, and an α-level of 0.05 was used to determine statistical significance. The Statistical Analysis Software (SAS) version 9.3 was used for all analyses (SAS Institute, Inc., Cary, NC)
2. Results
The 2713 participants from the DPP analyzed in this study had a mean (±SD) baseline age of 50.7 ± 10.7 years; 67% of the participants were women and 45% were nonwhite. At baseline, the mean BMI of the participants was 34.1 ± 6.7 kg/m2, and waist circumference was 105.4 ± 14.6 cm. The characteristics of the DPP participants at baseline divided into four groups for descriptive purposes based on the polygenic lipodystrophy GRS is listed in Table 2. The list of common genetic variants used in the GRS along with their published effect size for fasting insulin is shown in Table 1. This table also shows the frequencies of the risk alleles in the DPP participants by ethnicity as well as published frequencies reported in MAGIC. The distribution of the GRS across the DPP participants was normal with a mean score of 13.9 (SD ± 2.2) and a median of 14.0 without substantial differences across all three treatment groups. The baseline characteristics of the participants predicted by the GRS are presented in Table 3. At baseline, a higher GRS was associated with higher ALT levels, higher fasting insulin, and with lower ln ISI values after adjustment for age at randomization, sex, self-reported race/ethnicity, and waist circumference. We chose adjustment for waist circumference because central adiposity as measured by waist circumference instead of BMI is a better predictor of type 2 diabetes, metabolic syndrome, and cardiovascular disease and because this is the anthropometric measure most strongly associated with outcomes in the DPP [24]. In contrast, a higher GRS was associated with lower weight, lower BMI and lower waist circumference measurements. Figure 1a-1f demonstrates the relation of the some of the baseline variables with the GRS across four groups of genetic risk.
Table 3.
Baseline Characteristics Predicted by the Polygenic Lipodystrophy GRS
| Baseline Variable | Sample Size (n) | β Estimate per Allele | SE | P |
| Weight, kga | 2712 | −0.6341 | 0.1705 | <0.001 |
| BMI, kg/m2a | 2712 | −0.2342 | 0.0574 | <0.001 |
| Waist circumference, cma | 2712 | −0.4670 | 0.1288 | <0.001 |
| FI, μU/mL, ln | 2709 | 0.0116 | 0.0045 | 0.010 |
| Fasting glucose, mg/dL | 2712 | 0.1035 | 0.0734 | 0.159 |
| ISI, ln | 2709 | −0.0125 | 0.0046 | 0.007 |
| Systolic blood pressure (mm Hg) | 2712 | 0.0531 | 0.1271 | 0.676 |
| Diastolic blood pressure (mm Hg) | 2712 | 0.0334 | 0.0836 | 0.690 |
| HDL-cholesterol, mg/dL | 2707 | −0.0670 | 0.0983 | 0.496 |
| LDL-cholesterol, mg/dL | 2707 | 0.0554 | 0.3003 | 0.854 |
| Triglycerides, mg/dL, ln | 2707 | 0.0069 | 0.0046 | 0.128 |
| ALT, U/L, ln | 2709 | 0.0113 | 0.0047 | 0.015 |
| Visceral adipose tissue, cm2, L4-L5 ln | 791 | −0.0062 | 0.0048 | 0.196 |
| Subcutaneous adipose tissue, cm2, L4-5 ln | 791 | −0.0025 | 0.0035 | 0.480 |
All measures adjusted for age at randomization, sex, self-reported race/ethnicity and waist circumference except where indicated.
Abbreviation: FI, fasting insulin.
Adjusted for age at randomization, sex, and self-reported race/ethnicity.
Table 4 lists the association of the GRS with change in measured traits at one year. Over the first year, a higher GRS was associated with a smaller magnitude of weight loss, despite starting with lower weight and BMI measurements at baseline. Additionally, over one year, a higher GRS was associated with less improvement in the ln ISI after adjustment for baseline value, age at randomization, sex, self-reported race/ethnicity, waist circumference, and treatment group. These results remained noteworthy after adjusting for BMI in our analysis. There was no interaction of the GRS with the treatment arms on the change in ISI over the first year (P = 0.617).
Table 4.
Association of the GRS with Change in Traits at 1 Year
| Year 1-Baseline | Sample Size (n) | β Estimate per Allele | SE | P |
| Weight, kga | 2568 | 0.1234 | 0.0529 | 0.020 |
| BMI, kg/m2a | 2568 | 0.0461 | 0.0190 | 0.015 |
| Waist circumference, cma | 2568 | 0.0754 | 0.0593 | 0.204 |
| FI, μU/mL, ln | 2568 | 0.0130 | 0.0043 | 0.002 |
| Fasting glucose, mg/dL, lna | 2568 | 0.0003 | 0.0009 | 0.722 |
| ISI, lnb | 2506 | −0.0136 | 0.0047 | 0.004 |
| Systolic blood pressure, mm Hg | 2568 | −0.0874 | 0.1134 | 0.441 |
| Diastolic blood pressure, mm Hg | 2568 | 0.0277 | 0.0740 | 0.708 |
| HDL-cholesterol, mg/dL | 2559 | 0.0043 | 0.0591 | 0.942 |
| LDL-cholesterol, mg/dL | 2559 | 0.0067 | 0.2033 | 0.974 |
| Triglycerides, mg/dL, ln + | 2559 | 0.0047 | 0.0033 | 0.154 |
| ALT, U/L, ln+ | 1857 | 0.0801 | 0.1211 | 0.509 |
| Visceral adipose tissue, cm2, L4-5 | 618 | −0.0006 | 0.0017 | 0.721 |
| Subcutaneous adipose tissue, cm2, L4-5 | 618 | 0.4760 | 1.0300 | 0.644 |
All measures adjusted for baseline value, age at randomization, sex and self-reported race/ethnicity, waist circumference and treatment group except where indicated.
Abbreviation: FI, fasting insulin.
Adjusted for age at randomization, sex, and race/ethnicity
ln (Year 1 measurement) - ln (Baseline measurement)
We also evaluated the association of the GRS with diabetes incidence over the course of the main DPP trial using a Cox proportional hazards model with a mean follow-up of 3.2 years. We did not find interactions between either treatment arm and the GRS, therefore, we pursued our multivariable models in the full cohort adjusting for treatment arms. There was no association with diabetes incidence [hazard ratio = 1.03, P = 0.229, 95% CI (0.98 to 1.07)] with the model including adjustment for major risk factors for type 2 diabetes including age at randomization, sex, waist circumference, and self-reported race/ethnicity.
In secondary analyses, we tested the association of the GRS with cardiovascular disease risk factors in 2708 participants at baseline and at one year. Table 5 shows the association of the GRS with the cardiovascular disease risk factors tested at baseline. A higher GRS was associated with lower low-density lipoprotein (LDL) peak-particle size at baseline after adjusting for age, sex, self-reported race/ethnicity, and waist circumference (Fig. 1g). Over one year, the GRS was associated with lowering in the fibrinogen levels after adjustment for baseline value, age, sex, self-reported race/ethnicity, waist circumference, and treatment group. We found no association of the GRS with CAC scores at 14 years in the DPPOS follow-up.
Table 5.
Association of the Polygenic Lipodystrophy GRS With Cardiovascular Disease Risk Factors at Baseline
| Baseline Variable | Sample Size (n) | β Estimate per Allele | SE | P |
| Adiponectin, μg/mL, ln | 2691 | −0.0027 | 0.0033 | 0.407 |
| Fibrinogen, mg/dL, ln | 2704 | −0.0008 | 0.0019 | 0.661 |
| C-reactive protein, mg/dL, ln | 2708 | -0.0143 | 0.0085 | 0.094 |
| Tissue plasminogen activator, ng/mL, ln | 2698 | −0.0009 | 0.0032 | 0.789 |
| LDL particle size, Rf, ln | 2708 | −0.0022 | 0.0010 | 0.034 |
| ACC/AHA 10 y risk score ln | 2708 | −0.0009 | 0.0085 | 0.916 |
All measures adjusted for age at randomization, sex, self-reported race/ethnicity and waist circumference.
Abbreviations: ACC, American College of Cardiology; AHA, American Heart Association.
3. Discussion
With the use of a large-scale genetic study and the phenotypic accuracy of clinical trial data, we have extended the genetic link between various components of the metabolic syndrome resembling a lipodystrophy phenotype in a multiethnic cohort of individuals who are at high metabolic risk at baseline. In the DPP, a select cluster of common genetic variants associated with insulin sensitivity as captured by fasting insulin-based measures was associated with worse measures of insulin resistance and higher alanine transaminase levels but with lower weight, BMI, and waist circumference measurements at baseline. The association of the GRS with alanine transaminase levels is noteworthy despite the exclusion of participants with markedly elevated levels in the DPP [15]. These findings expand on previous results examining the genetics of insulin resistance from the DPP [25] and highlight the unique metabolic signature of this genetic risk score of common genetic variants linked to insulin resistance.
The DPP also allows us to examine how this GRS influences change in our variables of interest over one year of intervention with either metformin, intensive lifestyle intervention, or placebo. Despite starting at a lower weight and BMI, participants with a high genetic burden for lipodystrophic insulin resistance were less likely to lose weight or show improvement in their BMI. Additionally, despite having lower weight and BMI, participants with the highest genetic burden for the lipodystrophy phenotype were less likely to improve their insulin sensitivity after accounting for demographic characteristics including waist circumference. The change in one-year analyses for the all the end points tested took into account the baseline value. The lack of association of the GRS with CT-based adipose tissue measurements is likely because these measurements were only available in a small subset (n = 618) of the DPP participants, limiting the power of the analyses.
We have previously shown in the DPP, that a GRS of 17 established insulin resistance variants was associated with decreased insulin sensitivity at baseline and diminished improvement in the ISI over one year of the study [25]. This current study builds upon that body of work by examining a more refined GRS, based on 11 of the 17 insulin-resistance SNPs used in the previous study [25] but with the key difference being that this 11-SNP GRS (in contrast with the complete 17-SNP score) was associated with a lipodystrophy phenotype, capturing the critical feature of lower adiposity but higher metabolic disease risk. Our results, in comparison with the previous DPP report, highlight the relative importance of the subset of these 11 variants. Our results also indicate that in the DPP participants, the genetic burden of even a subset of insulin resistance variants was associated with diminished insulin sensitivity at baseline and over one year of interventions. We have also extended our previous exploration by characterizing the association of the new GRS against cardiovascular endophenotypes in the DPP. The genetic basis of the phenotype of high insulin resistance with low body adiposity was also recently examined by Lotte et al [26]. They combined GWAS results for fasting insulin, high-density lipoprotein (HDL) cholesterol, and triglyceride levels and identified 53 genomic regions associated with high fasting insulin, high triglyceride, and low HDL cholesterol levels, a subset of which have been previously implicated in insulin resistance. Because the DPP is a cohort of participants at risk for diabetes at baseline and is therefore enriched for insulin resistance, we chose to use a score that was purely derived from genetic variants associated with insulin resistance.
We have also previously shown that a GRS derived from known type 2 diabetes variants predicts diabetes incidence [27]. Because most of the risk alleles at loci associated with fasting insulin are not associated with type 2 diabetes in large population-based studies, it is not surprising that the polygenic lipodystrophy GRS was not associated with diabetes incidence in the current analyses. This is consistent with current evidence that common genetic variants associated with β-cell function have greater predictive power for type 2 diabetes [28], and it highlights the important role of β-cell secretory function in the pathogenesis of type 2 diabetes [1].
Metformin and intensive lifestyle modification both improved insulin sensitivity over one year in DPP participants irrespective of genetic risk burden. This is again consistent with previous results from the DPP and highlights the effectiveness of these preventive interventions across the gradient of genetic risk [25, 27] The lack of statistical significance achieved with the use of this GRS compared with previous scores in the DPP suggests pathophysiological differences in this particular subset of genetic variants and should be explored in future studies.
A major goal of diabetes prevention and treatment is to prevent microvascular and macrovascular events. In the DPP, those who did not develop diabetes had a lower prevalence of microvascular complications than those who did develop diabetes, supporting the importance of diabetes prevention [29]. However, there have not been a sufficient number of cardiovascular events in the DPPOS to permit meaningful analysis of macrovascular events [30]. Because this polygenic lipodystrophy GRS has previously been associated with coronary artery disease [8], we tested the GRS for association with cardiovascular disease risk factors in the DPP. These risk factors are quantitative laboratory-based measures that are heritable intermediate phenotypes and are related to the disease outcome of interest, in this case, cardiovascular disease. Our results show that among individuals who are at risk for diabetes, a higher GRS for insulin resistance with lower BMI was associated with higher cardiovascular disease risk profile, specifically smaller or more dense LDL peak particle measurements after adjustment for relevant demographic and anthropometric measurements [8]. LDL peak particle density is an emerging risk factor that seems to be an important predictor of cardiovascular events and progression of coronary artery disease [26]. Of note, the GRS was not associated with the CAC scores that predict total coronary atherosclerotic burden at year 14 in DPPOS. However, because we do not have baseline CAC data, our interpretation of results is based on the assumption that as a result of randomization to treatment groups, the distribution of CAC scores at baseline would have been similar among the treatment groups. Further evaluation of this GRS awaits results of cardiovascular events in the DPP, but our results suggest that this GRS may help characterize cardiovascular disease risk in a seemingly homogenous group of individuals at risk for diabetes.
One of the main strengths of our study is that the DPP randomized controlled trial design enabled extensive in-depth phenotyping and comprehensive longitudinal measurements as well as standardized therapeutic interventions to characterize the effects of genetic variants on various outcomes including diabetes incidence and response to in a multiethnic cohort with high metabolic risk at baseline. However, we recognize that our study also has limitations. The genetic variants in our score are associated with fasting insulin, which is not the most accurate measure of insulin sensitivity. However, more accurate clamp-based measures of insulin sensitivity are not feasible in a large-scale study such as the DPP. Also, our insulin measurements are posthepatic and do not reflect portal insulin secretion, which was not obtained in the DPP. DPP participants are adults ascertained by the presence of risk factors for the development of diabetes but do not have diabetes, therefore, their glycemic variables fall within a narrow range. Additionally, the GRS is based on variants that have been discovered in populations of European descent and therefore may not adequately capture genetic variation in individuals of non-European descent. For the analyses related to cardiovascular endophenotypes, we acknowledge the concern for multiple hypothesis testing, although the outcomes tested are closely related and likely not independent; we are therefore careful to interpret our findings related to these traits as emerging from exploratory analyses
In conclusion, our results provide further evidence for a genetic link between various risk factors that contribute to cardiovascular disease risk and highlight the potential association of adipose tissue dysfunction independent of BMI with worsening insulin resistance and potential increased cardiovascular disease risk. We hope that these results advance physiological understanding and will inform future functional studies to explore the underlying mechanisms behind these associations. Individuals at high risk of type 2 diabetes who had a high genetic burden of lipodystrophic insulin resistance had less improvement in their insulin sensitivity over time. We confirm that metformin treatment and intensive lifestyle modification are effective in improving insulin sensitivity regardless of genetic risk. A full evaluation of the effects of this GRS on cardiovascular disease awaits the accrual of hard cardiovascular event outcomes in the DPPOS.
Acknowledgments
The research group gratefully acknowledges the commitment and dedication of the participants of the DPP and DPPOS. During the DPP and DPPOS, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding to the clinical centers and the Coordinating Center for the design and conduct of the study, and collection, management, analysis, and interpretation of the data (U01 DK048489). The Southwestern American Indian Centers were supported directly by the NIDDK, including its Intramural Research Program, and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Funding was also provided by the National Institute of Child Health and Human Development, the National Institute on Aging, the National Eye Institute, the National Heart Lung and Blood Institute, the National Cancer Institute, the Office of Research on Women’s Health, the National Institute on Minority Health and Health Disparities, the Centers for Disease Control and Prevention, and the American Diabetes Association. Bristol-Myers Squibb and Parke-Davis provided additional funding and material support during the DPP, Lipha (Merck-Sante) provided medication and LifeScan Inc. donated materials during the DPP and DPPOS. This research was also supported, in part, by the intramural research program of the NIDDK. LifeScan Inc., Health O Meter, Hoechst Marion Roussel, Inc., Merck-Medco Managed Care, Inc., Merck and Co., Nike Sports Marketing, Slim Fast Foods Co., and Quaker Oats Co. donated materials, equipment, or medicines for concomitant conditions. McKesson BioServices Corp., Matthews Media Group, Inc., and the Henry M. Jackson Foundation provided support services under subcontract with the Coordinating Center. The sponsor of this study was represented on the Steering Committee and played a part in study design, how the study was done, and publication. The funding agency was not represented on the writing group, although all members of the Steering Committee had input into the report’s contents. All authors in the writing group had access to all data. The opinions expressed are those of the investigators and do not necessarily reflect the views of the funding agencies. A complete list of centers, investigators, and staff can be found on the DPPOS website at https://dppos.bsc.gwu.edu/web/dppos/appendix. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial Support: This work was supported by U01 DK048489 as well as R01 DK072041-12 to J.C.F. and K.A.J. S.S. was supported by NIH Training Grants 5T32GM007748-36 (Harvard Medical School/Department of Genetics) and 2T32DK007028-41(Massachusetts General Hospital/Endocrine Unit). The funding agencies were not involved in the design of the study; the collection, analysis, and interpretation of data; writing the report; or the decision to submit the report for publication.
Clinical Trial Information: ClinicalTrials.gov Identifier: NCT00004992 (registered 20 March 2000).
Author Contributions: S.S. wrote the article, helped conceive the analytic plans, led the interpretation of results, and edited and reviewed the article before submission. K.A.J. conducted statistical analyses, contributed to the interpretation of the results, and edited and reviewed the article before submission. W.C.K., S.D-J., S.K., E.J.B., G.A.B., E.S.H., R.G., and the DPP Research Group planned and conducted the clinical trial and obtained the phenotypic data. W.C.K., S.D-J., S.K., E.B., G.B., E.H., M-F.H., R.G., L.C., J.M., and M.H. contributed to the interpretation of the results, and edited and reviewed the article before submission. J.C.F. conceived the study design, contributed to the interpretation of the results, and edited and reviewed the article before submission. J.C.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Glossary
Abbreviations:
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BMI
body mass index
- CAC
coronary artery calcification
- DPP
Diabetes Prevention Program
- DPPOS
Diabetes Prevention Program Outcomes Study
- GRS
genetic risk score
- GWAS
Genome-wide association studies
- ISI
insulin sensitivity index
- SNP
single nucleotide polymorphism
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability:
DPP and DPPOS data are available in the NIDDK repository (www.niddkrepository.org/home/) and can be requested by any researcher. In accordance with the NIH Public Access Policy, we continue to provide all manuscripts to PubMed Central including this manuscript. DPP/DPPOS has provided the protocols and lifestyle and medication intervention manuals to the public through its public website (www.dppos.org). The DPPOS abides by the NIDDK data sharing policy and implementation guidance as required by the NIH/NIDDK.
References and Notes
- 1. Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of type 2 diabetes. Diabetologia. 2003;46(1):3–19. [DOI] [PubMed] [Google Scholar]
- 2. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk [published correction appears in: Nat Genet. 2010;42(5):464]. Nat Genet. 2010;42(2):105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga JJ, Ingelsson E, Jackson AU, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad MT, Montasser ME, Navarro P, Perry JR, Rasmussen-Torvik LJ, Salo P, Sattar N, Shungin D, Strawbridge RJ, Tanaka T, van Duijn CM, An P, de Andrade M, Andrews JS, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann JS, Beilby JP, Bellis C, Bergman RN, Blangero J, Boban M, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Borecki IB, Böttcher Y, Bouchard C, Brunner E, Budimir D, Campbell H, Carlson O, Chines PS, Clarke R, Collins FS, Corbatón-Anchuelo A, Couper D, de Faire U, Dedoussis GV, Deloukas P, Dimitriou M, Egan JM, Eiriksdottir G, Erdos MR, Eriksson JG, Eury E, Ferrucci L, Ford I, Forouhi NG, Fox CS, Franzosi MG, Franks PW, Frayling TM, Froguel P, Galan P, de Geus E, Gigante B, Glazer NL, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris TB, Hayward C, Heath S, Hercberg S, Hicks AA, Hingorani A, Hofman A, Hui J, Hung J, Jarvelin MR, Jhun MA, Johnson PC, Jukema JW, Jula A, Kao WH, Kaprio J, Kardia SL, Keinanen-Kiukaanniemi S, Kivimaki M, Kolcic I, Kovacs P, Kumari M, Kuusisto J, Kyvik KO, Laakso M, Lakka T, Lannfelt L, Lathrop GM, Launer LJ, Leander K, Li G, Lind L, Lindstrom J, Lobbens S, Loos RJ, Luan J, Lyssenko V, Mägi R, Magnusson PK, Marmot M, Meneton P, Mohlke KL, Mooser V, Morken MA, Miljkovic I, Narisu N, O’Connell J, Ong KK, Oostra BA, Palmer LJ, Palotie A, Pankow JS, Peden JF, Pedersen NL, Pehlic M, Peltonen L, Penninx B, Pericic M, Perola M, Perusse L, Peyser PA, Polasek O, Pramstaller PP, Province MA, Räikkönen K, Rauramaa R, Rehnberg E, Rice K, Rotter JI, Rudan I, Ruokonen A, Saaristo T, Sabater-Lleal M, Salomaa V, Savage DB, Saxena R, Schwarz P, Seedorf U, Sennblad B, Serrano-Rios M, Shuldiner AR, Sijbrands EJ, Siscovick DS, Smit JH, Small KS, Smith NL, Smith AV, Stančáková A, Stirrups K, Stumvoll M, Sun YV, Swift AJ, Tönjes A, Tuomilehto J, Trompet S, Uitterlinden AG, Uusitupa M, Vikström M, Vitart V, Vohl MC, Voight BF, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Wheeler E, Widen E, Wild SH, Willems SM, Willemsen G, Wilson JF, Witteman JC, Wright AF, Yaghootkar H, Zelenika D, Zemunik T, Zgaga L, Wareham NJ, McCarthy MI, Barroso I, Watanabe RM, Florez JC, Dupuis J, Meigs JB, Langenberg C; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Mägi R, Strawbridge RJ, Rehnberg E, Gustafsson S, Kanoni S, Rasmussen-Torvik LJ, Yengo L, Lecoeur C, Shungin D, Sanna S, Sidore C, Johnson PC, Jukema JW, Johnson T, Mahajan A, Verweij N, Thorleifsson G, Hottenga JJ, Shah S, Smith AV, Sennblad B, Gieger C, Salo P, Perola M, Timpson NJ, Evans DM, Pourcain BS, Wu Y, Andrews JS, Hui J, Bielak LF, Zhao W, Horikoshi M, Navarro P, Isaacs A, O’Connell JR, Stirrups K, Vitart V, Hayward C, Esko T, Mihailov E, Fraser RM, Fall T, Voight BF, Raychaudhuri S, Chen H, Lindgren CM, Morris AP, Rayner NW, Robertson N, Rybin D, Liu CT, Beckmann JS, Willems SM, Chines PS, Jackson AU, Kang HM, Stringham HM, Song K, Tanaka T, Peden JF, Goel A, Hicks AA, An P, Müller-Nurasyid M, Franco-Cereceda A, Folkersen L, Marullo L, Jansen H, Oldehinkel AJ, Bruinenberg M, Pankow JS, North KE, Forouhi NG, Loos RJ, Edkins S, Varga TV, Hallmans G, Oksa H, Antonella M, Nagaraja R, Trompet S, Ford I, Bakker SJ, Kong A, Kumari M, Gigante B, Herder C, Munroe PB, Caulfield M, Antti J, Mangino M, Small K, Miljkovic I, Liu Y, Atalay M, Kiess W, James AL, Rivadeneira F, Uitterlinden AG, Palmer CN, Doney AS, Willemsen G, Smit JH, Campbell S, Polasek O, Bonnycastle LL, Hercberg S, Dimitriou M, Bolton JL, Fowkes GR, Kovacs P, Lindström J, Zemunik T, Bandinelli S, Wild SH, Basart HV, Rathmann W, Grallert H, Maerz W, Kleber ME, Boehm BO, Peters A, Pramstaller PP, Province MA, Borecki IB, Hastie ND, Rudan I, Campbell H, Watkins H, Farrall M, Stumvoll M, Ferrucci L, Waterworth DM, Bergman RN, Collins FS, Tuomilehto J, Watanabe RM, de Geus EJ, Penninx BW, Hofman A, Oostra BA, Psaty BM, Vollenweider P, Wilson JF, Wright AF, Hovingh GK, Metspalu A, Uusitupa M, Magnusson PK, Kyvik KO, Kaprio J, Price JF, Dedoussis GV, Deloukas P, Meneton P, Lind L, Boehnke M, Shuldiner AR, van Duijn CM, Morris AD, Toenjes A, Peyser PA, Beilby JP, Körner A, Kuusisto J, Laakso M, Bornstein SR, Schwarz PE, Lakka TA, Rauramaa R, Adair LS, Smith GD, Spector TD, Illig T, de Faire U, Hamsten A, Gudnason V, Kivimaki M, Hingorani A, Keinanen-Kiukaanniemi SM, Saaristo TE, Boomsma DI, Stefansson K, van der Harst P, Dupuis J, Pedersen NL, Sattar N, Harris TB, Cucca F, Ripatti S, Salomaa V, Mohlke KL, Balkau B, Froguel P, Pouta A, Jarvelin MR, Wareham NJ, Bouatia-Naji N, McCarthy MI, Franks PW, Meigs JB, Teslovich TM, Florez JC, Langenberg C, Ingelsson E, Prokopenko I, Barroso I; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McFarlane SI, Banerji M, Sowers JR. Insulin resistance and cardiovascular disease. J Clin Endocrinol Metab. 2001;86(2):713–718. [DOI] [PubMed] [Google Scholar]
- 6. Langenberg C, Sharp SJ, Schulze MB, Rolandsson O, Overvad K, Forouhi NG, Spranger J, Drogan D, Huerta JM, Arriola L, de Lauzon-Guillan B, Tormo MJ, Ardanaz E, Balkau B, Beulens JW, Boeing H, Bueno-de-Mesquita HB, Clavel-Chapelon F, Crowe FL, Franks PW, Gonzalez CA, Grioni S, Halkjaer J, Hallmans G, Kaaks R, Kerrison ND, Key TJ, Khaw KT, Mattiello A, Nilsson P, Norat T, Palla L, Palli D, Panico S, Quirós JR, Romaguera D, Romieu I, Sacerdote C, Sánchez MJ, Slimani N, Sluijs I, Spijkerman AM, Teucher B, Tjonneland A, Tumino R, van der A DL, van der Schouw YT, Feskens EJ, Riboli E, Wareham NJ; InterAct Consortium. Long-term risk of incident type 2 diabetes and measures of overall and regional obesity: the EPIC-InterAct case-cohort study. PLoS Med. 2012;9(6):e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Semple RK, Savage DB, Cochran EK, Gorden P, O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocr Rev. 2011;32(4):498–514. [DOI] [PubMed] [Google Scholar]
- 8. Yaghootkar H, Scott RA, White CC, Zhang W, Speliotes E, Munroe PB, Ehret GB, Bis JC, Fox CS, Walker M, Borecki IB, Knowles JW, Yerges-Armstrong L, Ohlsson C, Perry JR, Chambers JC, Kooner JS, Franceschini N, Langenberg C, Hivert MF, Dastani Z, Richards JB, Semple RK, Frayling TM. Genetic evidence for a normal-weight “metabolically obese” phenotype linking insulin resistance, hypertension, coronary artery disease, and type 2 diabetes. Diabetes. 2014;63(12):4369–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dick DM, Jones K, Saccone N, Hinrichs A, Wang JC, Goate A, Bierut L, Almasy L, Schuckit M, Hesselbrock V, Tischfield J, Foroud T, Edenberg H, Porjesz B, Begleiter H. Endophenotypes successfully lead to gene identification: results from the collaborative study on the genetics of alcoholism. Behav Genet. 2006;36(1):112–126. [DOI] [PubMed] [Google Scholar]
- 10.The Diabetes Prevention Program. The Diabetes Prevention Program. Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care. 1999;22(4):623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Diabetes Prevention Program Research Group. The Diabetes Prevention Program: baseline characteristics of the randomized cohort. Diabetes Care. 2000;23(11):1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM; Diabetes Prevention Program Research Group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diabetes Prevention Program Research Group. Effects of withdrawal from metformin on the development of diabetes in the diabetes prevention program. Diabetes Care. 2003;26(4):977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Venditti EM, Bray GA, Carrion-Petersen ML, Delahanty LM, Edelstein SL, Hamman RF, Hoskin MA, Knowler WC, Ma Y; Diabetes Prevention Program Research Group. First versus repeat treatment with a lifestyle intervention program: attendance and weight loss outcomes. Int J Obes (Lond). 2008;32(10):1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krakoff J, Clark JM, Crandall JP, Wilson C, Molitch ME, Brancati FL, Edelstein SL, Knowler WC; Diabetes Prevention Program Research Group. Effects of metformin and weight loss on serum alanine aminotransferase activity in the diabetes prevention program. Obesity (Silver Spring). 2010;18(9):1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. [DOI] [PubMed] [Google Scholar]
- 17. Bray GA, Jablonski KA, Fujimoto WY, Barrett-Connor E, Haffner S, Hanson RL, Hill JO, Hubbard V, Kriska A, Stamm E, Pi-Sunyer FX; Diabetes Prevention Program Research Group. Relation of central adiposity and body mass index to the development of diabetes in the Diabetes Prevention Program. Am J Clin Nutr. 2008;87(5):1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mather KJ, Funahashi T, Matsuzawa Y, Edelstein S, Bray GA, Kahn SE, Crandall J, Marcovina S, Goldstein B, Goldberg R; Diabetes Prevention Program. Adiponectin, change in adiponectin, and progression to diabetes in the Diabetes Prevention Program. Diabetes. 2008;57(4):980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. RRID:AB_2801639, https://scicrunch.org/resolver/AB_2801639.
- 20.Diabetes Prevention Program Research Group. Lipid, lipoproteins, C-reactive protein, and hemostatic factors at baseline in the diabetes prevention program. Diabetes Care. 2005;28(10):2472–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goff DC Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, Greenland P, Lackland DT, Levy D, O’Donnell CJ, Robinson JG, Schwartz JS, Shero ST, Smith SC Jr, Sorlie P, Stone NJ, Wilson PW, Jordan HS, Nevo L, Wnek J, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Curtis LH, DeMets D, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Smith SC Jr, Tomaselli GF; American College of Cardiology/American Heart Association Task Force on Practice Guidelines. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129(25 Suppl 2):S49–S73. [DOI] [PubMed] [Google Scholar]
- 22. Goldberg RB, Aroda VR, Bluemke DA, Barrett-Connor E, Budoff M, Crandall JP, Dabelea D, Horton ES, Mather KJ, Orchard TJ, Schade D, Watson K, Temprosa M; Diabetes Prevention Program Research Group. Effect of long-term metformin and lifestyle in the Diabetes Prevention Program and Its Outcome Study on coronary artery calcium. Circulation. 2017;136(1):52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma S, Liu A, Carr J, Post W, Kronmal R. Statistical modeling of Agatston score in multi-ethnic study of atherosclerosis (MESA). PLoS One. 2010;5(8):e12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diabetes Prevention Program Research Group. Relationship of body size and shape to the development of diabetes in the diabetes prevention program. Obesity (Silver Spring). 2006;14(11):2107–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hivert MF, Christophi CA, Franks PW, Jablonski KA, Ehrmann DA, Kahn SE, Horton ES, Pollin TI, Mather KJ, Perreault L, Barrett-Connor E, Knowler WC, Florez JC; Diabetes Prevention Program Research Group. Lifestyle and metformin ameliorate insulin sensitivity independently of the genetic burden of established insulin resistance variants in Diabetes Prevention Program participants. Diabetes. 2016;65(2):520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lotta LA, Gulati P, Day FR, Ongen H, van de Bunt M, Gaulton KJ, Eicher JD, Sharp SJ, Luan J, De Lucia Rolfe E, Stewart ID, Wheeler E, Willems SM, Adams C, Yaghootkar H; EPIC-InterAct Consortium; Cambridge FPLD Consortium, Forouhi NG, Khaw KT, Johnson AD, Semple RK, Frayling T, Perry JR, Dermitzakis E, McCarthy MI, Barroso I, Wareham NJ, Savage DB, Langenberg C, O'Rahilly S, Scott RA. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49(1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hivert MF, Jablonski KA, Perreault L, Saxena R, McAteer JB, Franks PW, Hamman RF, Kahn SE, Haffner S, Meigs JB, Altshuler D, Knowler WC, Florez JC; DIAGRAM Consortium; Diabetes Prevention Program Research Group. Updated genetic score based on 34 confirmed type 2 diabetes Loci is associated with diabetes incidence and regression to normoglycemia in the diabetes prevention program. Diabetes. 2011;60(4):1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vassy JL, Hivert MF, Porneala B, Dauriz M, Florez JC, Dupuis J, Siscovick DS, Fornage M, Rasmussen-Torvik LJ, Bouchard C, Meigs JB. Polygenic type 2 diabetes prediction at the limit of common variant detection [published correction appears in Diabetes. 2014;63(6):2172]. Diabetes. 2014;63(6):2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nathan DM, Barrett-Connor E, Crandall JP, Edelstein SL, Goldberg RB, Horton ES, Knowler WC, Mather KT, Orchard TJ, Pi-Sunyer X, Schade D, Temprosa M; Diabetes Prevention Program Research Group. Long-term effects of lifestyle intervention or metformin on diabetes development and microvascular complications over 15-year follow-up: the Diabetes Prevention Program Outcomes Study. Lancet Diabetes Endocrinol. 2015;3(11):866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knowler WC, Fowler SE, Hamman RF, Christophi CA, Hoffman HJ, Brenneman AT, Brown-Friday JO, Goldberg R, Venditti E, Nathan DM; Diabetes Prevention Program Research Group. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet. 2009;374(9702):1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DPP and DPPOS data are available in the NIDDK repository (www.niddkrepository.org/home/) and can be requested by any researcher. In accordance with the NIH Public Access Policy, we continue to provide all manuscripts to PubMed Central including this manuscript. DPP/DPPOS has provided the protocols and lifestyle and medication intervention manuals to the public through its public website (www.dppos.org). The DPPOS abides by the NIDDK data sharing policy and implementation guidance as required by the NIH/NIDDK.