Abstract
CRISPR-Cas9-Cytidine deaminase fusion enzymes—termed “base editors”—allow targeted editing of genomic deoxycytidine to deoxythymidine (C:G→T:A) without the need for double-stranded break induction. Base editors represent a paradigm shift in gene editing technology due to their unprecedented efficiency to mediate targeted, single-base conversion. However, current analysis of base editing outcomes rely on methods that are either imprecise or expensive and time-consuming. To overcome these limitations, we developed a simple, cost-effective, and accurate program to measure base editing efficiency from fluorescence-based Sanger sequencing, termed “EditR.” We provide EditR as a free online tool or downloadable desktop application requiring a single Sanger sequencing file and guide RNA sequence. EditR is more accurate than enzymatic assays, and provides added insight to the position, type, and efficiency of base editing. Furthermore, EditR is likely amenable to quantify base editing from the recently developed adenosine deaminase base editors that act on either DNA (adenosine deaminase base editors [ABEs]) or RNA (REPAIRs) (catalyzes A:T→G:C). Collectively, we demonstrate that EditR is a robust, inexpensive tool that will facilitate the broad application of base editing technology, thereby fostering further innovation in this burgeoning field.
Introduction
Recently, several research groups have developed Cas9-Cytidine deaminase fusion enzymes for the purpose of gene editing with single base resolution.1–4 These base editors rely on the programmable specificity of the Cas9-guide RNA (gRNA) complex to localize a mutagenic cytidine deaminase enzyme to produce targeted deoxycytidine to deoxyuridine (C→U) mutations. Through DNA replication, deoxyuridine behaves like deoxythymidine, resulting in C→T mutations (antisense G→A). By leveraging disparate outcomes in DNA repair, some base editors preferentially induce C→T mutations (target mutations), while others were developed for random mutagenesis (non-target) of C→T, G, or A (antisense G→A, C, or T). The single nucleotide level resolution of base editing shows promise in gene therapy,5 agricultural engineering,6,7 and basic scientific research.8,9 Employing base editing in any laboratory setting requires the ability to quantify the efficiency, precision, accuracy, and reproducibility of base editing. Demonstrably, all work published on base editing to date includes a quantitative assessment of base editing efficiency.1–13
Of the several approaches used to measure base editing efficiency, all are limited by imprecision, high cost, or extended turnaround time. Rapid and cost-effective approaches for measuring base editing consist of enzymatic cleavage assays such as the Cel I, T7E, Surveyor, or Guide-it Resolvase assays.14,15 However, these assays are unable to discern the exact position and type of mutation because they only detect the presence of a mismatch bubble formed in heteroduplexes of stochastically annealed DNA.16 This approach is suboptimal for base editing where adjacent Cs may be edited or non-target C→T, G, or A mutations may occur,3–7,17 neither of which can be distinguished by enzymatic mismatch cleavage assay. As an alternative, bacterial colony sequencing of subcloned polymerase chain reaction (PCR) amplicons can elucidate the specific outcomes of base editing,2,7 but this is a time-consuming, laborious, and costly approach, making it impractical for medium- to high-throughput research. In comparison, the most informative method to measure base editing is next-generation deep sequencing (NGS) of the edited site.3,4,10,13 However, this is the most expensive and time-consuming method while also requiring bioinformatics expertise.
In the analysis of insertion-deletion (indels) mutations from CRISPR-Cas9* editing, bioinformatic approaches using fluorescent capillary Sanger sequencing provide rapid and affordable methods to measure and characterize editing efficiency, most notably with the free web tools Tracking of Indels by DEcomposition (TIDE; https://tide.nki.nl/) and Poly Peak Parser (http://yosttools.genetics.utah.edu/PolyPeakParser/).18,19 These programs analyze secondary Sanger sequencing traces to delineate the composition and frequency of indel mutations and have greatly reduced barriers by efficiently and accurately quantifying the outcomes of CRISPR-Cas9 gene editing. Inspired by these programs, we developed an accurate, fast, and low-cost method for the identification and quantification of base editing from fluorescent Sanger sequencing data. We provide this program, EditR (Edit deconvolution by inference of traces in R) as a free web tool (baseEditR.com) or an open-source R Shiny application that can run on a local desktop. EditR requires only a single Sanger sequencing file of a base-edited sample and the sequence of the gRNA protospacer to disentangle the outcomes of base editing.
Materials and Methods
Plasmids and gRNA design
The identity of all plasmids in this study was confirmed by Sanger sequencing and restriction enzyme digestion. All base editing was carried out using pCMV-BE3 developed by Dr. David Liu's Lab (Addgene # 73021).1 BE3 was the first published base editor and has arguably the most comprehensive examination of activity in vitro and in cell culture. Guide RNAs (gRNAs) for use with BE3 were designed to target the loci of interest using parameters outlined in previous publications, including size of the editing window, identity of preceding base, distance from the protospacer adjacent motif (PAM), and PAM specificity (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/crispr).1 gRNAs were ordered as complementary oligonucleotides: 5′-CACCG-protospacer-3′ and 5′-AAAC-reverse complement protospacer-C-3′ (Integrated DNA Technologies [IDT]). Complementary oligonucleotides were annealed and phosphorylated with T4 PNK (NEB) and 10 × T4 ligation buffer (NEB) in a thermocycler using the protocol: 30 min at 37°C, 5 min at 95°C, and step down to 25°C at 5°C/min. pENTR221-U6 stuffer vector was digested with BsmBI restriction enzyme, FastAP alkaline phosphatase (Fermentas), and 10 × Tango Buffer overnight at 37°C. Linearized pENTR221-U6 and 1:200 diluted annealed and phosphorylated oligonucleotides were ligated together with T4 DNA ligase and buffer (NEB) at room temperature for ≥1 h. Ligation reactions were transformed into DH10β Escherichia coli (Thermo Fisher Scientific) and grown on LB agar plates. Single colonies were chosen and cultured overnight after which plasmid DNA was extracted using a GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific). Plasmid identity was dually confirmed with HindIII-Hifi and PvuII-Hifi (NEB) restriction digest gel electrophoresis and Sanger sequencing of gRNA region (ACGT, Inc.). Confirmed plasmids were re-transformed, and plasmid DNA was extracted with a HiSpeed Plasmid Maxi Kit (Qiagen).
Cell line culturing and transfection
Cell lines were maintained at 37°C, 5% CO2, under 80% confluency and passaged 1:10 three times per week. HCT116 cells were maintained in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific), and human osteosarcoma (HOS) cells were maintained in Eagle's Minimum Essential Medium (ATCC). All cell culturing media were supplemented with 10% fetal bovine serum and 1× penicillin-streptomycin. Puromycin selection was performed using media containing 1 μg/mL of puromycin. HCT116 and HOS cells ≤80% confluent were electroporated using 1 μg of pENTR221-gRNA, 1 μg of pCMV-BE3, and 500 ng of pmaxGFP (Lonza) according to the manufacturer's protocol (Neon Transfection System, Life Technologies), and plated onto a polylysine-coated six-well plate. Twenty-four hours post electroporation, percent green fluorescent protein positive (GFP+) cells were observed to assess transfection efficiency qualitatively, and genomic DNA was isolated from cells harvested 72 h post electroporation.
Co-transposition and single colony isolation
Co-transposition was performed via electroporation of an additional 500 ng of PB-CG-Luciferase-EGFP (Puro) PiggyBac transposon and 500 ng of hyperactive PiggyBac transposase, as previously described,15 alongside the aforementioned pCMV-BE3 and pENTR221-gRNA plasmids. In principle, cells that obtain a transposition event integrating the puromycin resistance gene are also more likely to have taken up Cas9/BE3 and gRNA expression plasmids and are thus more likely to be edited. Twenty-four hours post electroporation, percent GFP+ cells were observed to assess transfection qualitatively, and genomic DNA was harvested from half of the cells 72 h post electroporation. The remaining cells were plated with puromycin supplemented media for single colony isolation in a 15 cm polylysine-coated dish or serially diluted on a 96-well-plate. Single colonies on 15 cm plates were allowed to grow for 14 days or until visible to the naked eye, and were isolated with colony isolators and Trypsin-EDTA (Thermo Fisher Scientific) or picked with a 10 μL pipette tip of Trypsin-EDTA and transferred to a 24-well dish. Once clones reached >90% confluency, genomic DNA was harvested to assess editing.
Surveyor nuclease assay
Primers were designed to produce amplicons approximately 300–400 bp in length, with the target site off-centered in the amplicon. Genomic DNA was PCR amplified with AccuPrime Taq DNA Polymerase, high fidelity (Invitrogen), 10× Accuprime buffer, and 5% dimethyl sulfoxide, and electrophoresed through a 1% agarose gel and gel extracted (QiaQuick Gel Extraction Kit; Qiagen) or PCR purified (PCR Purification Kit; Qiagen). PCR products were denatured and annealed in a thermocycler using the manufacturer's protocol (IDT). Three microliters of denatured PCR products were combined with 1 μL of 1× AccuPrime buffer II (Thermo Fisher Scientific), 0.7 μL of surveyor nuclease, and 0.7 μL of surveyor enhancer (IDT) before being incubated at 42°C for 20 min. Reactions were terminated with Ficoll loading dye and run on an agarose gel (2% m/v, 0.06 μL/mL ethidium bromide) in TAE buffer or a polyacrylamide gel in TBE buffer. Gel was imaged, and the fraction of amplicons edited was quantified in ImageJ with the formula FEdited = (b +c)/(a + b + c), where a is the integrated intensity of the undigested PCR band and b and c are the integrated intensities of each digested product band, as previously described.14
Sanger sequencing
Purified PCR product (1 ng/μL), primer (20 pmol/μL), and Big Dye Terminator v3.1 (4 μL) were brought to 12 μL in molecular H2O and sequenced using the protocol: 1 min at 95°C (30 s at 95°C, 30 s at 56°C, and 1 min at 60°C) × 24, and hold at 16°C. Sequencing reactions were analyzed on an Applied Biosystems 3730 DNA Analyzer.
Illumina NGS
Primers were designed using Primer3 and Primer-BLAST to 300–500 bp regions of interest, with Nextera universal adaptors flanking the site-specific primer (Supplementary Table S1). Genomic DNA was PCR amplified in one step using AccuPrime Taq DNA Polymerase, high fidelity, according to the manufacturer's protocol (Invitrogen). Samples were submitted to the University of Minnesota Genomics Center for subsequent amplification with indexed primers and sequencing on a MiSeq 2 × 300 bp run (Illumina). A minimum of 1,000 read-pairs were generated per sample.
Sequencing reads were demultiplexed using bcl2fastq2 (Illumina). FastQC v0.11.520 was used to assess the quality of the data. Overlapping read-pairs were assembled with Pear v0.9.10.21 Non-overlapping read-pairs and read-pairs with an assembled length 5 bp longer or shorter than the length of the amplicon reference sequence were discarded. Needle (EMBOSS v6.5.7)22 was used to generate optimal global sequence alignments between each assembled read and the amplicon reference sequence. The numbers of insertions, deletions, and substitutions at each base of the reference gRNA protospacer sequence were counted. Alignments of the 20 most common amplicon reads were visualized using MView v1.52.23
EditR software development
To determine if the measured percent editing was significant, we implemented a null hypothesis significance testing approach using a null distribution modeled from the background noise. The null distribution is generated by trimming the first 20 bases of the sequence and removing the 20 bases of the protospacer. Additionally, bases that fall within the 10th percentile of total area are removed, as small peaks are associated with poor initial primer binding and poor end extension.24 To account for the variability in sequencing, the user can manually select the region to model the null distribution in case the default trimming does not effectively remove low-quality sequencing. Next, the value of every “N” trace fluorescence under every non-“N” basecall (e.g., T fluorescence under A, C, or G peaks) is compiled to generate a sample of the noise distribution. The sample of the noise distribution for each base is fitted to a zero-adjusted gamma distribution (zΓ; Supplementary Fig. S1) using the package gamlss.25 We chose the zΓ distribution for three reasons: (1) it has a domain from 0 to +∞, (2) it is a continuous distribution allowing for non-integer values, and (3) it allows for a high proportion of zeros in the data, which accounted for 10% of the values in our data (Supplementary Fig. S1).25 Filliben's correlation coefficient (RF2) is calculated to assess the goodness of fit of the model given the data, where RF2 = 1 is a perfect fit. From this model, we can assign critical values using a default level of significance (α = 0.01), which the user can manually change on EditR's interface.
EditR was written in the R statistical programming environment v3.4.0. EditR requires a sample AB1 Sanger sequencing file (i.e., cells treated with base editor and gRNA) and a 15–24 nt character string of the edited region of interest (i.e., gRNA protospacer). Initial parameters for the program have set defaults that can be adjusted by the user under the advanced settings if desired. The EditR web app was written with R Shiny v1.0.1 and helped by incorporating design from TIDE and Poly Peak Parser.18,19 The former identifies simple indel mixtures from Sanger sequencing data, while the latter calculates the frequency and composition of complex indel mixtures.
The sample file is uploaded and read into EditR. The fluorescence area of all four bases at each base call is assigned, as measured by the software provided by the capillary electrophoretic instrument manufacturer and determined by the makeBaseCalls function of sangerseqR. The percent area of each base is calculated by dividing the total area of the focal base by the area of all the bases summed together. The guide sequence is then aligned to the primary sequence generated from the base calls using the ends-free overlap alignment algorithm in pairwiseAlignment() with type = “overlap” argument from the Biostrings package.26 Ends-free alignment was chosen, as it aligned to a local match while also being robust to changes in the first base of the guide, as well multiple base changes in the middle of the guide.
Results
EditR Workflow
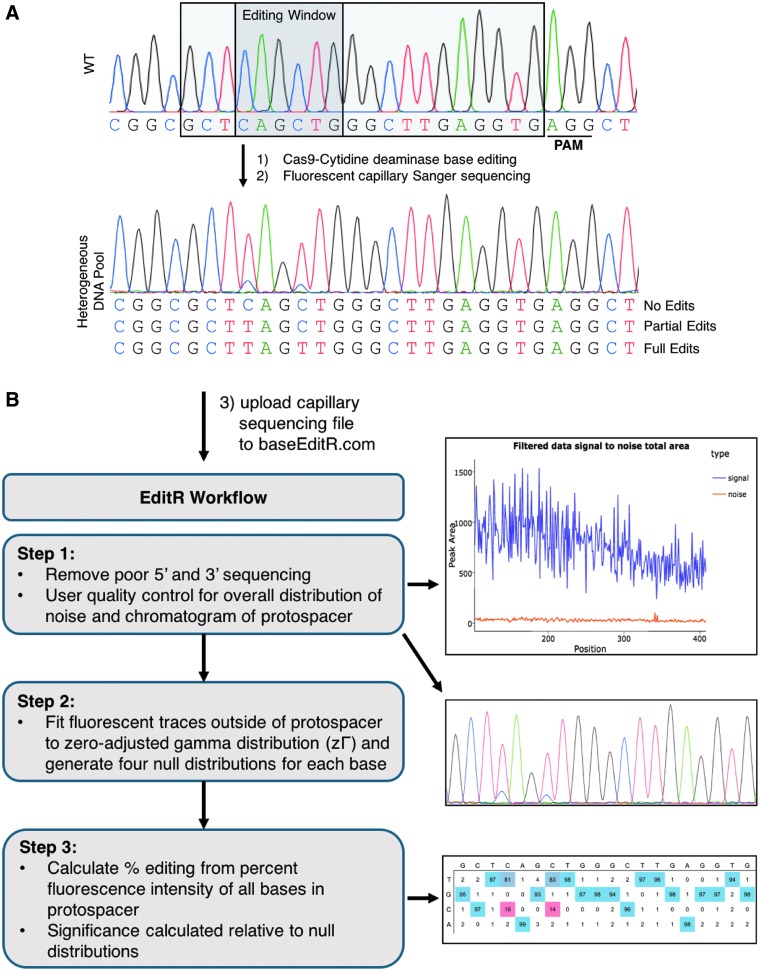
To analyze the mutation frequency, spectrum, and significance of BE3-treated cells, a 400–800 bp region encompassing the edited site is PCR amplified and sequenced by standard dideoxynucleotide chain termination based capillary electrophoresis (Sanger method). DNA isolated from BE3- and gRNA-treated cells with significant editing should demonstrate polymorphisms under C bases (antisense G) within the base editing window (∼5 bp of the protospacer with BE3 for example; Fig. 1A). Generally, these base edits are C→T (antisense G→A). However, there are several documented instances of non-target base editing (i.e., C→G or A), including our work here.2–4,10
FIG. 1.
Analysis of base editing by capillary Sanger sequencing trace quantification. (A) Following treatment with base editor and guide RNA (gRNA), Cs (antisense Gs) within the editing window are converted to Ts (antisense As), producing a heterogeneous population of edited cells. (B) Workflow of EditR steps with summary plots. (1) The first and last portions of the file are removed due to poor quality. The signal–noise plot allows users to visualize the amount of fluorescence at each basecall that is deemed signal (purple) versus noise (orange). A chromatogram of the protospacer is also produced for users to validate their results qualitatively. (2) A zero-adjusted gamma distribution is fit to the percent area noise of each trace (A, C, G, and T) to generate four null distribution to which the traces in the hypothesized sites of editing (i.e., the protospacer) are compared. (3) Percent composition of traces measured to be significantly different from noise are plotted in a colored heat map proportional to magnitude (red is low, blue is high).
EditR generates a graphic of the percent noise across the sequencing file, allowing the user to assess the sequencing quality (Fig. 1B, Step 1). If low-quality regions are not filtered out by default settings, users can modify the region used to generate the null distribution. A chromatogram of the protospacer is generated to determine if the gRNA is properly aligned to the sequencing file and to visualize if the predicted editing matches qualitative expectations (Fig. 1B, Step 1). The sequence traces within this region are compared to the traces in the rest of the sequencing file to quantify and determine the significance of base editing. EditR decomposes the trace at each basecall position into the percent fluorescence contribution of each of the four bases; A,C, G, and T. The value of each percent “N” fluorescence at every “non-N” basecall is used to model a zΓ distribution, resulting in one zΓ distribution for each nucleotide. From these zΓ distributions, a critical value is calculated, as determined by the level of significance, which serves as the threshold for calling an edit within the protospacer as significant (Fig. 1, Step 2, and Supplementary Fig. S1B). Percent editing is then calculated for traces within the protospacer that are above this threshold, the output of which is a heat-mapped table to visualize percent editing across the protospacer (Fig. 1B, Step 3). The pzΓ-value in this context is the probability of calling a fluorescent peak a significant edit when in fact that peak was merely noise rather than a base edit. On the EditR web app, users can download a report of the results and a summary of the operations performed on their data.
In vitro validation of EditR
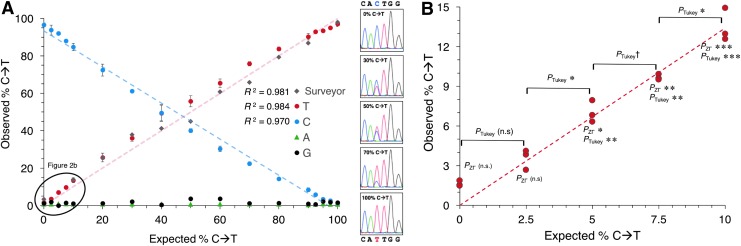
To determine if quantitative Sanger sequencing can accurately measure base editing under simulated conditions, we mixed together a WT PCR product with a fully edited PCR product containing a single C→T mutation. In three separate trials across multiple amplicons, samples were mixed in titrated amounts from 0% to 100% and subjected to capillary Sanger sequencing (Fig. 2A and Supplementary Figs. S2 and S3). The calculated percent C→T agreed well with the actual concentration of PCR products by measuring either C or T in all trials (R2 = 0.984, 0.979, and 0.970). As an additional analysis of our data, we performed a pairwise t-test to compare the observed and expected values of the titrations. Although we found that the observed and expected values were significantly different (p < 2.2 × 10–16, df = 491; Supplementary Fig. S5) with an average difference of −1.9% (95% confidence interval [CI] −2.2% to −1.6%; Supplementary Figure S4B–D), this difference is marginal when considering the mean ± 2SD, where 95% of observed values are expected to differ between −8.2% and 4.4% from the actual values (Supplementary Figure S4D).
FIG. 2.
In vitro validation of EditR as an accurate and sensitive method. (A) EditR data presented as the mean of independent triplicates ± 1 standard deviation. See Supplementary Figs. S2 and S3 for an additional titration series. Surveyor assay data presented as the mean of triplicate measurement ± 1 standard error of gel fluorescence densitometry, as previously described.15 When the expected editing rate is >50%, the observed editing is calculated as 100%–calculated editing, as the T-containing product shifted from the minor to major PCR product. See Supplementary Fig. S2 for gel image. (B) EditR can detect 2.5% differences in C→T down to 2.5% C→T. Individual red dots represent a replicate. †p < 0.05; *p < 0.01; **p < 0.001; ***p < 0.0001; n.s., not significant.
As a comparison to an alternative method of measuring base editing, titrations were also subjected to the Surveyor nuclease assay and quantified with fluorescence gel densitometry (Fig. 2A and Supplementary Figure S5). The calculated percent editing as calculated by the surveyor assay agreed well with the actual concentration of the PCR products (R2 = 0.981), showing that EditR is as accurate as the surveyor assay in measuring base editing efficiency (Fig. 2A).
To determine the precision and sensitivity of EditR, we performed statistical tests between differing titrations. Analysis of variance (ANOVA) with post hoc Tukey's HSD test of each titration compared to the WT titration (0% C→T) showed that titrations could be measured as significantly different from WT as low as 2.5% C→T (p < 0.01; Fig. 2B and Supplementary Figs. S2 and S3). One-way ANOVA followed by Tukey's HSD post hoc test demonstrated that triplicate samples could resolve incremental differences as small as 2.5% increments down to but not past 2.5% C→T (p < 0.05; Fig. 2B). By comparison, the EditR zΓ significance testing was able to resolve C→T editing from background noise down to 5.0% (p < 0.01; Fig. 2B). These results were mirrored with the percent C area at the 95% C→T end of the spectrum (Supplementary Figs. S2 and S3). Furthermore, EditR can distinguish 2.5% differences surrounding 50% editing (Supplementary Fig. S2B and C), even when two bases are edited. Collectively, these data demonstrates EditR is a sensitive and precise method for discerning and measuring even low-level mutations generated in base-edited cells.
Application of EditR to base-edited cells with target mutations (C→T)
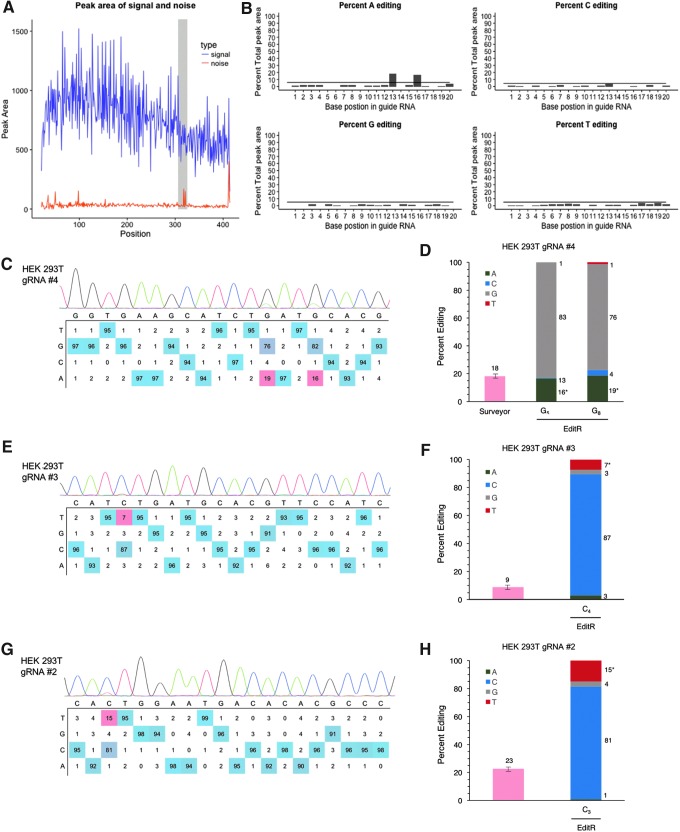
To assess the functionality of EditR in base-edited cells, we treated HEK 293T cells with pCMV-BE3 and pENTR221-U6-gRNA. As expected, PCR amplification and capillary Sanger sequencing of the target site demonstrated noisy initial sequencing followed by a several hundred base-pair span with a high percent signal (S/[S + N] ≥ 0.9; Fig. 3A). Informatively, the quality control plot generated by EditR showed two “noise” peaks within the highlighted protospacer region, which the editing quadplot (four-paneled graphic with percent base composition by each position) confirmed to be from base editing of C→T (antisense G→A editing; Fig. 3B). Percent editing as calculated by EditR was consistent with percent editing, as measured by the surveyor assay across three different targets, while in contrast to the surveyor assay, EditR was also able to distinguish the position and type of mutation (Fig. 3C–H). Importantly, EditR was also able to determine the discrete editing efficiency in a multiply base-edited sample (Fig. 3C) and measure editing as low as 7% (pzΓ < 0.01; Fig. 3C and E). These data show that EditR is able to measure target C→T and G→A mutations in base-edited cells.
FIG. 3.
Validation of EditR in base-edited cells with target mutations. (A) Output graphic showing the distribution of signal and noise in the sequencing file with peaks in the gRNA region. (B) Output plots of traces by base identity and significance. (C) Output editing table color-coded by proportionality showing editing of G→A at two bases. (D) Comparison of EditR to Surveyor assay, height of bars is the mean of triplicate measurement ± 1 standard error of gel fluorescence densitometry, as previously described.25 WT negative control not shown (0% editing). See Supplementary Fig. S2 for gel image. (E–H) Additional EditR-generated table plots of base-edited samples with target mutations.
Application of EditR to base-edited cells with non-target mutations (C→G or A)
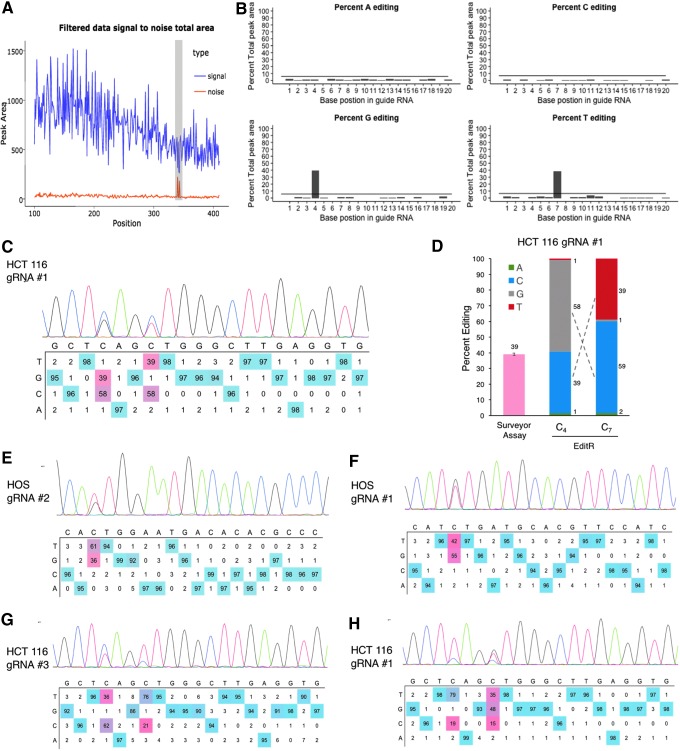
To assess the functionality of EditR in measuring the frequency of non-target mutations, which are regularly seen with the base editor BE3,10 we treated HOS and HCT116 cell lines with BE3 and gRNA using our previously published enrichment method that selects for highly edited cells.15 Sanger sequencing of cells treated with gRNA #1 was confirmed to be of high quality (S/[S + N] ≥ 97.5%; Fig. 4A) and demonstrated around 40% base editing of Cs at positions 4 and 7, with C4 exhibiting a non-target C→G mutation and C7 exhibiting a target C→T mutation (Fig. 4B and C). The surveyor assay yielded an editing efficiency of 39%, which was similar to the 39% of C4 and 39% of C7. Because the percent G at C4 was nearly identical to the percent C at C7, it is suggestive that C4–T7 are linked together and account for around 40% of the allelic pool, while G4–C7 are linked, accounting for the approximately 60% remainder. Further use of EditR shows its ability to resolve complex mixtures of non-target mutations in base-edited cells across multiple cell lines and target sites (Fig. 4E–H). This demonstrates EditR can measure the editing efficiency of non-target mutations while having the advantage over the surveyor assay in elucidating the discrete composition of non-target mutations.
FIG. 4.
Validation of EditR in base-edited cells with non-target mutations. (A) Output graphic showing the distribution of signal and noise in the sequencing file with peaks in the gRNA region. (B) Output plots of traces by base identity and significance. (C) Output editing table color-coded by proportionality showing significant C→T and C→G mutations. (D) Comparison of EditR to Surveyor assay, height of bars is the mean ± 1 standard deviation of fluorescence densitometry from independent surveyor assays. Similarity in the percent editing suggests linked mutations. See Supplementary Fig. S2 for gel image. (E–H) Additional EditR-generated table plots of base-edited samples with target mutations.
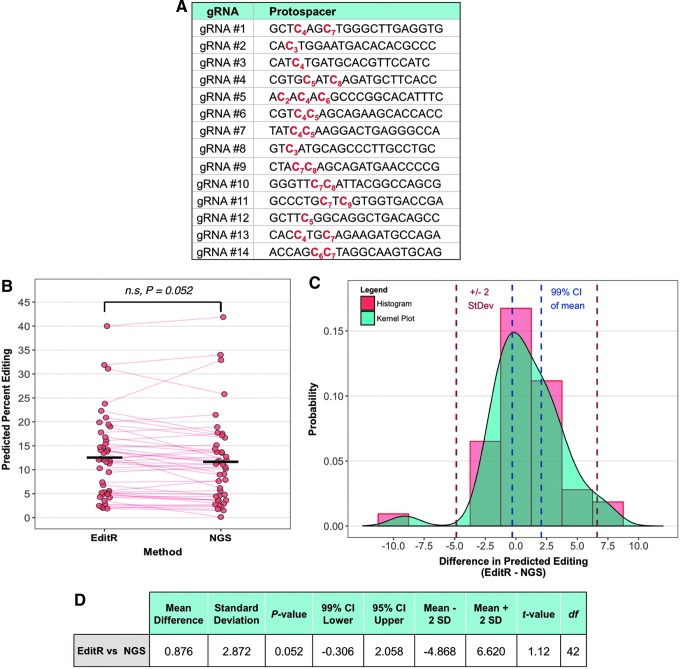
Comparison of EditR to NGS
To assess potential trade-offs of the ease of using EditR against the accuracy of its measurements, and to assess the accuracy of EditR in multiple sequence contexts, we compared EditR to NGS, which is the gold-standard for measuring base editing.1,3,4,10,17 HEK 293T cells were treated with BE3 and 14 different gRNAs that targeted one of nine unique genomic sites (Fig. 5A and Supplementary Table S1). Genomic DNA was harvested from treated cells, PCR amplified for the edited region of interest, and amplicons were concurrently Sanger sequenced and deep sequenced to compare EditR to NGS directly. EditR yielded measurements of base editing that were not significantly different from NGS by paired t-test (p = 0.052, df = 42; Fig. 5B and D), with an average difference of 0.9% (99% CI −0.6% to 2.1%; Fig. 5C and D) and standard deviation of 2.9%. Furthermore, samples were confirmed by NGS to possess non-target mutations spanning the spectrum of C→T, A, or G (Supplementary Fig. S6). While the non-significance of this difference is borderline with respect to a level of significance at α = 0.05, even if the difference between EditR and NGS were statistically significant, the implications of this difference would be marginal, given the small 99% confidence interval of the mean (Fig. 5D). This demonstrates that EditR is a robust method for measuring target and non-target base editing outcomes in diverse sequence contexts.
FIG. 5.
Validation of EditR compared to next-generation deep sequencing (NGS) across multiple guide sites. (A) Base editing guides used to compare EditR to NGS. (B) Comparison of measured editing by EditR and NGS. Solid black bars denote the mean of each group. Red lines drawn between points indicate which samples are paired. (C) Distribution of differences between EditR and NGS. Positive values indicate editing measured by EditR was larger. (D) Table summary of paired t-test performed on data. N = 43, comprised of 8 amplicons, 14 unique guides, 26 unique bases, and 21 unique edits with one or two independent replicates per guide. A single unique base may produce multiple unique edits via non-target editing, that is, one unique C edited to T, G, or A.
Discussion
Advantages of EditR
Cas9-Cytidine deaminase base editors are a new but rapidly expanding technology, with potential applications spanning the biomedical sciences. Notable recent advances in base editing also include the development of Cas9-adenosine deaminase base editors (ABEs) that edit A:T→G:C in DNA27 and Cas13-adenosine deaminase base editors (REPAIRs) that edit A→I in RNA.28 The rapidly expanding versatility of base editing is astronomical, requiring an equally adaptable method to analyze base editing outcomes. Here, we show that the Surveyor nuclease assay can accurately measure base editing mutations. However, it is unable to resolve the composition and position of base editing. While there are several other methods available to measure base editing efficiency, all suffer from poor accuracy or high costs, hindering access to base editing research.
Given the high requirement of resources needed to measure base editing accurately, this creates an accessibility barrier in base editing research. As an alternative, we developed EditR as a rapid, accurate, and inexpensive approach to measuring base editing efficiency. EditR takes advantage of the proportional change in the percent area of trace fluorescence as bases are edited. This percent area is compared to the background distribution of percent fluorescence noise to determine if significant editing is occurring. EditR enables researchers both to quantify base editing by position and to assess the composition of mutations at a particular base. EditR is a fraction of the cost of NGS, with results possible within a day. As such, EditR is a viable supplement or even alternative to NGS when an inexpensive and rapid analysis is desired, such as when identifying gRNAs with highest activity, or when screening cell populations for frequency of specific outcomes.
Comparison to other programs for base editing research
The resolution and accuracy of EditR are equal to that of other programs that quantify nucleotide polymorphisms (SNP) from Sanger sequencing such as QSVanalyser (http://dna.leeds.ac.uk/qsv/) and Mutation Surveyor® (http://www.softgenetics.com/mutationSurveyor.php; >5% resolution).29,30 While these programs are highly useful for analyzing discrete SNP or copy number variants, they are less suitable for base editing research. The algorithms of Mutation Surveyor® and QSVanalyser both rely on adjacent peaks as a reference to the base of interest when measuring editing efficiency.29 For example, QSVanalyser compares the intensity of the base of interest to the heights of the peaks between 5 and 10 bases upstream of the base of interest to measure the percentage of the minor SNP. This referencing method is powerful when looking at discrete single point mutations, but it is less amenable to base editing, as base editors are processive enzymes that will edit adjacent cytidines within the editing window. This issue is especially relevant when considering new generations of base editors, some of which have editing windows as large as 14 nucleotides.13 EditR overcomes these issues by comparing the trace within the protospacer against the background distribution of noise outside of the protospacer instead of adjacent peaks. Furthermore, EditR is accessible and intuitive as a free web application, or as open-source code that can be run locally as an R Shiny app on any major operating system.
Limitations of EditR
EditR is largely limited by the quality of the Sanger sequencing results, because EditR measures base editing by determining if trace fluorescence is due to editing or noise. As such, the baseline noise of chromatograms restricts the ability to detect edits of around 5% or more (Fig. 2B and Supplementary Fig. S1). Similarly, even a monoallelic sequence will not be called as 100%, given there will be some proportion of noise subtracting from calling a base as pure. To account for this, we recommend gel extracting or purifying PCR products with a commercial DNA isolation kit prior to sequencing. We advise using traces that have an average percent noise of ≤7.25% and modeled parameter μ of ≤2.5, as that is strongly correlated with EditR calling significance at p < 0.01 (Supplementary Fig. S7A and B). Furthermore, it is important that the zΓ models are properly fit in order to have sensitive detection of base editing. Thus, we recommend only using sequencing files with an RF2 of ≥0.95, as we found the vast majority of our chromatograms fall in this range (Supplementary Fig. S7C). As a note, even in files with a large proportion of noise, RF2 was still >0.9, showing that even in noisy samples the zΓ distribution effectively models the noise distribution (Supplementary Fig. S7D).
In considering the precision of EditR, we expect that 95% of samples analyzed via EditR will not deviate more than −4.7% to +6.6% (M ± 2SD; 0 ± ∼5.7%) from the percent editing as measured by NGS (Fig. 5B and D). This range of the precision of EditR is further reinforced by the pairwise analysis of the titration experiments (M ± 2SD; −8.2% to +4.4%; Supplementary Fig. S4B and D). Furthermore, the precision of EditR is similar to that of TIDE (M ± 2SD = −5.4% to +4.2%; Supplementary Fig. S8), which further supports the reliability and utility of using Sanger-based methods for quantifying gene editing events. To assess what may cause EditR to deviate from NGS values, future work needs to address how local sequence contexts may alter percent fluorescence area.
In fluorescent Sanger sequencing, the identity of the preceding base can affect the intensity of the subsequent base, but it is unclear how certain sequence contexts may affect calculations of editing efficiency.29,32 For example, EditR may be unable to measure base editing accurately in certain sequence contexts such as repetitive G-rich reads.29 Observationally, we have noticed that the height of any peak following a G appears to be less predictable than other motifs (e.g., TT motifs appear to have more consistent heights). This may be slightly problematic when measuring the first exon of protein coding genes, as these exons tend to be slightly more GC rich than subsequent exons (Supplementary Fig. S9 and Supplementary Script S1). Therefore, when choosing to sequence with either the forward or reverse primer, if possible, we recommend sequencing the strand that does not have a G immediately upstream of the base of interest. Future work will address which motifs are most reliably measured by EditR and develop algorithms that account for the local sequence context to quantify base editing more accurately.
Future applications of EditR
Here, we used the base editor BE3 as the basis of our work. However, we expect EditR could also measure the editing efficiency of the recently developed ABEs.27 We expect EditR will handle base edits produced by ABEs identically to how EditR handles base edits produced by cytidine deaminase base editors such as BE3, as ABEs edit in the reverse direction of BE3, that is, BE3 edits C:G→T:A, while ABEs edit to A:T→G:C. Therefore, the titration analyses (Fig. 2A and B and Supplementary Figs. S2A–E, S3, and S4) and comparisons to NGS (Fig. 5A–D) performed here are likely directly applicable to measuring ABE editing. Future work will examine the ability of EditR to measure ABE base editing, as well as any subsequent base editors. Ultimately, EditR is a resource-saving tool equipped to improve accessibility to the burgeoning field of base editing.
Supplementary Material
Acknowledgments
Thank you to Arianna Wegley and Nick Slipek for the helpful suggestions on the manuscript, Leah Hogdal for beta testing the web application, and Yaniv Brandvain for conversations around developing the statistical testing approach. This research was funded by the Sobiech Osteosarcoma Fund Award, The Jimmy V Foundation, and the Children's Cancer Research Fund. EditR is available at baseEditR.com/ and https://github.com/MoriarityLab/EditR7
Author Disclosure Statement
All authors declare no competing financial or non-financial interests.
Clustered Regularly Interspaced Short Palindromic Repeats.
References
- 1.Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;61:5985–5991. DOI: 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016;102:553–563. DOI: 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Zhang J, Yin W, et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 2016;13:1–9. DOI: 10.1038/nmeth.4027 [DOI] [PubMed] [Google Scholar]
- 4.Hess GT, Frésard L, Han K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 2016;13:1036–1042. DOI: 10.1038/nmeth.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim K, Ryu S-M, Kim S-T, et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol 2017;35:435–437. DOI: 10.1038/nbt.3816 [DOI] [PubMed] [Google Scholar]
- 6.Zong Y, Wang Y, Li C, et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Publ Gr 2017;35:438–440. DOI: 10.1038/nbt.3811 [DOI] [PubMed] [Google Scholar]
- 7.Shimatani Z, Kashojiya S, Takayama M, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 2017;35:441–443. DOI: 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- 8.Kuscu C, Parlak M, Tufan T, et al. CRISPR-STOP: gene silencing through nonsense mutations. Nat Publ Gr 2017;14:2–6. DOI: 10.1038/nmeth.4327 [DOI] [PubMed] [Google Scholar]
- 9.Billon P, Bryant EE, Joseph SA, et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell 2017;67:1068–1079.e4. DOI: 10.1016/j.molcel.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 2017;3eaao4774. DOI: 10.1126/sciadv.aao4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Sun Y, Du J, Zhao Y, et al. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 2016;1–4 DOI: 10.1016/j.molp.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 12.Kim D, Lim K, Kim S-T, et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol 2017;35:475–480. DOI: 10.1038/nbt.3852 [DOI] [PubMed] [Google Scholar]
- 13.Kim YB, Komor AC, Levy JM, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 2017;35:371–376. DOI: 10.1038/nbt.3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, Hsu PDP, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–2308. DOI: 10.1038/nprot.2013.143.Genome [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriarity BS, Rahrmann EP, Beckmann DA, et al. Simple and efficient methods for enrichment and isolation of endonuclease modified cells. PLoS One 2014;9:e9611–4.. DOI: 10.1371/journal.pone.0096114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Till BJ, Burtner C, Comai L, et al. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res 2004;32:2632–2641. DOI: 10.1093/nar/gkh599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keiji N, Takayuki A, Nozomu Y, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016;353:1248–1256. DOI: 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- 18.Brinkman EK, Chen T, Amendola M, et al. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014;42:1–8. DOI: 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill JT, Demarest BL, Bisgrove BW, et al. Poly peak parser: method and software for identification of unknown indels using Sanger sequencing of polymerase chain reaction products. Dev Dyn 2014;243:1632–1636. DOI: 10.1002/dvdy.24183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews S. FastQC: a quality control tool for high throughput sequence data. Available online at www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed June24, 2017)
- 21.Stamatakis A, Zhang J, Kobert K. Genome analysis PEAR: a fast and accurate Illumina Paired-End reAd mergeR. 2014;30:614–620. DOI: 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 2000;16:276–277 [DOI] [PubMed] [Google Scholar]
- 23.Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment. Bioinformatics 1998;14:380–381 [DOI] [PubMed] [Google Scholar]
- 24.Thornley DJ. Analysis of Trace Data from Fluorescence Based Sanger Sequencing [dissertation]. London: University of London, Imperial College of Science, Technology, and Medicine; 1997. [Google Scholar]
- 25.Rigby B, Stasinopoulos M. GAMLSS package: A flexible regression approach. Available online at https://cran.r-project.org/web/packages/gamlss/gamlss.pdf (accessed June24, 2017)
- 26.Aboyoun P, Gentleman R, Debroy S, Rmpi E. Package “Biostrings.” Available online at www.bioconductor.org/packages/3.7/bioc/manuals/Biostrings/man/Biostrings.pdf (accessed June24, 2017)
- 27.Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017;551:464–471. DOI: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science 2017;358:1019–1027. DOI: 10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr IM, Robinson JI, Dimitriou R, et al. Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics 2009;25:3244–3250. DOI: 10.1093/bioinformatics/btp583 [DOI] [PubMed] [Google Scholar]
- 30.Manion M, Ni S, Hulce D, Liu CSJ. Sanger sequencing traces with Mutation Surveyor software. Available online at https://softgenetics.com/mutationSurveyor.php (accessed June24, 2017)
- 31.Naue J, Sänger T, Schmidt U, et al. Factors affecting the detection and quantification of mitochondrial point heteroplasmy using Sanger sequencing and SNaPshot minisequencing. Int J Legal Med 2011;125:427–436. DOI: 10.1007/s00414-011-0549-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.