Japanese encephalitis virus (JEV) infection triggers endoplasmic reticulum (ER) stress and neuron apoptosis. ER stress sensor protein kinase R-like endoplasmic reticulum kinase (PERK) has been reported to induce apoptosis under acute or prolonged ER stress. However, whether the PERK pathway of ER stress response plays important roles in JEV-induced apoptosis and encephalitis remains unknown. Here, we found that JEV infection activates ER stress sensor PERK in neuronal cells and mouse brains. PERK activation induces apoptosis via the PERK-ATF4-CHOP apoptosis pathway upon JEV infection. Among the JEV proteins prM, E, NS1, NS2A, NS2B, and NS4B, only NS4B activates PERK. Moreover, activated PERK participates in apoptosis and encephalitis induced by JEV and NS4B. These findings provide a novel therapeutic approach for JEV-caused encephalitis.
KEYWORDS: apoptosis, ER stress, Japanese encephalitis virus, NS4B, PERK
ABSTRACT
Accumulated evidence demonstrates that Japanese encephalitis virus (JEV) infection triggers endoplasmic reticulum (ER) stress and neuron apoptosis. ER stress sensor protein kinase R-like endoplasmic reticulum kinase (PERK) has been reported to induce apoptosis under acute or prolonged ER stress. However, the precise role of PERK in JEV-induced apoptosis and encephalitis remains unknown. Here, we report that JEV infection activates the PERK-ATF4-CHOP apoptosis pathway both in vitro and in vivo. PERK activation also promotes the formation of stress granule, which in turn represses JEV-induced apoptosis. However, PERK inhibitor reduces apoptosis, indicating that JEV-activated PERK predominantly induces apoptosis via the PERK-ATF4-CHOP apoptosis pathway. Among JEV proteins that have been reported to induce ER stress, only JEV NS4B can induce PERK activation. PERK has been reported to form an active molecule by dimerization. The coimmunoprecipitation assay shows that NS4B interacts with PERK. Moreover, glycerol gradient centrifugation shows that NS4B induces PERK dimerization. Both the LIG-FHA and the LIG-WD40 domains within NS4B are required to induce PERK dimerization, suggesting that JEV NS4B pulls two PERK molecules together by simultaneously interacting with them via different motifs. PERK deactivation reduces brain cell damage and encephalitis during JEV infection. Furthermore, expression of JEV NS4B is sufficient to induce encephalitis via PERK in mice, indicating that JEV activates PERK primarily via its NS4B to cause encephalitis. Taken together, our findings provide a novel insight into JEV-caused encephalitis.
IMPORTANCE Japanese encephalitis virus (JEV) infection triggers endoplasmic reticulum (ER) stress and neuron apoptosis. ER stress sensor protein kinase R-like endoplasmic reticulum kinase (PERK) has been reported to induce apoptosis under acute or prolonged ER stress. However, whether the PERK pathway of ER stress response plays important roles in JEV-induced apoptosis and encephalitis remains unknown. Here, we found that JEV infection activates ER stress sensor PERK in neuronal cells and mouse brains. PERK activation induces apoptosis via the PERK-ATF4-CHOP apoptosis pathway upon JEV infection. Among the JEV proteins prM, E, NS1, NS2A, NS2B, and NS4B, only NS4B activates PERK. Moreover, activated PERK participates in apoptosis and encephalitis induced by JEV and NS4B. These findings provide a novel therapeutic approach for JEV-caused encephalitis.
INTRODUCTION
Endoplasmic reticulum (ER) is a cellular membrane organelle involved in protein synthesis, folding and secretion. Many factors, such as virus infection, ischemia, hypoxia, heat shock, and increased protein synthesis, impair the ER functions, resulting in ER stress. ER stress leads to the accumulation of unfolded and misfolded proteins in the ER, which is sensed by three ER transmembrane sensors: R-like endoplasmic reticulum kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1). These three sensors are maintained in inactive states through interactions with ER resident chaperone glucose regulated protein 78 (GRP78). When unfolded or misfolded proteins accumulate in the ER lumen, GRP78 dissociates from these three transducers, resulting in their activation and initiation of unfolded protein response (UPR). UPR functions to alleviate ER stress by reducing protein synthesis, inducing ER chaperones and activating ER-associated degradation. However, once the stress exceeds the capacity of the ER homeostatic machinery, cells undergo apoptosis (1).
PERK is an ER-resident transmembrane kinase. Under ER stress, it dissociates from GRP78, undergoes oligomerization and autophosphorylation to form active PERK. Active PERK phosphorylates eukaryotic translation initiation factor 2 (eIF2α) to reduce global protein synthesis, thus relieving ER stress. In addition, PERK selectively increases the translation of activating transcription factor-4 (ATF4) to facilitate cell survival (2). If ER stress exceeds the capacity of ER homeostatic machinery, activated PERK-eIF2α-ATF4 pathway promotes apoptosis by inducing the expression of CHOP (3).
Japanese encephalitis virus (JEV) is a positive-strand RNA virus belonging to the family of Flaviviridae. Its genome encodes three structural proteins (C, prM/M, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). JEV causes severe disease with encephalitis in humans and animals (4). Accumulating evidence indicates that JEV infection results in ER stress and neuron apoptosis (4–8). We have previously reported that the IRE1/JNK branch of ER stress response is involved in JEV-induced apoptosis in BHK-21 cells (6). However, whether the PERK pathway plays a role in JEV-induced apoptosis remains unknown. In this study, we found that JEV infection induces the activation of ER stress sensor PERK in neuronal cells and mouse brains. Moreover, PERK participates in neuron apoptosis and encephalitis induced by JEV via its NS4B.
RESULTS
JEV infection in vitro and in vivo activates PERK.
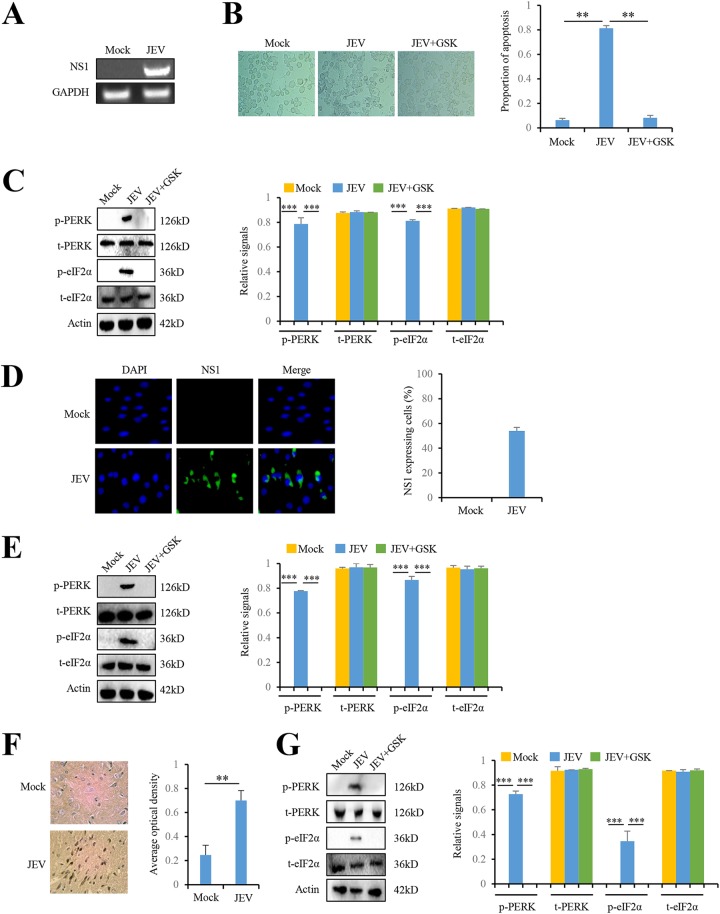
To assess whether JEV infection in vitro activated the PERK/eIF2α pathway, we analyzed the levels of phosphorylated PERK and eIF2α in JEV-infected Neuro-2a cells and BHK-21 cells. Viral NS1 RNA was detected in JEV-infected Neuro-2a cells (Fig. 1A), indicating the successful JEV infection in Neuro-2a cells. JEV infection induced significant cytopathy in Neuro-2a cells compared to mock-infected cells (Fig. 1B). JEV infection did not change the total levels of PERK and eIF2α, but significantly increased the levels of phosphorylated PERK and eIF2α in Neuro-2a cells (Fig. 1C). When JEV-infected Neuro-2a cells were treated with PERK inhibitor, GSK2606414, the phosphorylation levels of PERK and eIF2α were significantly reduced (Fig. 1C). We also investigated the effect of JEV infection on the PERK/eIF2α pathway in BHK-21 cells. JEV infection in BHK-21 cells was confirmed by immunofluorescence using JEV NS1 antibody (Fig. 1D). Consistent with the findings in Neuro-2a cells, JEV infection induced the phosphorylation of PERK and eIF2α in BHK-21 cells and PERK inhibitor GSK2606414 reduced JEV-induced phosphorylation of PERK and eIF2α (Fig. 1E). In Fig. 1B, we also noted that PERK inhibitor GSK2606414 reduced the cytopathy induced by JEV infection in Neuro-2a cells (Fig. 1B), implying that PERK could play a role in JEV-induced cytopathy.
FIG 1.
JEV activates the PERK pathway both in vitro and in vivo. (A to C) JEV infection activates PERK in Neuro-2a cells. Neuro-2a cells were infected with P3 strain at a multiplicity of infection (MOI) of 5. Mock-infected Neuro-2a cells were used as controls. After 3 days, the intracellular JEV NS1 RNA was determined by RT-PCR (A), and the cytopathic effect was determined by microscopy (B). Intracellular proteins, including PERK, phospho-PERK, eIF2α, phospho-eIF2α, and actin, were determined by immunoblot analysis (C). (D and E) JEV infection activates PERK in BHK-21 cells. BHK-21cells were infected with P3 strain at an MOI of 5. Mock-infected BHK-21 cells were used as controls. After 3 days, the cells were subjected to immunofluorescence using JEV NS1 antibody (D) and immunoblot analysis using PERK, phospho-PERK, eIF2α, phospho-eIF2α, and actin antibodies (E). (F and G) JEV infection activates PERK in mouse brains. The tissues from JEV-infected and mock-infected mice brains at 3 days postinfection were collected for IHC using antibody against JEV NS1 protein (F) and immunoblot analysis using antibodies against PERK, phospho-PERK, eIF2α, phospho-eIF2α, and actin (G). The left panels (B to G) show representative images; the right panels (B to G) show the quantitation data. Error bars indicate standard deviations (SD) of the means (n = 3). **, P < 0.01; ***, P < 0.001.
To assess whether JEV infection in vivo activates the PERK/eIF2α pathway, we analyzed the levels of phosphorylated PERK and eIF2α in JEV-infected mouse brains. We collected brain tissues from JEV-infected mice that showed clinical central nervous system symptoms such as paralysis. No neurological symptom was observed in mock-infected controls as expected. JEV NS1 protein was detected in the brain of JEV-infected mouse by immunohistochemistry (IHC) (Fig. 1F), confirming JEV infection in mouse brain. JEV infection did not change the total levels of PERK and eIF2α but increased the levels of phosphorylated PERK and eIF2α in mouse brains (Fig. 1G). Moreover, PERK inhibitor GSK2606414 reduced the phosphorylation of PERK and eIF2α induced by JEV, albeit without the effect on the levels of PERK and eIF2α protein in mouse brains (Fig. 1G). Collectively, these data indicate that JEV infection in vitro and in vivo activates the PERK/eIF2α pathway.
JEV infection induces apoptosis by activating the PERK-ATF4-CHOP pathway.
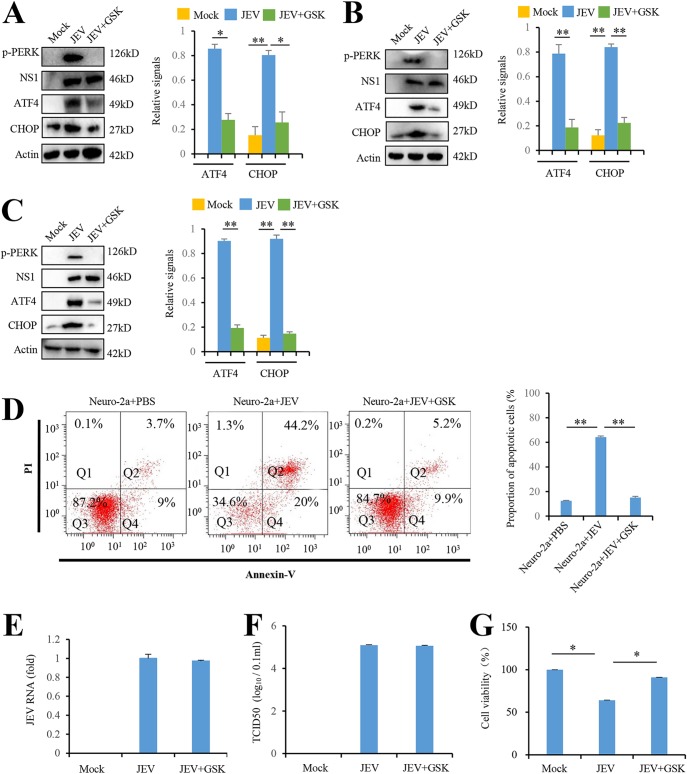
PERK has been reported to induce apoptosis by activating its downstream ATF4-CHOP pathway (3). Since JEV activates PERK and PERK inhibitor reduces the cytopathic effect triggered by JEV, we examined whether JEV-activated PERK increased the expression of the downstream ATF4 and CHOP. To address this question, we analyzed the effect of JEV infection on ATF4 and CHOP expression in Neuro-2a cells. JEV infection in Neuro-2a cells was confirmed by the presence of JEV protein NS1 (Fig. 2A). JEV-infected Neuro-2a cells had higher levels of ATF4 and CHOP than mock-infected Neuro-2a cells. Moreover, inhibiting PERK by its inhibitor GSK2606414 reduced the level of ATF4 and CHOP in JEV-infected Neuro-2a cells (Fig. 2A). Similar results were observed in BHK-21 cells (Fig. 2B), indicating that JEV infection in vitro activates the PERK-ATF4-CHOP apoptosis pathway. Moreover, we found the protein levels of ATF4 and CHOP were increased by JEV infection in mouse brain. In addition, induction of ATF4 and CHOP were compromised by PERK inhibitor treatment in JEV-infected mouse brain (Fig. 2C).
FIG 2.
JEV infection induces apoptosis by activating the PERK-ATF4-CHOP pathway. (A and B) JEV infection activates the PERK-ATF4-CHOP pathway in Neuro-2a cells (A) and BHK-21cells (B). Cells were infected with P3 strain at an MOI of 5 and treated with GSK2606414 (0.8 nM) as indicated. Mock-infected cells were used as controls. After 3 days, the cells were subjected to immunoblot analysis using JEV NS1, phospho-PERK, CHOP, ATF4, or actin antibodies. Mock-infected cells were used as controls. (C) JEV infection activates the PERK-ATF4-CHOP apoptosis pathway in mice brains. The tissues from JEV-infected and mock-infected mice brains at 3 days postinfection were collected and analyzed by immunoblotting with antibodies against JEV NS1, phospho-PERK, CHOP, ATF4, or actin. (D to G) JEV infection in Neuro-2a cells induces apoptosis via PERK. Neuro-2a cells were infected with P3 strain at an MOI of 5 and treated with GSK2606414 (0.8 nM) as indicated. Mock-infected Neuro-2a cells were used as controls. After 3 days, the cells were subjected to apoptosis analysis (D), virus replication analysis (E and F), and cell viability analysis (G). Apoptosis analysis was performed with an annexin V-EGFP/PI apoptosis detection kit and analyzed by flow cytometry. Q1, Q2, Q3, and Q4 represent PI-positive, annexin V-/PI-positive, annexin V/PI-negative, and annexin V-positive cells, respectively. Virus replication was analyzed by real-time RT-PCR using JEV-specific primers and a TCID50 assay. Cell viability was analyzed by an MTT assay. Left panels (A to D) show representative images; right panels (A to D) show quantitation data. Error bars indicate the SD of the mean (n = 3). *, P < 0.05; **, P < 0.01.
Next, we examined the role of PERK in JEV-induced apoptosis. Neuro-2a cells were infected with JEV P3 strain (MOI = 5) and treated with 0.8 nM PERK inhibitor GSK2606414 for 3 h as indicated. After 3 days, the cells were stained using an annexin V-EGFP/PI and analyzed by flow cytometry. Annexin V-EGFP stains early apoptotic cells by binding to membrane phospholipid phosphatidylserine exposed to external cellular environment. Propidium iodide (PI) is membrane impermeable and stains late apoptotic cells by binding to DNA. JEV infection significantly increased the percentage of apoptotic cells compared to mock infection. PERK inhibitor GSK2606414 in JEV-infected Neuro-2a cells remarkably reduced the percentage of apoptotic cells (Fig. 2D). Moreover, PERK inhibitor GSK2606414 did not alter the viral RNA level (Fig. 2E) and virus yield (Fig. 2F). However, PERK inhibitor enhanced cell viability (Fig. 2G), which is consistent with our previous data (Fig. 1B and Fig. 2D). All of these data indicate that JEV infection induces apoptosis by activating the PERK-ATF4-CHOP pathway.
JEV-activated PERK triggers the formation of stress granule.
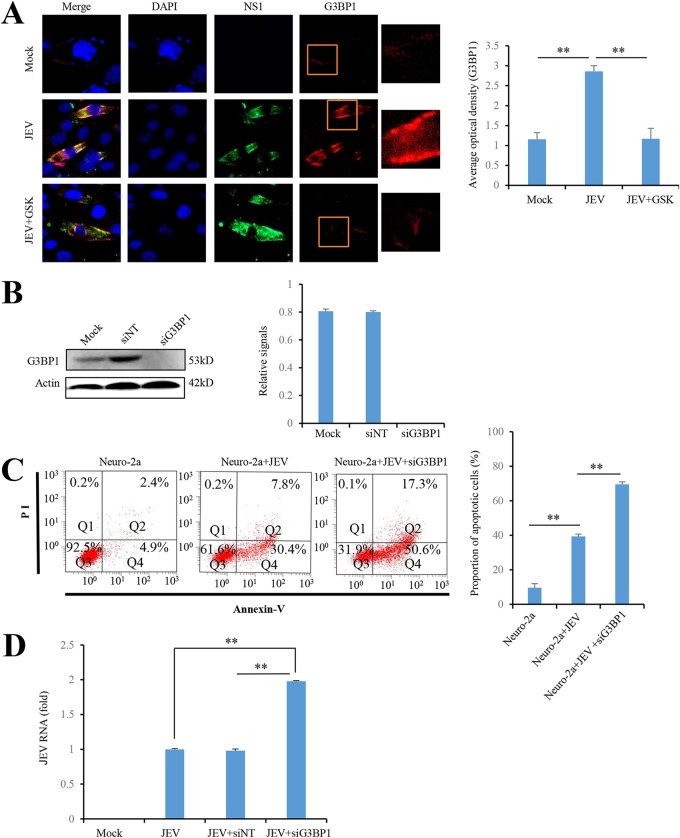
A stress granule (SG) is a discrete cytoplasmic protein-RNA structure that is quickly formed when eukaryotic cells are exposed to various stresses such as viral infection, oxidative stress, and hypoxia. PERK has been reported to initiate SG formation during viral infection (9). Therefore, we investigated whether JEV-activated PERK promotes SG formation by immunofluorescence analysis. Consistent with the previous report (10), cytoplasmic granules containing SG marker protein G3BP1 were markedly increased by JEV infection in Neuro-2a cells. However, PERK inhibitor GSK2606414 significantly reduced the formation of SGs in JEV-infected Neuro-2a cells (Fig. 3A), indicating that JEV-induced SG formation is dependent on PERK activation. In addition, G3BP1 knockdown significantly increased cell apoptosis (Fig. 3B and C) and JEV replication (Fig. 3B and D) in JEV-infected Neuro-2a cells. Taken together, these data show that JEV activates PERK to trigger the formation of SGs, which in turn reduce JEV replication and alleviate JEV-induced apoptosis.
FIG 3.
PERK activation upon JEV infection triggers the formation of SG inhibiting JEV-induced apoptosis. (A) PERK activation upon JEV infection triggers the formation of SG. Neuro-2a cells infected with JEV at an MOI of 5 were treated with 0.8 nM GSK2606414. Mock-infected Neuro-2a cells were used as controls. At 2 days postinfection, the cells were analyzed by immunofluorescence with rabbit anti-G3BP1 antibody and mouse anti-NS1 antibody, followed by Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 594-conjugated anti-rabbit IgG, respectively. The cell nucleus was stained with DAPI. The boxed areas show enlargements in the corresponding panels on the right. (B to D) Neuro-2a cells were infected with P3 strain at an MOI of 5 and transfected with siG3BP1 (25 nM) or siNT (25 nM) as indicated. At 2 days postinfection, the cells were subjected to immunoblot analysis with antibodies against G3BP1 and actin (B), flow cytometry analysis using an annexin V-EGFP/PI apoptosis detection kit (C), and real-time RT-PCR using JEV-specific primers (D). Left panels (A to C) show representative images; right panels (A to C) show quantitation data. Error bars indicate the SD of the mean (n = 3). **, P < 0.01.
JEV NS4B activates PERK by promoting its dimerization.
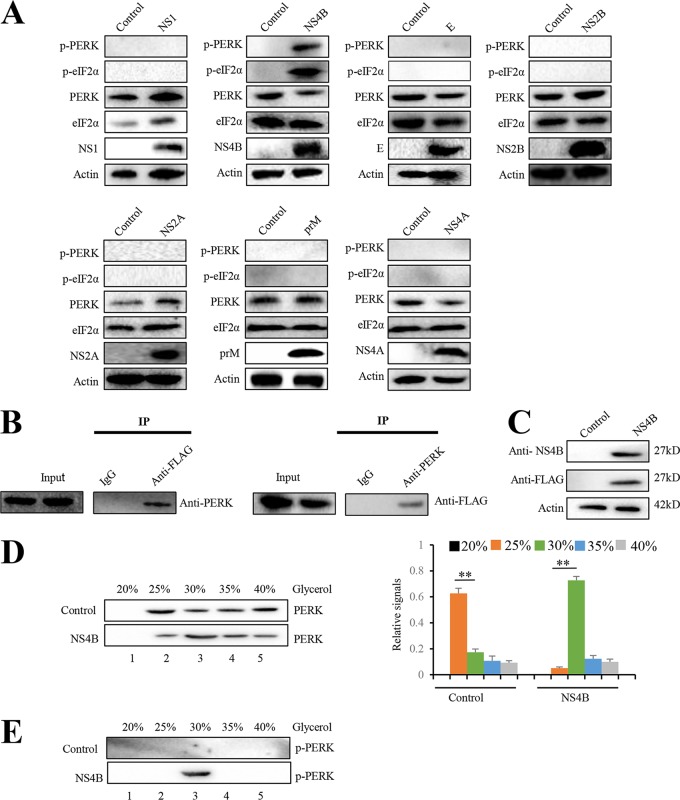
JEV proteins, including prM, E, NS1, NS2A, NS2B, and NS4B, have been reported to induce XBP1-mediated reporter gene expression in mouse neuroblastoma N18 cells (5). JEV NS4B is a hydrophobic protein which exhibits the membrane-modifying capability (7, 11). To identify which viral protein(s) activates PERK, Neuro-2a cells were individually transfected with plasmids that express JEV prM, E, NS1, NS2A, NS2B, NS4A, and NS4B individually. The expression of JEV proteins was confirmed by immunoblots analysis (Fig. 4A). Phosphorylation of PERK and eIF2α was induced by NS4B but not prM, E, NS1, NS2A, NS2B, and NS4A, indicating that only NS4B activates PERK (Fig. 4A).
FIG 4.
JEV NS4B activates PERK by promoting its dimerization. (A) Neuro-2a cells were transfected with plasmid expressing prM, E, NS1, NS2A, NS2B, NS4A, or NS4B. Empty vector-transfected Neuro-2a cells were used as negative controls. After 2 days, the cells were subjected to immunoblot analysis using PERK, eIF2α, phospho-PERK, phospho-eIF2α, FLAG, or actin antibody. Representative images from three independent experiments are shown. (B) Neuro-2a cells were transfected with the plasmid expressing NS4B-3FLAG. After 2 days, the cell lysates were immunoprecipitated with anti-FLAG antibody (left) or anti-PERK antibody (right). The resulting immunoprecipitates (IP) and whole-cell lysates used for immunoprecipitation (Input) were examined by an immunoblot assay with anti-FLAG antibody for detecting NS4B and anti-PERK antibody. The negative-control sample was generated by using control IgG antibody. Representative images from three independent experiments are shown. (C to E) Neuro-2a cells were transfected with plasmid expressing NS4B or empty vector. After 2 days, cellular lysates were subjected to immunoblot analysis with NS4B, FLAG, or actin antibody (C). For the analysis of PERK dimerization, cellular lysates were separated using a 20 to 40% glycerol gradient. PERK (D) and phosphorylated PERK (E) in gradient fractions were determined by immunoblotting. Fractions are numbered 1 to 5 in order from top to bottom (light to heavy), respectively. The left panel (D) shows representative images; the right panel (D) shows quantitation data. Error bars indicate the SD of the mean (n = 3). **, P < 0.01.
To explore the mechanism how NS4B activates PERK, we first examined whether NS4B interacts with PERK by coimmunoprecipitation assay. Neuro-2a cells were transfected with the plasmid expressing NS4B with a 3×FLAG tag and then subjected to immunoprecipitation with anti-FLAG resin. PERK was coimmunoprecipitated with NS4B (Fig. 4B, left panel). Reciprocal immunoprecipitation showed that NS4B was also coimmunoprecipitated with PERK (Fig. 4B, right panel). PERK has been reported to undergo dimerization and autophosphorylation to form active PERK following ER stress. To test whether NS4B induces PERK dimerization, we analyzed the multimeric state of PERK in Neuro-2a cells with or without NS4B expression. Glycerol gradient centrifugation has been used to analyze PERK dimerization (12). Dimerized PERK has a higher molecular weight and sediments more quickly than monomeric PERK. Therefore, PERK dimerization can be determined by PERK migration from the fraction with a low concentration of glycerol to the fraction with a high concentration of glycerol. We transfected Neuro-2a cells with NS4B expressing plasmid or empty vector. NS4B expression in transfected Neuro-2a cells was determined by immunoblots analysis (Fig. 4C). Lysates from Neuro-2a cells with or without NS4B expression were subjected to glycerol gradient centrifugation. PERK within each fraction was analyzed by immunoblotting. PERK in Neuro-2a cells without NS4B expression was enriched in the fraction with 25% glycerol. However, in Neuro-2a cells with NS4B expression, PERK enrichment occurred in the fraction with 30% glycerol, indicating that NS4B promotes PERK dimerization (Fig. 4D). Since PERK dimerization is required for its activation, we also examined the active form of PERK, phosphorylated PERK. Remarkably, phosphorylated PERK was observed in the fraction with 30% glycerol from cell lysate with NS4B expression. However, no phosphorylated PERK was detected in all fractions from cell lysate without NS4B expression (Fig. 4E). Taken together, these results demonstrate that NS4B induces PERK activation by promoting its dimerization.
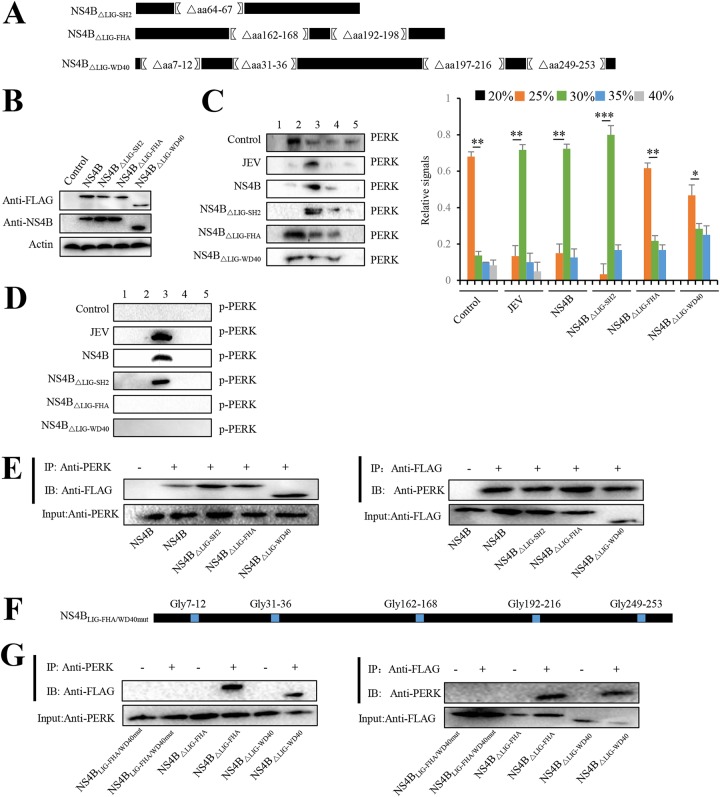
The question then arose is how NS4B induces dimerization by interacting with PERK. Based on the basic ELM prediction (http://elm.eu.org/), NS4B has motifs including LIG-SH2 (amino acids [aa] 64 to 67), LIG-FHA (aa 162 to 168 and aa 192 to 198), LIG-WD40 (aa 7 to 12, aa 31 to 36, aa 197 to 216, and aa 249 to 253), which can bind the SH2 domain, FHA domain, and WW domain within PERK, respectively. To demarcate the portions of NS4B required for PERK dimerization, we constructed plasmids that express NS4B deletion mutants, including NS4BΔLIG-SH2 without LIG-SH2 domain, NS4BΔLIG-FHA without LIG-FHA domain, and NS4BΔLIG-WD40 without LIG-WD40 domain (Fig. 5A) and then transfected them into Neuro-2a cells individually. The expression of NS4B mutants was confirmed by immunoblotting (Fig. 5B). The dimerization status of PERK in Neuro-2a cells expressing NS4B mutants was analyzed by glycerol gradient centrifugation. The higher-molecular-weight form of PERK was detected in Neuro-2a cells expressing NS4BΔLIG-SH2 but was rarely detected in Neuro-2a cells transfected with NS4BΔLIG-FHA or NS4BΔLIG-WD40 (Fig. 5C), suggesting that both the LIG-FHA and the LIG-WD40 domains within NS4B are required to induce PERK dimerization. Consistently, phosphorylated PERK was observed in Neuro-2a cells transfected with NS4BΔLIG-SH2 but not with NS4BΔLIG-FHA or NS4BΔLIG-WD40 (Fig. 5D). Together, these results demonstrate that NS4B induces PERK dimerization and subsequent phosphorylation by its LIG-FHA and LIG-WD40 domains.
FIG 5.
LIG-FHA and LIG-WD40 domains of JEV NS4B promote PERK dimerization. (A) Diagrams of NS4B constructs. Portions of NS4B that were deleted in frame are indicated by brackets with the deleted amino acid residues. (B to D) Neuro-2a cells were transfected with plasmids expressing NS4B, NS4BΔLIG-SH2, NS4BΔLIG-FHA, and NS4BΔLIG-WD40. After 2 days, cellular lysates were subjected to immunoblot analysis using NS4B, FLAG, or actin antibody (B) or separated using a 20 to 40% glycerol gradient. PERK (C) and phosphorylated PERK (D) in gradient fractions were determined by immunoblot analysis. JEV-infected and empty vector-transfected Neuro-2a cells were used as positive and negative controls, respectively. The left panel (C) shows representative images; the right panel (C) shows quantitation data. (E) Neuro-2a cells were individually transfected with plasmids expressing NS4B, NS4BΔLIG-SH2, NS4BΔLIG-FHA, and NS4BΔLIG-WD40. After 2 days, cell lysates were immunoprecipitated with anti-PERK antibody (left panel) or anti-FLAG antibody (right panel). The resulting immunoprecipitates (IP) and whole-cell lysates used for immunoprecipitation (Input) were examined by an immunoblot assay using anti-FLAG antibody or anti-PERK antibody. The negative-control sample was generated by using control IgG antibody. (F) Diagram of the NS4BLIG-FHA/WD40mut with mutated LIG-FHA and LIG-WD40 domains. Amino acids of LIG-FHA and LIG-WD40 domains were replaced with glycines (blue). (G) Neuro-2a cells were transfected with plasmid expressing NS4BΔLIG-FHA, NS4BΔLIG-WD40, or NS4BLIG-FHA/WD40mut. After 2 days, cell lysates were immunoprecipitated with anti-FLAG antibody (left) or anti-PERK antibody (right). The resulting immunoprecipitates (IP) and whole-cell lysates used for immunoprecipitation (Input) were examined by an immunoblot assay using anti-FLAG antibody and anti-PERK antibody. The negative-control sample was generated by using control IgG antibody. Representative images from three independent experiments are shown. Error bars indicate the SD of the mean (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As for how LIG-FHA and LIG-WD40 domains within NS4B contribute to PERK dimerization, we hypothesized that NS4B induces the formation of PERK dimer by simultaneously interacting with two PERK molecules via its LIG-FHA and LIG-WD40 domains. This speculation was supported by the finding that PERK was coimmunoprecipitated with both NS4BΔLIG-FHA and NS4BΔLIG-WD40 (Fig. 5E), implying that both LIG-FHA and LIG-WD40 domains of NS4B can interact with PERK. To further demonstrate that NS4B binds PERK via its LIG-FHA and LIG-WD40 domains, plasmid NS4BLIG-FHA/WD40mut, in which amino acids of LIG-FHA and LIG-WD40 domains of NS4B were mutated to glycines, was constructed (Fig. 5F). The NS4B mutants NS4BΔLIG-FHA and NS4BΔLIG-WD40, but not NS4BLIG-FHA/WD40mut, were coimmunoprecipitated with PERK (Fig. 5G, left panel), and PERK was coimmunoprecipitated with the NS4B mutants NS4BΔLIG-FHA or NS4BΔLIG-WD40 but not NS4BLIG-FHA/WD40mut (Fig. 5G, right panel), suggesting both LIG-FHA and LIG-WD40 domains of NS4B are required for interaction with PERK. Taken together, these results indicate that JEV NS4B pulls PERK molecules together by simultaneously interacting with different PERK molecules via different domains, resulting in PERK dimerization and subsequent activation.
PERK is involved in JEV-induced Japanese encephalitis in mice.
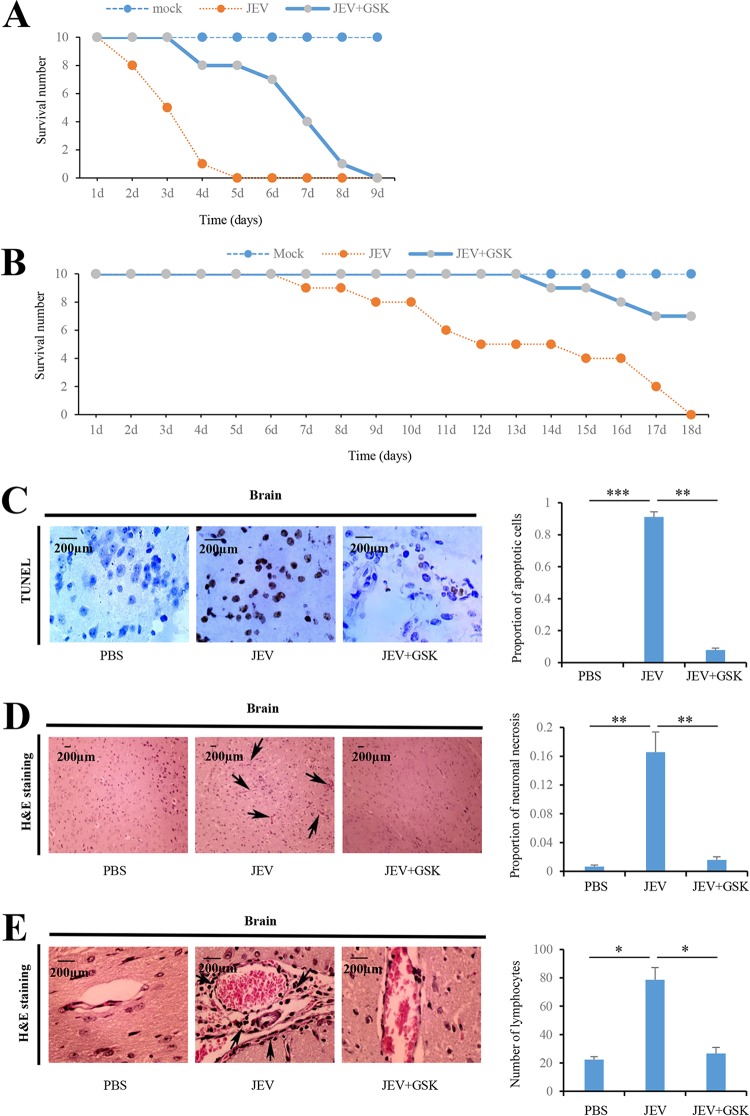
To examine whether PERK is involved in JEV-induced Japanese encephalitis in mice, we first investigated the role of PERK in JEV-induced lethality in vivo. PERK inhibitor GSK2606414 was introduced intracranially into mice at 3 h after JEV infection. All mock-infected mice survived until the end of experiments (Fig. 6A and B). However, mice inoculated intracranially at a dose of 104 PFU per mice died within 5 days after infection. A total of 80% of the JEV-infected mice that received GSK2606414 treatment survived at 5 days after infection (Fig. 6A). Mice inoculated subcutaneously at a dose of 102 PFU per mice died within 18 days after infection. However, 70% of the JEV-infected mice that received GSK2606414 treatment survived at 18 days after infection (Fig. 6B). Taken together, these results strongly suggest that administration of the PERK inhibitor can protect against JEV-induced mortality. Next, cell death in brain tissue was assessed with the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay. TUNEL-negative cells were significantly increased by GSK2606414 treatment in JEV-infected mice by day 3 postinfection (Fig. 6C), indicating that PERK deactivation reduces brain cell damage during JEV infection.
FIG 6.
JEV-activated PERK is involved in JEV-induced Japanese encephalitis in mice. (A) Ten mice inoculated intracranially with PBS were used as mock-infected controls. Ten mice were inoculated intracranially with JEV at a dose of 104 PFU per mice. Ten mice were inoculated intracranially with JEV at a dose of 104 PFU per mice, followed by intracranial injection with GSK2606414 (800 μg/kg [body weight]) 3 h after infection. The survival of mice in each group was monitored for 9 days after JEV inoculation. Data were collected and are shown as Kaplan-Meier survival curves (n = 10 mice). (B) Ten mice inoculated subcutaneously with PBS were used as mock-infected controls. Ten mice were inoculated subcutaneously with JEV at a dose of 102 PFU per mice. Ten mice were inoculated subcutaneously with JEV at a dose of 102 PFU per mice, followed by intracranial injection with GSK2606414 (800 μg/kg [body weight]) 3 h after infection. The survival of mice in each group was monitored for 18 days after JEV inoculation. Data were collected and are shown as Kaplan-Meier survival curves (n = 10 mice). (C to E) Mice were inoculated intracranially with PBS or JEV (104 PFU per mice), followed by intracranially injection with GSK2606414 (800 μg/kg [body weight]) as indicated. Brain tissues were collected 3 days after infection. The apoptotic cells in the brain sections were detected using the TUNEL assay kit (C). The histopathological changes of the mouse brain were observed by H&E staining (D and E). Meningitis (D) and perivascular cuffing (E) that appeared in the JEV-infected groups are indicated by arrows. Left panels (C, D, and E) show representative images; right panels (C, D, and E) show quantitation data. Error bars indicate the SD of the mean (n = 3). Scale bars, 200 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To examine the impact of PERK on encephalitis during JEV infection, histopathological changes in mouse brains were analyzed by hematoxylin and eosin (H&E) staining. At 3 days after infection, JEV infection significantly induced meningitis (Fig. 6D) and perivascular cuffing (Fig. 6E), which are indicators of severe encephalitis in JEV-infected mice. In contrast, these encephalitis indicators were significantly reduced in JEV-infected mice after receiving GSK2606414 treatment (Fig. 6D and E), indicating that PERK is involved in JEV-induced Japanese encephalitis in mice.
NS4B induces apoptosis and encephalitis via PERK.
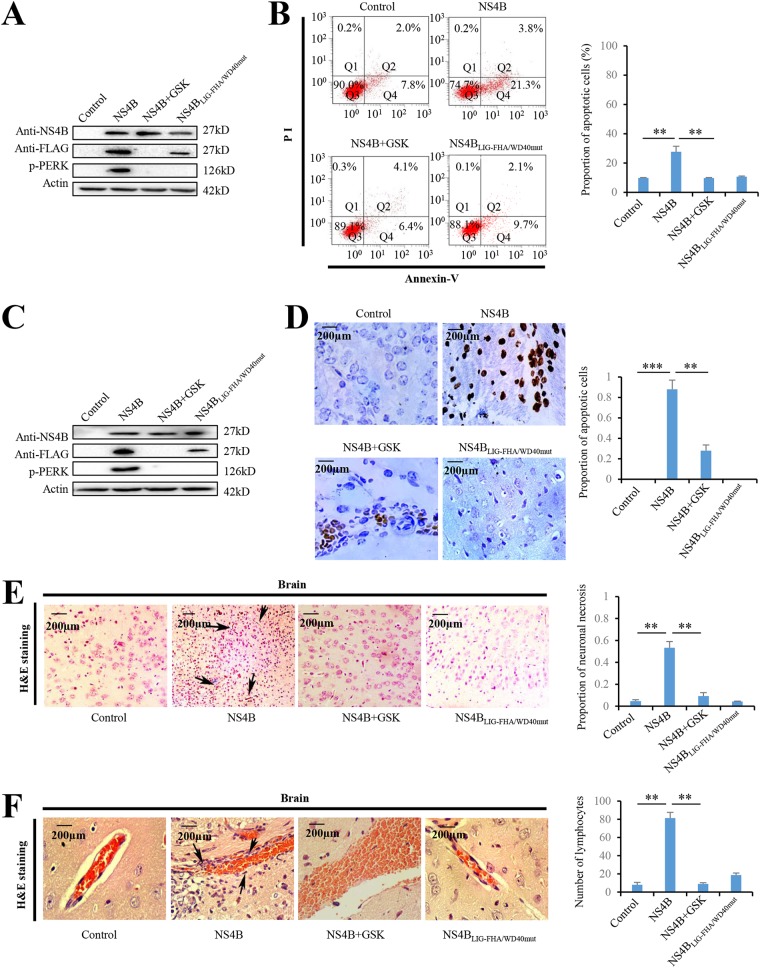
Since JEV NS4B activated PERK by promoting its dimerization in Neuro-2a cells (Fig. 4), we next examined the role of NS4B in PERK-caused apoptosis and encephalitis. Overexpression of NS4B in Neuro-2a cells activated PERK (Fig. 7A) and significantly induced cell apoptosis (Fig. 7B). Moreover, PERK inhibitor significantly reduced NS4B-induced apoptosis (Fig. 7B), suggesting that NS4B induced apoptosis via PERK in Neuro-2a cells. Next, we investigated whether NS4B induced encephalitis via PERK in vivo by injecting the plasmid expressing NS4B intracranially into the brains of mice. Expression of NS4B activates PERK in brains of mice (Fig. 7C). TUNEL assays showed that TUNEL-positive cells in brains were significantly increased by NS4B in brains of mice (Fig. 7D). Moreover, GSK2606414 dramatically reduced NS4B-induced cell apoptosis with no significant effect on NS4B expression in brains of mice (Fig. 7C and D), suggesting that NS4B promotes brain cell apoptosis by activating PERK in vivo. To determine the impact of NS4B on encephalitis in vivo, histopathological changes in mouse brains were analyzed by H&E staining. NS4B expression significantly induced meningitis (Fig. 7E) and perivascular cuffing (Fig. 7F) in the brains of mice, which were significantly reduced by GSK2606414 treatment (Fig. 7E and F). Taken together, these results showed that NS4B induces encephalitis by activating PERK. Since we have previously shown that JEV NS4B activates PERK by promoting its dimerization (Fig. 5), we also examined the effect of NS4B mutant (NS4BΔLIG-FHA/WD40mut), which lacks the domains to promote PERK dimerization, on apoptosis and encephalitis. As expected, the overexpression of NS4BLIG-FHA/WD40mut did not induce apoptosis (Fig. 7B and D) and encephalitis (Fig. 7E and F).
FIG 7.
JEV NS4B induces encephalitis via PERK in mice. (A and B) Neuro-2a cells were transfected with NS4B-expressing plasmid, NS4BLIG-FHA/WD40mut-expressing plasmid, or empty vector and treated with GSK2606414 (0.8 nM) as indicated. At 2 days posttransfection, all groups were subjected to immunoblot analysis using NS4B, FLAG, or actin antibody (A) and apoptotic analysis by flow cytometry (B). (C to F) Mice were injected intracranially with NS4B- or NS4BLIG-FHA/WD40mut-expressing plasmid (200 μg/kg [body weight]) and injected intracranially GSK2606414 (800 μg/kg [body weight]) as indicated. At day 5 after injection, brain tissues were collected and subjected to immunoblot analysis using NS4B, FLAG, or actin antibody (C), TUNEL assays using the TUNEL assay kit (D), and H&E staining (E and F). Meningitis (E) and perivascular cuffing (F) that appeared in the JEV-infected groups are indicated by arrows. Left panels (B, D to F) show representative images; right panels (B, D to F) show quantitation data. Error bars indicate the SD of the mean (n = 3). Scale bars, 200 μm. **, P < 0.01; ***, P < 0.001.
DISCUSSION
During productive virus infection, viruses produce a large amount of viral proteins accumulated in the ER lumen (13–15). Some viruses, such as hepatitis C virus (HCV) and dengue virus (DENV), also use the ER as their replication sites (16, 17). These viral activities often disrupt the homeostasis of the ER and cause ER stress, which can lead to a variety of prevalent diseases such as neurodegenerative diseases, renal disease, liver disease, ocular disease, and cancer (18–21). To attenuate ER stress, cells activate an ER-to-nucleus signaling pathway called the unfolded protein response (UPR), which restores the ER homeostasis (22, 23). However, if under unsolved or intense ER stress, cells would initiate the apoptotic cascades such as IRE1 and PERK-eIF2α-ATF4-CHOP pathways (3), which are primarily associated with virus pathogenesis (24).
ER stress is a common consequence in virus-infected cells. Numerous viruses such as DENV, prototype foamy virus (PFV), chikungunya virus (CHIKV), Newcastle disease virus (NDV), porcine circovirus 2 (PCV2), bluetongue virus (BTV), Venezuelan equine encephalitis virus (VEEV), HCV, salmon anemia virus (SAV), human cytomegalovirus (HCMV), and transmissible gastroenteritis virus (TGEV) have been reported to induce PERK activation or expression (25–27). Conversely, PERK activation is inhibited during influenza virus infection (28). PCV2 and NDV deploy the PERK pathway for its enhanced replication (29, 30). PERK activation contributes to DENV and BTV replication by inducing autophagy and promoting survivability of virus-infected cells (17, 31, 32). However, PFV infection activates PERK pathway, thereby inducing autophagy, which in turn represses PFV replication (33). PERK pathway also inhibits influenza virus replication by reducing viral protein synthesis (28). West Nile virus (WNV) in cells deficient for PERK shows increased replication and virus release compared to wild-type cells (34). These findings reveal that PERK upregulates or downregulates virus replication dependent on virus species. IRE1-dependent decay pathway of ER stress is activated during JEV infection and benefits viral replication (35). Our data showed that although JEV activates PERK, PERK has no significant effect on JEV replication.
NDV induces G0/G1 cell cycle arrest and apoptosis by activating the PERK-eIF2α-ATF4-CHOP signaling pathway (36, 37). VEEV induces apoptosis by activating the PERK arm of the UPR (38). CHIKV infection in HeLa cells leads to activation of the PERK/eIF2α branch of the UPR, the induction of autophagy and apoptosis, implying that PERK/eIF2α pathway could play a role in CHIKV-induced apoptosis (26). Interestingly, sustained activation of PERK axis of ER-stress during chronic HCV infection activates the oncogenic Nrf2 signaling that promotes hepatocyte survival (39). Our data strongly indicate that JEV infection in vivo and in vitro induces PERK activation by promoting its dimerization, resulting in cell apoptosis via the PERK-ATF4-CHOP pathway (Fig. 7). Meanwhile, JEV infection also triggers the formation of stress granule to inhibit apoptosis via PERK activation. However, stress granule plays a role in repressing JEV replication (Fig. 3D). Thus, JEV primarily induces apoptosis by activating the PERK-ATF4-CHOP pathway.
The Flaviviridae is a family of positive, single-stranded, enveloped RNA viruses. This family currently comprises over 100 species such as HCV, yellow fever virus (YFV), WNV, JEV, and DENV. NS4B proteins encoded by different flaviviruses, including YFV, WNV, JEV, and DENV, share the same predicted topology, with five integral transmembrane segments. Dimerization of DENV NS4B, as well as interaction between NS4B and NS4A of DENV-2 and JEV, has been reported. Dimerization motifs of HCV NS4B have been identified (40). Whether NS4B of JEV has dimerization motifs or forms homodimer remains to be determined. Our data showed that NS4B mutants NS4BΔLIG-FHA and NS4BΔLIG-WD40 can interact with PERK but cannot promote PERK dimerization (Fig. 5), indicating that both LIG-FHA and LIG-WD40 domains are required for PERK dimerization. It is highly likely that LIG-FHA binds one PERK molecule and LIG-WD40 binds another PERK molecule. Since NS4B contains both LIG-FHA and LIG-WD40 domains, one NS4B molecule is able to pull two PERK molecules together to form dimer. It is thus possible that JEV NS4B dimerization, if any, may not be required for PERK dimerization and activation.
Some viruses of Flaviviridae, such as HCV, WNV, JEV, and DENV, have been reported to regulate ER stress response (5, 6). The opposing functions of ER and cytosolic forms of HCV envelope proteins E1 and E2 have been reported depending on their subcellular localization (41). ER-targeting E1 and E2 of HCV are capable of eliciting the UPR. However, cytosolic targeting E1 and E2 bind PERK to dampen the UPR (41). The effects of DENV on the UPR are time-dependent and possibly cell type specific (40). WNV NS4B (together with NS2B and NS4A) activates the ATF6/IRE-1 pathway, resulting in Xbp-1 transcription and splicing. We demonstrate that JEV NS4B pulls two PERK molecules together by its LIG-FHA and LIG-WD40 domains, resulting in PERK dimerization and subsequent activation (Fig. 5). Moreover, apoptosis and encephalitis are induced by overexpression of JEV NS4B but not its mutant NS4BLIG-FHA/WD40mut lacking the domains to promote PERK dimerization (Fig. 7), indicating that JEV NS4B plays a critical role in JEV-induced apoptosis and encephalitis by inducing PERK dimerization and activation. Interestingly, NS4B of DENV decreased PERK activation in insect cells when treated with the ER stress inducer tunicamycin for 8 h. However, the decrease in PERK activation appears to be reversed after 24 h (42). NS4B of JEV and related flaviviruses appears to play an important role in flaviviral replication and pathogenesis (40). DENV NS4B enables virus replication by decreasing UPR-mediated immune effectors in insect cells treated with the ER stress inducer tunicamycin (42). Flaviviral NS4B has recently been shown to constitute an excellent target for the selective inhibition of flavivirus replication (40, 43, 44). Our data showed that JEV NS4B induces encephalitis via PERK in mice (Fig. 7), highlighting the prospect of NS4B as a molecular target for antiviral intervention.
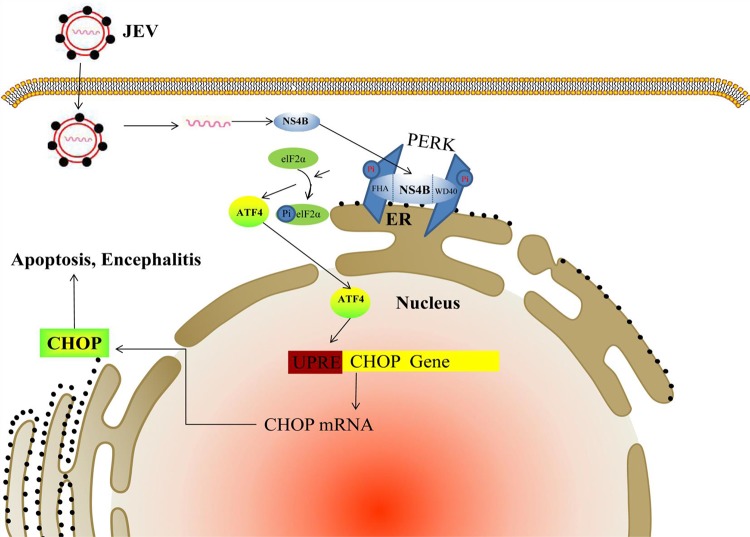
In conclusion, our study reveals that JEV infection induces dimerization and activation of the ER stress sensor PERK via its NS4B protein. Moreover, JEV-activated PERK induces neuron apoptosis and encephalitis through the PERK-eIF2α-ATF4-CHOP pathway (Fig. 8). These findings provide a novel therapeutic approach for JEV-caused encephalitis.
FIG 8.
Schematic diagram showing the molecular signaling pathway for JEV-induced apoptosis and encephalitis via NS4B. JEV particle enters cytoplasm and releases viral RNA, which directs the synthesis of NS4B protein. The synthesized NS4B protein puts two PERK molecules together with its LIG-FHA and LIG-WD40 domains, leading to dimerization and activation of PERK, which causes the phosphorylation of elF2α and subsequent increase of ATF4 translation. The ATF4 protein enters the nucleus and induces the transcription of CHOP gene by binding the UPR element (UPRE). CHOP then triggers apoptosis and encephalitis.
MATERIALS AND METHODS
Cell lines, viruses, and mice.
Baby hamster kidney (BHK) cells were used for virus titer measurement by 50% tissue culture infective dose (TCID50) assays (16). Neuro-2a cells or BALB/c mice were infected with the P3 strain of JEV as indicated. All animal experiments were approved by the Laboratory Animal Ethics Committee at Jiangxi Agricultural University, Nanchang, Jiangxi, China, and performed in accordance with the approved guidelines.
Plasmid construction and reagents.
To generate the plasmids to express JEV proteins, cDNA encoding individual viral proteins, including prM, E, NS1, NS2A, NS2B, NS4A, or NS4B, was amplified from the isolated RNA of JEV P3 strain (GenBank accession number U47032.1) and inserted into pcDNA3.1(–)-3×FLAG under the control of the cytomegalovirus promoter. NS4B deletion mutants, including NS4BΔLIG-SH2 (the LIG-SH2 domain is deleted), NS4BΔLIG-FHA (the LIG-FHA domain is deleted), NS4BΔLIG-WD40 (the LIG-WD40 domain is deleted), and NS4BLIG-FHA/WD40mut (the amino acids of the LIG-FHA and LIG-WD40 domains of NS4B were mutated to glycines), were cloned into the pcDNA3.1(–)-3×FLAG vector. Antibodies against JEV NS1, G3BP1, PERK, phospho-PERK, FLAG, eIF2a, and phospho-eIF2α were purchased from Abcam. Antibody against JEV NS4B was purchased from GeneTex. Antibodies against ATF4, CHOP, and actin were purchased from Proteintech. Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 594-conjugated anti-rabbit IgG were purchased from Invitrogen. Annexin V-FITC/PI was purchased from Beijing Zoman Biotechnology. PERK inhibitor GSK2606414 was purchased from MedChemExpress. The TUNEL assay kit was purchased from Beyotime Biotechnology.
Reverse transcription-PCR and immunoblot analysis.
Total RNA was extracted using TRIzol reagent and reverse transcription-PCR (RT-PCR) was performed as described previously (16). Specific primers for JEV NS1 and GADPH were used for RT-PCR. PCR products were electrophoresed on 1% agarose gel. For immunoblot analysis, total cell extracts were prepared as described previously (9). Protein samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with primary antibodies against JEV proteins, including NS1, NS4B, G3BP1, PERK, phospho-PERK, eIF2α, phospho-eIF2α, ATF4, CHOP, FLAG, or actin. Next, horseradish peroxidase-labeled IgG as secondary antibody was applied to the blots. The specific proteins were visualized by using an ECL detection kit (Amersham Biosciences).
Immunoprecipitation.
Immunoprecipitation was performed as described previously (9). Cells were washed with ice-cold phosphate-buffered saline (PBS) and suspended in lysis buffer (20 mM Tris-HCl [pH 7.4] containing 135 mM NaCl, 1% Triton X-100, and 10% glycerol) supplemented with 50 mM NaF, 5 mM Na3VO4, and EDTA-free Complete protease inhibitor cocktail (Roche). Cell lysates were sonicated for 10 min and then incubated for 30 min at 4°C, followed by centrifugation at 14,000 × g for 10 min. The supernatants were immunoprecipitated with anti-FLAG or anti-PERK antibody in the presence of protein A-agarose beads (Beyotime). The immunocomplexes with the beads were collected by centrifugation at 1,000 × g and then washed four times with lysis buffer. The proteins binding to the beads were boiled with SDS sample buffer and then subjected to SDS-PAGE.
Indirect immunofluorescence and apoptosis analysis.
Indirect immunofluorescence was performed as described previously (16). Briefly, cells were fixed in acetone-methanol and incubated with the primary antibodies. After three washes with PBS, the cells were incubated with Alexa Fluor 594- or 488-labeled secondary antibodies. Cell nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). The staining was observed with a fluorescence microscope (Nikon). The apoptotic Neuro-2a cells were subjected to flow cytometry analysis using annexin V-FITC/PI according to the manufacturer’s instructions. The apoptotic cells in the brain sections were detected using a TUNEL assay kit according to the manufacturer’s instructions.
RNA interference, DNA transfection, virus replication, and cell viability analysis.
The small interfering RNAs (siRNAs) were purchased from Sangon Biotech and transfected into the cells using Entranster-R4000 (Engreen Biosystem). siRNAs targeting G3BP1 (siG3BP1) included two strands (sense, 5′-GCAACGAUGACUUUGAUAATT-3′; antisense, 5′-UUAUCAAAGUCAUCGUUGCTT-3′). Nontargeting siRNA (siNT) was used as a negative-control. DNA transfection was performed using Lipofectamine 2000 transfection reagent (Thermo Fisher) according to the manufacturer’s instructions. Virus replication was analyzed by real-time RT-PCR and TCID50 assay as described previously (8). Cell viability was determined by MTT assays as described previously (8).
Glycerol gradient centrifugation.
Proteins in cell lysates were separated on a 5-ml exponential 20 to 40% glycerol gradient in cell lysis buffer (1% Triton X-100 in 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 100 mM NaF, 17.5 mM β-glycerolphosphate, 10% glycerol) supplemented with protease inhibitors (100 μM phenylmethylsulfonyl fluoride, 0.15 μM aprotinin, 1 μM leupeptin, and 1 μM pepstatin). The gradient was subjected to ultracentrifugation using a Beckman SW41 rotor at 39,000 rpm for 48 h at 4°C. Then, 1-ml fractions were collected from the gradients and separated by SDS-PAGE. The PERK levels were determined by immunoblot analyses using antibodies against PERK and phospho-PERK.
JEV challenge and PERK inhibitor administration.
Adult BALB/c mice (4 weeks old) were randomly assigned to six groups (n ≥ 10 per group). Mice in group 1 were inoculated intracranially with PBS. Mice in group 2 were inoculated intracranially with 104 PFU of JEV P3 strain. Mice in group 3 were inoculated intracranially with 104 PFU of JEV P3 strain (per mouse), followed by intracranial injection with GSK2606414 (800 μg/kg [body weight]) at 3 h after infection. Mice in group 4 were inoculated subcutaneously with PBS. Mice in group 5 were inoculated subcutaneously with 102 PFU of JEV P3 strain (per mouse). Mice in group 6 were inoculated subcutaneously with 102 PFU of JEV P3 strain (per mouse), followed by intracranial injection with GSK2606414 (800 μg/kg [body weight]) at 3 h after infection. The survival of mice in each group was monitored for 9 or 18 days after JEV inoculation, as indicated. Immunoblot, immunohistochemistry, TUNEL, and H&E staining assays were performed on brain samples obtained from three mice in the tested groups.
Administration of NS4B expression plasmid and PERK inhibitor.
Adult BALB/c mice (4 weeks old) were randomly assigned to four groups (n = 10 per group): group 1, the control group (empty vector-treated mice); group 2, the NS4B expression plasmid-treated mice; group 3, the NS4B expression plasmid- and GSK2606414-treated mice; and group 4, the NS4BLIG-FHA/WD40mut expression plasmid-treated mice. Mice in group 1 were injected intracranially with empty vector. Mice in group 2 were injected intracranially with NS4B expression plasmid (200 μg/kg [body weight]). Mice in group 3 were injected intracranially with NS4B expression plasmid (200 μg/kg [body weight]) and GSK2606414 (800 μg/kg [body weight]). Mice in group 4 were injected intracranially with NS4BLIG-FHA/WD40mut expression plasmid (200 μg/kg [body weight]). After 5 days, mice in each group were killed, and brain samples were collected for immunoblot (three mice per group), TUNEL (three mice per group), and H&E staining (three mice per group) assays.
H&E staining and immunohistochemistry.
The standard H&E staining protocol was followed for tissue staining. Immunohistochemistry (IHC) assay was performed as previously described (4). The tissues from JEV-infected and mock-infected mice brains were collected at 3 days postinfection, fixed in 10% formalin phosphate buffer solution (0.01 M PBS [pH 7.4]), and processed for paraffin embedding. JEV NS1 antibody was used as primary antibody for IHC according to the manufacturer’s instructions.
Statistical analysis.
Statistical analysis was performed using the Statistical Package Social Sciences (SPSS) program, version 11.5, by one-way analysis of variance, and significant differences among groups were determined using the least significant difference. The accepted level of statistical significance was P < 0.05.
ACKNOWLEDGMENTS
This study was funded by research grants from the National Natural Science Foundation of China (grants 31460667, 31160034, and 31860038) to L.K. and from Jiangxi Province (grants 20161BBF60084 and JXJG-16-3-8) to L.K.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Hisanaga S, Miyake M, Taniuchi S, Oyadomari M, Morimoto M, Sato R, Hirose J, Mizuta H, Oyadomari S. 2018. PERK-mediated translational control is required for collagen secretion in chondrocytes. Sci Rep 8:773. doi: 10.1038/s41598-017-19052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Kong L, Yu X. 2015. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol 41:150–164. doi: 10.3109/1040841X.2013.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye J, Zhang H, He W, Zhu B, Zhou D, Chen Z, Ashraf U, Wei Y, Liu Z, Fu ZF, Chen H, Cao S. 2016. Quantitative phosphoproteomic analysis identifies the critical role of JNK1 in neuroinflammation induced by Japanese encephalitis virus. Sci Signal 9:a98. [DOI] [PubMed] [Google Scholar]
- 5.Yu CY, Hsu YW, Liao CL, Lin YL. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol 80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Xu A, Wu X, Zhang Y, Guo Y, Guo F, Pan Z, Kong L. 2016. Japanese encephalitis virus induces apoptosis by the IRE1/JNK pathway of ER stress response in BHK-21 cells. Arch Virol 161:699–703. doi: 10.1007/s00705-015-2715-5. [DOI] [PubMed] [Google Scholar]
- 7.Su HL, Liao CL, Lin YL. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol 76:4162–4171. doi: 10.1128/jvi.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Yu X, Xu A, Xu J, Wang Q, Guo Y, Wu X, Tang Y, Ding Z, Zhang Y, Gong T, Pan Z, Li S, Kong L. 2018. Japanese encephalitis virus induces apoptosis by inhibiting Foxo signaling pathway. Vet Microbiol 220:73–82. doi: 10.1016/j.vetmic.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Fang L, Wang D, Cai K, Chen H, Xiao S. 2017. Porcine reproductive and respiratory syndrome virus infection induces stress granule formation depending on protein kinase R-like endoplasmic reticulum kinase (PERK) in MARC-145 cells. Front Cell Infect Microbiol 7:111. doi: 10.3389/fcimb.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, Kamitani W, Matsuura Y. 2013. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with caprin-1 and facilitates viral propagation. J Virol 87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang YS, Liao CL, Tsao CH, Chen MC, Liu CI, Chen LK, Lin YL. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J Virol 73:6257–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma K, Vattem KM, Wek RC. 2002. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem 277:18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 13.Jheng JR, Ho JY, Horng JT. 2014. ER stress, autophagy, and RNA viruses. Front Microbiol 5:388. doi: 10.3389/fmicb.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C, Qiu Y, Han P, Chen Y, Zhang L, Yuan Q, Zhang T, Cheng T, Yuan L, Huang C, Zhang S, Yin Z, Peng XE, Liang D, Lin X, Lin Y, Lin Z, Xia N. 2018. ER stress regulating protein phosphatase 2A-B56γ, targeted by hepatitis B virus X protein, induces cell cycle arrest and apoptosis of hepatocytes. Cell Death Dis 9:762. doi: 10.1038/s41419-018-0787-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Wang A. 2012. Virus-induced ER stress and the unfolded protein response. Front Plant Sci 3:293. doi: 10.3389/fpls.2012.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L, Fujimoto A, Nakamura M, Aoyagi H, Matsuda M, Watashi K, Suzuki R, Arita M, Yamagoe S, Dohmae N, Suzuki T, Sakamaki Y, Ichinose S, Suzuki T, Wakita T, Aizaki H. 2016. Prolactin regulatory element binding protein is involved in hepatitis C virus replication by interaction with NS4B. J Virol 90:3093–3111. doi: 10.1128/JVI.01540-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YR, Kuo SH, Lin CY, Fu PJ, Lin YS, Yeh TM, Liu HS. 2018. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci Rep 8:489. doi: 10.1038/s41598-017-18909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghemrawi R, Battaglia-Hsu SF, Arnold C. 2018. Endoplasmic reticulum stress in metabolic disorders. Cells 7:E63. doi: 10.3390/cells7060063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallazzini M, Pallet N. 2018. Endoplasmic reticulum stress and kidney dysfunction. Biol Cell 110:205–216. doi: 10.1111/boc.201800019. [DOI] [PubMed] [Google Scholar]
- 20.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 21.Kroeger H, Chiang WC, Felden J, Nguyen A, Lin JH. 2019. ER stress and unfolded protein response in ocular health and disease. FEBS J 286:399–412. doi: 10.1111/febs.14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernales S, Papa FR, Walter P. 2006. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 23.Schroder M, Kaufman RJ. 2005. The mammalian unfolded protein response. Annu Rev Biochem 74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 24.Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog 4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathore AP, Ng ML, Vasudevan SG. 2013. Differential unfolded protein response during chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2α phosphorylation. Virol J 10:36. doi: 10.1186/1743-422X-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khongwichit S, Wikan N, Abere B, Thepparit C, Kuadkitkan A, Ubol S, Smith DR. 2016. Cell-type specific variation in the induction of ER stress and downstream events in chikungunya virus infection. Microb Pathog 101:104–118. doi: 10.1016/j.micpath.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Xue M, Fu F, Ma Y, Zhang X, Li L, Feng L, Liu P. 2018. The PERK arm of the unfolded protein response negatively regulates transmissible gastroenteritis virus replication by suppressing protein translation and promoting type I IFN production. J Virol 92:e00431-18. doi: 10.1128/JVI.00431-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landeras-Bueno S, Fernandez Y, Falcon A, Oliveros JC, Ortin J. 2016. Chemical genomics identifies the PERK-mediated unfolded protein stress response as a cellular target for influenza virus inhibition. mBio 7:e16–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng JH, Sun YJ, Zhang FQ, Zhang XR, Qiu XS, Yu LP, Wu YT, Ding C. 2016. Newcastle disease virus NP and P proteins induce autophagy via the endoplasmic reticulum stress-related unfolded protein response. Sci Rep 6:24721. doi: 10.1038/srep24721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Qi B, Gu Y, Xu F, Du H, Li X, Fang W. 2016. Porcine circovirus 2 deploys PERK pathway and GRP78 for its enhanced replication in PK-15 cells. Viruses 8:E56. doi: 10.3390/v8020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou JN, Chen TH, Chiang YH, Peng JY, Yang TH, Cheng CC, Sofiyatun E, Chiu CH, Chiang-Ni C, Chen WJ. 2017. PERK signal-modulated protein translation promotes the survivability of dengue 2 virus-infected mosquito cells and extends viral replication. Viruses 9:E262. doi: 10.3390/v9090262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv S, Sun EC, Xu QY, Zhang JK, Wu DL. 2015. Endoplasmic reticulum stress-mediated autophagy contributes to bluetongue virus infection via the PERK-eIF2α pathway. Biochem Biophys Res Commun 466:406–412. doi: 10.1016/j.bbrc.2015.09.039. [DOI] [PubMed] [Google Scholar]
- 33.Yuan P, Dong L, Cheng Q, Wang S, Li Z, Sun Y, Han S, Yin J, Peng B, He X, Liu W. 2017. Prototype foamy virus elicits complete autophagy involving the ER stress-related UPR pathway. Retrovirology 14:16. doi: 10.1186/s12977-017-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol 85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya S, Sen U, Vrati S. 2014. Regulated IRE1-dependent decay pathway is activated during Japanese encephalitis virus-induced unfolded protein response and benefits viral replication. J Gen Virol 95:71–79. doi: 10.1099/vir.0.057265-0. [DOI] [PubMed] [Google Scholar]
- 36.Bu X, Zhao Y, Zhang Z, Wang M, Li M, Yan Y. 2016. Recombinant Newcastle disease virus (rL-RVG) triggers autophagy and apoptosis in gastric carcinoma cells by inducing ER stress. Am J Cancer Res 6:924–936. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Wang R, Li Y, Sun Y, Song C, Zhan Y, Tan L, Liao Y, Meng C, Qiu X, Ding C. 2018. Newcastle disease virus induces G0/G1 cell cycle arrest in asynchronously growing cells. Virology 520:67–74. doi: 10.1016/j.virol.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baer A, Lundberg L, Swales D, Waybright N, Pinkham C, Dinman JD, Jacobs JL, Kehn-Hall K. 2016. Venezuelan equine encephalitis virus induces apoptosis through the unfolded protein response activation of EGR1. J Virol 90:3558–3572. doi: 10.1128/JVI.02827-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aydin Y, Chedid M, Chava S, Danielle WD, Liu S, Hagedorn CH, Sumitran-Holgersson S, Reiss K, Moroz K, Lu H, Balart LA, Dash S. 2017. Activation of PERK-Nrf2 oncogenic signaling promotes Mdm2-mediated Rb degradation in persistently infected HCV culture. Sci Rep 7:9223. doi: 10.1038/s41598-017-10087-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Zmurko J, Neyts J, Dallmeier K. 2015. Flaviviral NS4b, chameleon and jack-in-the-box roles in viral replication and pathogenesis, and a molecular target for antiviral intervention. Rev Med Virol 25:205–223. doi: 10.1002/rmv.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egan PA, Sobkowiak M, Chan SW. 2013. Hepatitis C virus envelope protein E1 binds PERK and represses the unfolded protein response. Open Virol J 7:37–40. doi: 10.2174/1874357901307010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sepulveda-Salinas KJ, Ramos-Castaneda J. 2017. Participation of dengue virus NS4B protein in the modulation of immune effectors dependent on ER stress in insect cells. Cell Stress Chaperones 22:799–810. doi: 10.1007/s12192-017-0810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Liu Y, Guo J, Wang P, Zhang L, Xiao G, Wang W. 2017. Screening of FDA-approved drugs for inhibitors of Japanese encephalitis virus infection. J Virol 91:e01055-17. doi: 10.1128/JVI.01055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li XD, Ye HQ, Deng CL, Liu SQ, Zhang HL, Shang BD, Shi PY, Yuan ZM, Zhang B. 2015. Genetic interaction between NS4A and NS4B for replication of Japanese encephalitis virus. J Gen Virol 96:1264–1275. doi: 10.1099/vir.0.000044. [DOI] [PubMed] [Google Scholar]