Abstract
Background
The relationship between survivin and extranodal, nasal-type natural killer/T cell lymphoma (ENKTCL) was unclearly established yet. We here studied the potential prognostic roles of survivin and its implication as a target in ENKTCL therapy.
Methods
ENKTCL patients’ peripheral blood were collected and tested by ELISA. ENKTCL cell lines were cultured with or without survivin inhibitor and tested by MTT and Flow cytometry. According to the gene expression profiles from the ArrayExpress Archive under E-TABM-702, survivin co-regulated cluster was established by Coupled Two-way Clustering Algorithm.
Results
Seventeen point six percent of total 17 ENKTCL patients were serum survivin-positive. These patients had poorer outcome than that of negative cases (P<0.01). Analysis of survivin co-regulation genes in ENKTCL revealed that survivin was significantly involved in pluripotency, drug resistance, cell cycle and proliferation, indicating that it should be one of key regulators in ENKTCL and might be a latent therapeutic target. Our results just showed that YM155, a survivin inhibitor, had strong anti-tumor effect on ENKTCL cell lines in a dose dependent manner. It increased sub-G1 phase population and reduced G1- and G2-M phase populations (P<0.05). In addition, combining YM155 with DDP induced a larger decrease in cell viability than either agent alone and had a higher inhibition rate than Bliss index, suggesting their synergistic inhibition.
Conclusions
We concluded that survivin was a potential prognostic marker and a critical regulatory molecule in the pathological process of ENKTCL. It would be a promising target in drugs discovery for ENKTCL therapy.
Keywords: Extranodal, nasal-type natural killer/T cell lymphoma (ENKTCL); survivin; prognosis; therapeutic target
Introduction
Extranodal, nasal-type natural killer/T cell lymphoma (ENKTCL) is a special subtype of non-Hodgkin’s lymphoma with a geographical and ethnical predilection for Asian and South American populations. A conservative estimate in China is that 11.0% of malignant lymphomas newly diagnosed each year are ENKTCL, the second large proportion among all types of lymphoma (1). Up to 60–80% of patients were classified as Ann Arbor Stage I/II at the time of initial diagnosis. However, the traditional and classical chemotherapy CHOP was administrated in these stage I/II patients, the 5-year overall survival (OS) was less than 50%. For those patients of stage III/IV or relapsed and/or refractory disease, 1-year OS was just only 55% (2). Some new therapy regimens, composing of L-asparaginase, cisplatin, carboplatin, oxaliplatin, ifosfamide, methotrexate, etoposide, gemcitabine, and/or dexamethasone, showed a promising overall response rate (81–100%) and a 2–3-year OS (78–87.5%) in stage I/II patients (3-7). But 5-year OS was only about 60% and much less than that of other types of early lymphomas (8,9). Development of new effective drugs is thus desired to improve the survival of ENKTCL patients.
Survivin belongs to the family of inhibitors of apoptosis proteins (IAPs). In normal situation, survivin is prominently expressed during embryonic development and absently or little expressed in normal differentiated adult tissues. But survivin expression is abnormally upregulated in cells transformed into malignancy. Over-expression of survivin was closely associated with cancer cells proliferation, apoptosis, autophagy, anti-cancer-drugs resistance, and cancer angiogenesis (10-16). In addition, high level of survivin was detected in peripheral blood of patients with some kinds of cancers and might be a good marker for prognosis (17-20).
Due to the important roles of survivin in malignancies, drug development targeting survivin may yield cumulative benefits. YM155, as one of the low molecular weight antagonists, inhibits the gene transcription through binding with the Sp1 of survivin promoter. Some researches claimed that YM155 alone or combined with other anti-cancer drugs can inhibit proliferation, increase apoptosis, reverse drug resistance, and improve radiation sensitivity of cancer cells (21-24). A few phases I/II clinical trials were operated in advanced cancer patients to test the safety and tolerance of YM155 (25-28).
There were a couple of reports about the association between ENKTCL and survivin. The positive survivin expression in ENKTCL tissue samples from Singapore was 97% (22/23) (29). The positive rate of serum survivin in ENKTCL from Korea was 27.3% and significantly correlated with the level of EBV-DNA, tumor loading and OS (30). However, the roles of survivin in ENKTCL were not clearly understood yet. We here aimed to further study survivin as a therapy target for ENKTCL.
Methods
Patients and samples
All 17 patients received the unified treatment which was L-asparaginase-basic chemotherapy and local radiotherapy. After obtaining informed consent, we collected peripheral venous blood from ENKTCL patients at the time of initial diagnosis for testing serum survivin, from 09/2008 to 03/2014. For comparison, serum samples from healthy adults were collected in 03/2014 as well. The diagnosis of NKTCL was made according to the 2008 World Health Organization classification of tumors of the hematopoietic and lymphoid tissues. This study was approved by the West China Hospital of Sichuan University (ID of ethics approval: SCHX-2014-0302).
Cell line and reagents
The human NKTCL cell line SNK-10 was obtained from Fudan University (Shanghai) and cultured in RPMI1640 (Thermo Scientific) supplemented with 15% heat-inactivated human serum, 700 IU/mL of IL-2, 100 U/mL of penicillin sodium, 100 mg/mL of streptomycin sulfate, and 0.5 μg/mL amphotericin B at 37 °C in a humidified air atmosphere containing 5% CO2. The cell line was authenticated by testing membrane markers using flow cytometry (31). YM155 monobromide (YM155) was purchased from Selleck Chemicals Company and dissolved in dimethyl sulfoxide (DMSO), DDP from Qilu Pharmaceutical Co., Ltd.
ELISA
The blood was placed in room temperature for 4 hours for being fully solidified, and was centrifuged by 3,500 rpm/min. The serum samples were then sub-packaged and stored at −80 °C until tested for levels of survivin. An ELISA Kit (Human Survivin ELISA kit, Abcam) was used to determine the concentration of survivin in the serum samples. The instruction was carefully followed.
Establishment of BIRC5 co-regulated Cluster
Gene expression profiles of 51 samples were obtained from the ArrayExpress Archive under E-TABM-702, including 7 ENKTCL biopsies, 2 ENKTCL cell lines, 16 PTCL NOS biopsies, 6 normal NK-cells, and 20 normal B-cell samples (32). We removed those genes with both coefficient of variation and quartile coefficient of dispersion across all samples less than 0.15. Total 11,254 genes were left for following analysis.
MTT assay
In vitro cell proliferation was measured using MTT assay. Cells in the logarithmic phase of growth were seeded into 96-well culture plates at 1×104 cells per well. After treatment with different concentrations of YM155, or vehicle control, or different concentration combinations of YM155 with DDP, 100 µL of MTT solution (1 mg/mL) were added to each well, and the cells were further incubated at 37 °C for 4 hours. The supernatant was replaced with dimethyl sulfoxide (DMSO) to dissolve formazan production. The absorbance at wave length 570 nm was measured and 630 nm adjusted using micro-ELISA reader. The inhibition rates were calculated from 3 independent assays.
Flow cytometry
After YM155 treatment, cells were collected and fixed in chilled 70% ethanol, resuspended in PBS, and treated with RNaseA. Propidium iodide (PI) was added to cells, and samples were analyzed with flow cytometry in FACSCalibur (BD Biosciences). Cell-cycle profile analysis of DNA histograms of integrated red fluorescence was performed with ModFit LT software (VeritySoftware, Inc, Topsham). Mean values were obtained from three independent assays.
Analysis for synergy
The Bliss Additivism model was used to classify the effect of combining two agents as additive, synergistic, or antagonistic. A theoretical curve was calculated for combined inhibition using the equation: Bliss index Ebliss = EA + EB - EA×EB, where EA and EB are the fractional inhibitions obtained by drug A alone and drug B alone at specific concentrations. The combined effect of the two drugs was judged as follows: Synergistic if EA+B > Ebliss; Additive if EA+B = Ebliss; and antagonistic if EA+B < Ebliss.
Statistical analysis
Kaplan Meier plot was used to demonstrate survival time. Pearson coefficient was used to analyze the correlation. The coupled two-way clustering algorithm was used to obtain a stable cluster (33). The molecular signatures database was used for the gene set enrichment analysis (34), and the principle component analysis was used to visually and globally characterize the difference among various types of samples. Probit analysis (Logit model) was used to evaluate the half maximal inhibitory concentration (IC50) of agents. Inhibition rates were calculated as the means of at least three different experiments ± standard deviation (SD). The results were analyzed by one-way ANOVA, and P<0.05 was considered to be statistically significant.
Ethics approval
This study was conducted in State Key Laboratory of Biotherapy (SKLB), Sichuan University which was established by the Ministry of Sciences and Technology in China. SKLB is a highly-regarded, comprehensive and multidisciplinary research center in China. Through the seamless integration of basic research, preclinical development, translational and clinical medicine, an efficient and fully integrated technology chain for the discovery and development of innovative drug candidates has been established in a single institute. It has been exempted from ethics as a primary research project.
Results
Serum survivin level in ENKTCL patients
Peripheral venous blood of 17 patients, who were diagnosed as ENKTCLs with their primary sites in the upper aerodigestive tract according to the WHO criteria, were collected. The correlations between patients’ clinical characteristics and the level of serum survivin were shown in Table 1. Their averaged age was 44.9 years (ranged from 15 to 74 years). Three of them (17.6%) were positive of serum survivin. Their concentrations were 56.22, 13.89, and 21.03 pg/mL, respectively. The survivin level was negatively correlated with OS (R=−0.644, P=0.005), and positively correlated with the level of serum lactic dehydrogenase (LDH) (R=0.627, P=0.007). The serum survivin was negative in all 16 donators.
Table 1. Patients’ characteristics and their correlation with serum survivin.
| Characteristics | Percentage (%) | R& (correlated with serum survivin) |
|---|---|---|
| Stage | I, 23.5%; II, 58.8%; III, 5.9%; IV, 11.8% | 0.302 |
| Sex | Female, 41.2%; male, 58.8% | 0.074 |
| Age | Mean, 44.9 years (range, 15 to 74 years) | −0.394 |
| IPI scores | 0, 29.4%; 1, 35.3%; 2, 29.4%; 3, 5.9% | 0.413 |
| LDH | Increased, 35.3%; normal, 64.7% | 0.627** (P=0.007) |
| B symptom | Absent, 47.1%; present, 52.9% | 0.127 |
| ECOG scores | 0, 41.2%; 1, 470.1%; 2, 11.8% | 0.346 |
| OS | Mean, 3 years (range, 0.4 to 7.2 years) | −0.644** (P=0.005) |
| Survivin | Positive, 17.6%; negative, 82.4% | 1.0 |
**, correlation is significant at the 0.01 level (2-tailed); &, correlation coefficient. IPI, international prognosis index; LDH, Lactic dehydrogenase; ECOG, Eastern Cooperative Group performance score; OS, overall survival time.
Survival analysis
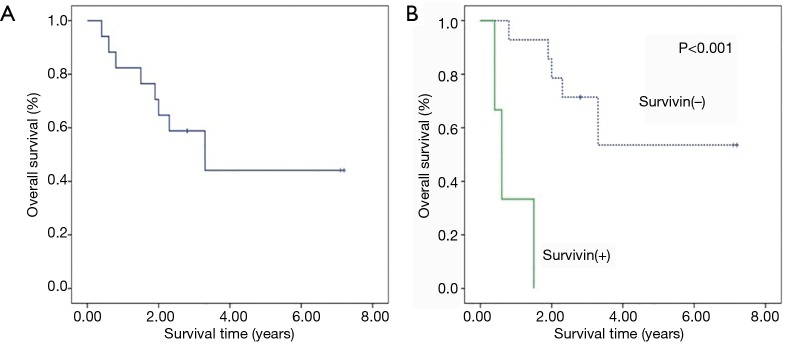
Up to the following-up deadline (12/2015), the median survival time of patients was 2.8 years with range from 0.4 to 7.2 years. For all patients, 2-year OS was 58.8%, and 3-year OS was 44.1% (Figure 1A). Among eight dead patients (47.1%), three of them had positive serum survivin. Their survival times were typically short, being 0.4, 0.6 and 1.5 years, respectively. In negative survivin group, 2- and 3-year OS was 71.4%, and 53.6% respectively, clearly higher than those of overall patients. The survival difference between survivin positive and negative patients was statistically significant (P<0.001), that was, survival time of patients with negative was significantly longer than that of positive patients (Figure 1B).
Figure 1.
Survival curve of ENKTCL patients. (A) Survival curve for all patients; (B) survival comparison between patients with survivin positive (survivin+) and patients with survivin negative (survivin−).
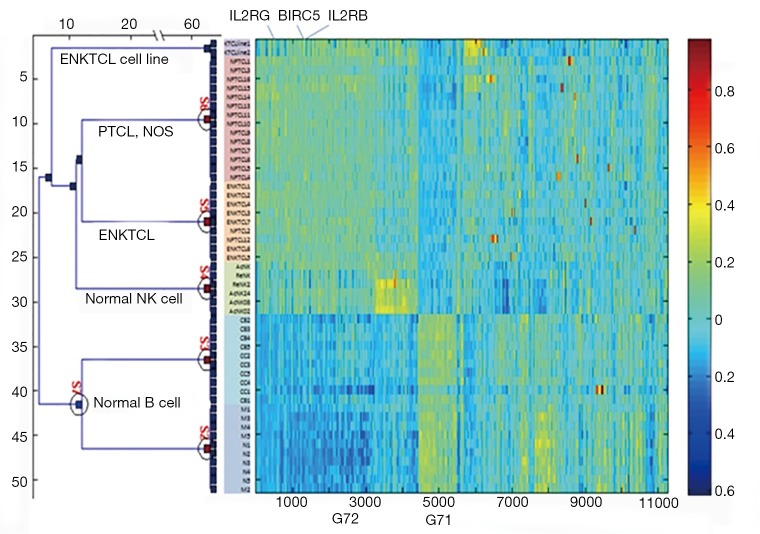
The role of survivin co-regulated gene set
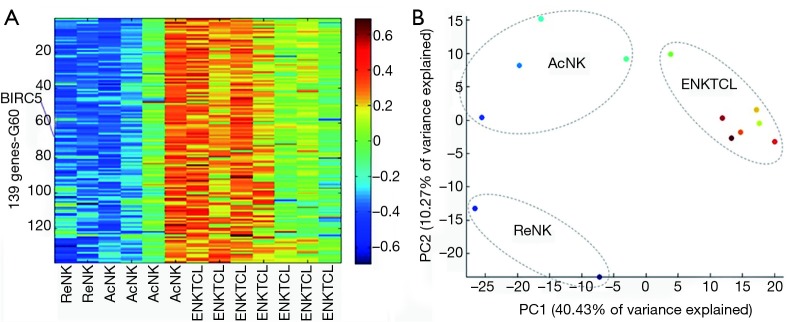
By using the coupled two-way clustering algorithm, we observed that BIRC5 (survivin gene) belonged to a large stable cluster (G72) with 4,419 genes, which could hardly differentiate between ENKTCL (S5) and normal NK-cell samples (S4) (Figure S1). To identify a systematic cluster of genes which are putatively co-regulated with survivin in ENKTCL, we considered only those ENKTCL biopsies and normal NK-cell samples. A stable 139-gene cluster (Table S1) was putatively survivin co-regulated because of their highly correlation of expressions (Figure 2A). By the principle component analysis, we noticed that ENKTCL was close to the activated NK-cells compared to the resting NK-cells (Figure 2B). Gene set enrichment analysis over this cluster also indicated that ENKTCLs were more activated, less differentiated, and earlier-effector-like than NK-cells (Table 2). According to the gene ontology analysis, this cluster was enriched with cell cycle and proliferation genes, consistent with the survivin functions. Additionally, it was significantly related to pluripotency and drug resistances. All of them suggested that survivin was involved in the process of ENKTCL (Table 2).
Figure S1.
Expression heatmap of 11,254 genes across 51 samples. PTCL, peripheral T-cell lymphoma; NOS, not otherwise specified.
Table S1. Categories in which 139 genes in the BIRC5 co-regulated cluster might be involved.
| Genes name | Cell Cycle | Proliferation | Pluripotency | Activation | Undifferentiation | Effector-like | Effector-like | Anti-drug favorite | Survival |
|---|---|---|---|---|---|---|---|---|---|
| THOC4 | + | ||||||||
| MAD2L1 | + | + | + | + | + | + | + | + | + |
| ASPM | + | + | + | + | |||||
| PBK | + | + | + | + | + | + | |||
| ANLN | + | + | + | + | + | + | |||
| CCNA2 | + | + | + | + | + | + | + | + | |
| FAM72A | |||||||||
| BUB1 | + | + | + | + | + | + | + | + | |
| CDKN3 | + | + | + | + | + | + | + | + | |
| CEP55 | + | + | + | + | |||||
| GAPDH | + | ||||||||
| E2F8 | + | + | + | + | + | + | + | ||
| MND1 | + | + | + | ||||||
| DLGAP5 | + | + | + | + | + | + | + | ||
| KIF14 | + | + | + | + | |||||
| TPX2 | + | + | + | + | + | + | + | ||
| HMMR | + | + | + | + | + | + | |||
| FANCI | + | + | + | + | + | + | |||
| CCNB1 | + | + | + | + | + | ||||
| PSRC1 | + | + | + | ||||||
| CENPA | + | + | + | + | + | + | + | ||
| WHSC1 | + | + | + | + | + | ||||
| KIF11 | + | + | + | + | + | + | + | ||
| TTK | + | + | + | + | + | + | + | + | |
| CKAP5 | + | + | + | + | |||||
| KIF23 | + | + | + | + | + | + | + | ||
| CDCA2 | + | + | + | ||||||
| NUF2 | + | + | + | + | |||||
| SPC25 | + | + | + | ||||||
| CDK1 | + | + | + | + | + | + | + | + | |
| KIF15 | + | + | + | + | + | ||||
| NEK2 | + | + | + | + | + | + | + | + | + |
| ECT2 | + | + | + | + | |||||
| NCAPG | + | + | + | + | + | + | + | ||
| SKA1 | + | + | + | + | |||||
| RRM2 | + | + | + | + | + | + | + | + | |
| DEPDC1B | + | + | |||||||
| GANAB | |||||||||
| KIF20A | + | + | + | + | + | + | |||
| MLF1IP | + | + | + | + | + | + | |||
| TPM4 | + | + | |||||||
| TMEM48 | + | + | + | + | + | ||||
| PRC1 | + | + | + | + | + | + | + | + | |
| KIAA0101 | + | + | + | + | + | + | + | ||
| CENPE | + | + | + | + | + | ||||
| CDC25C | + | + | + | + | + | + | + | + | |
| PLK1 | + | + | + | + | |||||
| CHEK1 | + | + | + | + | + | + | + | ||
| CKS1B | + | + | + | + | + | + | + | + | |
| SNRNP40 | + | + | |||||||
| BUB1B | + | + | + | + | + | + | |||
| KIF4A | + | + | + | + | + | ||||
| RRM1 | + | + | + | + | + | + | |||
| CASC5 | + | + | |||||||
| HJURP | + | + | + | ||||||
| CENPH | + | + | |||||||
| AURKB | + | + | + | + | + | + | + | ||
| EBP | + | ||||||||
| MKI67 | + | + | + | + | + | + | + | ||
| FBXO5 | + | + | + | + | |||||
| CCNB2 | + | + | + | + | + | + | + | + | |
| UBE2C | + | + | + | + | + | + | + | ||
| YWHAH | + | + | + | + | |||||
| KIF2C | + | + | + | + | + | + | + | + | |
| KIF18B | + | + | + | + | + | ||||
| CDCA3 | + | + | + | + | + | + | + | ||
| FOXM1 | + | + | + | + | |||||
| BIRC5 | + | + | + | + | + | + | + | + | + |
| CDCA8 | + | + | + | + | + | + | |||
| NCAPG2 | + | + | + | + | + | + | |||
| GTSE1 | + | + | + | + | + | + | |||
| PARVB | + | ||||||||
| NCAPH | + | + | + | + | + | + | + | ||
| TRIP13 | + | + | + | + | + | + | |||
| CKAP2L | + | + | |||||||
| NT5DC2 | + | ||||||||
| HN1 | + | + | + | + | + | ||||
| CDC20 | + | + | + | + | + | + | + | + | |
| ELMO1 | + | ||||||||
| MASTL | + | + | + | + | |||||
| MELK | + | + | + | + | |||||
| CDCA5 | + | + | + | + | + | ||||
| RAD54B | + | + | + | ||||||
| ERCC6L | + | + | + | + | |||||
| DHFR | + | + | + | + | |||||
| ARHGAP11A | + | + | + | ||||||
| TK1 | + | + | + | ||||||
| KPNA2 | + | + | + | + | + | + | + | + | + |
| STMN1 | + | + | + | + | + | + | + | ||
| SGOL1 | + | + | |||||||
| CCDC150 | + | ||||||||
| NUDCD2 | + | ||||||||
| GPSM2 | + | + | + | + | + | + | |||
| ESPL1 | + | + | + | + | + | ||||
| CDC45 | + | + | + | + | + | + | + | ||
| SKA3 | + | ||||||||
| DDX11 | + | + | + | ||||||
| KIF22 | + | + | + | + | + | ||||
| WDR90 | + | ||||||||
| GCAT | + | + | |||||||
| TACC3 | + | + | + | + | + | + | |||
| RBBP8 | + | + | + | + | + | + | |||
| CCNF | + | + | + | + | + | ||||
| CEP152 | + | ||||||||
| RCCD1 | + | + | |||||||
| COX18 | |||||||||
| TOMM22 | + | ||||||||
| E2F2 | + | + | + | ||||||
| ARHGAP19 | + | + | |||||||
| LOC100128191 | + | ||||||||
| CDCA4 | + | + | |||||||
| HMGB3L1 | |||||||||
| SRPK1 | + | + | + | ||||||
| POC1A | + | ||||||||
| AGBL5 | + | ||||||||
| RAD54L | + | + | + | + | + | + | + | + | |
| CENPM | + | + | + | + | |||||
| DNA2 | + | + | + | + | + | ||||
| CIT | + | + | + | ||||||
| CARHSP1 | + | + | + | + | |||||
| CHEK2 | + | + | + | ||||||
| SLC19A1 | + | + | |||||||
| PABPN1 | + | ||||||||
| TBC1D19 | + | ||||||||
| CCHCR1 | + | ||||||||
| UBE2T | + | + | + | + | + | ||||
| APOBEC3B | + | + | |||||||
| AURKA | + | + | + | + | + | + | + | + | + |
| CENPF | + | + | + | + | + | + | |||
| CKS2 | + | + | + | + | + | + | + | + | + |
| C6orf108 | + | + | |||||||
| PTTG1 | + | + | + | + | + | + | + | + | |
| SPAG5 | + | + | + | + | + | ||||
| EZH2 | + | + | + | + | + | + | + | ||
| MRPL37 | + | + | |||||||
| C15orf23 | + | + | + | ||||||
| HMGB3 | + | + | + | + | + | + | |||
| EIF4A3 | + | + | |||||||
| TRIP10 | + |
Figure 2.
Survivin co-regulated gene set. (A) Expression Heatmap of BIRC5-coregulated Cluster based on the coupled two-way clustering algorithm; (B) samples’ relationship by the principle component analysis (ReNK: Resting NK-cell; AcNK: Activated NK-cell).
Table 2. Gene sets enriched with genes in the BIRC5 co-regulated cluster.
| Corresponding functions | Gene set name | P value | Description |
|---|---|---|---|
| Cell cycle | CELL_CYCLE_GO_0007049 | <1E-6 | Genes annotated by the GO term GO:0007049 |
| CELL_CYCLE_PROCESS | <1E-6 | Genes annotated by the GO term GO:0022402 | |
| CELL_CYCLE_PHASE | <1E-6 | Genes annotated by the GO term GO:0022403 | |
| KEGG_CELL_CYCLE | <1E-6 | Cell cycle | |
| BENPORATH_CYCLING_GENES | <1E-6 | Genes showing cell-cycle stage-specific expression | |
| Proliferation | CELL_PROLIFERATION_GO_0008283 | <1E-6 | Genes annotated by the GO term GO:0008283 |
| BENPORATH_PROLIFERATION | <1E-6 | Set ‘Proliferation Cluster’: genes defined in human breast tumor expression data | |
| Pluripotency | MUELLER_PLURINET | <1E-6 | Genes constituting the PluriNet protein-protein network shared by the pluripotent cells |
| WONG_EMBRYONIC_STEM_CELL_CORE | <1E-6 | Core ESC-like gene module | |
| BHATTACHARYA_EMBRYONIC_STEM_CELL | <1E-6 | The stemness signature: genes up-regulated and common to 6 human embryonic stem cell lines tested | |
| BENPORATH_ES_1 | <1E-6 | Genes overexpressed in human embryonic stem cells according to 5 or more out of 20 profiling studies | |
| GSE27786_LSK_VS_NKCELL_UP | 0.000025 | Genes up-regulated in comparison of LSK versus NK cells | |
| GSE27786_LIN_NEG_VS_CD8_TCELL_UP | 0.000025 | Genes up-regulated in comparison of lineage negative versus CD8 T cells | |
| Treatment | WANG_RESPONSE_TO_GSK3_INHIBITOR_SB216763_DN | <1E-6 | Genes down-regulated in RS4;11 cells in response to SB216763 |
| CAFFAREL_RESPONSE_TO_THC_DN | <1E-6 | Genes down-regulated in EVSA-T cells treated THC | |
| ZERBINI_RESPONSE_TO_SULINDAC_DN | <1E-6 | Genes down-regulated in DU145 and PC-3 cells after treatment with sulindac | |
| MARTINEZ_RESPONSE_TO_TRABECTEDIN_DN | <1E-6 | Genes down-regulated in at least 8 of 11 sarcoma cell lines by trabectedin | |
| MITSIADES_RESPONSE_TO_APLIDIN_DN | <1E-6 | Genes down-regulated in the MM1S cells after treatment with aplidin | |
| KUMAMOTO_RESPONSE_TO_NUTLIN_3A_DN | <1E-6 | Genes down-regulated in response to nutlin-3a | |
| BLUM_RESPONSE_TO_SALIRASIB_DN | <1E-6 | Genes down-regulated in response to salirasib in a panel of cancer cell lines with constantly active HRAS | |
| SMIRNOV_RESPONSE_TO_IR_6HR_DN | <1E-6 | Genes down-regulated in B lymphocytes at 6 h after exposure to 10 Gy dose of ionizing radiation | |
| Activation | GSE7764_IL15_TREATED_VS_CTRL_NK_CELL_24H_UP | <1E-6 | Genes up-regulated in comparison of NK cells treated with IL15 versus untreated NK cells |
| GSE22886_UNSTIM_VS_IL2_STIM_NKCELL_DN | <1E-6 | Genes down-regulated in comparison of unstimulated NK cells versus those stimulated with IL2 at 16 h | |
| GSE22886_UNSTIM_VS_IL15_STIM_NKCELL_DN | <1E-6 | Genes down-regulated in comparison of unstimulated NK cells versus those stimulated with IL15 at 16 h | |
| Undifferentiation | LEE_EARLY_T_LYMPHOCYTE_UP | <1E-6 | Genes up-regulated at early stages of progenitor T lymphocyte maturation compared to the late stages |
| RHODES_UNDIFFERENTIATED_CANCER | <1E-6 | Genes commonly up-regulated in undifferentiated cancer relative to well-differentiated cancer | |
| GSE27786_LIN_NEG_VS_NKTCELL_UP | <1E-6 | Genes up-regulated in comparison of lineage negative versus NKT cells | |
| RODRIGUES_THYROID_CARCINOMA_POORLY_DIFFERENTIATED_UP | <1E-6 | Genes up-regulated in poorly differentiated thyroid carcinoma compared to normal thyroid tissue | |
| Effector-like | GSE3982_NKCELL_VS_TH1_DN | <1E-6 | Genes down-regulated in comparison of NK cells versus Th1 cells |
| GSE3982_NKCELL_VS_TH2_DN | <1E-6 | Genes down-regulated in comparison of NK cells versus Th2 cells | |
| GSE9650_NAIVE_VS_EFF_CD8_TCELL_DN | <1E-6 | Genes down-regulated in comparison of naive CD8 T cells versus effector CD8 T cells | |
| GSE9650_EFFECTOR_VS_MEMORY_CD8_TCELL_UP | <1E-6 | Genes up-regulated in comparison of effector CD8 T cells versus memory CD8 T cells | |
| KAECH_NAIVE_VS_DAY8_EFF_CD8_TCELL_DN | <1E-6 | Genes down-regulated in comparison of naive versus effector CD8 T cells (day 8) | |
| KAECH_DAY8_EFF_VS_DAY15_EFF_CD8_TCELL_UP | <1E-6 | Genes up-regulated in comparison of effector CD8 T cells (day 8) versus those (day 15) | |
| KAECH_DAY8_EFF_VS_MEMORY_CD8_TCELL_UP | <1E-6 | Genes up-regulated in comparison of effector CD8 T cells (day 8) versus memory CD8 T cells (day 40+) | |
| Survival | SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 | <1E-6 | Up-regulation of these genes in patients with non-small cell lung cancer predicts poor survival outcome |
Anti-tumor effect of YM155 alone and combined with Cisplatinum (DDP)
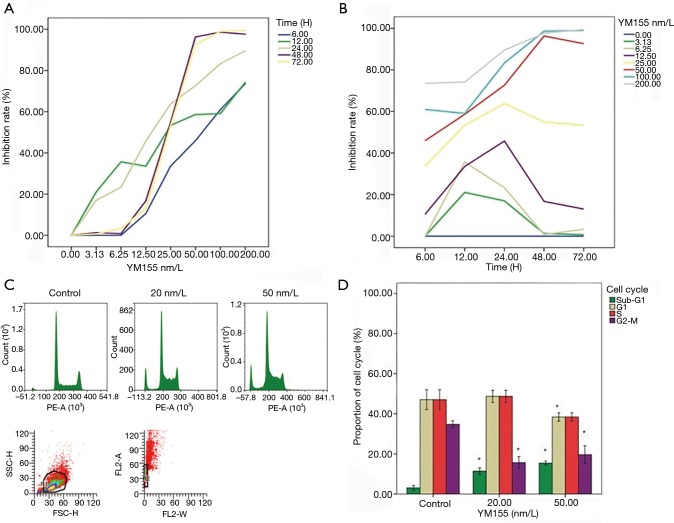
As shown in Figure 3A,B, YM155 alone had a strong anti-tumor effect in a dose dependent manner. Basically, the higher dose was applied, the higher inhibition rate was observed. When the dose went up to 50 nm/L or above, there was a big jump of inhibition rate following time extending, almost reaching 100% inhibition at 48 h. Statistically, both of time and dose were correlated to inhibition rate (P<0.001). Figure 3C,D showed that YM155 treatment increased sub-G1 phase population, and reduced G1- and G2-M phase populations significantly (P<0.05).
Figure 3.
Anti-tumor effect of YM155 alone. (A) The curve of inhibition rate followed by the varieties of YM155 concentrations (0, 3.125, 6.25, 12.5, 25, 50, 100 and 200 nm/L); (B) the curve of inhibition rate followed by time changes (6, 12, 24, 48 and 72 h); (C) after YM155 treatment for 24 h, cell-cycle distribution was detected via flow cytometry; (D) the difference of population in different cell-cycle stages compared with control (*, P<0.05).
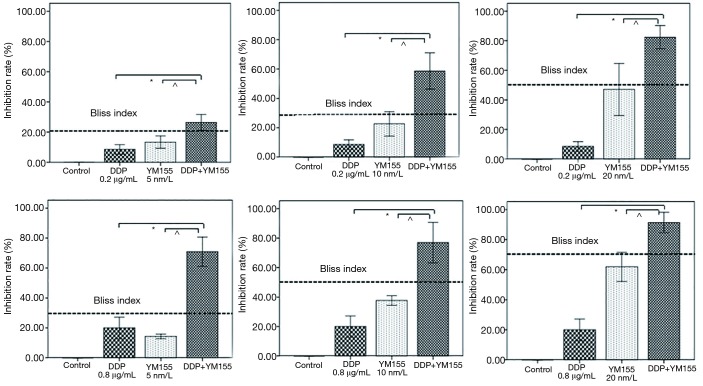
We then investigated the anti-tumor effect of YM155 combined with DDP. The IC50 was 17.47±4.82 nm/L for YM155 and 1.54±0.1 µg/mL for DDP. The dosages of YM155 combined with DDP were determined according to the IC50. The percentage of cell growth inhibition induced by combination of YM155 with DDP was much higher than either agent alone, even higher than the Bliss index (Figure 4), declaring that YM155 and DDP were synergistic.
Figure 4.
The effect after YM155 combined with DDP treatment for 24 h. Each bar represents the mean ± SD of inhibition rate from three separate experiments. The bliss index was showed as dashed line. DDP vs. DDP+YM155: *, P<0.001; YM155 vs. DDP+YM155: ^, P<0.01.
Discussion
Survivin plays an important role in many kinds of malignant tumors. For example, atypical survivin level was reported in peripheral blood of pancreatic cancer patients and might act as a diagnostic and prognostic marker (19). The patients were shown to have obviously higher survivin than that of normal adults, with a median concentration of 99.4 pg/mL in patients and 41.36 pg/mL in normal adults (P<0.001). Higher level of serum survivin was significantly associated with perineural invasion, venous invasion, lymph node status (N stage), cell differentiation, and recurrence. Median OS of normal serum survivin group was much longer than that of elevated group (26 vs. 9 months), and 5-year OS was 38.4% for the normal serum survivin group and 9.3% for the high serum survivin group, indicating that survivin was an independent prognostic factor for pancreatic cancer. Similar results were also reported in gallbladder cancer, breast cancer and ovarian cancer, with the median value of survivin being 4.1–112.4 pg/mL (17,18,20).
For ENKTCLs, Kim et al. reported that survivin was detected in peripheral blood and the positive rate was 27.3% in 66 patients (ranged from 0.8 to 165.0 pg/mL). In addition, serum survivin-positive patients showed a significant association with elevation of EBV DNA and stages reflecting high tumor burden and poorer prognosis (P<0.05) (30). In our study, the positive rate of survivin was 17.6% in ENKTCL patients’ serum (ranged from 13.9 to 56.22 pg/mL). And the level of serum survivin was also negatively correlated to OS. Patients with negative survivin had longer survival time than those with positive survivin. Otherwise, we found that serum survivin has significant association with serum LDH. Because high serum LDH is well known to be a worse factor for non-Hodgkin lymphoma’ prognosis, we inferred that survivin should be a factor for poor outcome of ENKTCL patients.
Above all, different concentration of serum survivin were reported by different researches over virous cancers (median value ranged from 4.1 to 176 pg/mL) and the positive detection rate was low, suggesting that survivin still needs to be further studied as a prognostic marker in clinic. A unified protocol and samples from multiple resources were desired for further researches.
At mRNA level, cell-free BIRC5 (survivin-encoded gene) was found to be significantly increased in serum of colorectal cancer (CRC), and closely correlated with tumor differentiation, regional lymph node metastasis and TNM stage. High serum BIRC5 mRNA expression corresponded to a poorer OS (35). These prompted that serum BIRC5 mRNA may be an interesting target for ENKTCL as well.
To further characterize the roles of survivin in ENKTCL, transcriptomic profiles were collected from the ArrayExpress Archive under E-TABM-702 (32). We obtained a stable 139-gene cluster which was supposed to be BIRC5 co-regulated. This cluster was enriched with genes related to pluripotency and drug resistances, in addition to the expected cell cycle and proliferation, strongly supporting that BIRC5 should be one of the targets critical for ENKTCL transformation and development. We also found that ENKTCL cells were maintained on a more activated, less differentiated, and earlier-effector-like stage compared to normal NK-cells. Therefore, BIRC5 might be involved in maintaining this pluripotency, activation, and undifferentiation within ENKTCL cells. BIRC5 inhibitor might be one of the potential drugs for ENKTCL treatment.
YM155, one of survivin inhibitors, inhibits survivin gene expression by suppressing promoter activity. It showed multiple functions such as anti-tumor proliferation and inducing apoptosis in cancer cell lines. It could also increase the sensitivity of radiotherapy and reverse drug-resistance. For example, YM155 in combination with bendamustine/rituximab showed a greater therapeutic efficacy than either agent alone, induced a greater sub-G1 phase population, and made a survival time extension in DLBCL xenografts (22). YM155 sensitized ovarian cancer to DDP, osteosarcoma to doxorubicin and CRC to 5-Fu (24,36,37). The safety and feasibility of YM155 were reported in phase I/II clinical trials in patients with refractory/recurrence lymphoma and solid malignances (25-28). But not ENKTCL yet. In this study, our results showed that YM155 resulted a greater sub-G1 population, depressed ENKTCL cells proliferation and induced apoptosis and necrosis. Our results demonstrated that the combination effect of YM155 and DDP was synergistic, suggesting that YM155 combined with DDP was potential regimen for advanced/refractory ENKTCL treatments.
Conclusions
This research has shown that survivin was a prognostic marker for the ENKTCL patients and a key regulatory molecule in the pathological process of ENKTCL. Survivin was also proved to be a promising target in the drug discovery for ENKTCL. In addition, the combined effect of YM155 and DDP was synergistic. Our results provide a starting point for future study about YM155 as a potent drug for ENKTCL therapy. However, this research just included a small sample. So, the results need to be further study. And now we are still collecting the blood samples and tissues samples and planning to establish animal model, doing more researches in ENKTCL.
Acknowledgments
We thank Tao Yin, Xiaowei Liu and Xiuran Zheng for providing technical assistance. We also thank Ling Gu for guiding cell line culture, all nurses working in cancer center of West China for helping at collecting blood. This work was supported by a grant from Sichuan University. This research funded by Sichuan University. The authors declare no potential conflicts of interest.
Ethical Statement: This study was approved by the West China Hospital of Sichuan University (ID of ethics approval: SCHX-2014-0302). Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images. The authors are accountable for all aspects of the work (if applied, including full data access, integrity of the data and the accuracy of the data analysis) in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Sun J, Yang Q, Lu Z, et al. Distribution of lymphoid neoplasms in China: analysis of 4,638 cases according to the World Health Organization classification. Am J Clin Pathol 2012;138:429-34. 10.1309/AJCP7YLTQPUSDQ5C [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi M, Kwong YL, Kim WS, et al. Phase II study of SMILE chemotherapy for newly diagnosed stage IV, relapsed, or refractory extranodal natural killer (NK)/T-cell lymphoma, nasal type: the NK-Cell Tumor Study Group study. J Clin Oncol 2011;29:4410-6. 10.1200/JCO.2011.35.6287 [DOI] [PubMed] [Google Scholar]
- 3.Kim SJ, Kim K, Kim BS, et al. Phase II trial of concurrent radiation and weekly cisplatin followed by VIPD chemotherapy in newly diagnosed, stage IE to IIE, nasal, extranodal NK/T-Cell Lymphoma: Consortium for Improving Survival of Lymphoma study. J Clin Oncol 2009;27:6027-32. 10.1200/JCO.2009.23.8592 [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi M, Tobinai K, Oguchi M, et al. Phase I/II study of concurrent chemoradiotherapy for localized nasal natural killer/T-cell lymphoma: Japan Clinical Oncology Group Study JCOG0211. J Clin Oncol 2009;27:5594-600. 10.1200/JCO.2009.23.8295 [DOI] [PubMed] [Google Scholar]
- 5.Jiang M, Zhang H, Jiang Y, et al. Phase 2 trial of "sandwich" L-asparaginase, vincristine, and prednisone chemotherapy with radiotherapy in newly diagnosed, stage IE to IIE, nasal type, extranodal natural killer/T-cell lymphoma. Cancer 2012;118:3294-301. 10.1002/cncr.26629 [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Wang ZH, Chen XQ, et al. First-line combination of gemcitabine, oxaliplatin, and L-asparaginase (GELOX) followed by involved-field radiation therapy for patients with stage IE/IIE extranodal natural killer/T-cell lymphoma. Cancer 2013;119:348-55. 10.1002/cncr.27752 [DOI] [PubMed] [Google Scholar]
- 7.Oh D, Ahn YC, Kim SJ, et al. Concurrent Chemoradiation Therapy Followed by Consolidation Chemotherapy for Localized Extranodal Natural Killer/T-Cell Lymphoma, Nasal Type. Int J Radiat Oncol Biol Phys 2015;93:677-83. 10.1016/j.ijrobp.2015.07.2267 [DOI] [PubMed] [Google Scholar]
- 8.Kim SJ, Yang DH, Kim JS, et al. Concurrent chemoradiotherapy followed by L-asparaginase-containing chemotherapy, VIDL, for localized nasal extranodal NK/T cell lymphoma: CISL08-01 phase II study. Ann Hematol 2014;93:1895-901. 10.1007/s00277-014-2137-6 [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Jiang M, Xie L, et al. Five-year analysis from phase 2 trial of "sandwich" chemoradiotherapy in newly diagnosed, stage IE to IIE, nasal type, extranodal natural killer/T-cell lymphoma. Cancer Med 2016;5:33-40. 10.1002/cam4.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu D, Liu S, Shi L, et al. Cleavage of survivin by Granzyme M triggers degradation of the survivin-X-linked inhibitor of apoptosis protein (XIAP) complex to free caspase activity leading to cytolysis of target tumor cells. J Biol Chem 2010;285:18326-35. 10.1074/jbc.M109.083170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu TK, Cheng Y, Ren X, et al. Interaction of Beclin 1 with survivin regulates sensitivity of human glioma cells to TRAIL-induced apoptosis. FEBS Lett 2010;584:3519-24. 10.1016/j.febslet.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling X, Calinski D, Chanan-Khan AA, et al. Cancer cell sensitivity to bortezomib is associated with survivin expression and p53 status but not cancer cell types. J Exp Clin Cancer Res 2010;29:8. 10.1186/1756-9966-29-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JL, Wang Y, Jiang J, et al. Inhibition of survivin expression and mechanisms of reversing drug-resistance of human lung adenocarcinoma cells by siRNA. Chin Med J (Engl) 2010;123:2901-7. [PubMed] [Google Scholar]
- 14.Wang Q, Chen Z, Diao X, et al. Induction of autophagy-dependent apoptosis by the survivin suppressant YM155 in prostate cancer cells. Cancer Lett 2011;302:29-36. 10.1016/j.canlet.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 15.Hu S, Qu Y, Xu X, et al. Nuclear survivin and its relationship to DNA damage repair genes in non-small cell lung cancer investigated using tissue array. PLoS One 2013;8:e74161. 10.1371/journal.pone.0074161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández JG, Rodríguez DA, Valenzuela M, et al. Survivin expression promotes VEGF-induced tumor angiogenesis via PI3K/Akt enhanced β-catenin/Tcf-Lef dependent transcription. Mol Cancer 2014;13:209. 10.1186/1476-4598-13-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan S, Bennit HF, Turay D, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014;14:176. 10.1186/1471-2407-14-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nigam J, Chandra A, Kazmi HR, et al. Expression of serum survivin protein in diagnosis and prognosis of gallbladder cancer: a comparative study. Med Oncol 2014;31:167. 10.1007/s12032-014-0167-5 [DOI] [PubMed] [Google Scholar]
- 19.Dong H, Qian D, Wang Y, et al. Survivin expression and serum levels in pancreatic cancer. World J Surg Oncol 2015;13:189. 10.1186/s12957-015-0605-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobrzycka B, Mackowiak-Matejczyk B, Terlikowska KM, et al. Prognostic significance of pretreatment VEGF, survivin, and Smac/DIABLO serum levels in patients with serous ovarian carcinoma. Tumour Biol 2015;36:4157-65. 10.1007/s13277-015-3050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasa T, Okamoto I, Suzuki M, et al. Radiosensitizing effect of YM155, a novel small-molecule survivin suppressant, in non-small cell lung cancer cell lines. Clin Cancer Res 2008;14:6496-504. 10.1158/1078-0432.CCR-08-0468 [DOI] [PubMed] [Google Scholar]
- 22.Kaneko N, Mitsuoka K, Amino N, et al. Combination of YM155, a survivin suppressant, with bendamustine and rituximab: a new combination therapy to treat relapsed/refractory diffuse large B-cell lymphoma. Clin Cancer Res 2014;20:1814-22. 10.1158/1078-0432.CCR-13-2707 [DOI] [PubMed] [Google Scholar]
- 23.Wang YF, Zhang W, He KF, et al. Induction of autophagy-dependent cell death by the survivin suppressant YM155 in salivary adenoid cystic carcinoma. Apoptosis 2014;19:748-58. 10.1007/s10495-013-0960-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mir R, Stanzani E, Martinez-Soler F, et al. YM155 sensitizes ovarian cancer cells to cisplatin inducing apoptosis and tumor regression. Gynecol Oncol 2014;132:211-20. 10.1016/j.ygyno.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 25.Tolcher AW, Mita A, Lewis LD, et al. Phase I and pharmacokinetic study of YM155, a small-molecule inhibitor of survivin. J Clin Oncol 2008;26:5198-203. 10.1200/JCO.2008.17.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giaccone G, Zatloukal P, Roubec J, et al. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J Clin Oncol 2009;27:4481-6. 10.1200/JCO.2008.21.1862 [DOI] [PubMed] [Google Scholar]
- 27.Lewis KD, Samlowski W, Ward J, et al. A multi-center phase II evaluation of the small molecule survivin suppressor YM155 in patients with unresectable stage III or IV melanoma. Invest New Drugs 2011;29:161-6. 10.1007/s10637-009-9333-6 [DOI] [PubMed] [Google Scholar]
- 28.Aoyama Y, Kaibara A, Takada A, et al. Population pharmacokinetic modeling of sepantronium bromide (YM155), a small molecule survivin suppressant, in patients with non-small cell lung cancer, hormone refractory prostate cancer, or unresectable stage III or IV melanoma. Invest New Drugs 2013;31:443-51. 10.1007/s10637-012-9867-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng SB, Selvarajan V, Huang G, et al. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol 2011;223:496-510. 10.1002/path.2823 [DOI] [PubMed] [Google Scholar]
- 30.Kim SJ, Hong M, Do IG, et al. Serum survivin and vascular endothelial growth factor in extranodal NK/T-cell lymphoma, nasal type: implications for a potential new prognostic indicator. Haematologica 2015;100:e106-9. 10.3324/haematol.2014.116087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Nagata H, Ikeuchi T, et al. Common cytological and cytogenetic features of Epstein-Barr virus (EBV)-positive natural killer (NK) cells and cell lines derived from patients with nasal T/NK-cell lymphomas, chronic active EBV infection and hydroa vacciniforme-like eruptions. Br J Haematol 2003;121:805-14. 10.1046/j.1365-2141.2003.04359.x [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, de Reyniès A, de Leval L, et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood 2010;115:1226-37. 10.1182/blood-2009-05-221275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Getz G, Levine E, Domany E. Coupled two-way clustering analysis of gene microarray data. Proc Natl Acad Sci U S A 2000;97:12079-84. 10.1073/pnas.210134797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Zhang X, Wang L, et al. Investigation of cell free BIRC5 mRNA as a serum diagnostic and prognostic biomarker for colorectal cancer. J Surg Oncol 2014;109:574-9. 10.1002/jso.23526 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Zhang Y, Lv J, et al. The survivin suppressant YM155 reverses doxorubicin resistance in osteosarcoma. Int J Clin Exp Med 2015;8:18032-40. [PMC free article] [PubMed] [Google Scholar]
- 37.Li WL, Lee MR, Cho MY. The small molecule survivin inhibitor YM155 may be an effective treatment modality for colon cancer through increasing apoptosis. Biochem Biophys Res Commun 2016;471:309-14. 10.1016/j.bbrc.2016.02.009 [DOI] [PubMed] [Google Scholar]