Abstract
Exonic circular RNAs (circRNAs) are covalently closed RNA molecules generated by a process named back‐splicing. circRNAs are highly abundant in eukaryotes, and many of them are evolutionary conserved. In metazoans, circular RNAs are expressed in a tissue‐specific manner, are highly stable, and accumulate with age in neural tissues. circRNA biogenesis can regulate the production of the linear RNA counterpart in cis as back‐splicing competes with linear splicing. Recent reports also demonstrate functions for some circRNAs in trans: Certain circRNAs interact with microRNAs, some are translated, and circRNAs have been shown to regulate immune responses and behavior. Here, we review current knowledge about animal circRNAs and summarize new insights into potential circRNA functions, concepts of their origin, and possible future directions in the field.
Keywords: circRNAs, circular RNAs, non‐coding RNAs, RNA processing, splicing
Subject Categories: RNA Biology
Introduction
“All truth passes through three stages. First, it is ridiculed. Second, it is violently opposed. Third, it is accepted as being self‐evident.” – Arthur Schopenhauer
From past to present—a summary
Circular RNAs (circRNAs) were first found in pathogens. In 1976, Sanger et al (1976) described viroids that contain “single‐stranded and covalently closed circular RNA molecules”. A second study published in 1979 by Hsu et al (1979) described RNAs that contained no free ends and their circularity was independent of associated proteins (Hsu & Coca‐Prados, 1979). These circRNAs were likely derived from viruses, but these findings were not followed up, probably due to the overwhelming evidence supporting the importance of linear RNAs and the excitement regarding the recent discovery of splicing.
After these initial reports, sporadic studies identified and characterized circRNAs generated from endogenous RNAs. The first of these reports, in 1991, described the serendipitous discovery of transcripts produced from non‐canonical splicing (“scrambled exons”) that originated from the deleted in colon cancer gene (DCC; Nigro et al, 1991). A year later, Cocquerelle et al (1992) reported similar findings for the human EST‐1 gene and linked the generation of those isoforms to the presence of large flanking introns (Cocquerelle et al, 1992).
A year later, two reports identified this type of non‐polyadenylated RNA with scrambled exons as covalently closed circular RNAs. The first study demonstrated the specific circularization of DCC and EST‐1 transcripts (Cocquerelle et al, 1993). Although the authors refer to the molecules as mis‐spliced, they raised the possibility that they might be functional. Cocquerelle et al (1993) also showed that the circularized RNAs were localized in the cytoplasm and were stable after a 48‐h treatment of cells with actinomycin D. In the second study, the authors determined that the Sry RNA scrambled products are indeed circular (Capel et al, 1993). Capel et al (1993) showed that this circular RNA is mainly cytoplasmic, tissue specific and is present in three different mice species.
In the following years, a few studies proposed mechanisms by which these molecules could be generated. These included the hypothesis that inverted repeats are necessary for Sry circularization (Dubin et al, 1995) and the finding that circRNAs can be produced in vitro from nuclear extracts (Pasman et al, 1996; Schindewolf et al, 1996).
During the late 1990s and early 2000s, additional circular RNAs were shown to be produced from the rat cytochrome P450 2C24 gene (Zaphiropoulos, 1996), the human cytochrome P450 gene, the rat androgen‐binding protein (ABP) gene (Zaphiropoulos, 1997), the human dystrophin gene (Surono et al, 1999), and human Inhibitor of cyclin‐dependent kinase 4 (INK4/ARF)‐associated non‐coding RNA (Burd et al, 2010). In addition, many studies classified other presumptive circular RNAs as scrambled‐exon, exon‐shuffling products, or just “non‐linear mRNAs” (Dixon et al, 2005; Al‐Balool et al, 2011). Although these early studies clearly documented the existence of circular RNA molecules, their potential impact was underappreciated.
Beginning in about 2010, the advancement of RNA‐seq technologies, together with the development of specialized computational pipelines, led to an explosion in circRNA research. Several studies in the early 2010s revealed that thousands of types of circRNAs are expressed in metazoans (Salzman et al, 2012; Hansen et al, 2013; Jeck et al, 2013; Memczak et al, 2013). Despite the low levels of most circRNA, some are highly abundant. Moreover, in many cases, like in the early discovered Sry gene (Capel et al, 1993), the circRNA can be the main product generated from the host gene (Salzman et al, 2012; Hansen et al, 2013; Jeck et al, 2013; Memczak et al, 2013). Moreover, two studies in 2013 not only identified thousands of circRNAs in different mammalian systems, but also showed that two circRNAs, CDR1as (also known as ciRS‐7) and circSry, can bind to and likely modulate activities of specific microRNAs (miRNAs; Hansen et al, 2013; Memczak et al, 2013). These and other works also showed that circRNAs are expressed in a tissue‐ and developmental stage‐specific manner in humans, mice, and flies (Westholm et al, 2014; Rybak‐Wolf et al, 2015; Gruner et al, 2016).
Further, these studies described novel methods for identifying and validating circRNAs. Among them was the analysis of non‐polyadenylated libraries enriched for circRNAs by pre‐treatment with RNase R (a 3′ exonuclease that only affects linear RNAs; Jeck et al, 2013; Memczak et al, 2013). This method allows enrichment for circRNAs as well as differentiation of real circRNAs from mRNAs with scrambled exons. Importantly, the presence of scrambled junctions can be due to other types of non‐canonical splicing (i.e., trans‐splicing), as well as common artifacts of the reverse transcriptase (i.e., template switching; Cocquet et al, 2006). Given the particular characteristics of the circRNA junction, circRNA identification and quantification requires specifically designed bioinformatic pipelines. Nowadays, there are a number of pipelines for annotating and quantifying circRNAs (reviewed by Hansen et al, 2016; Zeng et al, 2017; and compared by Hansen, 2018) as well as databases for circRNA reference (reviewed by Xu, 2017). It is worth mentioning that new circRNA detection methods and pipelines can also determine the existence of potentially internal alternative splicing of circRNAs (Gao et al, 2016; Data ref: Rahimi et al, 2019).
Despite their high abundance and cell specificity, the lack of additional insights raised questions about the functionality of circRNAs. For example, circRNAs could be side products of splicing. This hypothesis was backed up by the lack of evidence of translation of these molecules, as well as doubts about their cell specificity (Jeck et al, 2013; Guo et al, 2014). These conclusions were mainly based on the analysis of data collected from cell lines, which do not contain large amounts of circRNAs compared with tissues or primary cultured cells (Ashwal‐Fluss et al, 2014; Rybak‐Wolf et al, 2015). In recent years, the tissue‐specific and developmental stage‐regulated production of circRNAs has been confirmed. Four independent works showed that many circRNAs are highly abundant in the brain and that the levels of these RNAs increase during neuronal differentiation and development (Westholm et al, 2014; Rybak‐Wolf et al, 2015; Veno et al, 2015; You et al, 2015). In addition, circRNA production is modulated by neuronal activity and enriched in synaptosomes, the synaptic neuropil, and dendrites (You et al, 2015). The prevalence of circRNAs in neuronal tissue is even more evident in aging animals, which accumulate large amounts of circRNAs (Westholm et al, 2014; Gruner et al, 2016; Cortes‐Lopez et al, 2018). These findings were in line with previous work that had suggested a negative correlation between circRNA levels and cell division rates (Bachmayr‐Heyda et al, 2015). Along with functional experiments, these reports helped to remove doubts about the functionality of at least some circRNAs. Additional functional studies are essential to determine how many circRNAs have functions in vivo and to understand how common (or uncommon) this is.
Theoretically, circRNAs can act in cis or in trans. In 2014, Ashwal‐Fluss et al (2014) showed that circRNAs are produced co‐transcriptionally and in competition with regular splicing. Hence, biogenesis of circRNA results in reduced synthesis of mRNAs from the same locus. In this situation, production of the circRNA acts as an RNA trap for mRNA production. Several groups identified requirements for splicing and circularization of exons and demonstrated that circularization signals are located within the introns flanking the circularizable exons (Jeck et al, 2013; Ashwal‐Fluss et al, 2014; Liang & Wilusz, 2014; Wang et al, 2014; Zhang et al, 2014; Ivanov et al, 2015; Rybak‐Wolf et al, 2015; Starke et al, 2015; Veno et al, 2015; Sun et al, 2016; Zhang et al, 2016b). Ashwal‐Fluss et al (2014) also suggested the existence of a negative‐feedback loop for the regulation of circMbl production in flies and identified the first protein involved in exon circularization [the splicing factor muscleblind (MBL) in Drosophila and the vertebrate homolog muscleblind‐like protein 1 (MBNL1)]. Later works identified other RNA‐binding proteins (RBPs) that regulate exon circularization in different systems and organisms; these include adenosine deaminases acting on RNA (ADAR), quaking (QKI), FUS, nuclear factors NF90/NF110, DExH‐Box helicase 9 (DHX9), epithelial splicing regulatory protein 1 (ESRP1), and serine/arginine (SR)‐rich proteins (Conn et al, 2015; Ivanov et al, 2015; Kramer et al, 2015; Rybak‐Wolf et al, 2015; Aktas et al, 2017; Errichelli et al, 2017; Li et al, 2017a; Yu et al, 2017).
Finally, current works have uncovered the relevance of circRNAs in different systems. A subset of circRNA produces proteins in the fly brain and in murine and human cells (Legnini et al, 2017; Pamudurti et al, 2017; Yang et al, 2017), and others seem to be related to the immune response (Li et al, 2017a; Liu et al, 2019). Several reports demonstrated in vivo functions for circRNAs in the mouse and fly brains, as well as in bone marrow (Piwecka et al, 2017; Kleaveland et al, 2018; Xia et al, 2018; Data ref: Pamudurti et al, 2018). In addition, a number of studies suggest an association between circRNAs and cancer (reviewed in Kristensen et al, 2018; Patop & Kadener, 2018). These developments show a clear change of view in the scientific community toward circRNAs, representing a turning point in this exciting and rapidly developing field.
The making of a circRNA
The mechanism of back‐splicing
Exon‐derived circRNAs are generated by a particular type of splicing known as back‐splicing, in which a 5′ splice donor attacks an upstream 3′ splice site. This leads to a 3′–5′ phosphodiester bond that generates a circular RNA molecule (Fig 1.1). While circRNAs are produced by the spliceosome in most (if not all) eukaryotes, the exact mechanism seems to differ between yeast, plants, and metazoans. Contrary to animals, circRNAs in plants are produced from regions flanked by long introns but with very short complementary sequences or sequences with no complementarity at all (Ye et al, 2015; Sun et al, 2016). Interestingly, in Archaea the circularization process is independent of the spliceosome (Nielsen et al, 2003; Salgia et al, 2003) and results in a variety of circRNAs, of which only 16% derive from coding genes and a lesser portion from exons (Danan et al, 2012).
Figure 1. circRNAs at a glance.
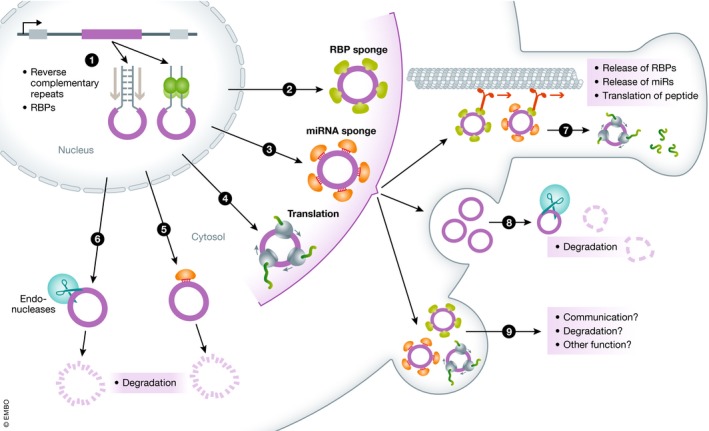
circRNA can be generated either with the help of reverse complementary repeats or RNA‐binding proteins and exported from the nucleus (1). In the cytoplasm, the circRNA might be bound by multiple factors. These can be RNA‐binding proteins (2), Argonaute proteins loaded with miRNAs as sponge or scaffold (3) or for direct degradation (5), ribosomes (4) or endonucleases that would cause degradation of the circRNA (6). From the non‐degradative binding, the circRNA‐factor complex might diffuse in the cytoplasm or been actively transported in into particular regions of the cell [e.g., the synapse (7)] where it can release its bound cargo or starts to be translated. The enclosure of circRNAs or circRNA factor complexes in vesicle that would be released into the extracellular space would remove circRNAs from the cytoplasm (8). However, protected by the vesicle, the circRNAs or circRNA complexes could reach other cells or tissues and therefore act as messenger molecules or fulfill other unknown functions (9).
In metazoans, previous studies showed that splice sites flanking circularizable exons are canonical and that back‐splicing is performed by the spliceosome (Cocquerelle et al, 1993; Zaphiropoulos, 1997; Ashwal‐Fluss et al, 2014; Guo et al, 2014; Westholm et al, 2014; Starke et al, 2015; Sun et al, 2016). Interestingly, circRNAs generally contain complete exons and are mostly generated from coding exons, particularly those located in the 5′ untranslated regions (UTRs) of protein‐coding genes (Guo et al, 2014; Westholm et al, 2014; Rybak‐Wolf et al, 2015). This results in back‐splice junctions formed by coding sequence to coding sequence regions (CDS‐CDS) and 5′ UTR‐CDS regions and tends to include the second exon of the gene (Salzman et al, 2012; Westholm et al, 2014). This might be related to their biogenesis, which requires longer and less efficiently spliced introns than average; both criteria are usually met with the first introns (Ashwal‐Fluss et al, 2014). While circRNA biogenesis might require inefficient canonical splicing, the introns within circRNAs seem to be mostly spliced out (Westholm et al, 2014; Guo et al, 2014; Gao et al, 2016; Data ref: Rahimi et al, 2019; Ji et al, 2019). These findings suggest that the inefficient splicing of the introns flanking circularizable exons is specific and not related to a poor processing of those transcripts in general.
In many cases, the production of circRNAs derives from intricate alternative splicing decisions (Zhang et al, 2016a; Gao et al, 2016). Since some genes produce dozens of alternative splicing isoforms and circRNAs, this suggests that back‐splicing and alternative splicing might be functionally related (Gao et al, 2016; Zhang et al, 2016a).
Sequence‐ vs. protein‐driven exon circularization
The production of exon‐derived circRNAs strongly depends on at least one of two mechanisms: introns with long inverted repeats or binding of RBPs. Both mechanisms bring the circRNA flanking introns in close proximity to each other. Introns flanking circRNAs are less efficiently spliced out than canonical splicing events at the junction sites, suggesting that linear splicing is the default splicing process (Zhang et al, 2016b). In addition, circularizable exons are flanked by long introns in fly, human, mouse, pigs, and also in plants (Jeck et al, 2013; Guo et al, 2014; Westholm et al, 2014; Veno et al, 2015; Ye et al, 2015; Zhao et al, 2017). Many of these introns display extensive reverse complementarity (Capel et al, 1993; Jeck et al, 2013; Liang & Wilusz, 2014; Zhang et al, 2014). Indeed, introns flanking exons that generate circRNAs are enriched in reverse complementary matches in Caenorhabditis elegans, human, mouse, and pig (Liang & Wilusz, 2014; Zhang et al, 2014; Ivanov et al, 2015; Rybak‐Wolf et al, 2015; Veno et al, 2015). Moreover, the presence of reverse complementary repeats in the introns can be used to predict which exons are likely to circularize (Ivanov et al, 2015). Reverse complementary elements show different motif enrichment depending on the analyzed species. In humans, 88% are ALU repeats, while in mouse, 22% are ALU‐like repeats, and in worms, only 11% are enriched for one specific motif (Jeck et al, 2013; Ivanov et al, 2015). Sequence alignment between these motifs points to a possible evolutionary relationship (Ivanov et al, 2015). Interestingly, ALU elements are believed to originate from transposons during evolution (Quentin, 1992). Moreover, the distribution of reverse elements among and within introns can have a strong influence on the amount and type of circRNAs produced (Zhang et al, 2014). While long inverted repeats in flanking introns promote exon circularization, the presence of additional repeats within one of these introns can inhibit inter‐intronic interactions in favor of intra‐intronic interactions. The latter tend to inhibit exon circularization, likely by competing with inter‐intronic secondary structures (Zhang et al, 2014).
Not all exons flanked by long introns are circularized, and there are many cases in which it is not possible to identify long stretches of complementing sequences. For instance, only 40% of circRNAs expressed in human fibroblasts are expressed from genes that contain ALU repeats in their flanking introns, from which 20% are also complementing (Jeck et al, 2013). In pigs, only approx. 30% of the circRNAs expressed in cortex have complementary short interspersed elements (SINEs, a group that includes the primate‐specific ALUs) in the introns flanking the circularizable exons (Veno et al, 2015). This proportion is similar to the 38% observed in C. elegans (Ivanov et al, 2015). The absence of inverted repeats in many introns flanking circularizable exons strongly suggests the presence of additional mechanisms of exon circularization. In fact, the splicing factor MBL mediates one alternative mechanism (Ashwal‐Fluss et al, 2014). MBL binds to several highly conserved intronic sites and promotes circularization of its own second exon. Interestingly, the Drosophila mbl gene produces mRNAs encoding several protein isoforms, not all of which promote exon circularization: Only MBL‐C (the predominant MBL isoform in fly heads) and MBL‐A do so. Conceivably, circMbl production is regulated in tissue and cell‐specific manner by the tissue‐specific MBL isoform production. Both fly MBL‐C and MBL‐A isoforms also promote circularization in the endogenous human MBNL1 locus, suggesting the existence of a highly conserved mechanism (Ashwal‐Fluss et al, 2014). circMbl also strongly binds to the MBL protein, pointing to the existence of a regulatory loop between these two molecules. Two of the MBL mammalian homologs (MBNL1 and MBNL2) generate circRNAs in mouse and humans (Salzman et al, 2012; Rybak‐Wolf et al, 2015). While these circRNAs are expressed at relatively low levels compared to the fly, they contain more than twice the number of binding sites for the MBL/MBNL1/MBNL2 protein, keeping the MB(N)L/circMbl(n)l ratio comparable between the different species (Ashwal‐Fluss et al, 2014). Interestingly, the introns flanking the second exon of mbl contain short inverted sequences that are likely to stabilize inter‐intron interactions, but may be too weak to promote exon circularization in the absence of MBL binding (Ashwal‐Fluss et al, 2014). This strongly suggests that MBL promotes circularization by binding to the flanking introns and promoting intron–intron interactions. The MBL molecules may dimerize, bringing the two ends of the exon together to allow circRNA formation (Yuan et al, 2007).
Other RBPs, such as QKI, FUS, and ESRP1, can also regulate exon circularization. QKI promotes global circRNA production during the epithelial‐to‐mesenchymal transformation in human cell culture by binding to intronic sequences surrounding the circularized exons (Conn et al, 2015). Interestingly, like MBL, QKI might dimerize to facilitate circularization (Teplova et al, 2013). FUS regulates circRNA formation in murine embryonic stem cell‐derived motor neurons by binding to specific exon–intron junctions (Errichelli et al, 2017). In addition, the splicing factor ESRP1 mediates circularization of circBIRC6 (derived from the baculoviral IAP repeat‐containing 6 gene) by binding to specific sites that are present in the introns flanking the circularizable exon (Yu et al, 2017). Last, the biosynthesis of a laccase‐2‐derived circRNA in Drosophila is regulated by a combination of different RBPs, such as heterogeneous nuclear ribonucleoproteins (hnRNPs) and SR proteins, suggesting that the circularization efficiency of a given exon can be the result of the integration of several signals (Kramer et al, 2015).
As mentioned, the presence of intronic long inverted repeats and the binding (and probably dimerizing) of RBPs facilitate intron–intron interactions leading to circRNA formation. This enhancement of circularization through intron–intron interaction could also be at least partially due to the steric inhibition of linear splicing. In this context, factors that promote or disrupt RNA structure, and hence putative intron–intron interactions, should alter circRNA biosynthesis. Indeed, recent work showed that RNA editing by the double‐stranded RNA (dsRNA)‐specific adenosine deaminase ADAR modulates circRNA biogenesis in human and mouse cell culture (Ivanov et al, 2015) and in fly heads (Rybak‐Wolf et al, 2015). Furthermore, the A‐to‐I editing signature of ADAR is enriched 200–600 nucleotides upstream and downstream of the splice sites in circRNA‐proximal introns in C. elegans (Ivanov et al, 2015). In addition, the RNA helicase DHX9 limits circRNA production by disrupting secondary structures like those based on ALU inverted repeats (Aktas et al, 2017). DHX9 interacts directly with an interferon‐inducible isoform of ADAR (p150), and this complex is responsible for disrupting RNA secondary structures, including many that could promote exon circularization. Interestingly, DHX9 downregulation doubles the population of uniquely detected circRNAs (~26,000 to ~50,000). This seems to be a correction mechanism to reduce the widespread production of circRNAs and suggests that some circRNAs are no more than “processing defects” or splicing noise. However, it also suggests a potential requirement for the circRNAs which are produced even in the presence of DHX9.
Particular physiological situations that involve the appearance of dsRNA structures can also alter circRNA biogenesis. For example, the immune response factors NF90 and NF110 (Patiño et al, 2015) regulate circRNA production. Interestingly, these proteins bind to viral RNA upon infection and interact with dsRNA structures formed during transcription. This union seems to stabilize such transient RNA duplexes and promotes back‐splicing of a subset of circRNAs (Li et al, 2017a). Interestingly, NF90‐binding sites are selectively enriched in ALU motifs of introns that bracket circRNA‐forming exons. Thus, circularization of these exons can also be subjected to ADAR and/or DHX9 control. Moreover, poly(I:C) (polyinosinic:polycytidylic acid) stimulation, which mimics viral infection, promotes the export of NF90/NF110 to the cytosol. Under these conditions, the circRNAs produced via the binding of NF90/NF110 are downregulated (Li et al, 2017a). NF90/NF110 binds not only to nascent circRNAs in the nucleus but also to at least two circRNAs in the cytoplasm (circPOLR2A and circDHX34). Upon poly(I:C) treatment or vesicular stomatitis virus infection, the interaction of both circRNAs with NF90/NF110 is reduced. This may also be the case for other circRNAs. Therefore, circRNAs might trigger the release of NF90/NF110 allowing it to bind to viral RNA upon viral infections (Li et al, 2017a). RIG‐1, a protein that detects dsRNAs and triggers antiviral cytokine production (Yoneyama & Fujita, 2008), alters circRNA production in HeLa cells as shown by siRNA treatment and qPCR of a reporter circRNA (Li et al, 2017a). This protein is at the core of a mechanism that allows the distinction between endogenous and foreign circRNAs (Chen et al, 2017). However, the magnitude of RIG‐1 activation due to exogenous circRNA was challenged by later reports (Wesselhoeft et al, 2018, 2019).
Our understanding of the mechanisms that promote, disturb, or modulate circRNA formation will very likely increase in the following years. It will be highly interesting to evaluate the degree of evolutionary conservation of mechanisms that generate circRNAs. Moreover, one wonders whether circRNAs that have functions in trans are generated by a specific type of mechanism.
Regulation of circRNA biosynthesis
circRNAs are transcribed by RNA polymerase II and generated by the spliceosome (Cocquerelle et al, 1993; Zaphiropoulos, 1997; Ashwal‐Fluss et al, 2014; Starke et al, 2015; Sun et al, 2016). Importantly, many of the exons forming circRNAs are not alternatively spliced (Aufiero et al, 2018). Hence, some highly abundant circRNAs regulate in cis the production of mRNAs from the host gene. Further evidence for the widespread importance of this cis‐regulation is the fact that in addition to splicing circRNA production is also linked to inefficient cleavage and polyadenylation (Liang et al, 2017).
Our understanding of circRNA biogenesis is hampered by the fact that circRNA production differs depending on the system used. In cell culture, circRNA biosynthesis seems to be post‐transcriptional (Liang & Wilusz, 2014; Zhang et al, 2014). This might constitute an idiosyncrasy of a system (cell culture) in which circRNAs are known to be produced at very low levels compared to neural tissue (Ashwal‐Fluss et al, 2014). If circRNA production is in competition with canonical splicing, changes in splicing efficiency should modulate the production of circRNAs. This could be achieved by modulation of trans‐acting splicing factors or by changes in the kinetics of RNA polymerase II transcription, which are known to modulate alternative splicing (Kadener et al, 2001, 2002; de la Mata et al, 2003). In this context, changes in circRNA production can be seen as a consequence of alterations in the efficiency of global RNA processing events that are influenced by variations in transcription rates. Indeed, downregulation of general splicing regulators (e.g., SR protein SF2) or core spliceosome components [e.g. small ribonucleoprotein particle U1 subunit 70K and C (snRNP‐U1‐70K, snRNP‐U1‐C), pre‐mRNA processing 8 (Prp8, Slu7), cell division cycle 40 (CDC40)] shifts the production from linear toward circular RNAs (Kramer et al, 2015; Liang et al, 2017). Moreover, inhibition of transcriptional termination increases circRNA biosynthesis (Liang et al, 2017). In some cases, circRNAs can be generated after read‐through transcription from upstream genes resulting, for example, from the inhibition of the upstream gene cleavage (Liang et al, 2017). Interestingly, chromatin structure could modulate all these co‐transcriptional processing events and it would not be surprising if changes in chromatin structure underlie changes in circRNA production in specific loci.
Degradation of circRNAs
circRNAs do not have free ends and therefore are not accessible to many canonical RNA decay pathways. An in vitro study of 60 circRNAs in cell culture using 4‐thiouridine metabolic labeling demonstrated that most of the assayed circRNAs have longer half‐lives (18.8–23.7 h) than their linear counterparts (4.0–7.4 h; Enuka et al, 2016). Other studies support this finding (Ashwal‐Fluss et al, 2014; Zheng et al, 2016; Liang et al, 2017). circRNAs might have even longer half‐life in vivo, particularly in cell types that do not divide. Indeed, the age‐dependent accumulation of circRNAs in the brain is likely due to the high stability of these molecules. Vice versa, it appears that circRNAs do not accumulate in tissues with a high proliferation rate (Bachmayr‐Heyda et al, 2015). This might be due to dilution of the circRNAs if the proliferation is higher than the production.
Very little is known about the mechanisms and rates at which circRNAs are degraded in vivo. In theory, circRNA degradation could be initiated by an endonuclease and followed through a combination of exo‐ and endonucleases. A first hint for circRNA degradation via endonuclease activity has been shown using RNase H and Rrp44 in vitro (Mackie, 1998; Schaeffer et al, 2009). In both cases, the authors tested artificial circular RNA constructs and cleavage of these circRNA species was very low. Small RNA‐mediated degradation of circRNAs is so far the best characterized circRNA degradation pathway. However, the only example until now, besides artificial shRNA/siRNA‐based systems (Jeck et al, 2013; Legnini et al, 2017; Pamudurti et al, 2017; Yu et al, 2017), is the degradation of CDR1as by miR‐671 (Hansen et al, 2011). Notably, the miRNA‐binding site in the circRNA is almost fully complementary to the miRNA. The amount of CDR1as is directly modulated by miR‐671 through AGO2‐mediated degradation (Hansen et al, 2013). CDR1as, miR‐671, and its binding site are highly conserved (Hansen et al, 2013), and the deletion of this site results in a significant increase in CDR1as levels (Kleaveland et al, 2018). Interestingly, CDR1as levels are modulated by miR‐7 likely through slicing, which is also dependent on miR‐671 (Kleaveland et al, 2018).
A recent study suggests that the RNA modification N6‐methylation of adenosine (m6A) facilitates the recruitment of endonucleases potentially able to degrade circRNAs (Park et al, 2019). Another study found global degradation of circRNAs upon poly(I:C) stimulation or encephalomyocarditis virus (EMCV) infection in HeLa cells (Liu et al, 2019). Both treatments lead to activation of the endoribonuclease RNase L and degradation of circRNAs. Interestingly, a subset of the circRNAs bind and inhibit PKR (protein kinase dsRNA‐activated), an activator for response to viral infections. Moreover, the authors found spontaneous RNase L activation, an increased phosphorylation of PKR and reduction of circRNAs in peripheral blood mononuclear cells derived from systemic lupus erythematosus patients (Liu et al, 2019).
In addition to degradation, circRNAs might be eliminated from cells by exocytosis. Several studies have detected circRNAs in exosomes (Li et al, 2015; Lasda & Parker, 2016; Preußer et al, 2018). However, it is not yet known whether the secretion of circRNAs contributes significantly to lowering their intracellular levels (Fig 1.8). Alternatively, circRNA secretion might constitute a communication mechanism (Fig 1.9). In sum, given the growing body of evidence that circRNAs are functional molecules, questions about their degradation and extracellular transport are of importance and should be the focus of future studies.
circRNA properties and features
Evolutionary conservation of circRNAs
circRNAs are present in most organisms, including Archaea, plants, yeast, and most metazoans (Danan et al, 2012; Wang et al, 2014; Broadbent et al, 2015; Lu et al, 2015; Sun et al, 2016; Ji et al, 2019). However, it is not clear how they have evolved, as they seem to be produced by different mechanisms. Understanding the details of their evolutionary conservation might shed light on both circRNA function and possible mechanisms of evolution.
CircRNA production can function in cis by balancing the production rates of their linear counterpart. As function and conservation are usually related, the first level of analysis is to determine whether orthologous (similar gene in different organisms) or paralogous loci (close related genes in the same organism) generate circRNAs. While the second approach focuses on specific circRNAs, the comparison of circRNAs derived from orthologous genes can be used more widely. For instance, 22% (457) of circRNAs detected in Hs68 cells (human fibroblast line) and murine testis are generated from homologous loci (Jeck et al, 2013). Another study compared circRNAs in heads of three different Drosophila species. The authors found that roughly 300 (between 30 and 40%) of these circRNAs were present in the orthologous genes and exons (Westholm et al, 2014). If an orthologous or paralogous locus is producing a circRNA, the next question is the level of circRNA production compared to its linear counterpart in each species (circRNA to linear RNA ratio). For those circRNAs with a function related to their production rate (cis regulatory function), we expect to see relatively constant circRNA/mRNA ratio across evolution, but not necessarily sequence conservation of the circRNA. In those cases, evaluating nascent transcripts would allow the analysis of production rates and putative mRNA‐trap‐like regulation by circRNAs. One such example is circRims2, which in mouse and human is approximately 20‐fold more abundant than the linear mRNA (Rybak‐Wolf et al, 2015). This form of conservation (presence of circRNAs in orthologous loci) also has been reported in more distant species: human TTBK2 (tau tubulin kinase 2), murine Ttbk2 and Drosophila asator, as well as human CELF2 (CUGBP elav‐like family member 2), murine Celf2 and Drosophila bru‐2 (bruno 2), also the MBNL1 and mbl in humans and flies (Rybak‐Wolf et al, 2015).
Some circRNAs are generated from the same or equivalent exons in multiple species. The conservation can extend in those cases to the particular splice sites flanking the circRNA. Splice site conservation of specific circRNAs was observed in flies, mice, macaque, and humans (Westholm et al, 2014; Rybak‐Wolf et al, 2015; Ji et al, 2019). While only 3% of all circRNAs share the exact splice junction in Hs68 cells and murine testis, this seems to be the exception rather than the rule (Jeck et al, 2013). A similar splice‐junction study analyzed circRNAs from brains of humans and mice by mapping the circularizing splice sites (Rybak‐Wolf et al, 2015). About one‐third (4,522 out of 15,849) of the detected circRNAs share both splice sites, another third (4,527) share one splice site, demonstrating a very high degree of conservation within the mammalian brain. The degree of conservation is even higher for circRNAs with higher expression (Rybak‐Wolf et al, 2015). However, these results have to be taken with caution, as highly expressed circRNAs mostly derive from exons close to the transcription start site and conservation is not homogeneous along the gene sequence (Rybak‐Wolf et al, 2015). Many of the introns flanking these conserved circRNAs contain inverse complementary repeats in both mouse and humans (Ivanov et al, 2015; Rybak‐Wolf et al, 2015). Many of these complementary sequences are repetitive elements, which usually evolve rapidly (Ivanov et al, 2015; Rybak‐Wolf et al, 2015). As the general assumption is that the introns flanking the circularizable exons are the main drivers of circRNA biogenesis, it is possible that transcriptional features of these loci lead to the generation of circRNAs without exact evolutionary conservation of the inverted repeats. Therefore, even if the sequences are not conserved, their presence among different species indicates evolutionary pressure for the generation of circRNAs from one particular region. This evolutionary conservation is likely linked to the conserved presence of short or long inverted repeats or binding sites for RBPs. The latter might reflect a more specific regulation. The conservation of specific sequences and/or RNA‐binding sites might indicate that the circularization has a particular adaptive importance (Fig 2).
Figure 2. Multiple levels of circRNAs’ evolutionary conservation.
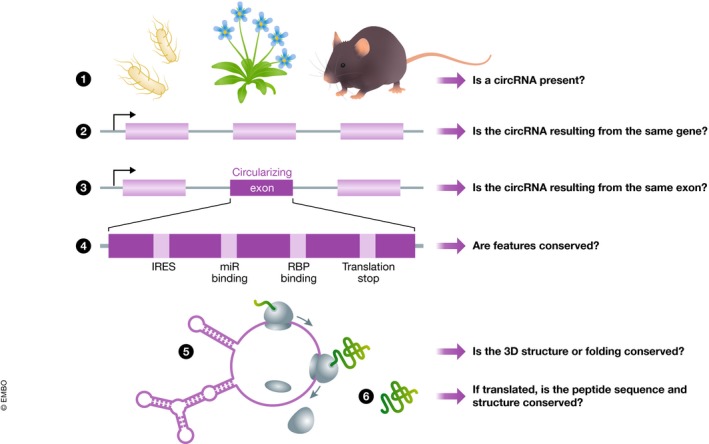
To determine the level of conservation of a circRNA between two or more organisms, we offer to ask several general questions: 1. Are circRNAs present in both organisms? 2. Is a circRNA produced from the same (homolog or orthologue) gene in both organisms? 3. Is a circRNA produced from the same exon(s)? 4. Are the potential features conserved? This would imply the potential presence of an RBP and microRNA‐binding sites, presence of IRES(s) and translational stop(s). 5. Is a potential 3D structure conserved? 6. If the circRNA is translated, is also the peptide sequence homolog?
The last level of evolutionary conservation is related to the functional elements within a given circRNA which could suggest a function in trans. These could include binding sites for RBPs, miRNAs, or elements necessary for functional secondary structures within the circRNA (Fig 2.4 and 2.5). For example, Rybak‐Wolf et al (2015) searched for miRNA‐binding sites within circRNA‐producing exons. The authors did not observe an enrichment of conserved miRNA‐binding sites compared to other short sequences. However, this study found that short sequences (some of which might be RBP‐binding sites) are enriched in exons present in circRNAs, pointing out a higher conservation level of circularized exons. Another study included 5′UTRs and confirmed a higher density of miRNA‐binding sites in circularized exons of flies (Westholm et al, 2014). The fact that Drosophila has more conserved miRNA target sites in their coding sequences than mammals might explain part of this difference (Schnall‐Levin et al, 2010). Further insights could be obtained by trying to determine evolutionary conservation in translated circRNAs, in particular to the regions surrounding the stop codons. Although stop codons of translated circRNAs are generally more evolutionarily conserved than other stop codons in the same 5′ UTR, their proximity to the splice site makes it impossible to determine whether the conservation is due to the importance of the splicing or the translation signal (Pamudurti et al, 2017). Another interesting feature is the secondary structure. Some human circRNAs might develop imperfect hairpins that are not detected in their linear counterparts (Liu et al, 2019). These hairpins are bound by RBPs responsible for intracellular immune response. Investigating if the hairpins are present in circRNA orthologues in other species and can still bind the same subset of RBPs could shed light not only on the conservation of circRNAs but also on the whole mechanism they are regulating.
Experimental validation of the putative interactions (i.e., with miRNAs) and/or translation capacity of circRNAs with putative conserved elements is necessary to fully understand the consequences of the conservation of one particular circRNA‐producing locus within different species (Fig 2.6). This is achievable by identifying translated circRNAs through ribosome footprinting, while protein–circRNA or circRNA–miRNA interactions can be tested by various methods as RIP, PAR‐CLIP (Photoactivatable Ribonucleoside‐Enhanced Crosslinking and Immunoprecipitation) and precipitation of the circRNA followed by protein identification and/or small RNA sequencing.
Tissue‐, developmental stage‐, and subcellular location‐specific expression
Genes hosting circRNAs are highly enriched for brain‐related genes (Ashwal‐Fluss et al, 2014; Westholm et al, 2014; Rybak‐Wolf et al, 2015; You et al, 2015). Hence, it is not surprising that circRNAs are highly enriched in neural tissue. This CNS‐specific enrichment seems to be a general feature of circRNAs in all the studied species (Guo et al, 2014; Westholm et al, 2014; Rybak‐Wolf et al, 2015; You et al, 2015). Notably, the expression of circRNAs within the brain is highly specific and, in many cases, independent of the expression of the linear isoform generated from the hosting gene (Rybak‐Wolf et al, 2015; Veno et al, 2015; You et al, 2015).
The notable enrichment of circRNAs in the CNS may be due to one or more factors. First, the brain and, more specifically, the neurons display the highest rates of alternative splicing in the body (Yeo et al, 2004; Pan et al, 2008), and circRNA biogenesis can be defined as a particular type of alternative splicing. Second, circRNAs are long‐lived, and, as neurons generally do not divide, circRNAs should in theory accumulate as the brain develops and ages even if produced at low rates (Westholm et al, 2014; Veno et al, 2015; Gruner et al, 2016). Indeed, circRNAs strongly accumulate with age in the brains of flies and mice (Westholm et al, 2014; Gruner et al, 2016), leading to the suggestion that circRNAs may be involved in age‐related brain diseases (reviewed in Hanan et al, 2017). As already mentioned, there is a strong anti‐correlation between the amount of circRNAs and cell replication rate (Bachmayr‐Heyda et al, 2015). Hence, it might seem that accumulation is the main reason behind the high levels of circRNAs in the brain. However, cardiomyocytes usually do not divide, but circRNAs do not accumulate in the heart as the animal ages (Gruner et al, 2016). A partial explanation might be that the heart displays a much smaller repertoire of alternatively spliced isoforms (Yeo et al, 2004). This suggests that the high levels of circRNAs observed in the CNS are related to the specific generation of these spliced isoforms, as well as accumulation due to low degradation and low or no cell division.
Another interesting feature of circRNAs is their subcellular localization. Unlike aberrantly spliced products, which localize near the site of transcription, circRNAs are predominantly cytoplasmic (Nigro et al, 1991; Capel et al, 1993; Cocquerelle et al, 1993; Salzman et al, 2012; Memczak et al, 2013; Werfel et al, 2016). This conclusion is mainly based on biochemical fractionation and in situ hybridization experiments (Hansen et al, 2013; Jeck et al, 2013). Moreover, reports show that subsets of circRNAs in neurons are localized to axons, dendrites, and synaptosomes (Rybak‐Wolf et al, 2015; You et al, 2015). Interestingly, some circRNA display a regulated switch in their nucleocytoplasmic localization during development (Veno et al, 2015).
A recent study identified Drosophila Hel25E (a DExH/D‐box helicase) and human UAP49/56 as key factors for nuclear export of circRNAs (Huang et al, 2018). Hel25E, UAP56, and UAP49 promote nuclear export in a manner that depends on the length of the circRNA. After depletion of the Drosophila helicase Hel25E in DL1 cells, circRNAs longer than 800 bases accumulate in the nucleus. Interestingly, this phenomenon is independent of the circRNA sequence. URH49 and UAP56 are human homologs with similar functions. URH49 seems responsible for the export of circRNAs shorter 356nts and UAP56 for circRNAs bigger than 1,298nts. For intermediate sizes, a more complex recognition mechanism might take part (Huang et al, 2018). In the future, exploring these intermediate length circRNAs for human cells and shorter circRNAs for fly cells will shed light over additional mechanism of circRNA export. It is unclear whether there is a direct or indirect interaction between the circRNAs and these proteins and whether other binding partners are involved. In most cases, the only features shared by circRNAs are their circularity, the presence of the exon junction complex, and the absence of the cap structure and the polyA tail. Therefore, recognition and export mechanisms have to be either highly specific for particular circRNAs or must recognize one or more of these features.
The localization of circRNAs to axons, dendrites, and synapses is also intriguing. It is still not clear whether the observed accumulation is due to a directed transport or to diffusion and retention of those molecules in these compartments (e.g., through binding to membrane proteins or just impossibility to come back). Further genetic and biochemical experiments should shed light on the mechanism that drives the subcellular localization of circRNAs within neurons.
As stated above, a large fraction of the genes hosting circRNAs encode for synaptic proteins and many brain circRNAs are strongly enriched in synaptosomes and in microdissected synaptic neuropil. As in many cases circRNAs and mRNAs share 5′ UTR regions (Pamudurti et al, 2017), it is possible that these sequences drive the localization of both the circular and linear forms produced from a given locus. Moreover, the mRNA and circRNA could compete for binding to transport and/or effector proteins. Competition for effector proteins might regulate translation and/or stability and could be a way in which circRNAs regulate gene expression in trans (Fig 1). To date, no studies have investigated circRNA production and transport using live‐cell imaging, and this type of approach will be key to test some of these hypotheses. Also, the field still lacks a precise description of the numbers and types of circRNA molecules in different cellular compartments.
circRNA as modulators of miRNA function
Some long non‐coding RNAs can regulate miRNA levels and/or activity by selective sponging (Cesana et al, 2011; Tay et al, 2011; Bitetti et al, 2018; Kleaveland et al, 2018). The initial observation that some circRNA possess many miRNA‐binding sites led to the speculation that these molecules could work as miRNA sponges. For example, CDR1as has 73 seed‐binding sites for miR‐7 (Hansen et al, 2013). Moreover, AGO2 CLIP data demonstrate that miR‐7 occupies many of these sites (Piwecka et al, 2017). As mentioned in previous sections, miR‐671 has one binding site with almost perfect complementarity to CDR1as. This suggests that while miR‐671 can mediate cleavage of CDR1as, this circRNA might regulate miR‐7 levels and/or activity.
The recent development of CDR1as knockout mice offers some insights into the functional consequences of miR‐7 binding to CDR1as (Piwecka et al, 2017). Piwecka et al (2017) showed that miR‐7 levels are modestly but significantly decreased and miR‐671 increased in the CDR1as knockout mice, suggesting that the presence of this circRNA stabilizes miR‐7 and destabilizes miR‐671. Moreover, the brains of CDR1as‐knockout animals show increased levels of several miR‐7 targets, again pointing out that this circRNA stabilizes rather than titrates miR‐7. Some of the miR‐7 targeted mRNAs are known to be induced by neuronal activity, and the knockout mice show specific excitatory and behavioral phenotypes (Piwecka et al, 2017). Thus, CDR1as may regulate storage and release of miR‐7 upon a specific signal. CDR1as could also transport and release miR‐7 to a particular cellular compartment, regulating miR‐7 function. This function might be harnessed in the future to deliver miRNA‐based therapeutics by taking advantage of the extreme stability of circRNA molecules.
Recent work showed that CDR1as is part of an even more complex regulatory circuit that involves the long intergenic non‐coding RNA (lincRNA) Cyrano (Kleaveland et al, 2018). By generating and using different CRISPR‐Cas9 engineered mice, Kleaveland et al (2018) show that Cyrano binds and targets miR‐7 for degradation. This effect of Cyrano on miR‐7 indirectly modulates the degradation of CDR1as by miR‐671. Thus, these non‐coding RNAs interact and regulate one another. However, as Cyrano−/− and miR‐7 double‐knockout mice had no observable phenotype, key details of this RNA network functionality remain unknown.
Although thorough inspection of circRNA sequences and analysis of AGO2 PAR‐CLIP data revealed that most circRNAs do not bind extensively to miRNAs (Guo et al, 2014), there are further examples of circRNAs that interact with miRNAs. For example, circSry has been shown to interact with miR‐138 (Hansen et al, 2013), but the physiological importance of this interaction has not been yet established. In addition, many other reports suggested interaction of circRNAs and miRNAs, but these putative contacts have not been biochemically demonstrated yet. For example, circHIPK3 (derived from the homeodomain interacting protein kinase 3 gene) has been described to interact with nine different miRNAs (miR‐124, miR‐152, miR‐193a, miR‐29a, miR‐29b, miR‐338, miR‐379, miR‐584, and miR‐654) and regulate cell growth (Li et al, 2017b). Also, circFOXO3 (derived from the forkhead box O3 gene) reportedly regulates cell growth by sponging specific miRNAs that regulate the production of FOXO3 mRNA (Du et al, 2016). circITCH (derived from the Itchy E3 ubiquitin‐protein ligase gene) might regulate the activity of oncogenic miR‐7 and miR‐214 in human cell lines (Huang et al, 2015), and it has been reported that circBIRC6 controls the pluripotency of human embryonic stem cells by sequestering miR‐34 and miR‐145 (Yu et al, 2017).
However, the circRNA‐miRNA sponge hypothesis must be seen critically. The abundance of specific circRNAs (besides MBL in flies or CDR1as in humans) is generally low. This matter contradicts possible sponge theories (Denzler et al, 2014; Jens & Rajewsky, 2015), since the miRNA might be more abundant than the total RNA in which the miRNA can bind and titrate, and there are more miRNA‐binding sites available than miRNA molecules. However, it is still possible that some circRNAs could act catalytically (i.e., by mobilizing, inactivating, and/or degrading miRNAs) but there are no indications that this is the case. Therefore, studies focusing on the sponge function of circRNAs would highly benefit from solid stoichiometric quantifications. The use of small RNA libraries and Argonaute‐RNA immunoprecipitation or crosslinking immunoprecipitation (AGO‐RIP/CLIP) techniques will be crucial to determine that there are direct interactions between the circRNAs and the regulated miRNAs. Generation of inducible knockdowns or knockout strains will be very helpful to determine the effect (if any) of the circRNAs on the putative miRNA function and levels.
Translation of circRNAs
In 2017, several groups reported that circRNAs are translated (Legnini et al, 2017; Pamudurti et al, 2017; Yang et al, 2017). Pamudurti et al (2017) utilized ribosome footprinting, mass spectrometry, and cellular assays to demonstrate translation of a subset of circRNAs in Drosophila heads and mammalian muscle cells. Interestingly, translated circRNAs tend to share a start codon with the hosting gene and have a stop codon that is evolutionary conserved and that is unique to the circular open reading frame. The same study found that circRNAs are translated by membrane‐associated ribosomes (Pamudurti et al, 2017). A simultaneously published study demonstrated the translation of circZNF609 (derived from the zinc finger protein 609 gene) in the context of murine and human skeletal muscle (Legnini et al, 2017). This study demonstrated the existence of a circRNA‐derived protein by polysome and Western blot assessments. Legnini et al (2017) also offered some insights into the function of the circRNA, as they showed that inhibition of the expression of circZNF609 caused a strong reduction in myoblast proliferation. In addition, Yang et al (2017) identified several small peptides generated from a subset of circRNAs in cancer cell lines and human fibroblasts (Yang et al, 2017). Interestingly, the authors found that RRACH motifs (R=G or A; H=A, C or U) upstream of the start codon enhance circRNA translation when the adenosine is methylated (Yang et al, 2017).
As circRNAs do not contain a 5′ cap, their translation needs to be cap‐independent. Indeed, Pamudurti et al (2017) showed that some of the translated circRNAs have internal ribosome entry sites (IRES) and can be translated in a cap‐independent way, both in vivo and in vitro (Pamudurti et al, 2017). Translation of circMbl in vivo is enhanced by signals that suppress cap‐dependent but not cap‐independent translation, like starvation or FOXO overexpression. Additional evidence of IRES‐mediated translation of circRNAs in vitro (Chen & Sarnow, 1998) and in cell culture experiments (Wang & Wang, 2015) was previously reported.
These studies do not provide much insight into the functions of the circRNA‐encoded protein products. A recently published report showed a potential molecular function of a circRNA‐encoded peptide in cancer (Yang et al, 2018). While encouraging, this study is based on circRNA overexpression. Forced expression of circRNAs in cell lines can result in concatemers (linear RNA molecules which contain several times the circRNA sequence) by trans‐splicing (e.g., see Liang & Wilusz, 2014; Pamudurti et al, 2017). Hence, definitive experiments will require exclusion of concatemer formation (i.e., by characterizing the RNA population following overexpression) as well as loss of function experiments (i.e., by generating cells with lower levels of the circRNA).
Interestingly, most of the predicted peptides are likely to be identical to the N‐terminal regions of the protein encoded from the circRNA hosting gene. These truncated proteins might act as competitors to their full‐length counterparts expressed from linear mRNAs. One possible case might be transcription factors like Mef2. Its N‐terminal DNA‐binding domain might be expressed from the circRNA (Legnini et al, 2017) and might compete for the DNA‐binding sites with the full‐length Mef2 or even have a function on its own (He et al, 2011; Legnini et al, 2017). The most straightforward hypothesis is that the circRNA‐encoded proteins have a function per se. Considering the rapid development of the field, we expect appearance of studies showing the physiological impact of circRNA translation and the resulting peptides in the next few years.
circRNAs as decoys, transporters, or scaffolds
As circRNAs are long‐lived and bind to RBPs, they can act as decoys or transporters for these factors. In some of the cases, there might be a direct or indirect crosstalk between the circRNA and the protein product of the host gene. This seems to be the case for circMbl, which might sequester/transport MBL protein. This is one component of a putative negative‐feedback loop in circMbl regulation (discussed in previous sections). Another circRNA, circPABPN1 (derived from the poly(A)‐binding protein nuclear 1 gene), binds the RBP HuR (ELAVL1 or Hu‐Antigen R). As HuR mediates the translation of the linear form of PABPN1 mRNA, the binding of circPABPN1 to HuR might inhibit it. Moreover, the interaction between HuR and the circRNA generally reduces translation (Abdelmohsen et al, 2017).
Another circRNA, circANRIL, is generally expressed at higher levels than the linear isoform and controls ribosomal RNA (rRNA) maturation by binding to PES1 (pescadillo ribosomal biogenesis factor 1; Holdt et al, 2016). Increased levels of circANRIL impair rRNA maturation and provoke nucleolar stress. circANRIL seems to form a stem loop structure that mimics rRNA and binds PES1, thus blocking its interaction with the PeBoW complex (Pes1, Bop1, WDR12), which is a key regulator of the 60S ribosome subunit biogenesis (Holdt et al, 2016).
In 2016, a study showed for the first time that circRNA can serve as a protein scaffold (Du et al, 2016). circFOXO3 was found to repress the transition through the cell cycle and to interact with both p21‐ and cyclin‐dependent kinase 2 (CDK2) in NIH3T3 murine fibroblasts. The formation of the circFOXO3‐p21‐CDK2 ternary complex arrests the function of CDK2 and consequently blocks the cell cycle progression. This ternary complex was validated by different complementary approaches, such as circRNA pulldown with specific probes, siRNA and RNase A protective assays (Du et al, 2016).
Some genomic studies indeed suggest that other circRNAs might regulate RBP function. For example, analysis of PAR‐CLIP data of 20 different RBPs suggested that circular exons have slightly higher cluster densities compared with their neighboring exons (Guo et al, 2014). However, this might not be a general phenomenon for all RBPs, as the motif density of 38 RBP expressed in the mouse brain is lower on exons that make circRNAs when compared with coding sequences and 3′ UTR (You et al, 2015).
Assessing the function of circRNAs in vivo
Two studies investigated the role of CDR1as in vivo (Piwecka et al, 2017; Kleaveland et al, 2018). Although this circRNA is embedded in a long non‐coding RNA locus (Barrett et al, 2017), the circularization of CDR1as is so efficient that previous studies could not detect any linear counterpart (Hansen et al, 2011, 2013; Memczak et al, 2013). Also, Piwecka et al (2017) could not detect any RNAs generated from the opposite strand and concluded that CDR1as is the only gene directly affected by the deletion (Piwecka et al, 2017). Deletion of CDR1as results in a behavioral phenotype that is associated with neuropsychiatric disorders (Piwecka et al, 2017). Kleaveland et al (2018) modulated CDR1as levels more mildly by doing a double knockout of Cyrano lincRNA (Cyrano −/−) and a mutant (Cyrano M3/M3) mice, which had a five nucleotides deletion in the miR‐7 site (Kleaveland et al, 2018). The authors also could increase CDR1as levels up to 4‐fold by disrupting the miR‐671 site within CDR1as. None of these mice, however, had evident phenotype.
A recent study evaluated mice deficient in a circRNA called cia‐cGAS (Cyclic GMP‐AMP synthase). The production of this circRNA was inhibited by deletion of the reverse complementary sequence in the downstream flanking intron using Cas‐9 (Xia et al, 2018). cia‐cGAS is normally highly expressed in the nuclei of long‐term hematopoietic stem cells. The authors claim that cia‐cGAS binds cGAS, blocking its activation. cia‐cGAS‐deficient mice had a reduced long‐term hematopoietic stem cell population compared with wild‐type mice. Also, the lack of cia‐cGAS causes elevated production of type I interferons in bone marrow leading to stem cell exhaustion (Xia et al, 2018).
A recent report investigates the consequences of knocking down the most abundant fly circRNA: circMbl (Data ref: Pamudurti et al, 2018). In this study, circMbl was knocked down using genetically encoded shRNAs directed against the back‐splice junction. This approach downregulates the circRNA of interest post‐transcriptionally without disturbing the linear RNA. Knocking down circMbl in the whole fly body leads to altered gene expression, male developmental lethality, behavioral defects, and defects in wing posture and flight. What is more, downregulation of circMbl in the fly central nervous system caused abnormal synaptic function. These phenotypes were recapitulated when using an additional shRNA, and no off targets were found. Interestingly, overexpression and knockdown of circMbl alter the expression of many common genes but in the opposite direction.
Potential additional functions of circRNAs
What other molecular functions could circRNAs have? A fascinating feature of circRNAs is their stability and accumulation over time. Hence, the population of circRNAs at a given time could provide the cell (or just us, the observer) a snapshot of the transcriptional history of a cell or even cell ancestry. From this perspective, circRNAs could serve as molecular memory molecules or “flight recorders” of the transcriptional history of the cell. This feature could potentially be exploited for researchers to determine exposure to stresses or specific situations. From the physiological point of view, long‐lived circRNAs could serve as reservoirs with protein‐coding potential. Considering that some circRNAs are known to be translated from IRES elements (Legnini et al, 2017; Pamudurti et al, 2017), upon developmental changes or stress these reservoirs could be translated into proteins that regulate stress response or physiological changes.
Local translation of circRNAs could be potentially important in synapses where other RNAs are translated (Bramham & Wells, 2007). Since some circRNAs bind RBPs (Du et al, 2016; Chen et al, 2017) as well as miRNAs, circRNAs might act by binding, delivering, and releasing their cargo at specific cellular compartments. On the other hand, circRNAs might compete in specific subcellular location for RBPs present in limiting amounts (i.e., translation factors or miRNAs). New biochemical, genetic, and imaging experiments are required to test these types of hypotheses, even more in vivo.
Going one step further and considering that circRNAs have been found in vesicles (Lasda & Parker, 2016; Preußer et al, 2018), they might also have a function as signaling molecules. Enclosed in vesicles they can be transported through the body, perhaps to specific receiver tissues. A circRNA loaded with one or several cargo molecules (miRNAs, RBPs) could be transported to an organ or a tissue. At its destination, the degradation of the circRNA or a different mechanism would release the cargo. However, it seems unlikely that this type of regulation could be very specific, as this would require very large amounts of circRNAs of a given type being secreted.
Conclusions and future directions
The studies described in this review suggest that circRNAs can act as scaffolds for proteins, can recruit other RNA species, and, through binding of miRNAs, can affect the transcriptional silencing, translation, and/or decay of specific mRNAs. The asymmetric distribution of circRNAs in neurons suggests the possibility of a directed transport of these RNAs across the cell.
circRNAs can encode proteins ranging from small peptides to proteins. Although physiological functions for most of these possible proteins have not yet been identified, it is likely that they will share some of the abilities of their full‐length protein counterparts encoded by the linear form of the transcript. As a side note, the coding ability and stability of circRNAs could be used in biotechnological applications that require production of peptides (Wesselhoeft et al, 2018, 2019).
As RNA technologies advance steadily, we foresee a great development of the circRNA field in the next years. Further understanding of circRNA localization, transportation, degradation in live cells, a completed circRNA interactome, and single‐cell profiling are some of the expected progress in the field.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank N. Reddy, J. A. Klaric, and M. Zaffagni for discussion and suggestions. This work was funded by the NIH R01 Grants: R01GM122406 and R01AG057700 to SK. SW was funded by the Alexander von Humboldt Foundation through the Feodor Lynen Research Fellowship.
The EMBO Journal (2019) 38: e100836
References
- Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A (2017) DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544: 115–119 [DOI] [PubMed] [Google Scholar]
- Al‐Balool HH, Weber D, Liu Y, Wade M, Guleria K, Nam PLP, Clayton J, Rowe W, Coxhead J, Irving J et al (2011) Post‐transcriptional exon shuffling events in humans can be evolutionarily conserved and abundant. Genome Res 21: 1788–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) CircRNA biogenesis competes with Pre‐mRNA splicing. Mol Cell 56: 55–66 [DOI] [PubMed] [Google Scholar]
- Aufiero S, van den Hoogenhof MMG, Reckman YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM, Creemers EE (2018) Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA 24: 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmayr‐Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner‐Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D (2015) Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 5: 8057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Parker KR, Horn C, Mata M, Salzman J (2017) ciRS‐7 exonic sequence is embedded in a long non‐coding RNA locus. PLoS Genet 13: e1007114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitetti A, Mallory AC, Golini E, Carrieri C, Carreño Gutiérrez H, Perlas E, Pérez‐Rico YA, Tocchini‐Valentini GP, Enright AJ, Norton WHJ et al (2018) MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat Struct Mol Biol 25: 244–251 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG (2007) Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 8: 776–789 [DOI] [PubMed] [Google Scholar]
- Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC (2015) Strand‐specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non‐coding RNA and circular RNA. BMC Genom 16: 454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF‐associated non‐coding RNA correlates with atherosclerosis risk. PLoS Genet 6: e1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell‐Badge R (1993) Circular transcripts of the testis‐determining gene Sry in adult mouse testis. Cell 73: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sarnow P (1998) Internal ribosome entry sites tests with circular mRNAs In Protein synthesis. Methods in molecular biology, vol 7, Martin R. (ed.), pp 355–364. Totowa, NJ: Springer; [DOI] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY (2017) Sensing self and foreign circular RNAs by intron identity. Mol Cell 67: 228–238 e225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B (1992) Splicing with inverted order of exons occurs proximal to large introns. EMBO J 11: 1095–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B (1993) Mis‐splicing yields circular RNA molecules. FASEB J 7: 155–160 [DOI] [PubMed] [Google Scholar]
- Cocquet J, Chong A, Zhang G, Veitia RA (2006) Reverse transcriptase template switching and false alternative transcripts. Genomics 88: 127–131 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 6: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Cortes‐Lopez M, Gruner MR, Cooper DA, Gruner HN, Voda AI, van der Linden AM, Miura P (2018) Global accumulation of circRNAs during aging in Caenorhabditis elegans . BMC Genom 19: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan M, Schwartz S, Edelheit S, Sorek R (2012) Transcriptome‐wide discovery of circular RNAs in Archaea. Nucleic Acids Res 40: 3131–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M (2014) Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 54: 766–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RJ, Eperon IC, Hall L, Samani NJ (2005) A genome‐wide survey demonstrates widespread non‐linear mRNA in expressed sequences from multiple species. Nucleic Acids Res 33: 5904–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 6: 2846–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H (1995) Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167: 245–248 [DOI] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas‐Chen A, Ulitsky I, Yarden Y (2016) Circular RNAs are long‐lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 44: 1370–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G et al (2017) FUS affects circular RNA expression in murine embryonic stem cell‐derived motor neurons. Nat Commun 8: 14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F (2016) Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 7: 12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner H, Cortés‐López M, Cooper DA, Bauer M, Miura P (2016) CircRNA accumulation in the aging mouse brain. Sci Rep 6: 38907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15: 409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanan M, Soreq H, Kadener S (2017) CircRNAs in the brain. RNA Biol 14: 1028–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J (2011) miRNA‐dependent gene silencing involving Ago2‐mediated cleavage of a circular antisense RNA. EMBO J 30: 4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Veno MT, Damgaard CK, Kjems J (2016) Comparison of circular RNA prediction tools. Nucleic Acids Res 44: e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB (2018) Improved circRNA identification by combining prediction algorithms. Front Cell Dev Biol 6: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ye J, Cai Y, Riquelme C, Liu JO, Liu X, Han A, Chen L (2011) Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly. Nucleic Acids Res 39: 4464–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A et al (2016) Circular non‐coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7: 12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, Coca‐Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280: 339–340 [DOI] [PubMed] [Google Scholar]
- Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X (2015) cir‐ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta‐catenin pathway. PLoS One 10: e0131225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, Wilusz JE (2018) A length‐dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C et al (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jens M, Rajewsky N (2015) Competition between target sites of regulators shapes post‐transcriptional gene regulation. Nat Rev Genet 16: 113–126 [DOI] [PubMed] [Google Scholar]
- Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, Cheng H, Yan J, Zhang S, Yang P et al (2019) Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep 26: 3444–3460.e5 [DOI] [PubMed] [Google Scholar]
- Kadener S, Cramer P, Nogues G, Cazalla D, de la Mata M, Fededa JP, Werbajh SE, Srebrow A, Kornblihtt AR (2001) Antagonistic effects of T‐Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J 20: 5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Fededa JP, Rosbash M, Kornblihtt AR (2002) Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc Natl Acad Sci USA 99: 8185–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP (2018) A network of noncoding regulatory RNAs Acts in the mammalian brain. Cell 174: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29: 2168–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Hansen TB, Venø MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R (2016) Circular RNAs co‐precipitate with extracellular vesicles: a possible mechanism for circrna clearance. PLoS One 11: e0148407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M et al (2017) Circ‐ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66: 22–37.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL (2017a) Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 67: 214–227.e7 [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F et al (2017b) CircHIPK3 sponges miR‐558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 18: 1646–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen L‐L, Cherry S, Wilusz JE (2017) The output of protein‐coding genes shifts to circular RNAs when the pre‐mRNA processing machinery is limiting. Mol Cell 68: 940–954.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C‐X, Li X, Nan F, Jiang S, Gao X, Guo S‐K, Xue W, Cui Y, Dong K, Ding H et al (2019) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177: 865–880.e21 [DOI] [PubMed] [Google Scholar]
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D et al (2015) Transcriptome‐wide investigation of circular RNAs in rice. RNA 21: 2076–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GA (1998) Ribonuclease E is a 5′‐end‐dependent endonuclease. Nature 395: 720–724 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR (2003) A slow RNA polymerase II affects alternative splicing in vivo . Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Fiskaa T, Birgisdottir AB, Haugen P, Einvik C, Johansen S (2003) The ability to form full‐length intron RNA circles is a general property of nuclear group I introns. RNA 9: 1464–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B (1991) Scrambled exons. Cell 64: 607–613 [DOI] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal‐Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez‐Hernandez D, Ramberger E et al (2017) Translation of CircRNAs. Mol Cell 66: 9–21.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Konakondla‐Jacob VV, Krishnamoorthy A, Ashwal‐Fluss R, Bartok O, Wust S, Seitz K, Maya R, Lerner N, Patop IL et al (2018) An in vivo knockdown strategy reveals multiple functions for circMbl. bioRxiv: 483271 10.1101/483271 [PREPRINT] [DOI] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high‐throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK (2019) Endoribonucleolytic cleavage of m6A‐containing RNAs by RNase P/MRP complex. Mol Cell 74: 494–507 [DOI] [PubMed] [Google Scholar]
- Pasman Z, Been MD, Garcia‐Blanco MA (1996) Exon circularization in mammalian nuclear extracts. RNA 2: 603–610 [PMC free article] [PubMed] [Google Scholar]
- Patiño C, Haenni AL, Urcuqui‐Inchima S (2015) NF90 isoforms, a new family of cellular proteins involved in viral replication? Biochimie 108: 20–24 [DOI] [PubMed] [Google Scholar]
- Patop IL, Kadener S (2018) circRNAs in cancer. Curr Opin Genet Dev 48: 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez‐Miranda LR, Memczak S, Wolf SA, Rybak‐Wolf A, Filipchyk A, Klironomos F, Jara CAC, Fenske P et al (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357: eaam8526 [DOI] [PubMed] [Google Scholar]
- Preußer C, Hung L‐H, Schneider T, Schreiner S, Hardt M, Moebus A, Santoso S, Bindereif A (2018) Selective release of circRNAs in platelet‐derived extracellular vesicles. J Extracell Vesicles 7: 1424473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y (1992) Origin of the Alu family: a family of Alu‐like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res 20: 3397–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi K, Venø MT, Dupont DM, Kjems J (2019) Nanopore sequencing of full‐length circRNAs in human and mouse brains reveals circRNA‐specific exon usage and intron retention. bioRxiv: 567164 10.1101/567164 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak‐Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal‐Fluss R et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58: 870–885 [DOI] [PubMed] [Google Scholar]
- Salgia SR, Singh SK, Gurha P, Gupta R (2003) Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA 9: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci USA 73: 3852–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez‐Rotunno M, Arraiano CM, van Hoof A (2009) The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindewolf C, Braun S, Domdey H (1996) In vitro generation of a circular exon from a linear pre‐mRNA transcript. Nucleic Acids Res 24: 1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall‐Levin M, Zhao Y, Perrimon N, Berger B (2010) Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Natl Acad Sci USA 107: 15751–15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Rep 10: 103–111 [DOI] [PubMed] [Google Scholar]
- Sun X, Wang L, Ding J, Wang Y, Wang J, Zhang X, Che Y, Liu Z, Zhang X, Ye J et al (2016) Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett 590: 3510–3516 [DOI] [PubMed] [Google Scholar]
- Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M (1999) Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 8: 493–500 [DOI] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F et al (2011) Coding‐independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, Patel DJ (2013) Structure‐function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev 27: 928–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J (2015) Spatio‐temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16: 245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9: e90859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S (2016) Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 98: 103–107 [DOI] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, Anderson DG (2018) Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun 9: 2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, Parker‐Hale FC, Huang Y, Bisaria N, Anderson DG (2019) RNA circularization diminishes immunogenicity and can extend translation duration in vivo . Mol Cell 74: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome‐wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age‐dependent neural accumulation. Cell Rep 9: 1966–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z (2018) A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS‐mediated exhaustion. Immunity 48: 688–701.e7 [DOI] [PubMed] [Google Scholar]
- Xu Y (2017) An overview of the main circRNA databases. Noncoding RNA Investig 1: 22 [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y et al (2017) Extensive translation of circular RNAs driven by N6‐methyladenosine. Cell Res 27: 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H et al (2018) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 110: 304–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015) Widespread noncoding circular RNAs in plants. New Phytol 208: 88–95 [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB (2004) Variation in alternative splicing across human tissues. Genome Biol 5: R74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T (2008) Structural mechanism of RNA recognition by the RIG‐I‐like receptors. Immunity 29: 178–181 [DOI] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C et al (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C‐Y, Li T‐C, Wu Y‐Y, Yeh C‐H, Chiang W, Chuang C‐Y, Kuo H‐C (2017) The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 8: 1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS (2007) Muscleblind‐like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res 35: 5474–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG (1996) Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA 93: 6536–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG (1997) Exon skipping and circular RNA formation in transcripts of the human cytochrome P‐450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 17: 2985–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Lin W, Guo M, Zou Q (2017) A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 13: e1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence‐mediated exon circularization. Cell 159: 134–147 [DOI] [PubMed] [Google Scholar]
- Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L (2016a) Diverse alternative back‐splicing and alternative splicing landscape of circular RNAs. Genome Res 26: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL (2016b) The biogenesis of nascent circular RNAs. Cell Rep 15: 611–624 [DOI] [PubMed] [Google Scholar]
- Zhao W, Cheng Y, Zhang C, You Q, Shen X, Guo W, Jiao Y (2017) Genome‐wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep 7: 5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7: 11215 [DOI] [PMC free article] [PubMed] [Google Scholar]


