Abstract
Background
MicroRNAs (miRNAs), a subset of small non-coding RNA molecules, play crucial roles in various pathophysiological processes. Studies increasingly indicate that dysregulated miRNAs are associated with bacterial infection. Nevertheless, little is known about miRNAs that respond to Brucella infection and their potential clinical value. Our research aimed to identify the serum miRNAs altered during Brucella infection.
Methods
We enrolled serum samples from 73 patients diagnosed with brucellosis and 65 age- and sex-matched control individuals. Illumina sequencing via synthesis (SBS) technology was performed for an initial screen of miRNAs expression profile in serum samples pooled from 29 patients and 29 controls, respectively. A quantitative real-time polymerase chain reaction (qRT-PCR) assay was conducted in the training and validation sets to confirm the concentrations of differentially expressed miRNAs in individual serum samples from 73 patients and 65 controls.
Results
The Illumina SBS technology identified 1,372 known miRNAs and 1,893 novel miRNAs in brucellosis patients. The three markedly upregulated miRNAs (miR-15a-3p, miR-7-2-3p, miR-103b) in brucellosis patients were subsequently validated by qRT-PCR assay, of which miR-103b was confirmed to be significantly and steadily increased in the brucellosis patients compared with the controls (>2-fold, P<0.001). The area under the receiver operating characteristic (ROC) curve (AUC) for miR-103b was 0.714 (95% CI, 0.624–0.804). Bioinformatics analysis predicted that some putative target genes of miR-103b are involved in immune regulation or the processes of apoptosis and autophagy in humans.
Conclusions
The serum miR-103b level markedly increases after Brucella infection and has the potential to serve as an auxiliary diagnostic indicator for Brucella infection that deserves further investigation.
Keywords: Brucella infection, brucellosis, serum, miR-103b, immune response
Introduction
MicroRNAs (miRNAs), a subset of non-coding RNA molecules of approximately 22 nucleotides in length, are believed to regulate gene expression by targeting mRNAs for posttranscriptional repression or mRNAs destabilization (1). Our group and others have recently highlighted that serum miRNAs can exist stably and that the special expression profiles of altered miRNAs are associated with many diseases by its special expression profiles (2-6). miRNAs, as an emerging area of investigation, are being paid increasing attention due to their altered expression profiles in numerous diseases, and the vital roles of miRNAs in microbial infection are gradually being uncovered. Accumulating evidence has demonstrated the functions of miRNAs in pathogens transmission and pathogenesis (7,8). The expression of host miRNAs can be regulated by pathogens, and the differentially expressed miRNAs can function as auxiliary indicators for diagnosing bacterial diseases (9,10). Herein, we speculated that the altered profile of serum miRNAs may furnish biomarkers for Brucella infection and may represent a promising new field for brucellosis research.
Brucellosis, a zoonotic infectious disease, is caused by a group of microorganisms belonging to the genus Brucella that are facultative intracellular Gram-negative bacteria (11). Brucella affects multiple systems throughout the body with a variety of clinical symptoms, including undulant fever, arthritis, endocarditis, osteoarthritis and meningitis in humans, as well as miscarriage or stillbirth in livestock (12). Approximately 500,000 individuals around the world are diagnosed with brucellosis annually, and the morbidity rate in some prevalent districts is as high as ten cases per 100,000 population, especially in Mediterranean countries, the Middle East and Latin America (13-15). In China, Inner Mongolia, Xinjiang, Qinghai, Ningxia and Henan Provinces are the main regions with severe epidemics (16). Additionally, Brucella infection not only causes serious public health problems in humans that mainly affect developing countries, but also results in significant economic losses in livestock (13,14,17,18). Although there are many studies on Brucella infection, few studies have been devoted to exploring the effects of brucellosis on serum miRNA expression. Since serum and plasma are accessed with relative ease, circulating biomarkers are one of the most promising means of diagnosis. Therefore, clarifying miRNA expression profile as a novel diagnostic indicator could provide some valuable information for the diagnosis of brucellosis.
In our study, to investigate the differentially expressed miRNA profile in brucellosis patients, we performed a comprehensive analysis of miRNA expression with Illumina SBS technology and subsequently confirmed candidates by qRT-PCR assay. The objective of our research was to identify the miRNAs with altered serum profiles and to explore their diagnostic value for Brucella infection.
Methods
Ethics approval
The present study was conducted in accordance with the principles of the World Medical Association Declaration of Helsinki. Meanwhile, we obtained written informed consent from all participants to allow the use of samples and clinical data for investigation. All collected samples were manipulated according to the protocols approved by the ethics committee of each participating institution.
Participants and processing of serum samples
For this case-control retrospective study, we enrolled 73 brucellosis patients, 64 of whom had acute infections and 19 of whom had chronic infections, at the Affiliated Hospital of Inner Mongolia Medical University or Zhungeer Qi Center for Disease Control and Prevention, Inner Mongolia, China, between 2016 and 2017. There were 48 males and 25 females, with average ages of 47.55±12.88 years and 45.51±9.01 years, respectively. Among them, 18 (75%) patients were positive on blood culture, and the others were diagnosed via serological tests. In addition, most of brucellosis patients (68/73, 94%) were Han Chinese. Meanwhile, we recruited 65 age- and gender-matched healthy Han Chinese (41 males and 24 females) as the parallel control group; these healthy controls underwent routine medical examinations at the Healthy Physical Examination Center of Jinling Hospital, Nanjing, China. The mean age of the control group was 48.00±14.18 years and 42.46±16.62 years in males and females, respectively. There was no significant difference in age (P=0.172) or gender (P=0.743) between the two groups.
All blood samples were collected in separating glue/coagulant tubes according to the standard operating procedure. All the collected samples were processed by centrifugation at 1,500 ×g for 10 min at room temperature, and the supernatants were subsequently centrifuged at 2,000 ×g for 10 min at 4 °C to remove the debris (19). The obtained serum samples were stored at −80 °C pending RNA extraction.
RNA extraction
For Illumina SBS technology, equal volumes of sera (20 mL) from 29 patients with brucellosis and 29 normal controls (0.69 mL each) were pooled separately to form case and control sample pools. Total RNAs were extracted from two pools using TRIzol reagent (Invitrogen, MA, USA) as previously described (3). The aqueous phase was subjected to 3 steps of acid phenol/chloroform purification to eliminate protein residues before isopropyl alcohol precipitation. The resulting RNA pellet was dissolved in 20 µL diethylpyrocarbonate (DEPC) water and stored at –80 °C until further analysis.
For the qRT-PCR assay, total RNA was extracted from 100 µL serum with a 1-step phenol/chloroform purification protocol as previously described (20); the serum was mixed with 300 µL deionized water, 200 µL acid phenol, and 200 µL chloroform. The mixture was vortex-mixed vigorously and incubated at room temperature for 15 min. After phase separation, the aqueous layer was blended with 1.5 volumes of isopropyl alcohol and 0.1 volumes of 3 mol/L sodium acetate (pH 5.3). This mixed solution was stored at −20 °C for at least 1 h. The RNA pellet was collected by centrifugation at 16,000 ×g for 20 min at 4 °C. We discarded the supernatant and washed the precipitate with 750 mL/L ethanol. The RNA pellet was dried for 10 min at room temperature. Finally, the pellet was dissolved in 30 µL DEPC water and stored at −80 °C until further analysis.
Illumina SBS technology
Illumina SBS technology (BGI Genomics Co., Shenzhen, China) was applied to RNA separately extracted from two pooled samples as previously described (21). Briefly, after PAGE purification of small RNA molecules (<50 nucleotides) and ligation of a pair of adaptors to the RNA 5' and 3' ends, RNA molecules were amplified using primers for the adaptor regions for 17 cycles. Fragments that were approximately 90 bp (small RNA + adaptors) in length were isolated from the agarose gel. The purified DNA was used for cluster generation and sequencing analysis on the Illumina sequencing system according to the manufacturer’s instructions. Image files were generated by the sequencer and processed to produce digital-quality data. The subsequent procedures performed with Illumina HiSeq 2000 were summarizing data production, evaluating sequencing quality, calculating length distribution of small RNA reads and filtrating reads contaminated by rRNA, tRNA, mRNA, snRNA, and snoRNA. Finally, clean reads were compared with miRBase 21.0. The experimental procedure of small RNA library construction was shown in Supplementary materials. After analysis, miRNAs with P<0.05 and fold change values >2 and <−2 were determined to be statistically significant between the groups.
Quantification of miRNAs by qRT-PCR analysis
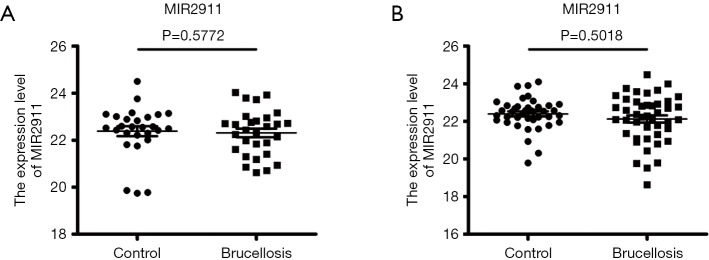
To normalize serum miRNAs, we added synthetic plant exogenous small non-coding RNA MIR2911 (5'-GGCCGG-GGGACGGGCUGGGA-3') during the RNA isolation process as an external reference to normalize the expression levels of the miRNAs (22). MIR2911 was added to each serum sample at an ultimate concentration of 106 fmol/L during RNA extraction, as perviously reported (20). There was no obvious difference between the two groups with regard to the CT values of MIR2911 (P>0.05) (Figure S1).
Figure S1.
The serum levels of MIR2911 in the training set (A) and the validation set (B) measured by qRT-PCR assay.
We performed a qRT-PCR assay according to the manufacturer’s instructions (Roche Light Cycler® 480 II, Roche Diagnostics Ltd., Rotkreuz, Switzerland) (3). Briefly, the reverse transcription reaction was conducted in a 10 µL volume: 2 µL of total RNA, 1 µL of 10 mmol/L dNTPs, 0.5 µL of AMV reverse transcriptase (TaKaRa, Dalian, China), 1 µL of a stem-loop RT primer (Applied Biosystems), 2 µL of 5× reverse transcription buffer and 3.5 µL of DEPC water. For the synthesis of cDNA, the reaction mixtures were incubated at 16 °C for 30 min, at 42 °C for 30 min, and at 85 °C for 5 min, and then they were stored at 4 °C. Real-time PCR was performed (1 cycle of 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min) with the Lightcycler® 480 II system (Roche). The reaction was performed with a final volume of 20 µL containing 1 µL of cDNA, 0.3 µL of Taq, 0.33 µL of hydrolysis probe (Applied Biosystems), 1.2 µL of 25 mmol/L MgCl2, 0.4 µL of 10 mmol/L dNTPs, 2 µL of 10× PCR buffer, and 14.77 µL of DEPC water. To ensure the reliability of the results, all reactions, including those involving non-template controls, were carried out in triplicate. The relative contents of the miRNAs were normalized to the level of MIR2911 and were calculated using the comparative cycle threshold (CT) method (2−∆CT). ∆CT was calculated by subtracting the CT values of MIR2911 from the CT values of the miRNAs. The concentrations of miRNAs relative to the concentration of MIR2911 were calculated using the equation 2−∆CT (23).
Data analysis
SPSS software 23.0 (IBM, NY, USA) and GraphPad Prism 6.0 (GraphPad Software, CA, USA) were used for the statistical analysis. Data are displayed as the mean ± SEM for miRNAs. The nonparametric Mann-Whitney U-test was selected to compare differences in variables between the two groups. For the differentially expressed miRNAs, we constructed ROC curves and calculated the area under the ROC curve (AUC) to assess their ability to distinguish brucellosis patients from control individuals. Binary logistic regression analysis was carried out to evaluate the risk of miR-103b on brucellosis. P<0.05 was considered statistically significant.
Results
Serum miRNAs profile analyzed by Illumina SBS technology
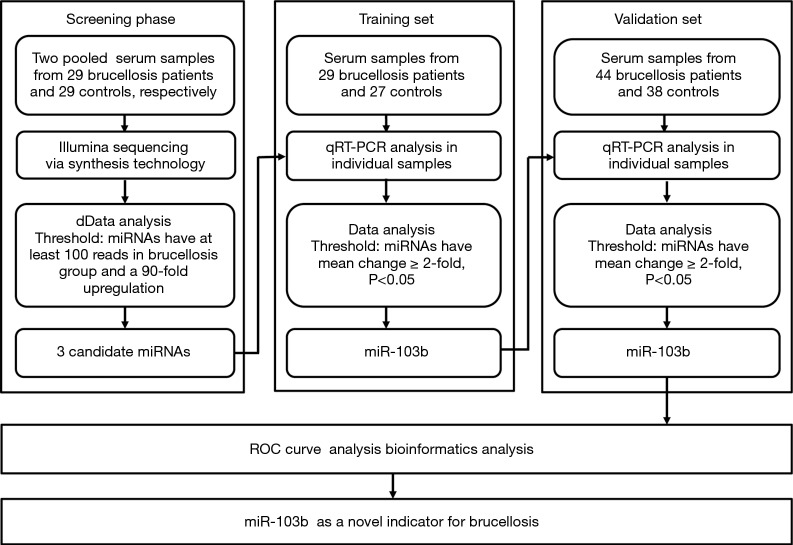
A multiphase, case-control study was designed to identify differentially expressed serum miRNAs (Figure 1). We first performed Illumina SBS technology to identify differentially expressed serum miRNAs between brucellosis patients and controls. The results indicated that the serum contained multiple and heterogeneous small RNA species (<50 nucleotides), as shown in Table 1. Among them, miRNA content was the most abundant (more than 40%) in the pooled serum samples (Figure S2). Pearson’s correlation scatter plots were used to compare the serum miRNA profile in brucellosis patients relative to healthy controls, in which the square of Pearson’s correlation coefficient (R2) value was 0.651 (P<0.0001) (Figure 2). In total, 3,265 miRNAs (1,372 known miRNAs and 1,893 novel miRNAs) were identified. Of the 1,372 known miRNAs, 1136 miRNAs were expressed differentially with more than a 2-fold change: 445 upregulated miRNAs and 691 downregulated miRNAs in the brucellosis sample compared with the control sample. A miRNA was regarded as markedly altered if the Illumina SBS technology detected more than 100 reads in either the patient or control group, and if the miRNA showed at least a 2-fold difference in expression level between the brucellosis and control groups. According to the above criteria, 354 known miRNAs exhibited a significant discrepancy between the two groups, with 106 upregulated (Table S1) and 248 downregulated (Table S2) miRNAs in brucellosis patients, respectively. To narrow down the candidates after global screening of the upregulated miRNAs, we therefore selected 3 miRNAs upregulated with the highest fold change values to validate the miRNA sequencing accuracy by qRT-PCR assay.
Figure 1.
Flow chart of the experimental design. qRT-PCR, quantitative real-time polymerase chain reaction; ROC, receiver operating characteristic.
Table 1. The categories of small RNAs in pooled serum respectively from controls and brucellosis patients by Illumina SBS technology.
| Variable | Controls (20 mL) | Brucellosis patients (20 mL) |
|---|---|---|
| Total (match genome) | 15,625,447 | 19,248,782 |
| intergenic | 9.05% | 4.34% |
| sRNA | 3.09% | 5.91% |
| snRNA | 0 | 0.01% |
| unmap | 13.1% | 2.47% |
| intron | 18.54% | 16.61% |
| piRNA | 2.2% | 1.15% |
| rRNA | 0.62% | 0.14% |
| snoRNA | 0.11% | 0.03% |
| precursor | 0.15% | 0.15% |
| repeat | 4.04% | 10.52% |
| exon | 7.25% | 2.33% |
| miRNA | 41.81% | 56.33% |
| tRNA | 0.04% | 0.01% |
Figure S2.
The length distribution of small non-coding RNAs screened by Illumina sequencing via synthesis (SBS) technology in control group (A) and brucellosis group (B).
Figure 2.
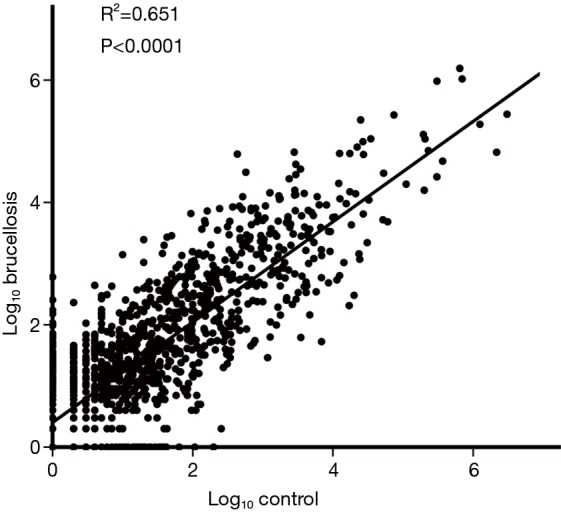
Pearson correlation scatter plot of serum miRNA levels. Pearson correlation scatter plot of serum miRNA levels in the brucellosis group and control group as determined by Illumina SBS technology, P<0.0001.
Table S1. Upregulated miRNAs in the serum sample from brucellosis determined by Illumina SBS technology.
| miRNA | Count (control) | Count (brucellosis) | Fold change (brucellosis/control) |
|---|---|---|---|
| miR-15a-3p | 0 | 198 | 10,500 |
| miR-7-2-3p | 2 | 255 | 96.57143 |
| miR-103b | 53 | 6,873 | 95.6168 |
| miR-642b-5p | 1 | 107 | 81 |
| let-7c-5p | 206 | 17,056 | 60.96156 |
| let-7i-5p | 303 | 20,046 | 48.71802 |
| miR-4732-5p | 62 | 3,483 | 41.39462 |
| miR-503-3p | 29 | 1,171 | 29.69856 |
| miR-1246 | 66,243 | 2,162,570 | 24.04372 |
| miR-142-5p | 145 | 4,673 | 23.7251 |
| miR-660-5p | 4 | 111 | 20.27586 |
| miR-98-3p | 5 | 129 | 19 |
| miR-1284 | 5 | 119 | 17.52778 |
| let-7f-5p | 542 | 11,398 | 15.48705 |
| miR-26a-2-3p | 1,178 | 24,099 | 15.06688 |
| miR-26b-3p | 63 | 1,272 | 14.883 |
| let-7g-5p | 636 | 12,445 | 14.41228 |
| let-7a-3p | 485 | 8,654 | 13.13979 |
| miR-7641 | 346 | 5,766 | 12.2743 |
| miR-331-5p | 7 | 114 | 12.08 |
| let-7a-2-3p | 1,440 | 23,519 | 12.02846 |
| miR-589-3p | 99 | 1,563 | 11.63624 |
| miR-3613-3p | 45 | 674 | 11.02778 |
| miR-194-3p | 50 | 738 | 10.86667 |
| miR-15b-5p | 55 | 771 | 10.32071 |
| miR-629-3p | 2,204 | 30,825 | 10.30059 |
| let-7f-1-3p | 1,039 | 14,329 | 10.15659 |
| miR-345-5p | 20 | 274 | 10.08333 |
| miR-3613-5p | 23 | 311 | 9.927711 |
| let-7b-3p | 15,806 | 200,789 | 9.356014 |
| let-7a-5p | 4,824 | 60,335 | 9.211533 |
| miR-16-1-3p | 144 | 1,781 | 9.111969 |
| miR-183-3p | 337 | 4,153 | 9.077526 |
| miR-16-5p | 215 | 2,641 | 9.049127 |
| miR-96-3p | 29 | 342 | 8.674641 |
| let-7f-2-3p | 589 | 6,860 | 8.577967 |
| let-7b-5p | 26,214 | 304,148 | 8.545214 |
| miR-4732-3p | 111 | 1,285 | 8.524406 |
| let-7g-3p | 262 | 2,979 | 8.372216 |
| miR-3184-3p | 276,762 | 3,048,029 | 8.111181 |
| miR-1290 | 5,205 | 51,656 | 7.30921 |
| miR-532-5p | 435 | 4,242 | 7.181412 |
| miR-101-3p | 336 | 3,197 | 7.008271 |
| miR-98-5p | 33 | 305 | 6.822785 |
| miR-182-3p | 420 | 3,884 | 6.810122 |
| miR-451b | 38 | 347 | 6.736264 |
| miR-502-3p | 252 | 2,169 | 6.337927 |
| miR-15b-3p | 194 | 1,642 | 6.234241 |
| miR-3074-5p | 500 | 4,135 | 6.091718 |
| miR-183-5p | 277 | 2,190 | 5.821464 |
| miR-92a-3p | 47,316 | 367,235 | 5.716203 |
| miR-6866-3p | 16 | 122 | 5.626087 |
| miR-130b-3p | 40 | 292 | 5.375 |
| miR-4525 | 14 | 101 | 5.29703 |
| miR-3605-3p | 52 | 360 | 5.101604 |
| miR-361-5p | 85 | 586 | 5.075163 |
| miR-486-3p | 189,535 | 1,250,919 | 4.860838 |
| miR-197-5p | 17 | 105 | 4.565574 |
| miR-3529-3p | 81 | 471 | 4.283019 |
| let-7d-5p | 624 | 3,615 | 4.266533 |
| miR-16-2-3p | 1,581 | 9,097 | 4.237915 |
| miR-25-3p | 19,904 | 110,993 | 4.107032 |
| miR-6723-5p | 39 | 210 | 3.960854 |
| miR-505-5p | 53 | 276 | 3.839895 |
| miR-128-3p | 1,354 | 6,914 | 3.760698 |
| miR-6815-3p | 24 | 122 | 3.739884 |
| miR-30c-5p | 69 | 344 | 3.668008 |
| miR-323a-3p | 32 | 149 | 3.434783 |
| miR-224-5p | 22 | 101 | 3.386076 |
| miR-942-5p | 64 | 294 | 3.37961 |
| miR-363-3p | 1,003 | 4,494 | 3.299626 |
| miR-21-5p | 1,210 | 5,342 | 3.251608 |
| miR-1304-3p | 96 | 411 | 3.153401 |
| miR-501-3p | 4,303 | 18,339 | 3.138918 |
| miR-421 | 32 | 136 | 3.134783 |
| miR-10b-5p | 680 | 2,855 | 3.092154 |
| miR-1294 | 238 | 999 | 3.091068 |
| miR-4488 | 6,475 | 27,090 | 3.081352 |
| miR-181a-5p | 432 | 1,773 | 3.022837 |
| miR-3138 | 113 | 460 | 2.99877 |
| miR-550a-3-5p | 55 | 224 | 2.997475 |
| miR-27b-3p | 1,687 | 6,844 | 2.987975 |
| miR-93-5p | 33 | 129 | 2.886076 |
| miR-3605-5p | 94 | 348 | 2.725258 |
| miR-503-5p | 75 | 277 | 2.718519 |
| miR-199b-3p | 81 | 289 | 2.627787 |
| miR-99a-3p | 582 | 2,075 | 2.625686 |
| miR-664a-3p | 239 | 832 | 2.563953 |
| miR-107 | 53 | 184 | 2.559055 |
| miR-450b-3p | 52 | 177 | 2.508021 |
| miR-6805-5p | 129 | 436 | 2.490302 |
| miR-20a-3p | 42 | 138 | 2.42053 |
| miR-340-3p | 45 | 147 | 2.404321 |
| miR-451a | 70,407 | 229,653 | 2.402303 |
| miR-550a-3p | 108 | 342 | 2.333333 |
| miR-4489 | 40 | 125 | 2.302083 |
| miR-3679-3p | 40 | 123 | 2.263889 |
| miR-103a-3p | 784 | 2,332 | 2.19089 |
| miR-30d-3p | 10,984 | 32,574 | 2.184147 |
| miR-5010-5p | 107 | 317 | 2.181818 |
| miR-877-3p | 249 | 719 | 2.126674 |
| miR-22-5p | 128 | 361 | 2.07709 |
| miR-181b-5p | 219 | 616 | 2.071701 |
| miR-7-1-3p | 90 | 252 | 2.061728 |
| miR-92b-3p | 618 | 1,717 | 2.046088 |
| let-7d-3p | 3,341 | 9,192 | 2.026284 |
Table S2. Downregulated miRNAs in the serum sample from brucellosis determined by Illumina SBS technology.
| miRNA | Count (controls) | Count (brucellosis) | Fold change (brucellosis/control) |
|---|---|---|---|
| miR-1249-3p | 604 | 0 | −43,470 |
| miR-4726-5p | 105 | 0 | −7,560 |
| miR-3653-3p | 255 | 1 | −367 |
| miR-1275 | 173 | 1 | −249 |
| miR-127-5p | 152 | 1 | −218.8 |
| miR-4433b-3p | 61,728 | 433 | −193.573 |
| miR-339-5p | 1,401 | 10 | −190.245 |
| miR-6852-5p | 2,464 | 20 | −167.292 |
| miR-4785 | 107 | 1 | −154 |
| miR-5187-3p | 441 | 4 | −151.143 |
| miR-2277-5p | 231 | 2 | −151.091 |
| miR-431-3p | 101 | 1 | −145.4 |
| miR-769-3p | 6,578 | 94 | −95.0622 |
| miR-4665-5p | 2,705 | 43 | −85.386 |
| miR-4665-3p | 427 | 7 | −83.0541 |
| miR-4668-5p | 298 | 5 | −79.4444 |
| miR-199a-5p | 2,860 | 50 | −77.6717 |
| miR-4433a-3p | 31,155 | 572 | −73.9512 |
| miR-1250-5p | 267 | 5 | −71.1852 |
| miR-378d | 1,045 | 20 | −70.9528 |
| miR-6763-5p | 311 | 6 | −69.9375 |
| miR-4433b-5p | 1,991 | 39 | −69.2222 |
| miR-134-3p | 7,254 | 161 | −61.2028 |
| miR-1260b | 1,462 | 37 | −53.6837 |
| miR-1181 | 106 | 3 | −47.6875 |
| miR-3155b | 177 | 5 | −47.1852 |
| miR-193b-5p | 5,048 | 148 | −46.3393 |
| miR-370-3p | 7,943 | 242 | −44.5557 |
| miR-758-3p | 157 | 5 | −41.8519 |
| miR-326 | 4,557 | 151 | −40.995 |
| miR-2355-3p | 145 | 5 | −38.6667 |
| miR-766-5p | 3,657 | 135 | −36.7584 |
| miR-765 | 131 | 5 | −34.9259 |
| miR-127-3p | 12,420 | 516 | −32.6823 |
| miR-1307-3p | 66,275 | 2,805 | −32.0806 |
| miR-4532 | 254 | 11 | −31.5172 |
| miR-3155a | 371 | 16 | −31.4118 |
| miR-483-5p | 8,452 | 372 | −30.8458 |
| miR-1307-5p | 504 | 24 | −28.5591 |
| miR-450a-5p | 104 | 5 | −27.7037 |
| miR-1260a | 396 | 20 | −26.8868 |
| miR-4443 | 432 | 22 | −26.5726 |
| miR-769-5p | 1,357 | 70 | −26.3235 |
| miR-1908-5p | 8,685 | 463 | −25.4707 |
| miR-129-2-3p | 156 | 9 | −23.3958 |
| miR-1268b | 384 | 24 | −21.7638 |
| miR-485-5p | 1,568 | 99 | −21.4952 |
| miR-3614-5p | 489 | 31 | −21.4573 |
| miR-939-5p | 3,525 | 226 | −21.1761 |
| miR-378i | 7,741 | 508 | −20.6873 |
| miR-4750-3p | 211 | 14 | −20.527 |
| miR-30c-1-3p | 965 | 64 | −20.4867 |
| miR-204-3p | 854 | 58 | −20.0195 |
| miR-139-3p | 4,900 | 338 | −19.6791 |
| miR-320d | 41,971 | 2,955 | −19.2851 |
| miR-4792 | 177 | 13 | −18.4638 |
| miR-6740-3p | 205 | 15 | −18.4375 |
| miR-4470 | 148 | 11 | −18.3621 |
| miR-450a-2-3p | 672 | 50 | −18.2491 |
| miR-642b-3p | 146 | 11 | −18.1207 |
| miR-409-3p | 4,028 | 313 | −17.4738 |
| miR-873-3p | 542 | 43 | −17.1096 |
| miR-4533 | 122 | 10 | −16.566 |
| miR-23a-5p | 1,487 | 123 | −16.4141 |
| miR-30b-3p | 1,413 | 119 | −16.1157 |
| miR-619-3p | 8,032 | 682 | −15.9906 |
| miR-3188 | 141 | 12 | −15.8594 |
| miR-193a-3p | 14,450 | 1,265 | −15.5101 |
| miR-185-3p | 14,332 | 1,256 | −15.4944 |
| miR-328-3p | 2,026 | 181 | −15.2044 |
| miR-1228-3p | 1,069 | 96 | −15.1159 |
| miR-4726-3p | 111 | 10 | −15.0755 |
| miR-3928-3p | 366 | 33 | −15.0514 |
| miR-129-1-3p | 278 | 25 | −15.0451 |
| miR-7704 | 230 | 21 | −14.9099 |
| miR-26a-5p | 7,354 | 682 | −14.6407 |
| miR-485-3p | 1,115 | 104 | −14.5644 |
| miR-181c-3p | 137 | 13 | −14.2899 |
| miR-6741-3p | 892 | 85 | −14.235 |
| miR-133a-3p | 420 | 41 | −13.9309 |
| miR-483-3p | 24,344 | 2,377 | −13.906 |
| miR-1273g-3p | 8,019 | 788 | −13.8166 |
| miR-4419a | 35,057 | 3,447 | −13.8089 |
| miR-5187-5p | 1,067 | 106 | −13.6637 |
| miR-4748 | 178 | 18 | −13.4842 |
| miR-6515-5p | 401 | 41 | −13.2995 |
| miR-6515-3p | 555 | 57 | −13.2252 |
| miR-4433a-5p | 28,431 | 2,952 | −13.0769 |
| miR-125b-1-3p | 241 | 25 | −13.0376 |
| miR-654-3p | 352 | 37 | −12.9235 |
| miR-27a-5p | 502 | 53 | −12.8577 |
| miR-6741-5p | 614 | 65 | −12.8087 |
| miR-296-3p | 1,340 | 142 | −12.8074 |
| miR-874-3p | 1,437 | 154 | −12.674 |
| miR-939-3p | 459 | 50 | −12.4642 |
| miR-5585-3p | 201 | 22 | −12.3675 |
| miR-151a-3p | 223,478 | 24,848 | −12.2116 |
| miR-3916 | 144 | 16 | −12.1882 |
| miR-1908-3p | 2,968 | 331 | −12.1779 |
| miR-345-3p | 412 | 47 | −11.9076 |
| miR-8485 | 847 | 100 | −11.5019 |
| miR-382-5p | 718 | 87 | −11.2082 |
| miR-6763-3p | 164 | 20 | −11.1321 |
| miR-3130-3p | 611 | 75 | −11.0477 |
| miR-193a-5p | 8,782 | 1,090 | −10.9386 |
| miR-6852-3p | 893 | 112 | −10.8199 |
| miR-1273d | 752 | 95 | −10.7381 |
| miR-4508 | 3,681 | 469 | −10.6565 |
| miR-7847-3p | 258 | 34 | −10.3167 |
| miR-574-5p | 12,816 | 1,687 | −10.3149 |
| miR-3184-5p | 12,792 | 1,699 | −10.2224 |
| miR-6884-3p | 339 | 45 | −10.2092 |
| miR-9-3p | 214 | 29 | −10 |
| miR-215-5p | 293 | 40 | −9.94811 |
| miR-6087 | 615 | 84 | −9.94607 |
| miR-382-3p | 6,201 | 849 | −9.91733 |
| miR-3158-3p | 554 | 79 | −9.51551 |
| miR-576-3p | 1,250 | 186 | −9.12373 |
| miR-3198 | 287 | 43 | −9.0614 |
| miR-1273f | 11,226 | 1,726 | −8.8308 |
| miR-134-5p | 11,504 | 1,770 | −8.82466 |
| miR-619-5p | 4,327 | 688 | −8.5388 |
| miR-671-3p | 611 | 98 | −8.47206 |
| miR-4750-5p | 266 | 45 | −8.00837 |
| miR-375 | 4,846 | 825 | −7.9753 |
| miR-1268a | 278 | 48 | −7.87795 |
| miR-4326 | 190 | 33 | −7.81143 |
| miR-148a-5p | 443 | 78 | −7.71913 |
| miR-4647 | 239 | 42 | −7.713 |
| miR-378c | 2,645 | 474 | −7.57803 |
| miR-744-5p | 15,854 | 2,858 | −7.53185 |
| miR-296-5p | 105 | 19 | −7.48515 |
| miR-3180-5p | 100 | 19 | −7.12871 |
| miR-7851-3p | 5,549 | 1,073 | −7.02233 |
| miR-744-3p | 5,083 | 997 | −6.92185 |
| miR-6734-3p | 2,431 | 477 | −6.92089 |
| miR-122-5p | 63,192 | 12,440 | −6.89719 |
| miR-3158-5p | 1674 | 332 | −6.84545 |
| miR-1273e | 2947 | 587 | −6.81742 |
| miR-422a | 112 | 23 | −6.60656 |
| miR-184 | 933 | 195 | −6.4942 |
| miR-28-3p | 13,167 | 2,835 | −6.30612 |
| miR-186-3p | 13,386 | 2,941 | −6.17987 |
| miR-493-3p | 309 | 69 | −6.0765 |
| miR-1273g-5p | 343 | 78 | −5.97821 |
| miR-4741 | 118 | 27 | −5.93706 |
| miR-24-2-5p | 249 | 58 | −5.83713 |
| miR-1273h-3p | 672 | 160 | −5.70283 |
| miR-760 | 2,735 | 659 | −5.63527 |
| miR-625-3p | 464 | 113 | −5.57429 |
| miR-125a-3p | 661 | 163 | −5.50579 |
| miR-566 | 105 | 26 | −5.47826 |
| miR-320e | 469 | 118 | −5.4 |
| miR-766-3p | 1,438 | 363 | −5.3789 |
| miR-4459 | 1,145 | 290 | −5.36109 |
| miR-6734-5p | 2,339 | 599 | −5.30205 |
| miR-3929 | 2,485 | 640 | −5.27241 |
| miR-99a-5p | 1,954 | 514 | −5.16263 |
| miR-1273a | 98,482 | 26,768 | −4.9954 |
| miR-589-5p | 730 | 199 | −4.98009 |
| miR-3648 | 220 | 60 | −4.97799 |
| miR-378f | 128 | 35 | −4.95161 |
| miR-146a-3p | 80,524 | 22,172 | −4.93115 |
| miR-3591-3p | 269,441 | 74,203 | −4.93027 |
| miR-378a-3p | 63,109 | 17,406 | −4.92288 |
| miR-5010-3p | 166 | 46 | −4.89754 |
| miR-221-3p | 14,041 | 3,896 | −4.89332 |
| miR-145-3p | 791 | 220 | −4.8825 |
| miR-6842-3p | 588 | 171 | −4.67108 |
| miR-339-3p | 1,920 | 588 | −4.43311 |
| miR-3180 | 328 | 102 | −4.36414 |
| miR-181b-3p | 302 | 94 | −4.36345 |
| miR-1254 | 1,291 | 407 | −4.30737 |
| miR-3922-3p | 219 | 69 | −4.30601 |
| miR-320b | 109,865 | 34,667 | −4.303 |
| miR-423-3p | 963,085 | 304,710 | −4.29148 |
| miR-197-3p | 271 | 86 | −4.27632 |
| miR-4429 | 2,865 | 936 | −4.15622 |
| miR-664b-5p | 128 | 42 | −4.13004 |
| miR-10a-5p | 1,496 | 497 | −4.0877 |
| miR-186-5p | 4,956 | 1,656 | −4.06334 |
| miR-1306-3p | 414 | 139 | −4.04342 |
| miR-92b-5p | 1,125 | 378 | −4.04042 |
| miR-3187-3p | 435 | 147 | −4.01926 |
| miR-1273h-5p | 626 | 214 | −3.97266 |
| miR-3691-3p | 159 | 55 | −3.91781 |
| miR-6729-3p | 104 | 36 | −3.91623 |
| miR-654-5p | 293 | 102 | −3.89834 |
| miR-10b-3p | 3,720 | 1,309 | −3.85889 |
| miR-6740-5p | 158 | 56 | −3.82828 |
| miR-1299 | 589 | 209 | −3.82581 |
| miR-330-3p | 723 | 259 | −3.78951 |
| miR-3913-3p | 493 | 178 | −3.76246 |
| miR-193b-3p | 823 | 306 | −3.65166 |
| miR-3665 | 400 | 150 | −3.62138 |
| miR-1538 | 161 | 62 | −3.5228 |
| miR-142-3p | 1,738 | 673 | −3.50659 |
| miR-342-3p | 5,847 | 2,283 | −3.4774 |
| miR-194-5p | 127 | 50 | −3.44906 |
| miR-143-3p | 7,336 | 2,901 | −3.4335 |
| miR-5095 | 181 | 72 | −3.41099 |
| miR-4516 | 2,313 | 926 | −3.39161 |
| miR-24-3p | 3,066 | 1,253 | −3.32219 |
| miR-146b-5p | 5,521 | 2,286 | −3.27919 |
| miR-320a | 1,554,360 | 644,710 | −3.27353 |
| miR-425-5p | 2,488 | 1,039 | −3.2515 |
| miR-4646-3p | 514 | 216 | −3.23057 |
| miR-150-5p | 298 | 127 | −3.18722 |
| miR-320c | 60,757 | 27,280 | −3.02399 |
| miR-3960 | 472 | 212 | −3.02224 |
| miR-4466 | 123 | 56 | −2.9798 |
| miR-433-3p | 348 | 159 | −2.97153 |
| miR-3180-3p | 216 | 100 | −2.93396 |
| miR-5096 | 663 | 307 | −2.93301 |
| miR-625-5p | 168 | 78 | −2.92736 |
| miR-425-3p | 4,662 | 2,168 | −2.9196 |
| miR-125b-2-3p | 294 | 137 | −2.9146 |
| miR-223-3p | 2,185 | 1,022 | −2.9029 |
| miR-30c-2-3p | 372 | 177 | −2.85394 |
| miR-532-3p | 3,089 | 1,472 | −2.8494 |
| miR-222-3p | 12,434 | 5,958 | −2.83363 |
| miR-100-3p | 957 | 467 | −2.78263 |
| miR-1292-5p | 129 | 63 | −2.77844 |
| miR-1291 | 155 | 77 | −2.73529 |
| miR-139-5p | 445 | 224 | −2.6984 |
| miR-324-3p | 282 | 148 | −2.58929 |
| miR-99b-5p | 1,793 | 953 | −2.55474 |
| miR-155-3p | 328 | 178 | −2.50371 |
| miR-424-3p | 670 | 369 | −2.46524 |
| miR-941 | 2,817 | 1,571 | −2.43473 |
| miR-1224-5p | 313 | 175 | −2.4278 |
| miR-129-5p | 173 | 97 | −2.42218 |
| miR-146b-3p | 2,817 | 1,580 | −2.42078 |
| miR-4669 | 232 | 131 | −2.40634 |
| miR-181a-2-3p | 4,421 | 2,497 | −2.40408 |
| miR-10a-3p | 1,402 | 808 | −2.35583 |
| miR-30d-5p | 6,051 | 3,570 | −2.30138 |
| miR-2110 | 20,462 | 12,102 | −2.29571 |
| miR-3656 | 7,432 | 4,451 | −2.26707 |
| miR-4446-3p | 388 | 235 | −2.24077 |
| miR-1-3p | 170 | 104 | −2.2196 |
| miR-1285-3p | 3,899 | 2,425 | −2.18306 |
| miR-93-3p | 541 | 339 | −2.16694 |
| miR-432-3p | 256 | 163 | −2.13194 |
| miR-664a-5p | 574 | 373 | −2.08953 |
| miR-7110-3p | 1,436 | 934 | −2.08746 |
| miR-423-5p | 1,044,294 | 696,892 | −2.03463 |
| miR-652-3p | 1,799 | 1,208 | −2.02202 |
Confirmation of Illumina SBS technology results by qRT-PCR analysis
Our previous studies have proven that circulating miRNAs can be efficiently extracted from serum and quantitatively detected by qRT-PCR assay which is reliable and reproducible (3,5). Therefore, we used qRT-PCR assay to verify the contents of the 3 markedly upregulated miRNAs (miR-15a-3p, miR-7-2-3p, miR-103b) in brucellosis patients. We first measured the expression levels of the 3 miRNAs in the training set composed of individual serum samples from 29 brucellosis patients and 27 healthy controls. Comparing the concentrations between the two groups, only the expression of miR-103b was significantly increased, with 2.4-fold upregulation and a P value <0.05 in the brucellosis group (Table 2). Therefore, miR-103b was selected for further verification.
Table 2. Relative concentrations of candidate miRNAs to MIR2911 in the serum samples from brucellosis group and control group determined by individual qRT-PCR assay in the training set and the validation seta.
| miRNA | Controls | Brucellosis | Fold change | P valueb |
|---|---|---|---|---|
| Training set | ||||
| Samples, n | 27 | 29 | ||
| miR-15a-3p | 52.53 (6.14) | 99.48 (18.27) | 1.89 | 0.1011 |
| miR-7-2-3p | 14.39 (1.70) | 27.91 (6.35) | 1.94 | 0.1378 |
| miR-103b | 58.65 (7.04) | 145.15 (30.27) | 2.47 | 0.0195 |
| Validation set | ||||
| Samples, n | 38 | 44 | ||
| miR-103b | 43.66 (3.99) | 132.90 (19.85) | 3.04 | 0.0003 |
a, miRNA data is presented as the mean ± SEM (×10−2). b, Brucellosis vs. Controls.
We examined the serum level of miR-103b in a larger cohort consisting of an additional 44 brucellosis patients and 38 matched controls (validation set). Consequently, consistent with the findings in the training set, the level of miR-103b also exhibited a significant increase (>3-fold, P<0.001) in the brucellosis patients in the validation set (Table 2). Figure S3 summarizes the differences in concentrations of miR-103b in all the patients and control individuals enrolled in the training and validation sets.
Figure S3.
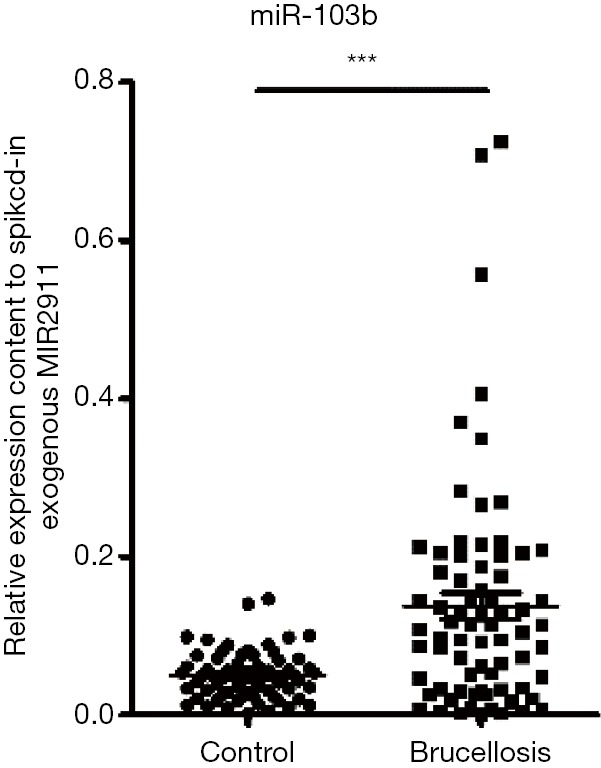
The relative levels of miR-103b in the combined two sets. The relative content of miR-103b was calculated using the 2−∆CT method. Each P Value was derived from a nonparametric Mann-Whitney U-test. ***, P<0.001.
ROC curve and binary logistic regression analysis
To distinguish brucellosis patients from healthy controls, ROC curve analysis was performed with miR-103b to assess the diagnostic value according to the AUC. We obtained an AUC of 0.714 (95% CI, 0.624–0.804) for the total serum samples (Figure 3). In addition, we performed univariate logistic regression analysis to further weigh the usefulness of miR-103b for the diagnosis of brucellosis. We defined brucellosis status as the dependent variable and controlled for other variables, including age and sex. The regression coefficient of miR-103 was 2.807, and the odds ratio was 16.557 (95% CI, 5.451–50.288), suggesting that miR-103b is a potential risk factor for Brucella infection.
Figure 3.
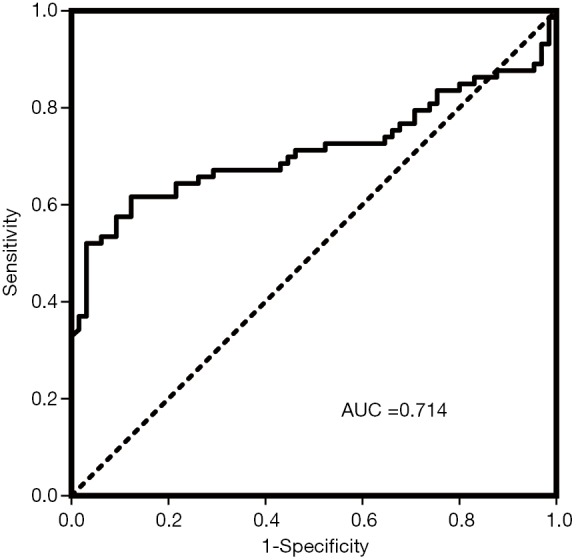
ROC curve analysis of miR-103b. ROC curve for the ability of miR-103b to distinguish brucellosis patients from normal controls in all subjects.
Target analysis of miR-103b
To explore the possible biological role of the upregulation of miR-103b in response to Brucella infection, we predicted the potential target genes associated with infection and immunity for miR-103b by bioinformatics analysis through several widely used target prediction databases, including TargetScan, miRDB and miRPathDB. The bioinformatics analysis revealed 72 potential target genes associated with miR-103b (Figure S4), of which cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), autophagy related 7 (ATG7), chemokine (C-X-C motif) ligand 9 (CXCL9) and caspase-7, apoptosis-related cysteine peptidase (CASP7), are involved in immune regulation or the regulation of apoptosis and autophagy in humans (Table 3).
Figure S4.
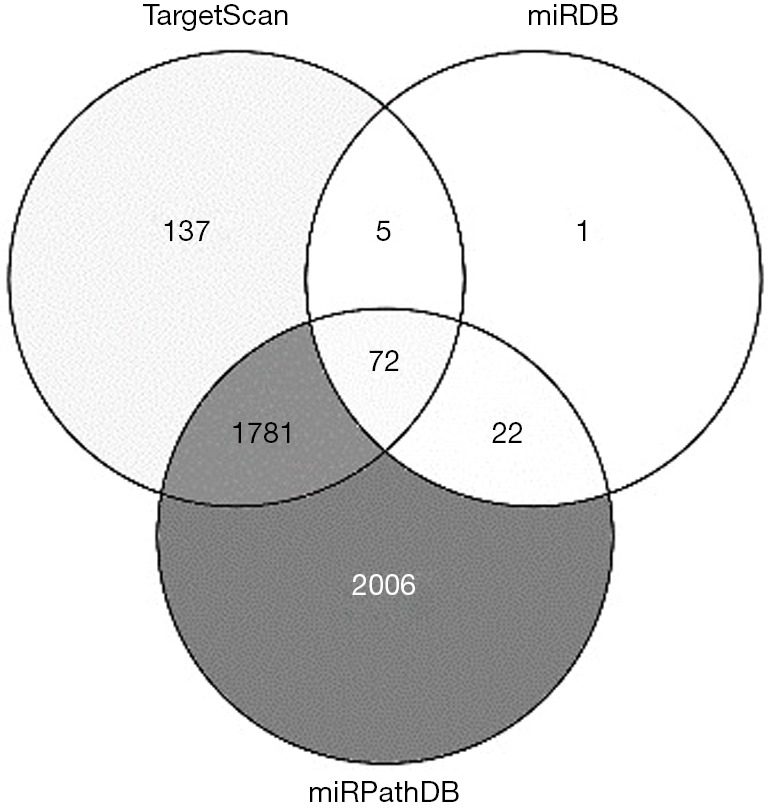
The predicted miR-103b target genes with online miRNA target prediction databases (TargetScan, miRDB, and miRPathDB).
Table 3. The target genes of miR-103b predicted by bioinformatics analysis.
| Target gene | Description | Validated or not |
|---|---|---|
| CTLA4 | Cytotoxic T-lymphocyte-associated protein 4 (24) | Yes |
| ATG7 | Autophagy related 7 | No |
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | No |
| CASP7 | Caspase-7, apoptosis-related cysteine peptidase | No |
Discussion
Human brucellosis is a multisystem disease with a large array of clinical symptoms but a lack of specific manifestations, which still presents a diagnostic challenge for clinicians owing to the limitations of detection methods (25,26). Recently, it has become promising to explore the pathogenesis and progress of infectious diseases from the molecular level with the application of high-throughput sequencing and PCR assays (27,28). In addition, a study has demonstrated that the optimal clinical specimen for brucellosis detection by PCR is serum and not whole blood, in that the sensitivity of the experiment is higher in the serum than that in the whole blood (29). Therefore, we attempted to investigate brucellosis from the perspective of serum small non-coding RNAs perspective. Some altered small non-coding RNAs might provide serological indicator candidates underlying Brucella infection.
It is well known that miRNAs are involved in numerous physiological and pathologic processes. Aberrantly expressed miRNAs have also been associated with various diseases, including cancers, infertility, aging and others (3,30,31). Meanwhile, mounting evidence indicates that miRNAs play a key role in bacterial infections by regulating inflammatory reactions (32-34). In the course of infection, Brucella mainly attacks the body's immune systems, modulating the innate and adaptive immunity, autophagy, apoptosis and possibly the expression of small non-coding RNAs in the serum (35). Considering its great potential in the regulation of the immune system, herein, we focused on the characteristic miRNA expression profile related to Brucella infection.
In this study, we first utilized Illumina SBS technology to comprehensively analyze the serum miRNA expression profile of brucellosis patients. We found that the miRNA expression profile in brucellosis patients was different from that in normal controls, with 1,136 dysregulated known miRNAs. After further validation of 3 miRNAs with highest fold upregulation by qRT-PCR in individual serum samples arranged in two independent cohorts, miR-103b was confirmed to be significantly and steadily upregulated in serum samples from brucellosis patients. In addition, ROC curve analysis showed that the corresponding AUC was 0.714, and the odds ratio was 16.557 larger than 1, indicating that miR-103b is a promising auxiliary serological index for brucellosis detection. We suspect that if serum miR-103b is detected in conjunction with other existing test indicators, it will be possible to improve diagnostic accuracy for brucellosis.
Until now, most existing studies have mainly concentrated on the levels of miRNAs in cells, mainly in RAW264.7 cells, CD8+ T cells and peripheral blood mononuclear cells (PBMCs) (11,12,36). A research group conducted a comprehensive analysis of miRNA expression profile in RAW264.7 cells with or without Brucella infection using high-throughput sequencing technology and found 57 differentially expressed miRNAs, of which eight altered miRNAs with high levels of abundance had putative target genes involved in the processes of apoptosis, autophagy and the immune response, indicating that Brucella may establish a stable infection by regulating miRNA expression (12). Furthermore, Budak et al. confirmed that differentially expressed miRNAs (miR-1238-3p, miR-494, miR-6069, and miR-139-3p) in PBMCs participate in the formation and progression of brucellosis (36). Previous studies have also shown that Brucella inhibits TNF-α to affect immune responses via Omp25 regulation of different miRNAs in macrophages (37). These findings fully demonstrate that miRNAs are closely related to human brucellosis, and they are crucial in pathogen-host interactions; however, no study to date has characterized the miRNA expression patterns in the sera of brucellosis patients, which we identified as being upregulated in the serum of brucellosis patients, may provide insight into the molecular mechanisms underlying Brucella infection.
Through bioinformatics analysis, we found that CTLA-4 was a potential target gene of miR-103b; CTLA-4 is a main component of the immune system and is expressed exclusively on activated CD4+ and CD8+ T cells and binds the same ligands (38). Eskandari-Nasab et al. demonstrated that CTLA-4 variants (CTLA-4 – 318C/T polymorphisms) act as risk factors for developing Brucella infection, facilitating the pathogenesis of disorders characterized by aberrant T-cell responses (24). Other predicted genes, such as ATG7, CXCL9 and CASP7, have also been linked to immune processes such as autophagy and apoptosis (39-41). The target genes associated with miR-103b and their functional linkage deserves further exploration.
In summary, for the first time, we investigated the serum miRNA expression profile in patients with brucellosis by Illumina SBS technology screening and confirmed it with qRT-PCR assays, and demonstrated that miR-103b is significantly upregulated in brucellosis patients. Further analysis suggests that miR-103b has potential to serve as an auxiliary detection indicator, combined with other existing laboratory tests such as blood culture and serological tests may improve the diagnostic accuracy for brucellosis. Further exploration and validation are required to evaluate the potential target genes of miR-103b and their relationship with the occurrence and development of brucellosis.
Acknowledgments
Funding: This work was supported by grants from National Natural Science Foundation of China (81472021, 81672102, 81772282 and 81401257), Fund of State Key Laboratory of Analytical Chemistry for Life Science (5431ZZXM1601), Fund of State Key Laboratory of Analytical Chemistry for Life Science (5431ZZXM1907), National Basic Research Program of China (2014CB542300), and Foundation of Jiangsu Provincial Medical Youth Talent (QNRC2016893).
Supplementary
Supplementary methods—small RNA library construction
Total RNAs were extracted from the two serum pools using TRIzol reagent (Invitrogen, MA, USA), and the RNA samples were then sent to BGI Genomics Co. in Shenzhen for further analysis. The detail information about library preparation are as follows:
First, the concentrations and purity of the total RNAs were measured using Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit) and NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific) to guarantee elimination of inorganic ions or polycarbonate contamination, and the qualified RNA samples were prepared for library construction. Subsequently, 0.2–1 µg of total RNA sample was used to separate RNA segment of different size by PAGE gel. The molecules of 18–50 nt RNA stripe were then recycled and ligated to adapters on the 5'- and 3'-terminals of the small RNAs. The reaction condition of adaptor ligation was performed according to the manufacture’s recommendation. Briefly, for 3′ adaptor system (TruSeq Small RNA Sample Pre Kit, Illumina), the reaction was performed under following cycling conditions: 70 °C for 2 min, 28 °C for 1 h and 28 °C for 15 min; For 5' adaptor mix system, the reaction was performed at 70 °C for 2 min and 28 °C for 1 h. Furthermore, the samples were subjected to RT-PCR for synthesis of cDNAs and amplification of cDNA fragments. In brief, reverse transcription was performed using First-Strand Master Mix and Super Script II (Invitrogen) for small RNAs at 70 °C for 2 min, followed by 50 °C for 1 h; PCR amplification with PCR Primer Cocktail and PCR Master Mix were performed to enrich the cDNA fragments under the reaction condition that initial denaturation for 30 s at 98 °C followed by 11 cycles of denaturation for 10 s at 98 °C, 30 s annealing at 60 °C, 15 s extension at 72 °C, and a final extension for 10 min at 72 °C. Finally, the PCR products were purified with 1% agarose gels and dissolved the recycled products in EB solution for Illumina SBS sequencing using Illumina HiSeq 2000.
Ethical Statement: The present study was conducted in accordance with the principles of the World Medical Association Declaration of Helsinki. Meanwhile, we obtained written informed consent from all participants to allow the use of samples and clinical data for investigation. All collected samples were manipulated according to the protocols approved by the ethics committee of each participating institution.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 3.Zhang C, Wang C, Chen X, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem 2010;56:1871-9. 10.1373/clinchem.2010.147553 [DOI] [PubMed] [Google Scholar]
- 4.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Y, Wang C, Chen X, et al. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin Chem 2013;59:658-66. 10.1373/clinchem.2012.195297 [DOI] [PubMed] [Google Scholar]
- 6.Han Z, Zhang L, Yuan L, et al. Change of plasma microRNA-208 level in acute myocardial infarction patients and its clinical significance. Ann Transl Med 2015;3:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D, Yi Z, Fu Y. Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by Mcl-1 upregulation. J Cell Biochem 2019;120:5889-96. 10.1002/jcb.27874 [DOI] [PubMed] [Google Scholar]
- 8.Zabaglia LM, Sallas ML, Santos MPD, et al. Expression of miRNA-146a, miRNA-155, IL-2, and TNF-alpha in inflammatory response to Helicobacter pylori infection associated with cancer progression. Ann Hum Genet 2018;82:135-42. 10.1111/ahg.12234 [DOI] [PubMed] [Google Scholar]
- 9.Wagh V, Urhekar A, Modi D. Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberculosis (Edinb) 2017;102:24-30. 10.1016/j.tube.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 10.Xiao B, Liu Z, Li BS, et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis 2009;200:916-25. 10.1086/605443 [DOI] [PubMed] [Google Scholar]
- 11.Budak F, Bal SH, Tezcan G, et al. MicroRNA Expression Patterns of CD8+ T Cells in Acute and Chronic Brucellosis. PLoS One 2016;11:e0165138. 10.1371/journal.pone.0165138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng K, Chen DS, Wu YQ, et al. MicroRNA expression profile in RAW264.7 cells in response to Brucella melitensis infection. Int J Biol Sci 2012;8:1013-22. 10.7150/ijbs.3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franco MP, Mulder M, Gilman RH, et al. Human brucellosis. Lancet Infect Dis 2007;7:775-86. 10.1016/S1473-3099(07)70286-4 [DOI] [PubMed] [Google Scholar]
- 14.Atluri VL, Xavier MN, de Jong MF, et al. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol 2011;65:523-41. 10.1146/annurev-micro-090110-102905 [DOI] [PubMed] [Google Scholar]
- 15.Al Dahouk S, Sprague LD, Neubauer H. New developments in the diagnostic procedures for zoonotic brucellosis in humans. Rev Sci Tech 2013;32:177-88. 10.20506/rst.32.1.2204 [DOI] [PubMed] [Google Scholar]
- 16.Lai S, Zhou H, Xiong W, et al. Changing Epidemiology of Human Brucellosis, China, 1955-2014. Emerg Infect Dis 2017;23:184-94. 10.3201/eid2302.151710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solera J, Solis Garcia Del Pozo J. Treatment of pulmonary brucellosis: a systematic review. Expert Rev Anti Infect Ther 2017;15:33-42. 10.1080/14787210.2017.1254042 [DOI] [PubMed] [Google Scholar]
- 18.Kassiri H, Amani H, Lotfi M. Epidemiological, laboratory, diagnostic and public health aspects of human brucellosis in western Iran. Asian Pac J Trop Biomed 2013;3:589-94; discussion 93-4. 10.1016/S2221-1691(13)60121-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding M, Wang C, Lu X, et al. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Analytical and Bioanalytical Chemistry 2018;410:3805-14. 10.1007/s00216-018-1052-4 [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Shi Y, Wang C, et al. Influence of a high-altitude hypoxic environment on human plasma microRNA profiles. Sci Rep 2015;5:15156. 10.1038/srep15156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding M, Wang C, Lu X, et al. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal Bioanal Chem 2018;410:3805-14. 10.1007/s00216-018-1052-4 [DOI] [PubMed] [Google Scholar]
- 22.Lu P, Wang F, Wu J, et al. Elevated Serum miR-7, miR-9, miR-122, and miR-141 Are Noninvasive Biomarkers of Acute Pancreatitis. Dis Markers 2017;2017:7293459. 10.1155/2017/7293459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y, Wang C, Zhou W, et al. Elevation of Circulating miR-210-3p in High-Altitude Hypoxic Environment. Front Physiol 2016;7:84. 10.3389/fphys.2016.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eskandari-Nasab E, Moghadampour M, Najibi H, et al. Investigation of CTLA-4 and CD86 gene polymorphisms in Iranian patients with brucellosis infection. Microbiol Immunol 2014;58:135-41. 10.1111/1348-0421.12119 [DOI] [PubMed] [Google Scholar]
- 25.Kose S, Serin Senger S, Akkoclu G, et al. Clinical manifestations, complications, and treatment of brucellosis: evaluation of 72 cases. Turk J Med Sci 2014;44:220-3. 10.3906/sag-1112-34 [DOI] [PubMed] [Google Scholar]
- 26.Andriopoulos P, Tsironi M, Deftereos S, et al. Acute brucellosis: presentation, diagnosis, and treatment of 144 cases. Int J Infect Dis 2007;11:52-7. 10.1016/j.ijid.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Xu D, Shen Z, et al. Differential expression of miRNA in Carassius auratus gibelio in response to cyprinid herpesvirus 2 infection. Dev Comp Immunol 2018;82:1-6. 10.1016/j.dci.2017.12.027 [DOI] [PubMed] [Google Scholar]
- 28.Cao Y, Wang D, Li S, et al. Identification and analysis of differentially expressed microRNAs in rainbow trout (Oncorhynchus mykiss) responding to infectious hematopoietic necrosis virus infection. Dev Comp Immunol 2018;88:28-36. 10.1016/j.dci.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 29.Zerva L, Bourantas K, Mitka S, et al. Serum is the preferred clinical specimen for diagnosis of human brucellosis by PCR. J Clin Microbiol 2001;39:1661-4. 10.1128/JCM.39.4.1661-1664.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Y, Wang C, Fu Z, et al. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci Rep 2016;6:24229. 10.1038/srep24229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Yang H, Zhang C, et al. Investigation of microRNA expression in human serum during the aging process. J Gerontol A Biol Sci Med Sci 2015;70:102-9. 10.1093/gerona/glu145 [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Li X, Wu M. miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther 2018;3:14. 10.1038/s41392-018-0006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang T, Ge B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett 2018;431:22-30. 10.1016/j.canlet.2018.05.028 [DOI] [PubMed] [Google Scholar]
- 34.Keck J, Gupta R, Christenson LK, et al. MicroRNA mediated regulation of immunity against gram-negative bacteria. Int Rev Immunol 2017;36:287-99. 10.1080/08830185.2017.1347649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed W, Zheng K, Liu ZF. Establishment of Chronic Infection: Brucella's Stealth Strategy. Front Cell Infect Microbiol 2016;6:30. 10.3389/fcimb.2016.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budak F, Bal SH, Tezcan G, et al. Altered Expressions of miR-1238-3p, miR-494, miR-6069, and miR-139-3p in the Formation of Chronic Brucellosis. J Immunol Res 2016;2016:4591468. 10.1155/2016/4591468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X, Zhang X, Wu X, et al. Brucella Downregulates Tumor Necrosis Factor-alpha to Promote Intracellular Survival via Omp25 Regulation of Different MicroRNAs in Porcine and Murine Macrophages. Front Immunol 2018;8:2013. 10.3389/fimmu.2017.02013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmune diseases--a general susceptibility gene to autoimmunity? Genes Immun 2000;1:170-84. 10.1038/sj.gene.6363655 [DOI] [PubMed] [Google Scholar]
- 39.Delcour C, Amazit L, Patino LC, et al. ATG7 and ATG9A loss-of-function variants trigger autophagy impairment and ovarian failure. Genet Med 2019;21:930-8. [DOI] [PubMed] [Google Scholar]
- 40.Ruiduo C, Ying D, Qiwei W. CXCL9 promotes the progression of diffuse large B-cell lymphoma through up-regulating beta-catenin. Biomed Pharmacother 2018;107:689-95. 10.1016/j.biopha.2018.07.171 [DOI] [PubMed] [Google Scholar]
- 41.Lindner AU, Lucantoni F, Vareslija D, et al. Low cleaved caspase-7 levels indicate unfavourable outcome across all breast cancers. J Mol Med (Berl) 2018;96:1025-37. 10.1007/s00109-018-1675-0 [DOI] [PubMed] [Google Scholar]