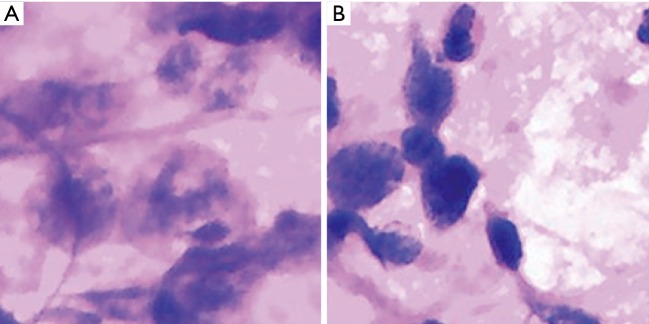
Abstract
Background
In this study, we exploited the Inception-v3 deep convolutional neural network (DCNN) model to differentiate cervical lymphadenopathy using cytological images.
Methods
A dataset of 80 cases was collected through the fine-needle aspiration (FNA) of enlarged cervical lymph nodes, which consisted of 20 cases of reactive lymphoid hyperplasia, 24 cases of non-Hodgkin’s lymphoma (NHL), 16 cases of squamous cell carcinoma (SCC), and 20 cases of adenocarcinoma. The images were cropped into fragmented images and divided into a training dataset and a test dataset. Inception-v3 was trained to make differential diagnoses and then tested. The features of misdiagnosed images were further analysed to discover the features that may influence the diagnostic efficiency of such a DCNN.
Results
A total of 742 original images were derived from the cases, from which a total of 7,934 fragmented images were cropped. The classification accuracies for the original images of reactive lymphoid hyperplasia, NHL, SCC and adenocarcinoma were 88.46%, 80.77%, 89.29% and 100%, respectively. The total accuracy on the test dataset was 89.62%. Three fragmented images of reactive lymphoid hyperplasia and three fragmented images of SCC were misclassified as NHL. Three fragmented images of NHL were misclassified as reactive lymphoid hyperplasia, one was misclassified as SCC, and one was misclassified as adenocarcinoma.
Conclusions
In summary, after training with a large dataset, the Inception-v3 DCNN model showed great potential in facilitating the diagnosis of cervical lymphadenopathy using cytological images. Analysis of the misdiagnosed cases revealed that NHL was the most challenging cytology type for DCNN to differentiate.
Keywords: Deep convolutional neural network (DCNN), Inception v3, cervical lymphadenopathy, cytological images, fine-needle aspiration (FNA), liquid-based cytology
Introduction
Palpable cervical lymph nodes are a common presentation in the clinic in the head and neck surgery department and can be involved in all types of lymphadenopathy, such as reactive lymphoid hyperplasia, lymphoma and metastases from various neoplastic lesions. Determining the nature of an enlarged lymph node is the most important step of differential diagnosis, especially when there are no other symptoms present. Fine-needle aspiration (FNA) cytology is a reliable and minimally invasive diagnostic method; although FNA cannot replace histopathology, it can provide evidence on the pathology type in a more rapid and cost-effective way. However, cytomorphologic diagnosis can be challenging in some cases, such as the discrimination of low-grade lymphoma from reactive lymphoid hyperplasia or lymphoma from metastases, particularly small round cell tumours, such as peripheral neuroectodermal tumours, rhabdomyosarcoma, and small cell carcinoma (1). Recently, machine learning (ML) has been introduced into cytological diagnosis, with promising results (2,3).
ML is defined as a set of methods for automatically detecting patterns in data and then utilizing the identified patterns to predict future data or enable decision making under uncertain conditions (4). Deep convolutional neural networks (DCNNs) are a type of ML and a special type of artificial neural network that resembles the multilayered human cognition system. Most recently, an automated DCNN scheme was developed to classify adenocarcinoma, squamous cell carcinoma (SCC), and small cell carcinoma in lung cancer using cytological images, and 70% of lung cancer cells were classified correctly (2). Momeni-Boroujeni et al. reported a study using a multilayer perceptron neural network (MNN) to distinguish benign from malignant pancreatic nodules using cytological images, which achieved 100% accuracy. Most impressively, this technology can categorize atypical cases as either benign or malignant with 77% accuracy (3).
In this study, we employed the Inception-v3 DCNN model to classify the four major pathology types of cervical lymphadenopathy: reactive lymphoid hyperplasia, non-Hodgkin’s lymphoma (NHL), SCC and adenocarcinoma. Inception-v3 has been proven to achieve better performance than other deep learning networks do on image classification tasks. To our knowledge, Inception-v3 has not previously been applied to cytological images of cervical lymphadenopathy for diagnosis.
Methods
Patients and cytological images
Eighty FNA samples from head and neck masses obtained between December 2016 and December 2017 were analysed retrospectively. They consisted of 20 cases of reactive lymphoid hyperplasia, 24 cases of NHL, 16 cases of SCC, and 20 cases of adenocarcinoma. In our study, the reactive lymphoid hyperplasia cases were confirmed as germinal centre B cell hyperplasia by flow cytometry immunophenotyping, the NHL cases were confirmed by follow-up histological biopsy, and the SCC and adenocarcinoma cases were confirmed by ancillary cell block and immunocytochemistry. The NHL cases consisted of 12 cases of diffuse large B cell lymphoma, 5 cases of follicular lymphoma, 2 cases of marginal zone lymphoma, 2 cases of mantle cell lymphoma, 1 case of peripheral T cell lymphoma, 1 case of angioimmunoblastic T cell lymphoma and 1 case of small lymphocytic lymphoma. The SCC cases consisted of 8 cases of lung cancer, 2 cases of oesophageal cancer, 1 case of cervical cancer, 1 case of renal cancer, and 4 cases of unknown origin. All cases of adenocarcinoma were proven to be metastases from lung cancer.
This work was approved by the ethical committee of Fudan University Shanghai Cancer Centre (FUSCC) (ID: 050432-4-1212B), and all patients were required to sign informed consent forms before the FNA procedures, which were performed with a 22-gauge needle by experienced cytopathologists. The direct smears, fixed with 95% ethanol and stained with haematoxylin and eosin (H&E) staining, were used for preliminary morphological evaluation. If the initial diagnosis indicated the possibility of reactive lymphoid hyperplasia or NHL, one or more additional needles were required to obtain sufficient cells for further flow cytometry immunophenotyping. For SCC or adenocarcinoma requiring definite diagnosis by ancillary immunocytochemical examination, the remaining specimens were placed in 10% formalin after routine smear preparation. After fixing for 24 to 48 hours, the centrifuged materials were combined with liquid 1% agarose to process the cell block. Paraffin-embedded cell block sections were stained with H&E for morphologic evaluation, and unstained sections were used for immunostaining.
The main features that distinguish reactive lymphoid hyperplasia from lymphoma are a mixed population of lymphoid cells representing the whole range of lymphocyte transformation from small lymphocytes to immunoblasts and plasma cells, a predominance of small lymphocytes, centroblasts and centrocytes associated with dendritic reticulum cells, and tangible body macrophages derived from germinal centres. The FNA diagnosis of NHL is based largely on the monomorphism of smear populations with a disproportionate representation of cell types. The populations of lymphoid cells are usually monotonous, heterogeneous and pleomorphic. The malignant cells of well-differentiated SCC are usually disperse, and necrosis is a common accompaniment. Single keratinizing cells are the most reliable indicators of squamous differentiation. Non-keratinizing tumours usually present as irregular solid cohesive fragments, elongated or spindle-shaped nuclei, and variable chromatin density in adjacent cells. The cellular morphology of adenocarcinoma is usually described as tumour cells with delicate cytoplasm, rosettes, acinar or cell clusters/cell balls, and round to oval eccentric nuclei with larger solitary nucleoli.
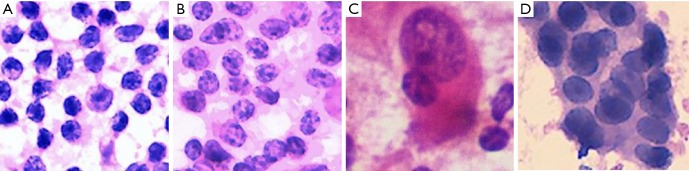
All photographs were collected with a digital still camera (DP27, Olympus, Tokyo, Japan) with a ×40 objective lens attached to a microscope (BX45, Olympus) and were saved in JPEG format. A total of 184 images of reactive lymphatic hyperplasia, 182 images of lymphoma, 196 images of SCC, and 180 images of adenocarcinoma were collected. The initial matrix size of each JPEG image was 1,388×1,036 pixels. Figure 1 shows examples of fragmented images from each cytology type of cervical lymphadenopathy.
Figure 1.
Fragmented images of cervical lymphadenopathy. (A) Reactive lymphoid hyperplasia (H&E, 40×); (B) NHL (H&E, 40×); (C) SCC (H&E, 20×); (D) adenocarcinoma (H&E, 40×). NHL, non-Hodgkin’s lymphoma; SCC, squamous cell carcinoma.
Data augmentation
Each image in the dataset was manually cropped into several 224×224 fragments that contained several cells, resulting in a total of 7,934 224×224 fragmented images: 2,750 of reactive lymphoid hyperplasia, 2,266 of NHL, 1,472 of SCC and 1,446 of adenocarcinoma. As shown in Table 1, we split the dataset randomly into training data and test data for each cytology type according to a ratio of approximately 6:1; meanwhile, we guaranteed that there was no overlapping of the original images between the two datasets.
Table 1. Numbers of images in the training and test groups.
| Cytological diagnosis | Training | Test | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Original | Fragmented | Original | Fragmented | Original | Fragmented | |||
| Reactive lymphoid hyperplasia | 158 | 2,410 | 26 | 340 | 184 | 2,750 | ||
| NHL | 156 | 1,969 | 26 | 297 | 182 | 2,266 | ||
| SCC | 168 | 1,308 | 28 | 164 | 196 | 1,472 | ||
| Adenocarcinoma | 154 | 1,257 | 26 | 189 | 180 | 1,446 | ||
NHL, non-Hodgkin’s lymphoma; SCC, squamous cell carcinoma.
We augmented the training data by flipping and rotating. Each image fragment was flipped horizontally and rotated by 0º, 90º, 180º and 270º. By flipping and rotating, we increased the size of the training dataset by a factor of 8. If we had directly augmented the training data, the storage space needed for the training data would have been 8 times as large as before. Therefore, to save storage space, we did not augment the training data in advance but instead augmented it only during the training process. In each iteration of the training process, we fetched a batch of images from the training dataset. For each image in the batch, we randomly flipped and rotated it. Each time, we had 8 transformation options and randomly applied only one of them to the image.
Network architecture
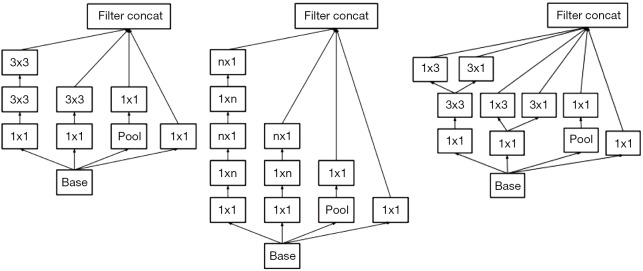
Inception-v3 (5) was the DCNN model used in our experiment. The layers of Inception-v3 are shown in Table 2. There are three kinds of Inception modules in Inception-v3, as shown in Figure 2: from left to right, Inception A, Inception B and Inception C. The Inception modules are well-designed convolution modules that can both generate discriminatory features and reduce the number of parameters. Each Inception module is composed of several convolutional layers and pooling layers in parallel. Small convolutional layers, such as 3×3, 1×3, 3×1, and 1×1 layers, are used in the Inception modules to reduce the number of parameters. In Inception-v3, 3 Inception A modules, 5 Inception B modules and 2 Inception C modules are stacked in series. The default input image size of Inception-v3 is 299×299; however, the image size in the dataset was 224×224. We did not resize the images to 299×299 when training and testing Inception-v3. This did not change the number of channels but instead changed only the size of the feature maps generated during the procedure, and the result was satisfactory. After the convolutional layers and Inception modules, the feature map dimensions were 5×5 with 2,048 channels. Then, we added 3 fully connected layers to the end of the Inception modules to allow us to utilize the pretrained model and finetune the parameters for our own task. Finally, a softmax layer was added as a classifier outputting a probability for each class, and the one with the highest probability was chosen as the predicted class.
Table 2. Inception-v3 network.
| Layer | Patch size/stride | Input size |
|---|---|---|
| Conv | 3×3/2 | 224×224×3 |
| Conv | 3×3/1 | 111×111×32 |
| Conv padded | 3×3/1 | 109×109×32 |
| Pool | 3×3/2 | 109×109×64 |
| Conv | 3×3/1 | 54×54×64 |
| Conv | 3×3/2 | 52×52×80 |
| Conv | 3×3/1 | 25×25×192 |
| Inception A ×3 | – | 25×25×288 |
| Inception B ×5 | – | 12×12×768 |
| Inception C ×2 | – | 5×5×1,280 |
| Fc | 51,200×1,024 | 5×5×2,048 |
| Fc | 1,024×1,024 | 1,024 |
| Fc | 1,024×4 | 1,024 |
| Softmax | Classifier | 4 |
Inception-v3 network. Conv denotes a convolutional layer, pool denotes a pooling layer, and fc denotes a fully connected layer. All layers before fc form the base network of Inception-v3. The patch size is the kernel size of the convolutional, pooling or fully connected layer. The stride is the gap between two operations. The input size is the size of the feature map input to the layer, and the output size of each layer is the input size of the next layer.
Figure 2.
The Inception modules of Inception-v3: Inception modules A, B and C (from left to right). Each Inception module is composed of several convolutional layers and pooling layers. Pool denotes a pooling layer, and n × m denotes a convolutional layer, where n and m specify the dimensions of the convolutions.
The output of the original Inception-v3 network contains 1,000 classes, but we had only 4 classes, namely, reactive lymphoid hyperplasia, NHL, SCC and adenocarcinoma; therefore, we changed the number of output channels of the last layer from 1,000 to 4. We also applied dropout with a dropout rate of 50% during the training process. Dropout randomly discards some inputs to a layer and is a commonly used trick to avoid over-fitting.
We used the pretrained model offered by TensorFlow and finetuned it using our cytological images. It was pretrained on the ImageNet dataset and can be found in the TensorFlow-Slim image classification library. We initialized the parameters from the pretrained model because ImageNet contains approximately 14,000,000 images, whereas we had only 7,934 images. It would be difficult to train a deep network with such a small number of images because of the large number of network parameters. Pretraining can also speed up the convergence of the network.
Statistical analysis
Categorical data were summarized with frequencies and percentages. Inter observer agreement were assessed with Cohen’ kappa analysis with 95% confidence interval (CI). Statistical analyses were performed using the SPSS 22.0 for Windows (SPSS Inc., Chicago, IL, USA).
Results
Table 3 shows the classification accuracies for the original and fragmented images. The classification accuracies for the original images of reactive lymphoid hyperplasia, NHL, SCC, and adenocarcinoma were 88.46%, 80.77%, 89.29% and 100%, respectively. The total accuracy on the test dataset was 89.62%. Table 4 shows the confusion matrix of the classification results, the agreement between cytopathologists and DCNN was calculated by Cohen’ kappa, which was 0.862±0.076. Three fragmented images of reactive lymphoid hyperplasia and three fragmented images of SCC were misclassified as NHL. Three fragmented images of NHL were misclassified as reactive lymphoid hyperplasia, one was misclassified as SCC, and one was misclassified as adenocarcinoma.
Table 3. Classification accuracies in the test group.
| Cytological diagnosis | Classification accuracy (%) | |
|---|---|---|
| Original | Fragmented | |
| Reactive lymphoid hyperplasia | 23/26 (88.46) | 337/340 (99.12) |
| NHL | 21/26 (80.77) | 292/297 (98.32) |
| SCC | 25/28 (89.29) | 161/164 (98.17) |
| Adenocarcinoma | 26/26 [100] | 189/189 [100] |
| Total | 95/106 (89.62) | 979/990 (98.89) |
NHL, non-Hodgkin’s lymphoma; SCC, squamous cell carcinoma.
Table 4. Confusion matrix of the classification results.
| Cytology | DCNN, n (%) | |||
|---|---|---|---|---|
| Reactive lymphoid hyperplasia | NHL | SCC | Adenocarcinoma | |
| Reactive lymphoid hyperplasia | 23 (88.46) | 3 (11.54) | 0 | 0 |
| NHL | 3 (11.54) | 21 (80.77) | 1 (3.85) | 1 (3.85) |
| SCC | 25 (89.29) | 3 (10.71) | 0 | 0 |
| Adenocarcinoma | 0 | 0 | 0 | 26 [100] |
DCNN, deep convolutional neural network; NHL, non-Hodgkin’s lymphoma; SCC, squamous cell carcinoma.
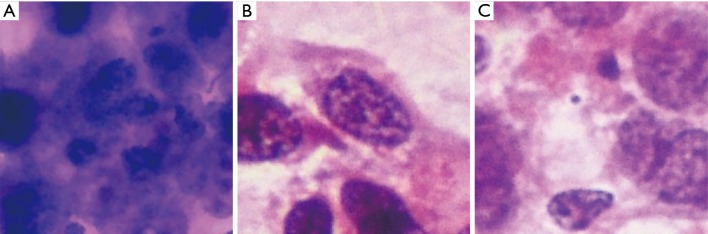
We further investigated the misdiagnosed images to analyse the reasons for failure. Figure 3 shows the fragmented images of reactive lymphoid hyperplasia that were misdiagnosed as NHL. Figure 4 shows the fragmented images of NHL that were misdiagnosed as reactive lymphoid hyperplasia. Figure 5 shows the fragmented images of NHL that were misdiagnosed as SCC and adenocarcinoma. Figure 6 shows the fragmented images of SCC that were misdiagnosed as NHL. The analyses of the images by cytopathologists are described in the figure legends.
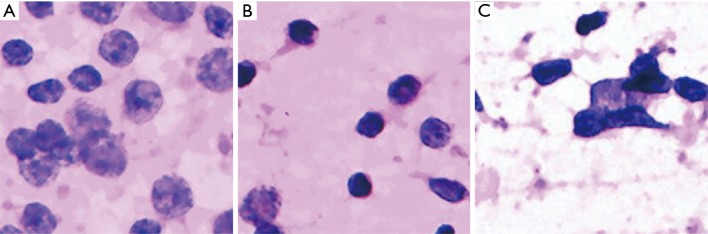
Figure 3.
Fragmented images of reactive lymphoid hyperplasia that were misdiagnosed as NHL (H&E, 40×). The analyses by cytopathologists are as follows: (A) hyperplasia of immunoblast or centroblast cells may have led to the misdiagnosis as lymphoma; (B) several lymphocytes around the small lymphocyte are relatively large and consistent, which may be the cause of misdiagnosis; (C) hyperchromasia and irregular nuclear membranes may be the reason for misdiagnosis as lymphoma. NHL, non-Hodgkin’s lymphoma.
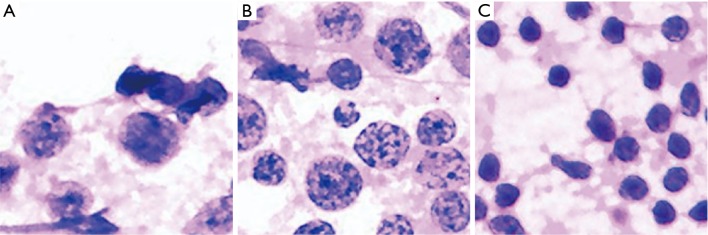
Figure 4.
Fragmented images of NHL that were misdiagnosed as reactive lymphoid hyperplasia (H&E, 40×). The analyses by cytopathologists are as follows: (A) follicular lymphoma, with the large cells showing obvious degeneration and the three adjacent small lymphocytes being close to normal lymphocytes in size, causing them to be easily misdiagnosed as normal lymphocytes; (B) marginal zone lymphoma, which presents with variable cell sizes and thus may be prone to misdiagnosis as reactive lymphoid hyperplasia; (C) follicular lymphoma, involving mainly small cells that may be misdiagnosed as normal lymphocytes. NHL, non-Hodgkin’s lymphoma.
Figure 5.
Fragmented images of NHL that were misdiagnosed as SCC (A) and as adenocarcinoma (B) (H&E, 40×). The analyses by cytopathologists are as follows: (A) diffuse large B cell lymphoma, where cell degeneration with mechanical damage resulting in unclear chromatin details may be the cause of misdiagnosis; (B) diffuse large B cell lymphoma, where the unfocused, blurry image may have resulted in unclear chromatin details. NHL, non-Hodgkin’s lymphoma.
Figure 6.
Fragmented images of SCC that were misdiagnosed as lymphoma (H&E, 20×). The analyses by cytopathologists are as follows: (A): the unfocused, blurry image may have resulted in unclear chromatin details; (B) the unfocused, blurry image may have resulted in unclear chromatin details; (C) the scattered tumour cells are poorly differentiated, and the cytoplasm is not prominent. SCC, squamous cell carcinoma.
Discussion
FNA cytology has been advocated as an integral part of the initial diagnosis and management of patients with lymphadenopathy due to its simplicity, the early availability of results, and the fact that it results in minimal trauma with few complications. The reported accuracy of FNA based on cytomorphology alone ranges from 85% to 94.4% (6-10). With the Inception-v3 model and selected patients, our DCNN model achieved 98.89% accuracy on fragmented images and an accuracy rate of 89.62% on the original images, which was quite satisfactory. Since this was a pilot study, the shortage of this study is that the number of cases was limited and all images had typical cell morphologies and arrangements. However, in clinical practice, more challenging cases arise, and further studies should be conducted to test the performance of DCNN diagnosis on random cases to compare its diagnostic efficiency with that of human practitioners. It is too soon to conclude that DCNN models could be used independently in clinical practice, but the most promising feature of this technique is that there is no observer bias when using DCNN.
In this study, 11 of the fragmented images were misdiagnosed, and those images were further analysed by cytopathologists to determine the main causes of misdiagnosis. Three fragmented images of reactive lymphoid hyperplasia were misdiagnosed as NHL. According to our analysis, the hyperplasia of immunoblast or centroblast cells and the presence of large and consistent lymphocytes, hyperchromasia, and irregular nuclear membranes may have been the causes. Mendon concluded that if the aspiration of the reactive node is derived from the large germinal centre, then the proportion of large cells (centroblasts and dendritic cells) might be high enough to suggest malignant lymphoma (11). Landgren et al. reported that the number of immature lymphoid cells might increase in reactive lymphoid hyperplasia due to the presence of the hyperplasia of lymphoid cells and, consequently, cell division; therefore, lymphoma should be diagnosed when these immature cells account for greater than 50% of the cell population (12). Because fragmented images focus on only a small proportion of the smear, the DCNN analysis could be misled by these immature cells; therefore, further training should be conducted to enable the integration of fragmented images.
In this study, NHL was the most challenging cytology type for the DCNN to differentiate, and there were five fragmented images of lymphoma that were misdiagnosed. The variety of subtypes of NHL may be the main cause of this phenomenon. The three NHL images that were misdiagnosed as reactive lymphoid hyperplasia consisted of two cases of follicular lymphoma and one case of marginal zone lymphoma. Alam et al. concluded that low-grade NHL, including follicular lymphoma grade 1 and grade 2, with minimal cytomorphological atypia, remain very difficult to evaluate cytologically and are usually misdiagnosed as reactive lymphoid hyperplasia (10). The two cases of NHL that were misdiagnosed as SCC and adenocarcinoma were both diffuse large B cell lymphoma. In some cases of diffuse large B cell lymphoma, extensive necrosis may yield necrotic material with sparsely suspicious lymphoid cells that might be degenerated and crushed (13).
There were also three fragmented images of SCC that were misdiagnosed as NHL, and they were all considered to have unclear chromatin details caused by unfocused, blurry images and mechanical damage. Alam et al. (10) found that the dissociation pattern that can occur in metastatic carcinoma cases may be indistinguishable from diffuse large cell NHL or anaplastic large cell lymphoma. In addition to being trained to recognize the disease itself, the DCNN should be trained to exclude such low-quality images, or more qualified images should be used.
This study has proven the capacity of DCNNs to be applied in the cytology of cervical lymphadenopathy, and there is still unexcavated potential that could be tapped by training the model with more images. Additionally, incorporating flow cytometry may assist with the diagnosis of NHL. Future studies should expand the training pool and integrate cytomorphology with molecular biomarkers.
Conclusions
In summary, after training with a large dataset, the Inception-v3 DCNN model showed great potential in facilitating the diagnosis of cervical lymphadenopathy using cytological images. Analysis of the misdiagnosed cases revealed that lymphoma was the most challenging cytology type for the DCNN to differentiate.
Acknowledgments
Funding: This study was funded by Shanghai municipal planning commission of science and research fund for young scholar (award number 20154Y0050).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This work was approved by the ethical committee of Fudan University Shanghai Cancer Centre (FUSCC) (ID: 050432-4-1212B), and all patients were required to sign informed consent forms before the FNA procedures.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Kocjan G. Best Practice No 185. Cytological and molecular diagnosis of lymphoma. J Clin Pathol 2005;58:561-7. 10.1136/jcp.2004.019133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teramoto A, Tsukamoto T, Kiriyama Y, et al. Automated Classification of Lung Cancer Types from Cytological Images Using Deep Convolutional Neural Networks. Biomed Res Int 2017;2017:4067832. 10.1155/2017/4067832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Momeni-Boroujeni A, Yousefi E, Somma J. Computer-assisted cytologic diagnosis in pancreatic FNA: An application of neural networks to image analysis. Cancer Cytopathol 2017;125:926-33. 10.1002/cncy.21915 [DOI] [PubMed] [Google Scholar]
- 4.Murphy KP. Machine learning: a probabilistic perspective. 1st ed. Cambridge: The MIT Press, 2012. [Google Scholar]
- 5.Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the Inception Architecture for Computer Vision. IEEE Conference on Computer Vision and Pattern Recognition, 2016:2818-26. [Google Scholar]
- 6.Shakya G, Malla S, Shakya KN, et al. A study of fine needle aspiration cytology of cervical lymph nodes. J Nepal Health Res Counc 2009;7:1-5. 10.3126/jnhrc.v7i1.2267 [DOI] [Google Scholar]
- 7.Jeffers MD, Milton J, Herriot R, et al. Fine needle aspiration cytology in the investigation on non-Hodgkin's lymphoma. J Clin Pathol 1998;51:189-96. 10.1136/jcp.51.3.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Alwan NA, Al Hashimi AS, Salman MM, et al. Fine needle aspiration cytology versus histopathology in diagnosing lymph node lesions. East Mediterr Health J 1996;2:320-5. [Google Scholar]
- 9.Khajuria R, Goswami KC, Singh K, et al. Pattern of Lymphadenopathy on Fine Needle Aspiration cytology in Jammu. JK Sci 2006;8:157-9. [Google Scholar]
- 10.Alam K, Maheshwari V, Haider N, et al. Fine needle aspiration cytology (FNAC), a handy tool for metastatic lymphadenopathy. Internet J Pathol 2010;10(2).
- 11.Mendon ME. Fine needle aspiration cytology of lymph nodes. Prog Diagn Cytol 1999;32:453-6. [Google Scholar]
- 12.Landgren O, Porwit MacDonald A, Tani E, et al. A prospective comparison of fine-needle aspiration cytology and histopathology in the diagnosis and classification of lymphomas. Hematol J 2004;5:69-76. 10.1038/sj.thj.6200316 [DOI] [PubMed] [Google Scholar]
- 13.Ciatto S, Brancato B, Risso G, et al. Accuracy of fine needle aspiration cytology (FNAC) of axillary lymph nodes as a triage test in breast cancer staging. Breast Cancer Res Treat 2007;103:85-91. 10.1007/s10549-006-9355-0 [DOI] [PubMed] [Google Scholar]