This cross-sectional study investigates the association of surgical skin markings in dermoscopic images with the diagnostic performance of a deep learning convolutional neural network in 3 image sets of 130 melanocytic lesions each.
Key Points
Question
Are surgical skin markings in dermoscopic images associated with the diagnostic performance of a trained and validated deep learning convolutional neural network?
Findings
In this cross-sectional study of 130 skin lesions, skin markings by standard surgical ink markers were associated with a significant reduction in the specificity of a convolutional neural network by increasing the melanoma probability scores, consequently increasing the false-positive rate of benign nevi by approximately 40%.
Meaning
This study suggests that the use of surgical skin markers should be avoided in dermoscopic images intended for analysis by a convolutional neural network.
Abstract
Importance
Deep learning convolutional neural networks (CNNs) have shown a performance at the level of dermatologists in the diagnosis of melanoma. Accordingly, further exploring the potential limitations of CNN technology before broadly applying it is of special interest.
Objective
To investigate the association between gentian violet surgical skin markings in dermoscopic images and the diagnostic performance of a CNN approved for use as a medical device in the European market.
Design and Setting
A cross-sectional analysis was conducted from August 1, 2018, to November 30, 2018, using a CNN architecture trained with more than 120 000 dermoscopic images of skin neoplasms and corresponding diagnoses. The association of gentian violet skin markings in dermoscopic images with the performance of the CNN was investigated in 3 image sets of 130 melanocytic lesions each (107 benign nevi, 23 melanomas).
Exposures
The same lesions were sequentially imaged with and without the application of a gentian violet surgical skin marker and then evaluated by the CNN for their probability of being a melanoma. In addition, the markings were removed by manually cropping the dermoscopic images to focus on the melanocytic lesion.
Main Outcomes and Measures
Sensitivity, specificity, and area under the curve (AUC) of the receiver operating characteristic (ROC) curve for the CNN’s diagnostic classification in unmarked, marked, and cropped images.
Results
In all, 130 melanocytic lesions (107 benign nevi and 23 melanomas) were imaged. In unmarked lesions, the CNN achieved a sensitivity of 95.7% (95% CI, 79%-99.2%) and a specificity of 84.1% (95% CI, 76.0%-89.8%). The ROC AUC was 0.969. In marked lesions, an increase in melanoma probability scores was observed that resulted in a sensitivity of 100% (95% CI, 85.7%-100%) and a significantly reduced specificity of 45.8% (95% CI, 36.7%-55.2%, P < .001). The ROC AUC was 0.922. Cropping images led to the highest sensitivity of 100% (95% CI, 85.7%-100%), specificity of 97.2% (95% CI, 92.1%-99.0%), and ROC AUC of 0.993. Heat maps created by vanilla gradient descent backpropagation indicated that the blue markings were associated with the increased false-positive rate.
Conclusions and Relevance
This study’s findings suggest that skin markings significantly interfered with the CNN’s correct diagnosis of nevi by increasing the melanoma probability scores and consequently the false-positive rate. A predominance of skin markings in melanoma training images may have induced the CNN’s association of markings with a melanoma diagnosis. Accordingly, these findings suggest that skin markings should be avoided in dermoscopic images intended for analysis by a CNN.
Trial Registration
German Clinical Trial Register (DRKS) Identifier: DRKS00013570
Introduction
Incidence rates of malignant melanoma are increasing in many countries of the world.1 Despite much progress being made regarding public awareness, basic research, and clinical care for treating malignant melanoma, mortality rates are still high.2 Therefore, there is a continuous need for improvements in the methods for the early detection of malignant melanoma. When diagnosed early, melanoma may be cured by surgical excision, whereas the prognosis of more advanced cases is limited. In clinical routine, a high sensitivity for the detection of melanoma is of utmost importance; nevertheless, the number of excised benign nevi should be limited.3 Dermoscopy was shown to significantly improve the diagnostic sensitivity and specificity compared with that obtained by naked eye examination.4,5,6 Various dermoscopic features have been associated with the diagnoses of melanoma,7 and a number of simplified algorithms have been defined and validated to support dermatologists in deciding which lesions to excise.8,9,10
As in other fields of medicine, automated and computerized deep learning systems are emerging for the diagnosis of skin cancer.11 Deep learning is defined as a form of machine learning in which large data sets (eg, dermoscopic images) and corresponding classification labels (eg, diagnoses of nevi or melanomas) are fed into a neural network for training purposes. Within the network, which is composed of many sequential layers, input images are assessed on a pixel level for the presence of “good representations” (here, dermoscopic features) of the input classification. With the increasing number of training images, the network assembles and weights image features that are useful for differentiating nevi from melanomas. Therefore, deep learning could be described as a hierarchical feature learning. Deep learning convolutional neural networks (CNNs) form a subcategory of deep learning algorithms that have shown strong performance in image classification. To date, deep learning CNNs have demonstrated a diagnostic performance at the level of experienced physicians in the evaluation of medical images from the fields of dermatology,12,13,14 radiology,15 ophthalmology,16 and pathology.17
While a single physician with a low diagnostic performance in the detection of melanoma may cause serious harm, the effect of a broadly applied neural network with inherent “diagnostic gaps” or unknown pitfalls would be even more detrimental. In dermoscopic images, artifacts such as air bubbles, hair, or overlayed rulers have previously been reported to present some of the difficulties in automated image evaluation.11 Because suspicious lesions are often routinely marked with gentian violet surgical skin markers, our study investigated whether highlighting lesions with a skin marker may alter the evaluation scores of a computerized deep learning CNN for melanoma recognition.
Methods
This noninterventional study was approved by the ethics committee of the medical faculty of the University of Heidelberg, Heidelberg, Germany, and performed in accordance with the Declaration of Helsinki18 principles. Informed consent of patients was waived by the ethics committee because all images were acquired as part of clinical routine procedures and only deidentified data were used. The study was conducted from August 1, 2018, to November 30, 2018. A pretrained CNN architecture (Inception-v4; Google)19 was used that was additionally trained with more than 120 000 dermoscopic images and corresponding labels (Moleanalyzer-Pro; FotoFinder Systems GmbH). Details on the CNN architecture and training have been described earlier.12
For the present study, 3 image sets were created, with each including 130 melanocytic lesions (107 benign nevi and 23 melanomas). Dermoscopic images of nevi with and without skin markings were prospectively and sequentially acquired in clinical routine with a mobile digital dermatoscope attached to a smartphone (Handyscope; FotoFinder Systems GmbH). The diagnoses of benign nevi were not based on histopathologic findings but rather on the absence of any melanoma-associated clinical and dermoscopic features in combination with an uneventful follow-up over the past 2 years. Skin markings included variable dots, streaks, or circles made with a gentian violet skin marker (Devon Surgical Skin Marker; Cardinal Health or pfm medical skin marker; pfm medical ag) to the skin adjacent to the nevi. All nevi were first imaged as unmarked lesions, after which they were marked in vivo and imaged again as marked lesions (Figure 1). Melanoma images without markings were randomly selected from the image library of the Department of Dermatology, University of Heidelberg. All melanoma cases were validated by histopathologic analysis with additional information on localization, Breslow thickness, and patient data being available. To allow for corresponding analyses of melanomas, the skin markings were digitally superimposed on the melanoma images with the use of photograph manipulation software (Photoshop CS6, version 13.0.1 x32; Adobe Inc). For a statistical comparison, 20 nevi from the test set were used to demonstrate that electronically superimposed markings provide comparable results to in vivo markings. In 20 unmarked benign nevi, the CNN’s mean melanoma probability score was 0.15 (95% CI, 0.01-0.29). Melanoma probability scores can range from 0 to 1; higher scores indicate a higher probability of the measured lesion being a melanoma. In vivo markings increased the mean score to 0.52 (95% CI, 0.31-0.74), whereas electronically superimposed markings led to a comparable mean score of 0.59 (95% CI, 0.39-0.79). The Mann-Whitney test did not reveal a significant difference between in vivo and electronically marked nevi (P = .78). Moreover, in each of the 20 nevi, the CNN classification of in vivo and electronically marked lesions showed consistent results. For more details, refer to the eMethods and eFigures 1 and 2 in the Supplement. All dermoscopic images were then cropped to reduce the background and to focus solely on the melanocytic lesions. The aforementioned steps resulted in 3 complete sets of the same 130 dermoscopic images, namely, set 1 with unmarked lesions, set 2 with marked lesions, and set 3 with cropped images.
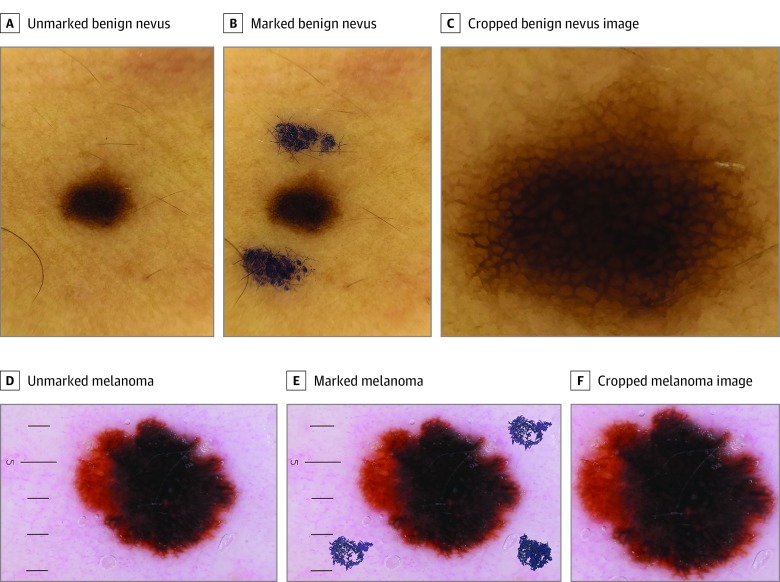
Figure 1. Convolutional Neural Network (CNN) Classification and Melanoma Probability Scores for Dermoscopic Images of Unmarked, Marked, and Cropped Benign Nevus and Melanoma.
A gentian violet surgical skin marker was used to highlight the marked examples. A, CNN classification: benign; score, 0.001. B, CNN classification: malignant; score, 0.981. C, CNN classification: benign; score, 0.001. D, CNN classification: malignant; score, 0.999. E, CNN classification: malignant; score, 0.999. F, CNN classification: malignant; score, 0.999.
Heat Maps
Deep learning CNNs do not provide any information about why a certain classification decision was reached. There are many different interpretability approaches that may help to more clearly visualize the information “learned” by the model.20
Heat maps were created to identify the most important pixels for the CNN’s diagnosis to better explain how much each pixel of the image contributes to the diagnostic classification. These heat maps were derived by vanilla (meaning “basic”) gradient descent backpropagation.21
Statistical Analysis
The primary outcome measures were sensitivity, specificity, and area under the curve (AUC) of receiver operating characteristic (ROC) curves for the diagnostic classification of lesions by the CNN. The CNN accorded a malignancy probability score between 0 and 1, and a validated a priori cutoff greater than 0.5 for the dichotomous classification of malignant vs benign lesions was applied. Descriptive statistical measures, such as frequency, mean, range, and SD, were used. Mann-Whitney tests were performed to assess the differences in the melanoma probability scores between the 3 sets of images. A 2-sample McNemar test was performed to compare the sensitivities and specificities attained by the CNN.22 Results were considered statistically significant at the P < .05 level (2-sided). All analyses were carried out using SPSS version 24 (IBM).
Results
Characteristics of Imaged Lesions
In all, 130 melanocytic lesions (107 benign nevi and 23 melanomas) were imaged. Of the 23 imaged melanomas, 18 (78.3%) were localized on the trunk and extremities, 3 (13.0%) on the facial skin, 1 (4.3%) on the scalp, and 1 (4.3%) on the palmoplantar skin (eTable in the Supplement). Nineteen melanomas (82.6%) were invasive (mean thickness, 1 mm [range, 0.2-5.6 mm]) and 4 (17.4%) in situ. The analysis of melanoma subtypes revealed the following subtypes: 15 superficial spreading melanomas, 2 lentigo maligna melanomas, 1 nodular melanoma, and 1 acrolentiginous melanoma. Of the 4 in situ melanomas, 1 was classified as lentigo maligna (eTable in the Supplement). The 123 imaged benign nevi showed no clinical or dermoscopic criteria associated with the presence of melanoma and had an uneventful follow-up for at least 2 years (Figure 1).
CNN’s Melanoma Probability Scores
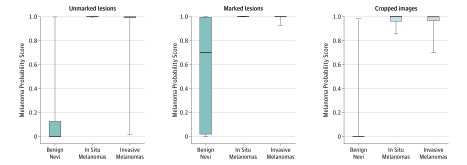
Box plots in Figure 2 show the distribution of the CNN melanoma probability scores for the 3 different sets of images (unmarked, marked, and cropped). Skin markings significantly increased the mean melanoma probability scores of the classifier in benign nevi from 0.16 (95% CI, 0.10-0.22) to 0.54 (95% CI, 0.46-0.62) (P < .001). Figure 3 and Figure 4 show heat maps of representative unmarked and marked nevi in which the most important pixels for the CNN’s diagnostic classifications were identified by vanilla gradient descent backpropagation.21 In nevi images that were cropped to reduce the background, the mean melanoma probability scores were significantly reduced to 0.03 (95% CI, 0-0.06) compared with those in unmarked (0.16; 95% CI, 0.10-0.22) and marked (0.54; 95% CI, 0.46-0.62) images (P < .001). In melanoma images we also observed an increase of the mean melanoma probability scores in unmarked vs electronically marked images from 0.94 (95% CI, 0.85-1.00) to 1.00 (95% CI, 0.99-1.00). However, as unmarked melanoma images already showed mean scores close to the maximum score of 1, the induced changes did not reach statistical significance (P = .10). Irrespective of markups or cropping, the statistical differences in melanoma probability scores between benign nevi vs melanomas remained significant across all image sets. At the same time, no significant difference was observed between the melanoma probability scores of in situ melanomas vs invasive melanomas across all image sets.
Figure 2. Box Plots Representing Convolutional Neural Network (CNN)’s Melanoma Probability Scores for Benign Nevi, In Situ Melanomas, and Invasive Melanomas.
Probability scores are presented for unmarked lesions, lesions marked by a gentian violet ink surgical pen, and cropped lesion images. Probability scores range from 0 to 1; scores closer to 1 indicate a higher probability of melanoma. The top and bottom borders of the boxes indicate the 75th and 25th percentiles, respectively, while the horizontal line in the box represents the median. The whiskers indicate the full range of the probability scores. Statistical analyses revealed significantly different melanoma probability scores when comparing benign lesions with in situ or invasive melanomas (P < .001).
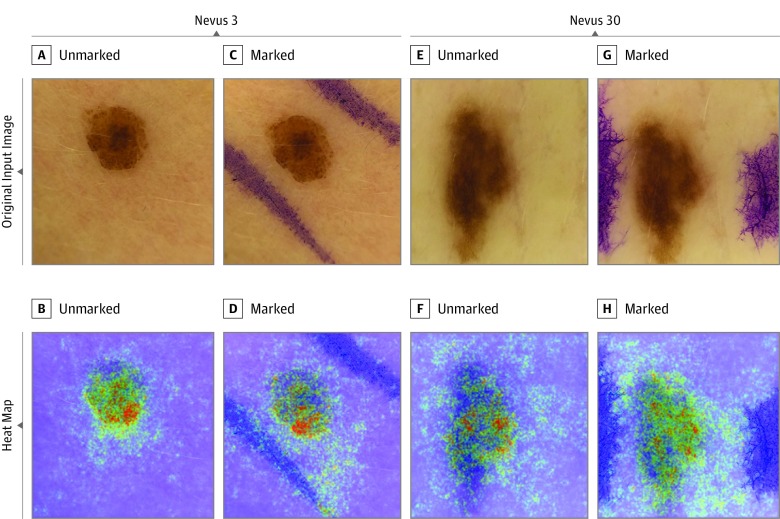
Figure 3. Heat Maps of 2 Benign Nevi With Unchanged Melanoma Probability Scores After Addition of In Vivo Skin Markings.
The heat maps were created by vanilla (meaning basic) gradient descent backpropagation. A and E, Unmarked input images. B and F, Heat maps reveal relevant pixels for the convolutional neural network’s (CNN’s) prediction of benign nevi. C and G, Marked input images. D and H, Heat maps reveal that skin markings are “ignored” by the CNN, thus leaving the CNN’s prediction of benign nevi unchanged.
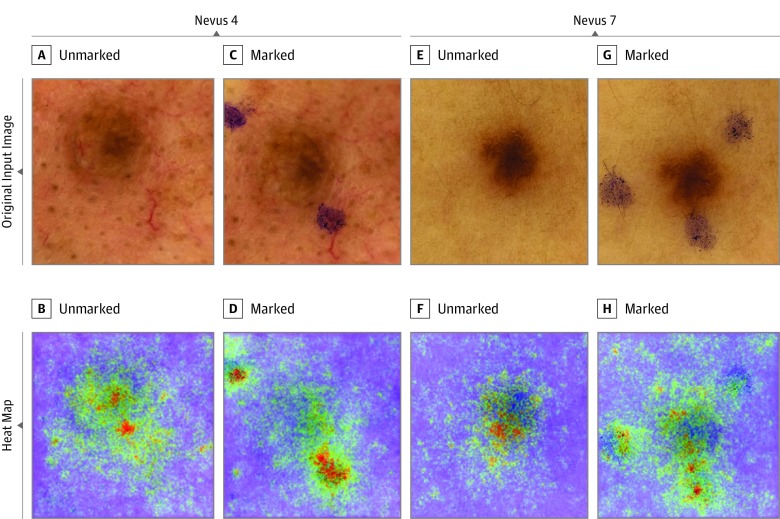
Figure 4. Heat Maps of 2 Benign Nevi With Major Increase in Melanoma Probability Scores After Addition of In Vivo Skin Markings.
The heat maps were created by vanilla (meaning basic) gradient descent backpropagation. A and E, Unmarked input images. B and F, Heat maps reveal relevant pixels for the convolutional neural network’s (CNN’s) prediction of benign nevi. C and G, Marked input images. D and H, Heat maps reveal that skin markings are of high relevance for CNN’s changed prediction of malignant melanomas, while the nevus itself is mostly ignored.
CNN’s Sensitivity, Specificity, and ROC AUC
At the a priori operation point of 0.5, the sensitivity of the CNN in the unmarked image set was 95.7% (95% CI, 79%-99.2%) and the specificity was 84.1% (95% CI, 76%-89.8%). When lesions were marked, the sensitivity changed to 100% (95% CI, 85.7%-100%) and the specificity to 45.8% (95% CI, 36.7%-55.2%). In cropped images, the CNN showed a sensitivity of 100% (95% CI, 85.7%-100%) and a specificity of 97.2% (95% CI, 92.1%-99%). A pairwise comparison of the CNN’s sensitivities in unmarked, marked, or cropped images revealed no significant differences. A pairwise comparison of the specificities showed significant differences between unmarked and marked images (84.1%; 95% CI, 76.0%-89.8% vs 45.8%; 95% CI, 36.7%-55.2%; P < .001), unmarked and cropped images (84.1%; 95% CI, 76.0%-89.8% vs 97.2%; 95% CI, 92.1%-99.0%; P = .003), and marked and cropped images (45.8%; 95% CI, 36.7%-55.2% vs 97.2%; 95% CI, 92.1%-99.0%; P < .001).
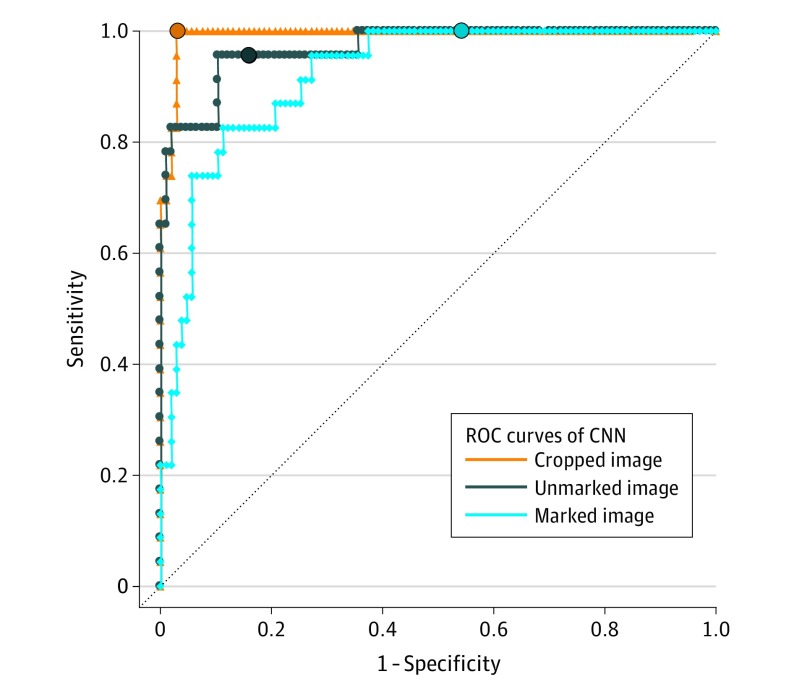
The ROC AUC in unmarked images was 0.969 (95% CI, 0.935-1.000), in marked images was 0.922 (95% CI, 0.871-0.973), and in cropped images was 0.993 (95% CI, 0.984-1.000). All 3 ROC curves that were calculated for the 3 image sets are depicted in Figure 5 and illustrate a significant reduction in specificity of nearly 40% in marked vs unmarked lesions as well as the outperformance of the CNN when using cropped lesions.
Figure 5. Receiver Operating Characteristic (ROC) Curves of the Performance of the Convolutional Neural Network (CNN) Diagnostic Classification .
Shown are ROC curves of the CNN in the original unmarked image set vs marked and cropped images (dichotomous classification). The sensitivity and specificity of the CNN for each image set at the a priori operation point are indicated by a circle on the curve and were as follows: (1) unmarked images: sensitivity, 95.7%; specificity, 84.1%; area under the curve (AUC), 0.969; (2) marked images: sensitivity, 100%; specificity, 45.8%; AUC, 0.922; and (3) cropped images: sensitivity, 100%; specificity, 97.2%; AUC, 0.993.
Discussion
Deep learning CNNs have recently been applied to different diagnostic tasks in medical image recognition and classification (eg, ophthalmology,16 radiology,15 histopathology,17 and dermatology23). Several landmark studies compared human and machine accuracy in skin cancer detection.24,25 Two recent publications reported an expert dermatologist-level classification of dermoscopic images of benign melanocytic nevi and cutaneous melanomas,12,13 and a first deep learning CNN for classification of skin neoplasms has gained market access in Europe as a medical device (Moleanalyzer-Pro). While these achievements represent major successes, further exploring the limitations of deep learning CNNs is important before considering a broader application worldwide.
It has previously been shown that artifacts in dermoscopic images, such as dark corners (caused by viewing through the tubular lens of the dermatoscope), gel bubbles, superimposed color charts, overlayed rulers, and occluding hair, may impede image segmentation and classification by automated algorithms.11,26 Various methods have been reported for the removal of such artifacts,27,28 and strategies for preprocessing of images were described to improve the classification outcomes of CNNs.29 However, the removal of artifacts by image preprocessing may ultimately alter the original image and itself be prone to error. Therefore, a major advantage of deep learning CNNs is that the raw RGB dermoscopic image may be used as an input, thus bypassing preprocessing.30
This study investigated the possible association of surgical skin markers as artifacts in dermoscopic images with the classification outcomes by a deep learning CNN. In clinical routine, suspicious lesions are frequently marked before being excised or photographed. Our attention was drawn to this issue when evaluating dermoscopic images of benign nevi under sequential digital dermoscopy follow-up. We observed that sequentially imaged benign nevi, although largely unchanged, were frequently labeled as being malignant by the CNN when ink markers were visible at the periphery of the dermoscopic image. To systematically and prospectively investigate our observation, 3 sets of dermoscopic images (unmarked, marked, and cropped) of the same 130 melanocytic lesions were created. Our assessments of these images with the CNN showed that skin markings at the periphery of benign nevi were associated with an increase in the melanoma probability scores that increased the false-positive rate by approximately 40%. To prove that this association may be attributed solely to the dermoscopic background and not the melanocytic lesion itself, the dermoscopic images were cropped manually. This procedure reversed the negative association of skin markings with the diagnostic performance of the CNN. Overall, image preprocessing by manually cropping images led to the best diagnostic performance of the CNN, achieving a sensitivity of 100%, specificity of 97.2%, and ROC AUC of 0.993. The CNN’s specificity in the cropped images (97.2%) was significantly improved compared with that in the unmarked images (84.1%). However, cropping was done manually by experienced dermatologists, and the results may deteriorate with automated cropping by a formal preprocessing step using border segmentation algorithms.
When reviewing the open-access International Skin Imaging Collaboration database, which is a source of training images for research groups, we found that a similar percentage of melanomas (52 of 2169 [2.4%]) and nevi (214 of 9303 [2.3%]) carry skin markings. Nevertheless, it seems conceivable that either an imbalance in the distribution of skin markings in thousands of other training images that were used in the CNN tested herein or the assignment of higher weights to blue markings only in lesions with specific (though unknown) accompanying features may induce a CNN to associate skin markings with the diagnosis of melanoma. The latter hypothesis may also explain why melanoma probability scores remained almost unchanged in many marked nevi while being increased in others.
The fact that blue markings are associated with changes in melanoma probability scores while the underlying mechanisms remain unclear highlights the lack of transparency in the classification process of neural network models. Thus, although not being dependent on manmade criteria for classification has opened a new level of performance, it may impede the insights into a mechanistic understanding. The CNN tested in this study applies the melanoma probability score as a softmax output classifier. Recently, content-based image retrieval has been shown to provide results comparable to softmax classifiers.14 In this alternative approach, the CNN generates several images that are visually similar to the input image along with the corresponding diagnoses. The displayed output images are retrieved from the compiled training images based on overlapping features identified by the neural network. This strategy has been hypothesized to increase the explainability for clinicians.
There are several approaches to the problem of bias induced by skin markings. Avoiding markings in images that are intended for analysis seems the most straightforward solution for the CNN tested in our study. Avoiding markings in training images (eg, by cropping images before training) is logical with regard to future algorithms. In contrast, teaching the CNN to ignore parts of the image that may or may not be artificial skin markings appears rather difficult. Because there are many more types of artifacts in images other than blue surgical skin markers, some artifacts may still be undetected. At the same time, other parts of images may erroneously be interpreted as artifacts that preclude them from analysis by the CNN. Moreover, as stated above, automated segmentation with border detection of the lesion of interest may be another option to improve evaluation.27
Limitations
Our study has some limitations. First, benign melanocytic nevi were not excised for histologic verification, but rather were selected from patients under follow-up and showed no changes during the past 2 years. Second, dermoscopic images of melanomas were extracted from a validated database; thus, skin markings could not be added in vivo. Alternatively, skin markings were electronically duplicated from digital images and superimposed on the melanoma background. This procedure and its association with changes in the classification by the CNN were extensively tested with images of benign nevi. In all these cases, no differences were found between the melanoma probability scores attained with the CNN in images with “in vivo” markings vs images with electronically superimposed markings. Third, most images included in this study were derived from fair-skinned patients residing in Germany; therefore, the findings may not be generalized for lesions of patients with other skin types and genetic backgrounds.
Conclusions
In summary, the results of our investigation suggest that skin markings at the periphery of dermoscopic images are significantly associated with the classification results of a deep learning CNN. Melanoma probability scores of benign nevi appear to be significantly increased by markings causing a strong increase in the false-positive rate. In clinical routine, these lesions may have been sent for unnecessary excisions. Therefore, we recommend to avoid skin markings in dermoscopic images intended for analysis by a deep learning CNN.
eMethods. Supplemental Methods
eFigure 1. Technical Approach of Creating Electronically Superimposed Markings in Melanoma Images
eFigure 2. Technical Approach for Validating the Comparability of Electronically Superimposed and In Vivo Markings
eTable. Characteristics of Melanomas Including Localization, Invasiveness, and Histopathology
References
- 1.Arnold M, Holterhues C, Hollestein LM, et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. J Eur Acad Dermatol Venereol. 2014;28(9):1170-1178. doi: 10.1111/jdv.12236 [DOI] [PubMed] [Google Scholar]
- 2.Shellenberger R, Nabhan M, Kakaraparthi S. Melanoma screening: a plan for improving early detection. Ann Med. 2016;48(3):142-148. doi: 10.3109/07853890.2016.1145795 [DOI] [PubMed] [Google Scholar]
- 3.Argenziano G, Cerroni L, Zalaudek I, et al. Accuracy in melanoma detection: a 10-year multicenter survey. J Am Acad Dermatol. 2012;67(1):54-59. doi: 10.1016/j.jaad.2011.07.019 [DOI] [PubMed] [Google Scholar]
- 4.Bafounta M-L, Beauchet A, Aegerter P, Saiag P. Is dermoscopy (epiluminescence microscopy) useful for the diagnosis of melanoma? results of a meta-analysis using techniques adapted to the evaluation of diagnostic tests. Arch Dermatol. 2001;137(10):1343-1350. doi: 10.1001/archderm.137.10.1343 [DOI] [PubMed] [Google Scholar]
- 5.Vestergaard ME, Macaskill P, Holt PE, Menzies SW. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol. 2008;159(3):669-676. doi: 10.1111/j.1365-2133.2008.08713 [DOI] [PubMed] [Google Scholar]
- 6.Kittler H, Pehamberger H, Wolff K, Binder M. Diagnostic accuracy of dermoscopy. Lancet Oncol. 2002;3(3):159-165. doi: 10.1016/S1470-2045(02)00679-4 [DOI] [PubMed] [Google Scholar]
- 7.Pehamberger H, Steiner A, Wolff K. In vivo epiluminescence microscopy of pigmented skin lesions, I: pattern analysis of pigmented skin lesions. J Am Acad Dermatol. 1987;17(4):571-583. doi: 10.1016/S0190-9622(87)70239-4 [DOI] [PubMed] [Google Scholar]
- 8.Stolz W. ABCD rule of dermatoscopy: a new practical method for early recognition of malignant melanoma. Eur J Dermatol. 1994;4:521-527. [Google Scholar]
- 9.Menzies SW, Ingvar C, Crotty KA, McCarthy WH. Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features. Arch Dermatol. 1996;132(10):1178-1182. doi: 10.1001/archderm.1996.03890340038007 [DOI] [PubMed] [Google Scholar]
- 10.Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol. 1998;134(12):1563-1570. doi: 10.1001/archderm.134.12.1563 [DOI] [PubMed] [Google Scholar]
- 11.Okur E, Turkan M. A survey on automated melanoma detection. Eng Appl Artif Intell. 2018;73:50-67. doi: 10.1016/j.engappai.2018.04.028 [DOI] [Google Scholar]
- 12.Haenssle HA, Fink C, Schneiderbauer R, et al. ; Reader Study Level-I and Level-II Groups . Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29(8):1836-1842. doi: 10.1093/annonc/mdy166 [DOI] [PubMed] [Google Scholar]
- 13.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115-118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tschandl P, Argenziano G, Razmara M, Yap J. Diagnostic accuracy of content-based dermatoscopic image retrieval with deep classification features [published online September 12, 2018]. Br J Dermatol. doi: 10.1111/bjd.17189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611-629. doi: 10.1007/s13244-018-0639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du X-L, Li W-B, Hu B-J. Application of artificial intelligence in ophthalmology. Int J Ophthalmol. 2018;11(9):1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Binder A Comparison of deep learning architectures for H&E histopathology images. Paper presented at: IEEE Conference on Big Data and Analytics (ICBDA); November 16, 2017; Kuching, Malaysia. https://ieeexplore.ieee.org/document/8284105. Accessed November 7, 2018.
- 18.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 19.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. Paper presented at: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); June 8, 2015; Boston, MA. https://arxiv.org/pdf/1409.4842.pdf. Accessed November 7, 2018.
- 20.Montavon G, Samek W, Müller K-R. Methods for interpreting and understanding deep neural networks. Digit Signal Process. 2018;73:1-15. doi: 10.1016/j.dsp.2017.10.011 [DOI] [Google Scholar]
- 21.Simonyan K, Vedaldi A, Zisserman A Deep inside convolutional networks: visualising image classification models and saliency maps. https://arxiv.org/abs/1312.6034. Published December 20, 2013. Accessed April 5, 2019.
- 22.Xiang JX. On two-sample McNemar test. J Biopharm Stat. 2016;26(2):217-226. doi: 10.1080/10543406.2014.1000548 [DOI] [PubMed] [Google Scholar]
- 23.Brinker TJ, Hekler A, Utikal JS, et al. Skin cancer classification using convolutional neural networks: systematic review. J Med Internet Res. 2018;20(10):e11936. doi: 10.2196/11936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchetti MA, Codella NCF, Dusza SW, et al. ; International Skin Imaging Collaboration . Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. 2018;78(2):270-277.e1. doi: 10.1016/j.jaad.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. 2018;138(7):1529-1538. doi: 10.1016/j.jid.2018.01.028 [DOI] [PubMed] [Google Scholar]
- 26.Mishra NK, Celebi ME An overview of melanoma detection in dermoscopy images using image processing and machine learning. https://arxiv.org/ftp/arxiv/papers/1601/1601.07843.pdf. Published January 27, 2016. Accessed November 7, 2018.
- 27.Jafari MH, Karimi N, Nasr-Esfahani E, et al. Skin lesion segmentation in clinical images using deep learning. Paper presented at: 2016 23rd International Conference on Pattern Recognition (ICPR); December 4, 2016; Cancun, Mexico. https://ieeexplore.ieee.org/document/7899656. Published April 24, 2017. Accessed November 7, 2018.
- 28.Salido JAA, Ruiz C Jr. Using deep learning to detect melanoma in dermoscopy images. Int J Mach Learn Comput. 2018;8(1):61-68. [Google Scholar]
- 29.Yoshida T, Celebi ME, Schaefer G, Iyatomi H Simple and effective pre-processing for automated melanoma discrimination based on cytological findings. Paper presented at: IEEE International Conference on Big Data; December 6, 2016; Washington, DC. https://ieeexplore.ieee.org/document/7841005. Published February 6, 2017. Accessed November 7, 2018.
- 30.Sultana NN, Puhan N Recent deep learning methods for melanoma detection: a review. In: Ghosh D, Giri D, Mohapatra R, Savas E, Sakurai K, Singh L, eds. Mathematics and Computing: ICMC 2018: Communications in Computer and Information Science Singapore: Springer; 2018;834:118-132. https://www.researchgate.net/publication/324502578_Recent_Deep_Learning_Methods_for_Melanoma_Detection_A_Review. Published April 2018. Accessed November 7, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Supplemental Methods
eFigure 1. Technical Approach of Creating Electronically Superimposed Markings in Melanoma Images
eFigure 2. Technical Approach for Validating the Comparability of Electronically Superimposed and In Vivo Markings
eTable. Characteristics of Melanomas Including Localization, Invasiveness, and Histopathology