Abstract
Background
This study aimed to assess gene expression changes associated with hypoxia pathway on bone marrow stem cells (BMSCs) and explore effects of bone mass index (BMI) on hypoxia pathway of osteoporosis (OP) patients.
Methods
Human BMSCs were isolated from bone marrow. Subjects were divided into healthy control group and OP group which was further divided into BMI <25 OP subgroup and BMI ≥25 OP subgroup.
Results
The genes downregulated in OP patients were involved in hypoxia pathway. Furthermore, those genes were even downregulated in OP patients BMI ≥25 subgroup than OP patients BMI <25 subgroup. The genes were expressed in response to decreased oxygen levels, and their functions are related to photoperiodism, positive regulation of myoblast differentiation, and entrainment of circadian clock by gene ontology (GO) analysis.
Conclusions
The expression of genes associated with hypoxia pathway on BMSCs in OP patients are lower than healthy subjects, and the expression of genes related to carbohydrate metabolism are lower in overweight OP patients than in normal weight OP patients. These results need further research.
Keywords: Bone marrow stem cells (BMSCs), osteoporosis (OP), hypoxia pathway, bone mass index (BMI)
Introduction
Aging bone loss and osteoporosis (OP) are characterized by decreased bone mass which is caused by the disruption of the balance between bone formation and bone resorption. Studies showed that decreased bone formation was partially caused by bone marrow mesenchymal stem cells (BMSCs), a common precursor of osteoblasts and bone marrow adipocytes, which inclined to differentiate into adipocytes and caused increased marrow fat. Many factors would affect the BMSC differentiation in bone marrow cavity (1), among which hypoxia inducible factor (HIF), a member of Helix-loop-helix family, is a pivotal one and plays a key role in BMSC differentiation.
The bone marrow cavity is one of the most hypoxic microenvironment of human body. Low oxygen level within the bone marrow would promote hypoxia signaling pathways such as HIF, which is regulated by oxygen requiring prolyl hydroxylases (PHDs) and von Hippel-Lindau (VHL) tumor suppressor (2). These hypoxia signaling pathways have profound effects on bone development and homeostasis (3). It is well-known that bone marrow is desaturated and contains oxygen concentration ranging from 1% to 7% despite its high degree of vascularization (4). The levels of O2 consumption by leukocytes as well as blood speed in the sinusoids were contributed to the generation of a gradient of oxygenation (2). Healthy bone marrow can provide balanced hypoxic microenvironments. It has been confirmed by direct measurements of PO2 in the bone marrow of mice that hypoxic conditions existed in perivascular areas even at a 5-cell distance from a blood sinus (5).
Dong-Feng et al. (6) found that down-regulated the expression of HIF1α could reduce the adhesion and secretion function of hBMSC in acute leukemia, which indicates that hypoxia signaling pathway plays an important role in regulating hBMSC function (7). Furthermore, with aging, oxidative stress and inflammation occurred, adipocyte differentiation increased in bone marrow with a disruption of hypoxic osteoblastic niches (8). And it remains unclear whether adipocytes respond to the same bone marrow microenvironment in a possibly opposite manner in regulating BMSC proliferation and differentiation. HIF and its target genes from hBMCSs were found up-regulated in patients with psoriasis (9), which may be caused by aberrant immunoregulatory and chemotactic function of BMSCs. HIF also can be mediated by inflammation (10). OP was reported to related to inflammation and immunity (11,12), thus BMSCs function was investigated from OP patients. Consequently, it is logical to study hypoxia pathway gene expression on the BMSCs from OP patients.
Besides, obesity is a potent risk factor for metabolic diseases at the population level. At the individual patient level, however, correlations between body mass index and bone mineral density are not always straightforward due, in part, to differences among adipose tissue depots with respect to the overall rate of adipocyte dysfunction, local degree of inflammation, and tissue vascularization (13). Hypoxia has been proposed as a key underlying mechanism triggering tissue dysfunction (14). The root cause of obesity-related adipose tissue dysfunction is viewed as oxygen deficiency or “hypoxia”, pushing the tissue toward a proinflammatory environment (15,16). It remains unclear whether obesity could further influence hypoxic pathway gene expression on BMSCs of OP patients.
In this study, the Qiagen Human Hypoxia Signaling Pathway PCR Array was firstly used to screen for differently expressed genes associated with hypoxia pathway on hBMSCs between normal controls and OP patients which were further divided into bone mass index (BMI) <25 OP subgroup and BMI ≥25 OP subgroup separately. Based on information above, Real-time PCR, western blot and immunohistochemistry were further performed to verify the expression level of certain hypoxia genes on BMSCs of OVX-induced OP mice model. The aim of this study was to compare the expression of hypoxia pathway genes between normal controls and OP patients of different BMI and provide a view of the potential regulation of hypoxia signaling pathway on OP bone marrow microenvironment and bone metabolism.
Methods
Participants
This study was approved by the Ethics Committee of Shanghai Tongji Hospital (KYSB-2015-20). A total of 6 patients with OP (marked decline in BMD, spine T-score ≤−2.5) and 3 control subjects were recruited from Endocrinology Department of Shanghai Tongji Hospital between March 2014 and June 2014. Patients with OP were further divided into 2 subgroups according to their BMI: BMI <25 (n=3) and BMI ≥25 (n=3). Informed consent was provided by all participants.
All participants were questioned of their medication history and verified no serious complications existed such as cancer, cardio-pulmonary diseases and all diseases that may cause secondary OP. Totally, 6 patients with OP participated in this study prior to the start of any treatment. Both the spine and femoral neck T-score was measured for each participant. T-scores of spine (L1-L4) and femoral neck were obtained by using a dual energy X-ray absorptiometry scanner (Lunar DXA, USA). The hBMSCs from anterior superior iliac crest of all participants were investigated in this study.
Isolation hBMSCs from human anterior superior iliac crest
Fresh human bone marrow was harvested from the anterior superior iliac crest of 6 patients and 3 volunteers during bone marrow biopsy (Shanghai Tongji Hospital, Shanghai, China). Patients were provided informed consent using guidelines approved by the Shanghai Tongji Hospital. From bone marrow aspirates, the mononuclear cell fraction was harvested for MSC isolation using conventional density gradient centrifugation with a density gradient solution (Ficoll-Paque PREMIUM, Sigma-Aldrich, MO, USA). The resulting cells were collected and isolated (17).
Microarray analysis
To analyze the genes expression changes associated with hypoxia pathway on BMSCs, the PAHS-032Z Human Hypoxia Signaling Pathway PCR Arrary (Qiagen) was used. All the gene information was described in Table S1.
OP mice model
Thirty 8-week-old C57 female mice were randomly equally divided into SHAM group and OVX group. All the mice were housed under 12 h light/12 h dark cycles, temperature 20.0±2.0 °C and relative humidity of 55%±10%. Food and water were available ad libitum. All procedures were performed according to the National Institutes of Health for the care and use of laboratory animals (NIH publication No 80-23) and were approved by the Animal Ethics Committee of Shanghai Tongji Hospital. Despite that OVX group mice received complete resection on both sides of ovarian while equal amount of fat was resected in SHAM group, other procedures were all the same. All the mice were killed 12 weeks after surgery and both sides of femur and tibia were separated under aseptic condition. Right femur and tibia were used to get original mBMSCs; left femur was used for immunohistochemical staining.
Cell culture
mBMSCs were isolated according to the following method. In brief, femurs and tibias from mice were removed. Muscle and extraossial tissue were trimmed. The ends of bones were cut and the marrow was flushed using a needle and syringe. The whole BM cells were cultured in α-MEM (Gibco) supplemented with 10% FBS and penicillin (100 U/mL)/streptomycin (100 µg/mL) at 37 °C in humid air with 5% CO2 and then nonadherent cells were removed thoroughly 24 h later. Afterwards, replace the cell medium every 48 h to remove non-adherent cells to get purified mBMSCs.
RNA preparation and quantitative real-time PCR
Total RNA was isolated using TriZol (Invitrogen, CA, USA) and purified from cell pellet using RNeasy mini kit as recommended by the manufacturer (Qiagen). Using the Primescript RT Reagent kit (Takara, Japan), 1 µL of total RNA was reverse-transcribed into 20 µL cDNA. Quantitative Real-time PCR was performed in a 10 µL reaction mixture in ABI Prism 7900HT system (Applied Biosystems, CA, USA) using SYBR Premix Ex TaqTM II (Takara, Japan) according to the manufacturer’s instructions. The 10 µL PCR reaction mixture contained 5 µL SYBR, 0.4 µL primer, 0.2 µL ROX, 3.4 µL ddH2O and 1 µL cDNA. The primers used were presented in the following: 5'-CAGCCTTCCTTCTTGGGTAT-3' and 3'-GGCATAGAGGTCTTTACGG-5' for β-actin; 5'-GAAAGGATCGGCGCAATTAA-3' and 3'-CATCATCCGAAAGCTGCATC-5' for BHLHE40; 5'-CGAGCATGACGAAGAGATCAT-3' and 3'-TCGAAGGTTGGCCTATCTGA-5' for PIM1.
Western blot
Protein lysates of mBMSC were separated using 8% SDS-PAGE gel electrophoresis and then transferred to PVDF membrane and blocked with 5% BSA. The primary antibodies for β-actin (1:1,000, Abcam) and Bhlhe40 (1:50, Genetex, CA, USA) were incubated on shaking bed overnight at 4 °C. Secondary antibody was incubated at room temperature for 1 h. Developed films were digitized by scanning and the optical densities were analyzed by AlphaView SA software (V3.3.0, Cell Biosciences).
Histology
Tibias were removed and fixed in PLP fixative (2% paraformaldehyde containing 0.075 M lysine and 0.01 M sodium periodate) overnight at 4 °C and processed histologically as described (18). Tibias were decalcified in ethylene diamine tetraacetic acid (EDTA)-glycerol solution for 5 to 7 days at 4 °C. Decalcified tibias were dehydrated and embedded in paraffin, after which 5 mm sections were cut on a rotary microtome (RM2235, Leica, Germany). These sections were stained with hematoxylin and eosin (HE) or histochemically for total collagen or immunohistochemical staining as described in the next section.
Immunohistochemical staining
Immunohistochemical staining for Bhlhe40 was performed using the avidin-biotin-peroxidase complex technique with primary antibody rabbit polyclonal anti-Bhlhe40 (1:50, Genetex, CA, USA). Briefly, endogenous peroxidase was blocked with 3% H2O2 and sections were treated overnight at 4 °C with primary antibodies. Sections were then incubated with the secondary antibody (KIT-5010, Max Version, Maixin.Bio, China) for 1 h at room temperature, followed by coloration with 3,3-diaminobenzidine (DAB, Sigma, Germany) and hematoxylin counterstaining. The sections were washed with 0.01 mol/L PBS between each step.
Data analysis
Calculate the ΔCt for each pathway-focused gene in each treatment group.
ΔCt (group 1) = average Ct – average of HK genes’ Ct for group 1 array;
ΔCt (group 2) = average Ct – average of HK genes’ Ct for group 2 array.
Calculate the ΔΔCt for each gene across two PCR arrays (or groups).
ΔΔCt = ΔCt (group 2) – ΔCt (group 1).
Where group 1 is the control and group 2 is the experimental.
Calculate the fold-change for each gene from group 1 to group 2 as 2-ΔΔCt.
If the fold-change is greater than 1, then the result may be reported as a fold up-regulation. If the fold-change is less than 1, then the negative inverse of the result may be reported as a fold down-regulation. Statistical analyses also were performed and P value correction to account for a high false-positive rate was performed by using the false discovery rate method.
Statistical analysis
Measurement data were expressed as mean ± standard deviation (SD). Significance of the difference between two groups was analyzed using two-tailed Student’s t-test. All statistical analyses were performed with SPSS 20.0 software (IBM, Armonk, NY, USA). P<0.05 was considered statistically significant. Statistical analyses were performed and P value correction to account for a high false-positive rate was performed by using the false discovery rate method with the R package limma in Bioconductor. Volcano Plot and GO analysis of differentially expressed mRNAs and between group statistical analysis were performed.
Results
Demographic characteristics of participants
The demographic characteristics of the participants are summarized (Table 1).
Table 1. Demographic characteristics of participants.
| Item | Healthy control (n=3) | Osteoporosis | ||
|---|---|---|---|---|
| BMI <25 (n=3) | BMI ≥25 (n=3) | P value | ||
| Age (years) | 52.67±1.15 | 59±3.61 | 52.67±5.86 | 0.186 |
| BMI (kg/m2) | 20.91±1.62 | 19.8±3.87 | 26.37±0.90 | 0.046 |
| HbA1c (%) | 4.97±0.55 | 5.07±0.21 | 6.27±1.25 | 0.176 |
| TC (mmol/L) | 2.24±1.91 | 4.37±0.78 | 4.92±1.20 | 0.591 |
| TG (mmol/L) | 2.26±1.40 | 1.99±1.52 | 2.62±1.09 | 0.544 |
| ALT (U/L) | 16±4.58 | 16.67±2.52 | 22.33±9.50 | 0.374 |
| AST (U/L) | 13.67±2.30 | 17.67±3.51 | 17.67±6.66 | 1.000 |
| HB (g/L) | 158.67±15.70 | 126.33±9.87 | 127.67±17.04 | 0.912 |
| Cr (µmol/L) | 86±9.85 | 63.33±3.51 | 68.33±28.57 | 0.779 |
Data are expressed as the mean ± SD. There was no significant difference between osteoporosis subgroups except BMI. No significant differences existed between healthy control group and osteoporosis group (data are not shown). BMI, body mass index.
One female and 5 males with OP were investigated in this study. Mean age of 6 patients was 55 years. Healthy controls were comprised of 3 healthy males with a normal BMD (Table 2). Except BMD, no significant differences existed between healthy control group and OP group (data are not shown). These results do not change much when the data were removed from the female patients. The OP patients can be further divided into two subgroups according to their BMI, except which no other significant differences existed between these two subgroups (Table 3).
Table 2. BMD of participants recruited for the study.
| BMD | Healthy control (n=3) | Osteoporosis (n=6) | P value |
|---|---|---|---|
| L1 BMD (g/cm3) | 0.91±0.02 | 0.63±0.06 | 0.000125 |
| L2 BMD (g/cm3) | 0.96±0.04 | 0.68±0.09 | 0.001876 |
| L3 BMD (g/cm3) | 1.00±0.08 | 0.70±0.07 | 0.000717 |
| L4 BMD (g/cm3) | 1.00±0.08 | 0.71±0.07 | 0.001377 |
| Total BMD (g/cm3) | 0.97±0.06 | 0.69±0.07 | 0.000462 |
| Neck BMD (g/cm3) | 0.84±0.05 | 0.60±0.08 | 0.003156 |
| T score | −0.8±0.15 | −3.33±0.51 | – |
Data are expressed as the mean ± SD. BMD was significantly decreased in osteoporosis group compared with healthy control group.
Table 3. BMD of osteoporosis subgroups recruited for the study.
| BMD | Osteoporosis | P value | |
|---|---|---|---|
| BMI <25 (n=3) | BMI ≥25 (n=3) | ||
| L1 BMD (g/cm3) | 0.63±0.06 | 0.65±0.08 | 0.553 |
| L2 BMD (g/cm3) | 0.68±0.09 | 0.73±0.09 | 0.211 |
| L3 BMD (g/cm3) | 0.70±0.07 | 0.72±0.10 | 0.610 |
| L4 BMD (g/cm3) | 0.72±0.08 | 0.71±0.09 | 0.960 |
| Total BMD (g/cm3) | 0.69±0.07 | 0.71±0.09 | 0.071 |
| Neck BMD (g/cm3) | 0.60±0.08 | 0.67±0.08 | 0.071 |
| T score | −3.23±0.5 | −3.43±0.50 | – |
Data are expressed as the mean ± SD. No significant differences existed between osteoporosis subgroups. BMI, body mass index.
Normoxic downregulation of hypoxia related genes from hBMSCs in OP patients
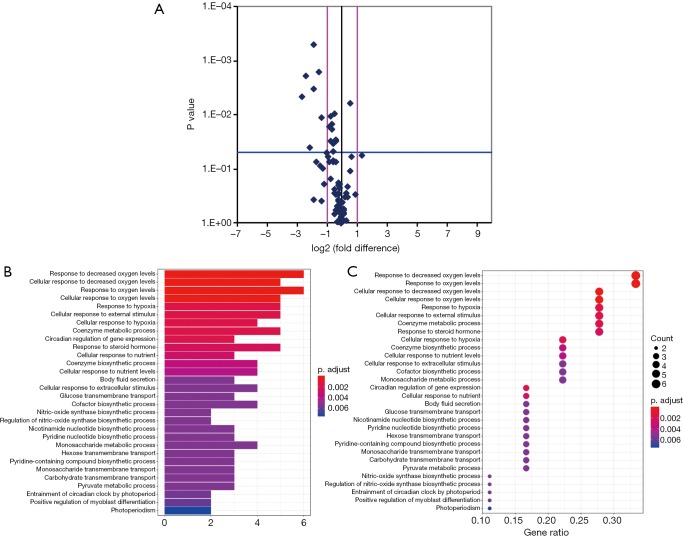
Among the hypoxia signaling pathway genes, the expression levels of 7 genes (BHLHE40, HIF1AN, MAP3K1, MET, PER1, IL1B, PFKFB3and PIM1) were down-regulated when compared with control group. Particularly, PER1 was significantly down-regulated. No gene expression was found be up-regulated in OP group compared with control group (Table 4). Some important downregulated GO functions may be related to response to decreased oxygen levels, photoperiodism, positive regulation of myoblast differentiation and entrainment of circadian clock by photoperiod (Figure 1).
Table 4. Differential genes from osteoporosis group and control group.
| Gene symbol | Fold up-/down-regulation, osteoporosis/control | t-test, P value | FDR, Q value |
|---|---|---|---|
| BHLHE40 | −2.85 | 0.001576 | 0.04727 |
| HIF1AN | −2.52 | 0.010969 | 0.06581 |
| MAP3K1 | −3.69 | 0.003010 | 0.04963 |
| MET | −4.28 | 0.038899 | 0.12964 |
| PER1 | −6.39 | 0.004551 | 0.05461 |
| PFKFB3 | −3.76 | 0.000486 | 0.02918 |
| PIM1 | −5.33 | 0.001885 | 0.03769 |
Figure 1.
Volcano plot of significantly differential experiment genes and significant gene ontology (GO) analyses and pathways of differentially expressed genes. (A) Analyzing the differentially expressed genes by t-test, log2 (fold change) was taken as abscissa and negative logarithm-log10 (P value) of P value was taken as ordinate. (B) The significant GO of differentially expressed genes. (C) The significant function of differentially expressed genes. The y axis shows the GO or pathway category and the x axis shows the negative logarithm of the P value (–LgP). A larger –LgP indicated a smaller P value for the difference.
Screening for differential genes from BMI <25 OP subgroup and control group
Among the hypoxia signaling pathway genes, the expression levels of 8 genes (ADM, BHLHE40, MAP3K1, LOX, TXNIP, NAMPT, PFKFB3 and PIM1) were down-regulated in BMI <25 OP subgroup when compared with the control group. Particularly, NAMPT was significantly down-regulated. Two genes (ANGPTL4, SLC16A3) expression were found to be up-regulated in BMI <25 OP subgroup compared with control group (Table 5).
Table 5. Differential genes from BMI <25 osteoporosis subgroup and control group.
| Gene symbol | Fold up-/down-regulation, osteoporosis/control | t-test, P value | FDR, Q value |
|---|---|---|---|
| ADM | −2.14 | 0.011382 | 0.08473 |
| BHLHE40 | −2.19 | 0.029789 | 0.09504 |
| LOX | −2.20 | 0.037472 | 0.10916 |
| MAP3K1 | −2.04 | 0.022416 | 0.08348 |
| NAMPT | −4.66 | 0.001422 | 0.04765 |
| PFKFB3 | −2.53 | 0.011585 | 0.07762 |
| PIM1 | −3.16 | 0.036125 | 0.11001 |
| TXNIP | −2.12 | 0.012865 | 0.07836 |
| SLC16A3 | 5.26 | 0.006528 | 0.08748 |
| ANGPTL4 | 3.36 | 0.023373 | 0.08700 |
BMI, body mass index.
Screening for differential genes from BMI ≥25 OP subgroup and control group
Among the hypoxia signaling pathway genes, the expression levels of 15 genes (ANXA2, BHLHE40, DDIT4, DNAJC5, GYS1, HIF1AN, MAP3K1, P4HB, PER1, PFKFB3, PFKL, PIM1, SLC2A1, TP53 and USF2) were down-regulated in BMI ≥25 OP subgroup when compared with the control group. Particularly, PFKL was significantly down-regulated. No gene expression was found be up-regulated in BMI ≥25 OP subgroup compared with control group (Table 6).
Table 6. Differential genes from BMI ≥25 osteoporosis subgroup and control group.
| Gene symbol | Fold up-/down-regulation, osteoporosis/control | t-test, P value | FDR, Q value |
|---|---|---|---|
| ANXA2 | −3.27 | 0.010590 | 0.05703 |
| BHLHE40 | −3.73 | 0.009262 | 0.05403 |
| DDIT4 | −8.55 | 0.028992 | 0.10681 |
| DNAJC5 | −2.54 | 0.017724 | 0.07298 |
| GYS1 | −7.48 | 0.007312 | 0.05119 |
| HIF1AN | −3.40 | 0.017200 | 0.07525 |
| MAP3K1 | −6.70 | 0.000017 | 0.08717 |
| P4HB | −2.64 | 0.006077 | 0.06077 |
| PER1 | −8.20 | 0.035503 | 0.01183 |
| PFKFB3 | −5.59 | 0.001887 | 0.04402 |
| PFKL | −16.04 | 0.000010 | 0.08109 |
| PIM1 | −8.99 | 0.006255 | 0.05473 |
| SLC2A1 | −4.92 | 0.004637 | 0.08114 |
| TP53 | −6.38 | 0.006409 | 0.04984 |
| USF2 | −2.13 | 0.016859 | 0.07867 |
BMI, body mass index.
Screening for differential genes from BMI <25 OP subgroup and BMI ≥25 OP subgroup
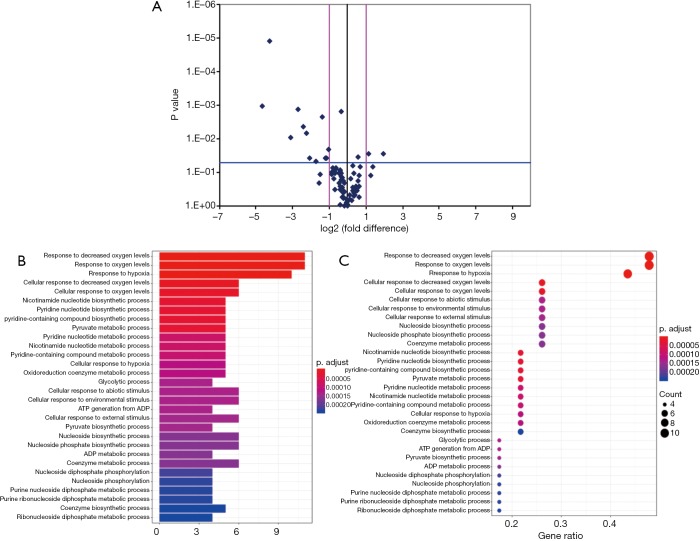
Among the hypoxia signaling pathway genes, the expression levels of 12 genes (ANXA2, CA9, DDIT4, GYS1, MAP3K1, P4HB, PFKFB3, PFKL, PGF, SLC16A3, SLC2A1 and TP53) were down-regulated in BMI ≥25 OP subgroup when compared with the BMI <25 OP subgroup. Particularly, PFKL and SLC16A3 were significantly down-regulated. Two genes (NAMPT, NCOA1) expression were found to be up-regulated in BMI ≥25 OP subgroup compared with BMI <25 OP subgroup (Table 7). Some important downregulated GO functions may be related to metabolic and glycolytic genes (Figure 2).
Table 7. Differential genes from BMI <25 osteoporosis subgroup and BMI ≥25 osteoporosis subgroup.
| Gene symbol | Fold up-/down-regulation, BMI ≥25/BMI <25 | t-test, P value | FDR, Q value |
|---|---|---|---|
| ANXA2 | −2.64 | 0.002236 | 0.03131 |
| CA9 | −4.20 | 0.038368 | 0.16786 |
| DDIT4 | −6.66 | 0.001337 | 0.03119 |
| GYS1 | −8.61 | 0.009239 | 0.07186 |
| MAP3K1 | −3.29 | 0.047246 | 0.19454 |
| P4HB | −2.38 | 0.037994 | 0.18997 |
| PFKFB3 | −2.21 | 0.038362 | 0.17902 |
| PFKL | −19.15 | 0.000013 | 0.00091 |
| PGF | −2.07 | 0.021402 | 0.14982 |
| SLC16A3 | −25.84 | 0.001075 | 0.03763 |
| SLC2A1 | −4.76 | 0.006630 | 0.06630 |
| TP53 | −5.31 | 0.004376 | 0.05106 |
| NAMPT | 3.75 | 0.027735 | 0.16179 |
| NCOA1 | 2.14 | 0.027111 | 0.17253 |
BMI, body mass index.
Figure 2.
Volcano plot of significantly differential experiment genes and significant gene ontology (GO) analyses and pathways of differentially expressed genes. (A) Analyzing the differentially expressed genes by t-test, log2 (fold change) was taken as abscissa and negative logarithm-log10 (P value) of P value was taken as ordinate. (B) The significant GO of differentially expressed genes. (C) The significant function of differentially expressed genes. The y axis shows the GO or pathway category and the x axis shows the negative logarithm of the P value (–LgP). A larger –LgP indicated a smaller P value for the difference.
Expression of hypoxia pathway genes on mBMSCs
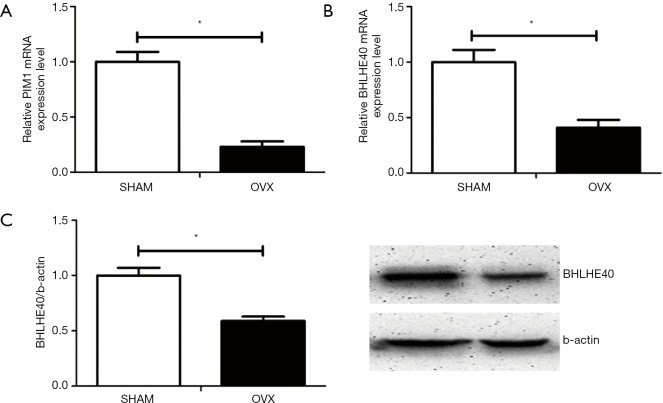
Based on previous microarray screening on differently expressed hypoxia signaling pathway genes between OP patients and normal controls, real-time PCR and Western blot were performed to verify the expression level of certain hypoxia genes on mBMSCs of OVX and SHAM mice model. Compared with mBMSCs of SHAM mice, the expression of PIM1 on mRNA level (Figure 3A) and the expression of BHLHE40 on both mRNA (Figure 3B) and protein level (Figure 3C) decreased significantly on mBMSCs of OVX mice, which was consistent with the results of previous microarray screening.
Figure 3.
Expression of BHLHE40 and PIM1 on mBMSCs of mice model. (A) Relative expression of PIM1 on mRNA level; (B) relative expression of BHLHE40 on mRNA level; (C) relative expression of Bhlhe40 on protein level. All experiments in this figure were repeated at least three times, and data were expressed as mean ± SD. *, P<0.05 compared with SHAM group.
Evaluation of mice model and expression of BHLHE40 in femur bone marrow of mouse model
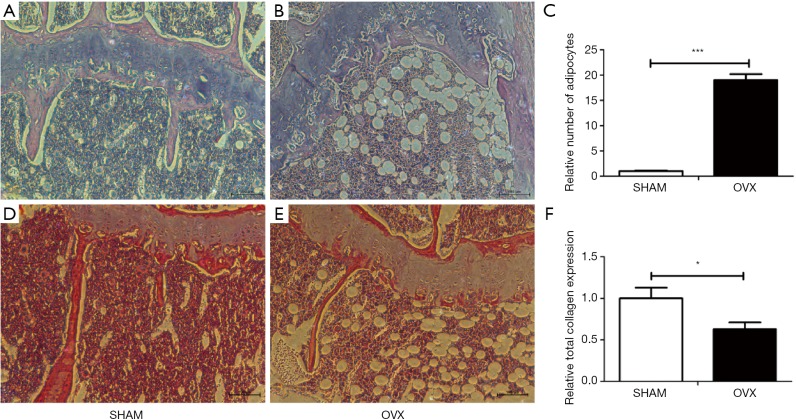
Morphologic and histological analyses of proximal femora were performed with HE staining and histochemistry for total collagen (Figures 4 and 5). Compared with SHAM mice, the number, thickness and length of trabecular bone (Figure 4A,B) and the expression of total collagen (Figure 4D,E) in OVX mice decreased significantly with significantly increased number of adipocytes in bone marrow (Figure 4C), which indicated the OVX mice models were successfully built.
Figure 4.
Evaluation of mice model by HE staining and histochemistry for total collagen. (A,D) HE staining and total collagen expression of SHAM mice (10×10); (B,E) HE staining and total collagen expression of OVX mice (10×10); (C) relative number of adipocytes in bone marrow of SHAM and OVX mice; (F) relative total collagen expression of SHAM and OVX mice. All experiments in this figure were repeated at least three times, and data were expressed as mean ± SD. *, P<0.05; ***, P<0.001 compared with SHAM group.
Figure 5.
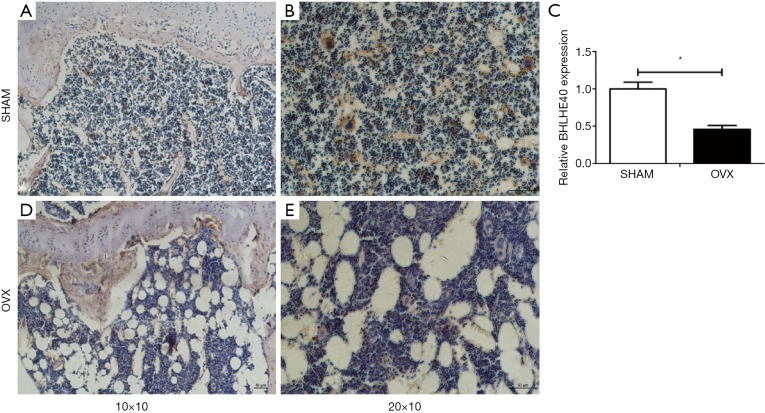
Expression of Bhlhe40 of SHAM and OVX mice by immunohistochemistry. (A) Expression of Bhlhe40 of SHAM mice (10×10); (B) expression of Bhlhe40 of SHAM mice (20×10); (D) expression of Bhlhe40 of OVX mice (10×10); (E) expression of Bhlhe40 of OVX mice (20×10); (C) relative expression of Bhlhe40 between two groups. All experiments in this figure were repeated at least three times, and data were expressed as mean ± SD. *, P<0.05 compared with SHAM group.
The BHLHE40 expression was analyzed by immunohistochemistry in this study. It showed that the BHLHE40 expression of OVX mice decreased markedly compared with that of the SHAM group (Figure 5).
Discussion
Hypoxia is a critical factor for stem cells in bone marrow by protecting them from ROS-mediated damage, which allows them to maintain normal function and self-renewal potential. The roles of hypoxia in the differentiation of MSCs remain controversial. MSCs under reduced oxygen conditions were believed to preserve their stemness and remain undifferentiated (19). However, it was also reported that hypoxia enhances mesoderm lineage differentiation, including adipogenic, osteogenic or chondrogenic differentiation (20,21). Studies comparing fracture incidence in obese and non-obese individuals have demonstrated that obesity, defined on the basis of body mass index (BMI), is associated with increased risk of fracture at some sites but seems to be protective at others (22). Extensive research using genetic models has revealed that hypoxia signaling is a key mechanism in adipose tissue dysfunction, leading to adipose tissue fibrosis, inflammation and insulin resistance (23,24). Consequently, it is quite significant to study hypoxia pathway gene expression on the BMSCs from OP patients, furthermore, whether the adipose tissue exert effect on hBMSC function.
Data in this study indicated that hypoxic pathway genes were down-regulated in the BMSCs from OP patients. Genes were found down-regulated when compared with the control group. Some important downregulated GO functions may be related to photoperiodism, positive regulation of myoblast differentiation and entrainment of circadian clock by photoperiod, among which PER1 was significantly down-regulated. PER1 encodes the period circadian protein homolog 1 protein in human and studies have demonstrated that the PER1 polymorphisms were singly and in combination related to the lumbar spine BMD (25). These data further explained the potential important function of PER1 on hypoxia pathway and OP. Basic helix-loop-helix transcription factor BHLHE40 (DEC1) promotes chondrogenic differentiation of MSC and myoblast differentiation in early and terminal stage and modulated the osteogenic differentiation of MSC (26,27). Disruption of hypoxic microenvironment in bone marrow of OP patients causes decreased DEC1 expression, which might affect the differentiation of BMSCs, further leading to decreased bone formation. Thus, DEC1 may play an important role in both hypoxia pathway and OP, but further studies are still needed. The serine/threonine kinase Pim1 is an important regulator of cell proliferation and survival, cell metabolism and transcriptional activity (28), which has not been studied in OP yet. Besides, studies have demonstrated its importance in anti-proinflammation (18). This study may shed new light on the important function of PIM1 in OP. PIM1 and BHLHE40 expression was further checked in mice OP model, which also down-regulated than control group.
This study further confirmed the expression of PIM1 and BHLHE40 decreased significantly on mBMSCs of OVX mice, which was consistent with the results of previous microarray screening in human BMSCs. Basic helix-loop-helix transcription factor BHLHE40 (DEC1) promotes chondrogenic differentiation of MSC in early and terminal stage and modulated the osteogenic differentiation of MSC (26). Disruption of hypoxic microenvironment in bone marrow of OP patients causes decreased DEC1 expression, which might affect the differentiation of BMSCs, further leading to decreased bone formation. Thus, DEC1 may play an important role in both hypoxia pathway and OP, but further studies are still needed. The serine/threonine kinase Pim1 is an important regulator of cell proliferation and survival, cell metabolism, and transcriptional activity (28), which has not been studied in OP yet. Besides, studies have demonstrated its importance in anti-proinflammation (18). This study may shed new light on the important function of PIM1 in OP.
Transcription factor HIF-1 has been reported to play a major role in the induction of hypoxia-inducible genes including many glycolytic enzymes, VEGF, and erythropoietin (29). However, in this study, it was found that HIF-1 expression was down-regulated without statistical significance (data were not shown) while HIF-1 inhibitor FIH was down-regulated dramatically. This may partly attribute to unstable expression of HIF-1 under the normoxia environment and the limitation by the sample size as the genes downstream of HIF-1 were all down-regulated. The other reason might be that the balance between HIF-1 and FIH declined due to increased oxidative stress in bone marrow cavity.
The gene expression levels were studies in BMI <25 OP subgroup. Eight genes (ADM, BHLHE40, MAP3K1, LOX, TXNIP, NAMPT, PFKFB3 and PIM1) were down-regulated and two genes (ANGPTL4, SLC16A3) were up-regulated in BMI <25 OP subgroup compared with control group. ADM acts as a survival factor in osteoblastic cells via a CGRP1 receptor-MEK-ERK pathway, which provides further understanding on the physiological function of ADM in osteoblasts (30). Lysyl oxidase (LOX) is a critical mediator of bone marrow cell recruitment to form the premetastatic niche (31). Thioredoxin-interacting protein (TXNIP), which is induced by oxidative stress, is a known regulator of intracellular ROS, which confirmed the relationship between ROS and OP. Nicotinamide phosphoribosyltransferase (Nampt) affects the lineage fate determination of mesenchymal stem cells (32) which could be a possible cause for decreased osteogenesis and increased adipogenesis in elder individuals. Angptl4 is up-regulated under inflammatory conditions in the bone marrow of mice (33), which might play an important role in the inflammatory condition in OP bone marrow. SLC16A3 is one of carbohydrate transporters which have not been reported concerned with OP.
Abnormal adiposity is associated with many metabolic diseases, where the link seems to be the mild and chronic inflammation occurred in obesogenic conditions. One of the mechanisms triggering inflammation has been associated with adipose tissue hypoxia (34). Studies have confirmed that hypoxia, mainly due to hypoperfusion, exists in adipose tissue no matter in obesity mice or human (15,35). Oxygen stress plays a pivotal role in normal human development and physiology, so disturbance of oxygen balance by abnormal adiposity in bone marrow may cause tissue dysfunction and affect a series of genes expression such as HIF-1α, IL-6, GLUT-1, PPAR-γ, etc., pushing the tissue toward a pro-inflammatory environment. All the above can disrupt the balanced microenvironment in bone marrow and result in BMSCs dysfunction, which may play an important role in pathophysiology of OP. Therefore, it was later investigated the influence of obesity on hypoxia pathway of BMSCs from OP patients. Hypoxia-mediated HIF-1α-activated downstream target genes were down-regulated in BMI ≥25 OP subgroup, including glycolytic enzymes (Pfkl, GYS1), carbohydrate transport and metabolism genes (CA9, SLC2A1, SLC16A3, NAMPT). AnxA2 is also expressed in cells of the osteoblast lineage and chondrocytes and may play a role in matrix mineralization (36). P4HB is involved in hydroxylation of prolyl residues in precollagen. These results demonstrated that BMI ≥25 had more negative effect on metabolic and glycolytic genes expression of BMSCs rather than VEGF which was viewed as another important downstream of HIF-1. In summary, these findings shed new light on the hypoxia pathway in OP and have implications for future researches.
Table S1. Gene data.
| Position | Unigene | GeneBank | Symbol | Description | Gene name |
|---|---|---|---|---|---|
| A01 | Hs.441047 | NM_001124 | ADM | Adrenomedullin | AM |
| A02 | Hs.167046 | NM_000676 | ADORA2B | Adenosine A2b receptor | ADORA2 |
| A03 | Hs.513490 | NM_000034 | ALDOA | Aldolase A, fructose-bisphosphate | ALDA, GSD12, MGC10942, MGC17716, MGC17767 |
| A04 | Hs.9613 | NM_001039667 | ANGPTL4 | Angiopoietin-like 4 | ANGPTL2, ARP4, FIAF, HFARP, NL2, PGAR, pp1158 |
| A05 | Hs.508154 | NM_181726 | ANKRD37 | Ankyrin repeat domain 37 | Lrp2bp, MGC111507 |
| A06 | Hs.511605 | NM_004039 | ANXA2 | Annexin A2 | ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV |
| A07 | Hs.73722 | NM_080649 | APEX1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | APE, APE1, APEN, APEX, APX, HAP1, REF1 |
| A08 | Hs.632446 | NM_001668 | ARNT | Aryl hydrocarbon receptor nuclear translocator | HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 |
| A09 | Hs.271791 | NM_001184 | ATR | Ataxia telangiectasia and Rad3 related | FRP1, MEC1, SCKL, SCKL1 |
| A10 | Hs.728782 | NM_003670 | BHLHE40 | Basic helix-loop-helix family, member e40 | BHLHB2, DEC1, FLJ99214, HLHB2, SHARP-2, STRA13, Stra14 |
| A11 | Hs.716515 | NM_000057 | BLM | Bloom syndrome, RecQ helicase-like | BS, MGC126616, MGC131618, MGC131620, RECQ2, RECQL2, RECQL3 |
| A12 | Hs.144873 | NM_004052 | BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | NIP3 |
| B01 | Hs.131226 | NM_004331 | BNIP3L | BCL2/adenovirus E1B 19kDa interacting protein 3-like | BNIP3a, NIX |
| B02 | Hs.255935 | NM_001731 | BTG1 | B-cell translocation gene 1, anti-proliferative | – |
| B03 | Hs.63287 | NM_001216 | CA9 | Carbonic anhydrase IX | CAIX, MN |
| B04 | Hs.13291 | NM_004354 | CCNG2 | Cyclin G2 | – |
| B05 | Hs.491912 | NM_006837 | COPS5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | CSN5, JAB1, MGC3149, MOV-34, SGN5 |
| B06 | Hs.517076 | NM_000308 | CTSA | Cathepsin A | GLB2, GSL, NGBE, PPCA, PPGB |
| B07 | Hs.523012 | NM_019058 | DDIT4 | DNA-damage-inducible transcript 4 | Dig2, FLJ20500, REDD-1, REDD1, RP11-442H21.1, RTP801 |
| B08 | Hs.164419 | NM_025219 | DNAJC5 | DnaJ (Hsp40) homolog, subfamily C, member 5 | CSP, DKFZp434N1429, DKFZp761N1221, DNAJC5A, FLJ00118, FLJ13070 |
| B09 | Hs.511899 | NM_001955 | EDN1 | Endothelin 1 | ET1, HDLCQ7, PPET1 |
| B10 | Hs.444450 | NM_022051 | EGLN1 | Egl nine homolog 1 (C. elegans) | C1orf12, DKFZp761F179, ECYT3, HIFPH2, HPH2, PHD2, SM20, ZMYND6 |
| B11 | Hs.515417 | NM_053046 | EGLN2 | Egl nine homolog 2 (C. elegans) | DKFZp434E026, EIT6, FLJ95603, HIF-PH1, HIFPH1, HPH-1, HPH-3, PHD1 |
| B12 | Hs.326035 | NM_001964 | EGR1 | Early growth response 1 | AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 |
| C01 | Hs.411641 | NM_004095 | EIF4EBP1 | Eukaryotic translation initiation factor 4E binding protein 1 | 4E-BP1, 4EBP1, BP-1, MGC4316, PHAS-I |
| C02 | Hs.517145 | NM_001428 | ENO1 | Enolase 1, (alpha) | ENO1L1, MPB1, NNE, PPH |
| C03 | Hs.2303 | NM_000799 | EPO | Erythropoietin | EP, MGC138142, MVCD2 |
| C04 | Hs.592304 | NM_014584 | ERO1L | ERO1-like (S. cerevisiae) | ERO1-alpha, ERO1A |
| C05 | Hs.361463 | NM_000504 | F10 | Coagulation factor X | FX, FXA |
| C06 | Hs.62192 | NM_001993 | F3 | Coagulation factor III (thromboplastin, tissue factor) | CD142, FLJ17960, TF, TFA |
| C07 | Hs.728789 | NM_005252 | FOS | FBJ murine osteosarcoma viral oncogene homolog | AP-1, C-FOS |
| C08 | Hs.436062 | NM_000158 | GBE1 | Glucan (1,4-alpha-), branching enzyme 1 | GBE |
| C09 | Hs.466471 | NM_000175 | GPI | Glucose-6-phosphate isomerase | AMF, DKFZp686C13233, GNPI, NLK, PGI, PHI, SA-36, SA36 |
| C10 | Hs.386225 | NM_002103 | GYS1 | Glycogen synthase 1 (muscle) | GSY, GYS |
| C11 | Hs.597216 | NM_001530 | HIF1A | Hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha, HIF1, HIF1-ALPHA, MOP1, PASD8, bHLHe78 |
| C12 | Hs.500788 | NM_017902 | HIF1AN | Hypoxia inducible factor 1, alpha subunit inhibitor | DKFZp762F1811, FIH1, FLJ20615, FLJ22027 |
| D01 | Hs.420830 | NM_152794 | HIF3A | Hypoxia inducible factor 3, alpha subunit | HIF-3A, IPAS, MOP7, PASD7, bHLHe17 |
| D02 | Hs.406266 | NM_000189 | HK2 | Hexokinase 2 | DKFZp686M1669, HKII, HXK2 |
| D03 | Hs.517581 | NM_002133 | HMOX1 | Heme oxygenase (decycling) 1 | HO-1, HSP32, bK286B10 |
| D04 | Hs.116462 | NM_178849 | HNF4A | Hepatocyte nuclear factor 4, alpha | FLJ39654, HNF4, HNF4a7, HNF4a8, HNF4a9, HNF4alpha, MODY, MODY1, NR2A1, NR2A21, TCF, TCF14 |
| D05 | Hs.591785 | NM_003897 | IER3 | Immediate early response 3 | DIF-2, DIF2, GLY96, IEX-1, IEX-1L, IEX1, PRG1 |
| D06 | Hs.450230 | NM_000598 | IGFBP3 | Insulin-like growth factor binding protein 3 | BP-53, IBP3 |
| D07 | Hs.514505 | NM_015167 | JMJD6 | Jumonji domain containing 6 | KIAA0585, PSR, PTDSR, PTDSR1 |
| D08 | Hs.2795 | NM_005566 | LDHA | Lactate dehydrogenase A | GSD11, LDH1, LDHM |
| D09 | Hs.531081 | NM_002306 | LGALS3 | Lectin, galactoside-binding, soluble, 3 | CBP35, GAL3, GALBP, GALIG, L31, LGALS2, MAC2 |
| D10 | Hs.102267 | NM_002317 | LOX | Lysyl oxidase | MGC105112 |
| D11 | Hs.657756 | NM_005921 | MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 | MAPKKK1, MEKK, MEKK1 |
| D12 | Hs.132966 | NM_000245 | MET | Met proto-oncogene (hepatocyte growth factor receptor) | AUTS9, HGFR, RCCP2, c-Met |
| E01 | Hs.407995 | NM_002415 | MIF | Macrophage migration inhibitory factor (glycosylation-inhibiting factor) | GIF, GLIF, MMIF |
| E02 | Hs.297413 | NM_004994 | MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | CLG4B, GELB, MANDP2, MMP-9 |
| E03 | Hs.501023 | NM_005962 | MXI1 | MAX interactor 1 | MAD2, MGC43220, MXD2, MXI, bHLHc11 |
| E04 | Hs.489615 | NM_005746 | NAMPT | Nicotinamide phosphoribosyltransferase | 1110035O14Rik, DKFZp666B131, MGC117256, PBEF, PBEF1, VF, VISFATIN |
| E05 | Hs.596314 | NM_003743 | NCOA1 | Nuclear receptor coactivator 1 | F-SRC-1, KAT13A, MGC129719, MGC129720, RIP160, SRC1, bHLHe42, bHLHe74 |
| E06 | Hs.372914 | NM_006096 | NDRG1 | N-myc downstream regulated 1 | CAP43, CMT4D, DRG1, GC4, HMSNL, NDR1, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 |
| E07 | Hs.654408 | NM_003998 | NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | DKFZp686C01211, EBP-1, KBF1, MGC54151, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 |
| E08 | Hs.707978 | NM_000603 | NOS3 | Nitric oxide synthase 3 (endothelial cell) | ECNOS, eNOS |
| E09 | Hs.467701 | NM_002539 | ODC1 | Ornithine decarboxylase 1 | ODC |
| E10 | Hs.500047 | NM_000917 | P4HA1 | Prolyl 4-hydroxylase, alpha polypeptide I | P4HA |
| E11 | Hs.464336 | NM_000918 | P4HB | Prolyl 4-hydroxylase, beta polypeptide | DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB, PO4DB, PO4HB, PROHB |
| E12 | Hs.470633 | NM_002610 | PDK1 | Pyruvate dehydrogenase kinase, isozyme 1 | – |
| F01 | Hs.445534 | NM_002616 | PER1 | Period homolog 1 (Drosophila) | MGC88021, PER, RIGUI, hPER |
| F02 | Hs.195471 | NM_004566 | PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | FLJ37326, IPFK2, PFK2 |
| F03 | Hs.476217 | NM_004567 | PFKFB4 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 | – |
| F04 | Hs.255093 | NM_002626 | PFKL | Phosphofructokinase, liver | DKFZp686G1648, DKFZp686L2097, FLJ30173, FLJ40909, PFK-B |
| F05 | Hs.26010 | NM_002627 | PFKP | Phosphofructokinase, platelet | FLJ40226, FLJ44165, PFK-C, PFKF |
| F06 | Hs.592599 | NM_002629 | PGAM1 | Phosphoglycerate mutase 1 (brain) | PGAM-B, PGAMA |
| F07 | Hs.252820 | NM_002632 | PGF | Placental growth factor | D12S1900, PGFL, PLGF, PlGF-2, SHGC-10760 |
| F08 | Hs.78771 | NM_000291 | PGK1 | Phosphoglycerate kinase 1 | MGC117307, MGC142128, MGC8947, MIG10, PGKA |
| F09 | Hs.81170 | NM_002648 | PIM1 | Pim-1 oncogene | PIM |
| F10 | Hs.534770 | NM_002654 | PKM2 | Pyruvate kinase, muscle | CTHBP, MGC3932, OIP3, PK3, PKM, TCB, THBP1 |
| F11 | Hs.77274 | NM_002658 | PLAU | Plasminogen activator, urokinase | ATF, UPA, URK, u-PA |
| F12 | Hs.479396 | NM_005349 | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region | CBF1, IGKJRB, IGKJRB1, KBF2, MGC61669, RBP-J, RBPJK, RBPSUH, SUH, csl |
| G01 | Hs.515846 | NM_006666 | RUVBL2 | RuvB-like 2 (E. coli) | ECP51, INO80J, REPTIN, RVB2, TIH2, TIP48, TIP49B |
| G02 | Hs.414795 | NM_000602 | SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | PAI, PAI-1, PAI1, PLANH1 |
| G03 | Hs.500761 | NM_004207 | SLC16A3 | Solute carrier family 16, member 3 (monocarboxylic acid transporter 4) | MCT 3, MCT 4, MCT-3, MCT-4, MCT3, MCT4, MGC138472, MGC138474 |
| G04 | Hs.473721 | NM_006516 | SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | DYT17, DYT18, GLUT, GLUT1, GLUT1DS, MGC141895, MGC141896, PED |
| G05 | Hs.419240 | NM_006931 | SLC2A3 | Solute carrier family 2 (facilitated glucose transporter), member 3 | FLJ90380, GLUT3 |
| G06 | Hs.529618 | NM_003234 | TFRC | Transferrin receptor (p90, CD71) | CD71, TFR, TFR1, TRFR |
| G07 | Hs.654481 | NM_000546 | TP53 | Tumor protein p53 | FLJ92943, LFS1, P53, TRP53 |
| G08 | Hs.524219 | NM_000365 | TPI1 | Triosephosphate isomerase 1 | MGC88108, TIM, TPI |
| G09 | Hs.533977 | NM_006472 | TXNIP | Thioredoxin interacting protein | EST01027, HHCPA78, THIF, VDUP1 |
| G10 | Hs.454534 | NM_003367 | USF2 | Upstream transcription factor 2, c-fos interacting | FIP, bHLHb12 |
| G11 | Hs.519320 | NM_003374 | VDAC1 | Voltage-dependent anion channel 1 | MGC111064, PORIN, VDAC-1 |
| G12 | Hs.73793 | NM_003376 | VEGFA | Vascular endothelial growth factor A | MGC70609, MVCD1, VEGF, VPF |
| H01 | Hs.520640 | NM_001101 | ACTB | Actin, beta | PS1TP5BP1 |
| H02 | Hs.534255 | NM_004048 | B2M | Beta-2-microglobulin | – |
| H03 | Hs.592355 | NM_002046 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | G3PD, GAPD, MGC88685 |
| H04 | Hs.412707 | NM_000194 | HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | HGPRT, HPRT |
| H05 | Hs.546285 | NM_001002 | RPLP0 | Ribosomal protein, large, P0 | L10E, LP0, MGC111226, MGC88175, P0, PRLP0, RPP0 |
| H06 | N/A | SA_00105 | HGDC | Human genomic DNA contamination | HIGX1A |
| H07 | N/A | SA_00104 | RTC | Reverse transcription control | RTC |
| H08 | N/A | SA_00104 | RTC | Reverse transcription control | RTC |
| H09 | N/A | SA_00104 | RTC | Reverse transcription control | RTC |
| H10 | N/A | SA_00103 | PPC | Positive PCR control | PPC |
| H11 | N/A | SA_00103 | PPC | Positive PCR control | PPC |
| H12 | N/A | SA_00103 | PPC | Positive PCR control | PPC |
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (Grant No. 81570799).
Ethical Statement: This study was approved by the Ethics Committee of Shanghai Tongji Hospital (KYSB-2015-20). Informed consent was provided by all participants. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Devlin MJ, Cloutier AM, Thomas NA, et al. Caloric restriction leads to high marrow adiposity and low bone mass in growing mice. J Bone Miner Res 2010;25:2078-88. 10.1002/jbmr.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson RW, Schipani E, Giaccia AJ. HIF targets in bone remodeling and metastatic disease. Pharmacol Ther 2015;150:169-77. 10.1016/j.pharmthera.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Provot S, Zinyk D, Gunes Y, et al. Hif-1alpha regulates differentiation of limb bud mesenchyme and joint development. J Cell Biol 2007;177:451-64. 10.1083/jcb.200612023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison JS, Rameshwar P, Chang V, et al. Oxygen saturation in the bone marrow of healthy volunteers. Blood 2002;99:394. 10.1182/blood.V99.1.394 [DOI] [PubMed] [Google Scholar]
- 5.Spencer JA, Ferraro F, Roussakis E, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 2014;508:269-73. 10.1038/nature13034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong-Feng Z, Ting L, Cheng C, et al. Silencing HIF-1alpha reduces the adhesion and secretion functions of acute leukemia hBMSCs. Braz J Med Biol Res 2012;45:906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell 2010;7:150-61. 10.1016/j.stem.2010.07.007 [DOI] [PubMed] [Google Scholar]
- 8.Chinn IK, Blackburn CC, Manley NR, et al. Changes in primary lymphoid organs with aging. Semin Immunol 2012;24:309-20. 10.1016/j.smim.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou R, Liu R, Niu X, et al. Biological characteristics and gene expression pattern of bone marrow mesenchymal stem cells in patients with psoriasis. Exp Dermatol 2014;23:521-3. 10.1111/exd.12446 [DOI] [PubMed] [Google Scholar]
- 10.Johnson AR, Milner JJ, Makowski L. The inflammation highway: metabolism accelerates inflammatory traffic in obesity. Immunol Rev 2012;249:218-38. 10.1111/j.1600-065X.2012.01151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frommer KW, Neumann E, Lange U. Inflammation and bone: Osteoimmunological aspects. Z Rheumatol 2016;75:444-50. 10.1007/s00393-016-0101-7 [DOI] [PubMed] [Google Scholar]
- 12.Kuhn H, O'Donnell VB. Inflammation and immune regulation by 12/15-lipoxygenases. Prog Lipid Res 2006;45:334-56. 10.1016/j.plipres.2006.02.003 [DOI] [PubMed] [Google Scholar]
- 13.Despres JP, Arsenault BJ, Cote M, et al. Abdominal obesity: the cholesterol of the 21st century? Can J Cardiol 2008;24 Suppl D:7D-12D. [DOI] [PMC free article] [PubMed]
- 14.Hodson L. Adipose tissue oxygenation: Effects on metabolic function. Adipocyte 2014;3:75-80. 10.4161/adip.27114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosogai N, Fukuhara A, Oshima K, et al. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes 2007;56:901-11. 10.2337/db06-0911 [DOI] [PubMed] [Google Scholar]
- 16.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest 2003;112:1785-8. 10.1172/JCI20514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JS, Park JC, Kim TW, et al. Human bone marrow stem cells cultured under hypoxic conditions present altered characteristics and enhanced in vivo tissue regeneration. Bone 2015;78:34-45. 10.1016/j.bone.2015.04.044 [DOI] [PubMed] [Google Scholar]
- 18.de Vries M, Hesse L, Jonker MR, et al. Pim1 kinase activity preserves airway epithelial integrity upon house dust mite exposure. Am J Physiol Lung Cell Mol Physiol 2015;309:L1344-53. 10.1152/ajplung.00043.2015 [DOI] [PubMed] [Google Scholar]
- 19.Jiang C, Sun J, Dai Y, et al. HIF-1A and C/EBPs transcriptionally regulate adipogenic differentiation of bone marrow-derived MSCs in hypoxia. Stem Cell Res Ther 2015;6:21. 10.1186/s13287-015-0014-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valorani MG, Montelatici E, Germani A, et al. Pre-culturing human adipose tissue mesenchymal stem cells under hypoxia increases their adipogenic and osteogenic differentiation potentials. Cell Prolif 2012;45:225-38. 10.1111/j.1365-2184.2012.00817.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SY, He YB, Deng SY, et al. Exercise affects biological characteristics of mesenchymal stromal cells derived from bone marrow and adipose tissue. Int Orthop 2017;41:1199-209. 10.1007/s00264-017-3441-2 [DOI] [PubMed] [Google Scholar]
- 22.Fassio A, Idolazzi L, Rossini M, et al. The obesity paradox and osteoporosis. Eat Weight Disord 2018;23:293-302. 10.1007/s40519-018-0505-2 [DOI] [PubMed] [Google Scholar]
- 23.Berry DC, Jiang Y, Arpke RW, et al. Cellular Aging Contributes to Failure of Cold-Induced Beige Adipocyte Formation in Old Mice and Humans. Cell Metab 2017;25:481. 10.1016/j.cmet.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 24.Lin Q, Yun Z. The Hypoxia-Inducible Factor Pathway in Adipocytes: The Role of HIF-2 in Adipose Inflammation and Hypertrophic Cardiomyopathy. Front Endocrinol (Lausanne) 2015;6:39. 10.3389/fendo.2015.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H, Koh H, Ku SY, et al. Association between polymorphisms in period genes and bone density in postmenopausal Korean women. Climacteric 2014;17:605-12. 10.3109/13697137.2014.905527 [DOI] [PubMed] [Google Scholar]
- 26.Iwata T, Kawamoto T, Sasabe E, et al. Effects of overexpression of basic helix-loop-helix transcription factor Dec1 on osteogenic and adipogenic differentiation of mesenchymal stem cells. Eur J Cell Biol 2006;85:423-31. 10.1016/j.ejcb.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 27.Chang HC, Kao CH, Chung SY, et al. Bhlhe40 differentially regulates the function and number of peroxisomes and mitochondria in myogenic cells. Redox Biol 2019;20:321-33. 10.1016/j.redox.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cen B, Xiong Y, Song JH, et al. The Pim-1 protein kinase is an important regulator of MET receptor tyrosine kinase levels and signaling. Mol Cell Biol 2014;34:2517-32. 10.1128/MCB.00147-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev 1998;8:588-94. 10.1016/S0959-437X(98)80016-6 [DOI] [PubMed] [Google Scholar]
- 30.Uzan B, Villemin A, Garel JM, et al. Adrenomedullin is anti-apoptotic in osteoblasts through CGRP1 receptors and MEK-ERK pathway. J Cell Physiol 2008;215:122-8. 10.1002/jcp.21294 [DOI] [PubMed] [Google Scholar]
- 31.Erler JT, Bennewith KL, Cox TR, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 2009;15:35-44. 10.1016/j.ccr.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, He X, Li Y, et al. Nicotinamide phosphoribosyltransferase (Nampt) affects the lineage fate determination of mesenchymal stem cells: a possible cause for reduced osteogenesis and increased adipogenesis in older individuals. J Bone Miner Res 2011;26:2656-64. 10.1002/jbmr.480 [DOI] [PubMed] [Google Scholar]
- 33.Schumacher A, Denecke B, Braunschweig T, et al. Angptl4 is upregulated under inflammatory conditions in the bone marrow of mice, expands myeloid progenitors, and accelerates reconstitution of platelets after myelosuppressive therapy. J Hematol Oncol 2015;8:64. 10.1186/s13045-015-0152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Muniesa P, Quintero P, De Andres J, et al. Hypoxia: a consequence of obesity and also a tool to treat excessive weight loss. Sleep Breath 2015;19:7-8. 10.1007/s11325-014-0972-2 [DOI] [PubMed] [Google Scholar]
- 35.Virtanen KA, Lonnroth P, Parkkola R, et al. Glucose uptake and perfusion in subcutaneous and visceral adipose tissue during insulin stimulation in nonobese and obese humans. J Clin Endocrinol Metab 2002;87:3902-10. 10.1210/jcem.87.8.8761 [DOI] [PubMed] [Google Scholar]
- 36.Genetos DC, Wong A, Watari S, et al. Hypoxia increases Annexin A2 expression in osteoblastic cells via VEGF and ERK. Bone 2010;47:1013-9. 10.1016/j.bone.2010.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]