Many flaviviruses are significant human pathogens that frequently cause outbreaks and epidemics around the world. Development of novel vaccine platforms against these pathogens is a public health priority. Using WNV as a model, we developed a new vaccine platform for flaviviruses. WNV containing a NS1 deletion (WNV-ΔNS1) could be efficiently trans complemented in Vero cells that constitutively expressed WT NS1 protein. A single-dose immunization with WNV-ΔNS1 elicited robust immune responses in mice. The immunized animals were fully protected against pathogenic WNV infection. No adverse effects related to the WNV-ΔNS1 vaccination were observed. The results have demonstrated the potential of the NS1 complementation system as an alternative platform for flavivirus vaccine development, especially for highly pathogenic flaviviruses.
KEYWORDS: flavivirus, NS1, West Nile virus, vaccine
ABSTRACT
We previously produced a replication-defective West Nile virus (WNV) lacking NS1 (WNV-ΔNS1) that could propagate at low levels (105 infectious units [IU]/ml) in a 293T cell line expressing wild-type (WT) NS1. This finding indicates the potential of developing WNV-ΔNS1 as a noninfectious vaccine. To explore this idea, we developed an NS1-expressing Vero cell line (VeroNS1) that significantly improved the yield of WNV-ΔNS1 (108 IU/ml). We evaluated the safety and efficacy of WNV-ΔNS1 in mice. WNV-ΔNS1 appeared to be safe, as no replicative virus was found in naive Vero cells after continuous culturing of WNV-ΔNS1 in VeroNS1 cells for 15 rounds. WNV-ΔNS1 was noninfectious in mice, even when IFNAR−/− mice were administered a high dose of WNV-ΔNS1. Vaccination with a single dose of WNV-ΔNS1 protected mice from a highly lethal challenge with WT WNV. The antibody response against WNV correlated well with the protection of vaccinated mice. Our study demonstrates the potential of the NS1 trans complementation system as a new platform for flavivirus vaccine development.
IMPORTANCE Many flaviviruses are significant human pathogens that frequently cause outbreaks and epidemics around the world. Development of novel vaccine platforms against these pathogens is a public health priority. Using WNV as a model, we developed a new vaccine platform for flaviviruses. WNV containing a NS1 deletion (WNV-ΔNS1) could be efficiently trans complemented in Vero cells that constitutively expressed WT NS1 protein. A single-dose immunization with WNV-ΔNS1 elicited robust immune responses in mice. The immunized animals were fully protected against pathogenic WNV infection. No adverse effects related to the WNV-ΔNS1 vaccination were observed. The results have demonstrated the potential of the NS1 complementation system as an alternative platform for flavivirus vaccine development, especially for highly pathogenic flaviviruses.
INTRODUCTION
West Nile virus (WNV) is an important mosquito-transmitted human pathogen. WNV causes no illness or a mild, self-limiting, febrile illness in most cases, but it causes more severe disease in elderly and immunocompromised individuals. Since its emergence in New York in 1999, WNV has remained an important public health threat in the United States (1). The virus has now been reported in many other regions, including Africa, Europe, and West Asia (2, 3), indicating that WNV may be a global public health threat.
WNV belongs to the genus Flavivirus in the family Flaviviridae (4). The Flavivirus genus also includes many other important human pathogens, such as dengue virus, yellow fever virus, Zika virus, and tick-borne encephalitis virus. The flavivirus genome is a single-stranded, positive-sense RNA with approximately 11,000 nucleotides, comprising a 5′ untranslated region (UTR), a single open reading frame (ORF), and a 3′ UTR. The single ORF encodes three structural proteins (capsid, membrane, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (5–7). The structural proteins play essential roles in virus entry, fusion, and assembly, while the nonstructural proteins are important for viral replication, virion assembly, and evasion of the innate immune response.
NS1 is a highly conserved glycoprotein, with a molecular mass of about 46 to 50 kDa, that plays important roles in viral replication and immune responses (8–11). The nascent NS1 is hydrophilic and water soluble. Following its cleavage from a polyprotein in the endoplasmic reticulum (ER), NS1 undergoes glycosylation and homodimerization. After dimerization, NS1 acquires partial hydrophobicity, traffics to the replication complex, and plays an essential role in the early stage of viral RNA replication (12, 13). The protein functions in viral RNA synthesis by modulating different viral and host factors. For example, NS1 was found to interact genetically or physically with viral transmembrane proteins NS4A and NS4B, which are responsible for replication complex formation (8); such interactions allow the lumen-resident NS1 to regulate viral replication that occurs on the cytoplasmic side of the ER (12, 14). Mutations of NS1 decrease viral replication, and the replication defect can be restored in trans by ectopic expression of wild-type (WT) NS1 (11, 15–18).
Since there are no licensed antivirals against WNV infection, vaccination is a good means to prevent infections and disease. It has been well demonstrated that humoral immunity is sufficient to control WNV infection (19, 20), although WNV-specific T cells also contribute to protection and the clearance of WNV (20). The envelope of WNV is the main viral antigen. Mapping of human and murine antibody repertoires against the envelope has revealed important insights into the protective immune response against the virus (19). A number of vaccine strategies have been explored to combat WNV infection (21, 22), including inactivated vaccine, subunit vaccine (23), live-attenuated or chimeric vaccines (24–27), DNA-based vaccine (28–32), and virus-like particle vaccine (33). Among these different types of vaccines, live-attenuated vaccines are superior to other vaccines as they can elicit robust humoral and cellular adaptive immune responses. However, the safety of live-attenuated vaccine is a major concern, particularly for highly pathogenic pathogens.
In this study, higher titers of replication-defective WNV with a NS1 deletion (WNV-ΔNS1) were produced in a selected Vero cell line that expresses WT NS1 protein (VeroNS1). WNV-ΔNS1 was evaluated as a vaccine candidate in mice. WNV-ΔNS1 was generated with a WNV infectious clone by deleting most coding sequences of NS1. WNV-ΔNS1 was not able to replicate in naive Vero cells unless NS1 protein was provided in trans (11). In animals, we demonstrated the safety of WNV-ΔNS1 in IFNAR−/− mice, a susceptible animal model deficient in type I interferon (IFN) receptor (IFNAR). A single dose of WNV-ΔNS1 immunization protected C57BL/6 mice from a lethal challenge with WT WNV. The antibody response against WNV correlated well with the protection efficacy of WNV-ΔNS1. Our study demonstrates the potential of the NS1 complementation system as an alternative platform for flavivirus vaccine development.
RESULTS
High-titer production of WNV-ΔNS1 in VeroNS1 cell line.
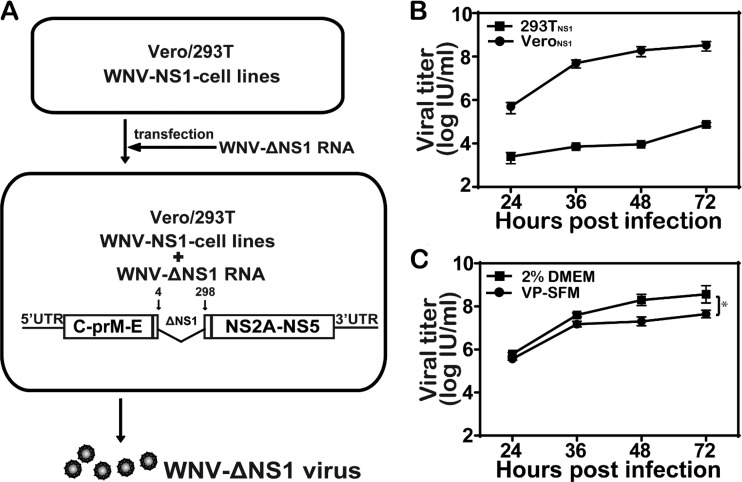
The goal of this study was to examine whether a replication-defective WNV-ΔNS1 could elicit protective immunity against WNV infection and serve as a safe vaccine platform for highly pathogenic flaviviruses. Although WNV-ΔNS1 was genetically stable after serial passaging in 293TNS1 cells without recombination (11), the viral titer of WNV-ΔNS1 was low (around 105 infectious units [IU]/ml), which was not ideal for vaccine production. In the present study, we selected a new NS1 helper cell line, using Vero cells (VeroNS1) to produce WNV-ΔNS1 (Fig. 1A). In VeroNS1 cells, WNV-ΔNS1 replicated efficiently with a mean peak viral titer of 1 × 108 IU/ml (Fig. 1B) at 72 h postinfection (hpi), about 1,000-fold higher than that observed in the previously reported 293TNS1 cells (11). In addition, WNV-ΔNS1 could propagate efficiently in serum-free medium in the VeroNS1 cell line, although the viral titer was about 5- to 10-fold lower than that in serum-containing medium (Fig. 1C).
FIG 1.
High-titer production of WNV-ΔNS1 in VeroNS1 cell line. (A) Method for production of WNV-ΔNS1. WNV-ΔNS1 was generated by transfection of WNV-ΔNS1 RNA (containing a deletion of residues 4 to 298 in the NS1 coding sequence) into the puromycin-resistant Vero/293T cell line stably expressing WNV-NS1 protein. (B) Comparison of growth kinetics of WNV-ΔNS1 in VeroNS1 and 293TNS1 cells. VeroNS1 and 293TNS1 cells were infected with WNV-ΔNS1 virus at an MOI of 0.1. The supernatants were harvested at the indicated time points, and viral titers were determined by IFA in VeroNS1 cells, as described in Materials and Methods. Two independent experiments were performed in triplicate. (C) Comparison of growth kinetics of WNV-ΔNS1 in serum-free medium and serum-containing medium. VeroNS1 cells were infected with WNV-ΔNS1 virus at an MOI of 0.1 and incubated in virus production serum-free medium (VP-SFM) or serum-containing medium. The supernatants were harvested at the indicated time points, and viral titers were determined as described above. Data represent the mean ± standard deviation of triplicate measurements in a representative experiment. The asterisks denote statistical differences between the indicated groups. *, P < 0.05.
Characterization of WNV-ΔNS1.
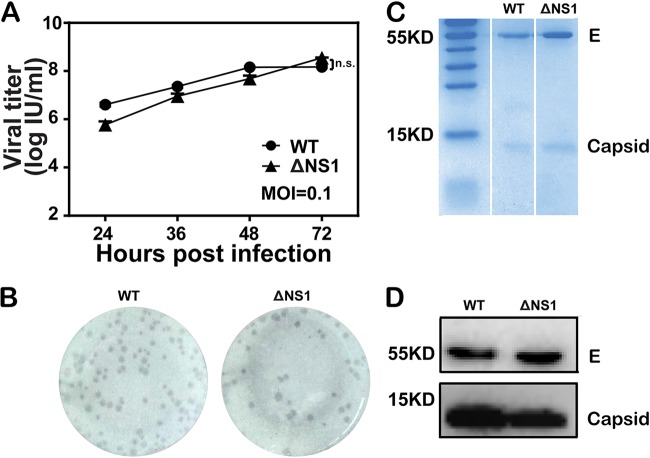
To characterize WNV-ΔNS1, the viral growth and plaque morphology were compared with those of WT WNV (Fig. 2). Similar viral growth curves were observed for these two viruses in the VeroNS1 cell line at a multiplicity of infection (MOI) of 0.1 (Fig. 2A), with the highest titer being about 1 × 108 IU/ml at 72 hpi. The replication foci of WNV-ΔNS1 and WT viruses were also similar when detected using an immunostaining-based focus-forming assay (Fig. 2B). After concentration and purification through polyethylene glycol 8000 (PEG8000) precipitation and ultracentrifugation, both viruses were analyzed by SDS-PAGE with Coomassie brilliant blue staining. Two bands, at about 55 and 15 kDa, corresponding to viral envelope (E) and capsid (C) proteins, respectively, were observed in both WNV-ΔNS1 and WT samples (Fig. 2C). The identities of the E and C bands were further confirmed by Western blotting with specific antibodies (Fig. 2D).
FIG 2.
Characterization of WNV-ΔNS1. (A) Comparison of growth kinetics between WT WNV and WNV-ΔNS1 virus in VeroNS1 cells. VeroNS1 cells were infected with either WT WNV or WNV-ΔNS1 virus at an MOI of 0.1. The supernatants were harvested at the indicated time points, and viral titers were determined as described above. Data represent the mean ± standard deviation of triplicate measurements in a representative experiment. n.s., no statistical difference. (B) Comparison of immunostained foci between WT WNV and WNV-ΔNS1 virus in VeroNS1 cells. The plaque morphology of WNV-ΔNS1 was analyzed by a standard immunostaining-based focus-forming assay and compared with that of WT virus. (C) Coomassie brilliant blue staining analysis of purified WT WNV and WNV-ΔNS1 virions. Following concentration and purification through PEG8000 precipitation and ultracentrifugation, equal amounts of purified WT and ΔNS1 virions were loaded onto a SDS-polyacrylamide gel, and protein bands were visualized by Coomassie brilliant blue staining. The bands corresponding to E and capsid proteins are indicated. (D) Western blotting analysis of purified WT WNV and WNV-ΔNS1 virions. The virus antigens described above were analyzed by Western blotting with an anti-E monoclonal antibody and anti-C polyclonal antibody.
To examine the antigenicity of WNV-ΔNS1, we performed neutralization assays in VeroNS1 cells using two different monoclonal neutralization antibodies against WNV envelope. Dose-dependent decreases in viral infection were observed for WT WNV and WNV-ΔNS1 (Fig. 3A and B). In addition, the antisera against WNV could recognize WNV-ΔNS1 coated on enzyme-linked immunosorbent assay (ELISA) plates, suggesting that the WNV-ΔNS1 virion remains antigenically intact (Fig. 3C). These results indicate that the WNV-ΔNS1 virion has antigenic properties similar to those of WT WNV.
FIG 3.
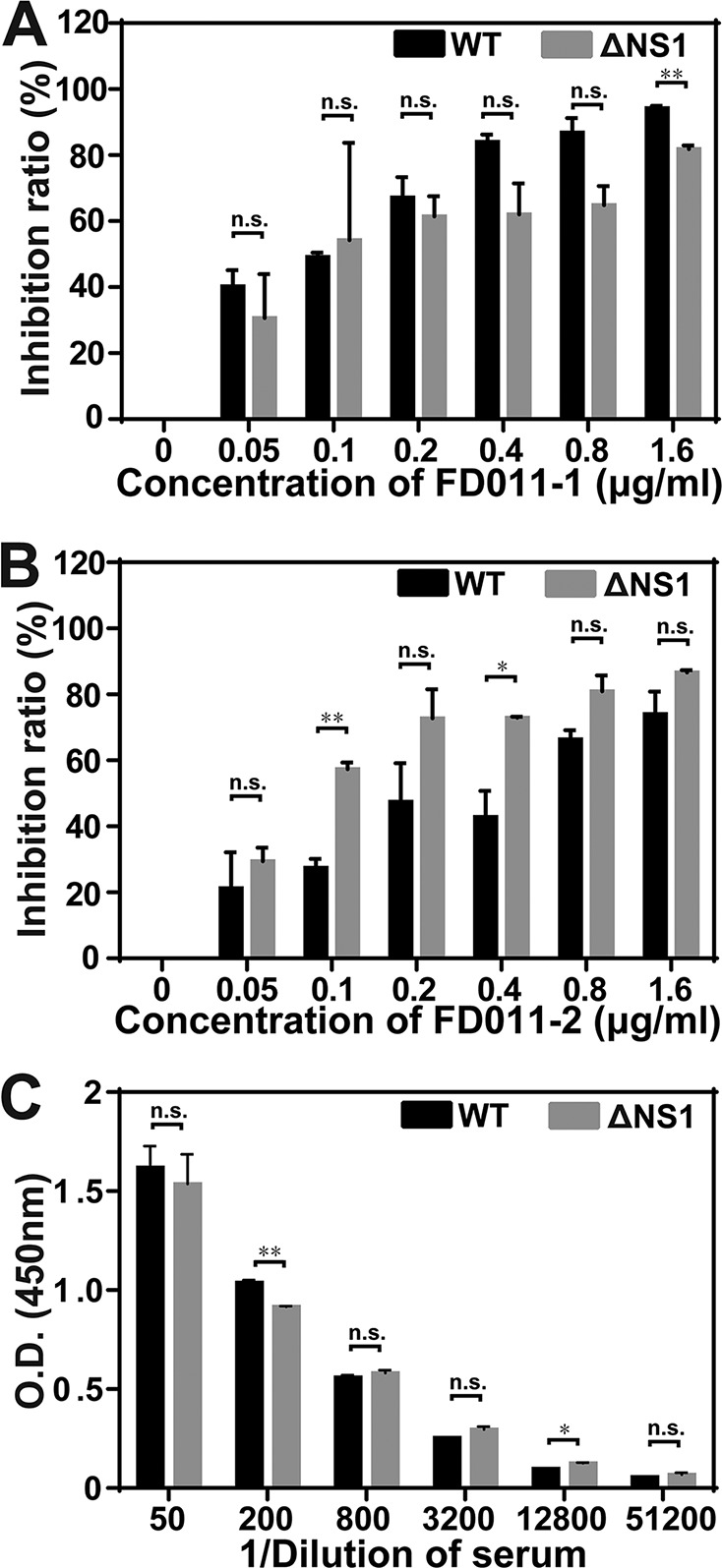
Immunogenicity of WNV-ΔNS1. (A and B) PRNT50 assay. PRNT50 testing against both WT WNV and WNV-ΔNS1 was performed with two different neutralizing antibodies against WNV envelope, i.e., FD011-1 (A) and FD011-2 (B). Serial 2-fold dilutions of neutralizing antibodies were incubated with WT WNV or WNV-ΔNS1 viruses. The inhibition rates were calculated by defining no-antibody treatment of WT WNV (0 μg/ml) as 0%. (C) ELISA of the antisera against WNV and WNV-ΔNS1. Two independent experiments were performed in triplicate. Data represent the mean ± standard deviation of triplicate measurements in a representative experiment. The asterisks denote statistical differences between the indicated groups. *, P < 0.05; **, P < 0.01; n.s., no statistical difference.
Stability and safety of WNV-ΔNS1.
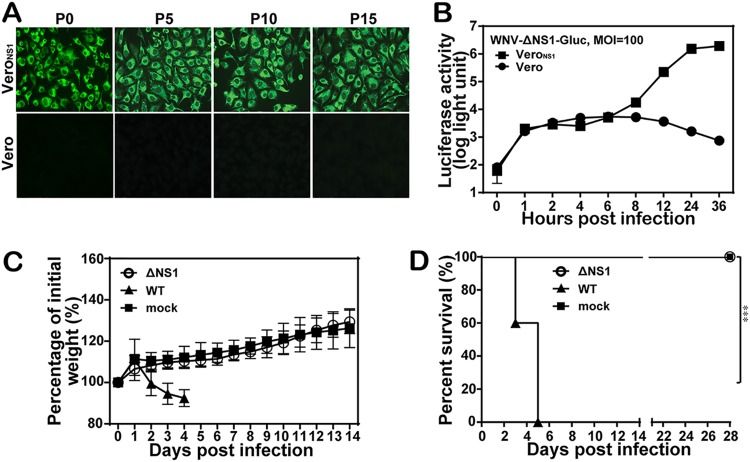
For stability testing, WNV-ΔNS1 was passaged serially on VeroNS1 cell lines for 15 rounds (passages 0 to 15). The resulting passage 0 to passage 15 WNV-ΔNS1 viruses were used to infect VeroNS1 and naive Vero cells. All passaged WNV-ΔNS1 viruses produced robust indirect immunofluorescence assay (IFA)-positive signals in VeroNS1 cells, whereas they did not generate any IFA-positive signals in naive Vero cells (Fig. 4A), suggesting that WNV-ΔNS1 was genetically stable and did not recombine with the WT NS1 sequence in the VeroNS1 cells. To further confirm the replication defect of WNV-ΔNS1, we infected both naive Vero and VeroNS1 cells with the Gaussia luciferase (Gluc) reporter WNV-ΔNS1-Gluc at an MOI of 100. As shown in Fig. 4B, viral translations were observed in both cell lines at 1 to 6 hpi with similar luciferase activities, whereas only VeroNS1 cell lines produced significantly increased luciferase activities after 8 hpi (representing viral replication). The results demonstrated that WNV-ΔNS1-Gluc could replicate only in VeroNS1 cells and not in naive Vero cells. To further evaluate the biosafety of WNV-ΔNS1, IFNAR−/− mice were inoculated intraperitoneally (i.p.) with 2 × 108 IU of WNV-ΔNS1 or 102 IU of WT WNV (Fig. 4C). The mice infected with WNV-ΔNS1 showed no signs of illness, with steadily increasing body weight (Fig. 4C), and survived during the entire observation period (Fig. 4D). In contrast, WT WNV caused disease in mice, such as ruffled fur and weight loss from day 2 postinfection (Fig. 4C), and all mice died by day 5 (Fig. 4D). These data demonstrate that WNV-ΔNS1 is not pathogenic in IFNAR−/− mice. Overall, the results showed that WNV-ΔNS1 is genetically stable in VeroNS1 cells and safe in IFNAR−/− mice.
FIG 4.
Stability and safety of WNV-ΔNS1. (A) IFA detection of WNV-ΔNS1 virus at different passages using the 4G2 monoclonal antibody. WNV-ΔNS1 viruses were blind passaged for 15 rounds in VeroNS1 cells. The viruses at passages 0, 5, 10, and 15 were used to infect naive VeroNS1 cells and Vero cells. (B) Detection of the Gluc activities in cell lysates at different time points after WNV-ΔNS1-Gluc infection. Naive VeroNS1 cells and Vero cells were infected with WNV-ΔNS1-Gluc reporter virus at an MOI of 100, and the cell lysates collected at the indicated time points were subjected to the luciferase assay. Data represent the mean ± standard deviation of triplicate measurements in a representative experiment. (C and D) Morbidity (C) and survival (D) analyses of IFNAR−/− mice after inoculation with WT and WNV-ΔNS1 viruses. Six-week-old IFNAR−/− female mice were inoculated i.p. with 102 IU of WT WNV or 2 × 108 IU of WNV-ΔNS1. All mice (n = 5 for each group) were monitored for morbidity and death for 28 days. Two experiments were performed independently. The asterisks denote statistical differences between the indicated groups. ***, P < 0.001.
Protection of C57BL/6 mice against WNV challenge.
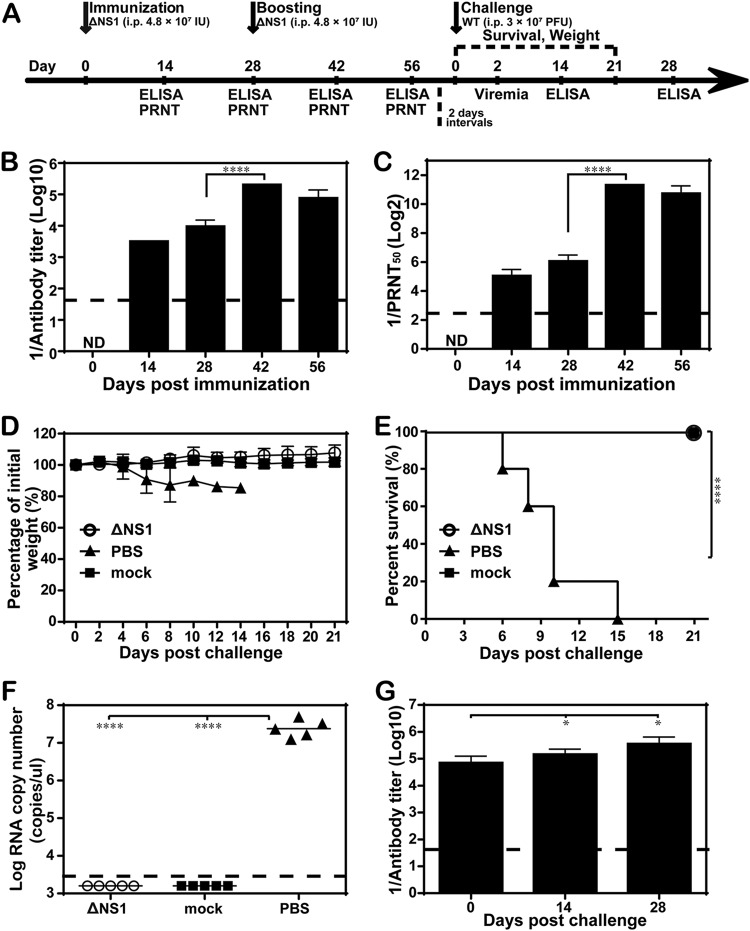
To assess the potential of WNV-ΔNS1 as a vaccine, we examined the antibody response induced by WNV-ΔNS1 infection in immunocompetent C57BL/6 mice. Four-week-old C57BL/6 mice (n = 5 in each group) were immunized with 4.8 × 107 IU of WNV-ΔNS1, with a booster with the same dose 28 days after the first immunization (Fig. 5A). As shown in Fig. 5B, WNV-ΔNS1 immunization induced robust ELISA antibody titers on day 14 (1:3,200) and day 28 (1:9,600); the booster on day 28 resulted in an additional increase in antibody titers to 1:204,800 on day 42 and afterward (Fig. 5B). In agreement with the ELISA titers, neutralizing antibody titers reached 1:33, 1:66, 1:2,560, and 1:1,706 on days 14, 28, 42, and 56, respectively (Fig. 5C). The results demonstrated that WNV-ΔNS1 could induce a robust antibody response.
FIG 5.
Antibody responses of mice immunized with WNV-ΔNS1 virus. (A) Animal experimental schedule. Four-week-old C57BL/6 female mice (n = 5 for each group) were i.p. immunized with 4.8 × 107 IU of WNV-ΔNS1, followed by the same dose as a booster immunization 28 days later. (B and C) ELISA and PRNT50 assay. Sera were harvested from mice at the indicated time points to determine the IgG antibody and neutralizing antibody titers against WNV, using the ELISA (B) and PRNT50 assay (C), respectively. ND, not detected. (D and E) Morbidity and survival analyses. All mice were i.p. injected with 3 × 107 PFU of WT WNV 30 days after the booster immunization, and weight loss (D) and survival (E) were monitored for 21 days after the challenge. (F) Viremia levels. Serum viremia levels were measured by real-time RT-PCR on day 2 postchallenge. Mice without any treatment (mock) were used as a negative control. (G) Postchallenge IgG antibody titers. On day 14 and day 28 after challenge, IgG antibody titers in sera were quantified using ELISA. The dashed lines in panels B, C, F, and G represent the limits of detection. Data represent the mean ± standard deviation of 5 mice at each time point in each group. The asterisks denote statistical differences between day 28 and day 42, which indicated that the booster dose resulted in enhanced antibody titers. *, P < 0.05; ****, P < 0.0001; n.s., no statistical difference. Two independent experiments were performed, and data from one experiment are presented.
To determine the protective efficacy of WNV-ΔNS1 virus, both phosphate-buffered saline (PBS)- and WNV-ΔNS1-immunized mice were i.p. challenged with 3 × 107 PFU of WT WNV at 30 days after booster immunization (Fig. 5A). As expected, all PBS-immunized mice started to lose weight on day 6 postchallenge (Fig. 5D), displayed signs of disease (such as ruffled fur and lameness), and succumbed to infection by 15 days postchallenge (Fig. 5E). In contrast, the WNV-ΔNS1-vaccinated mice survived (Fig. 5E) without any signs of sickness or body weight loss (Fig. 5D) in 21 days. The viremia levels were measured on day 2 postchallenge (Fig. 5F). All PBS-immunized mice developed a high level of viremia (viral RNA copy numbers of >107 copies/μl), compared to no detectable viral RNA from the WNV-ΔNS1-vaccinated group (Fig. 5F). IgG antibody titers against WNV increased 1.6-fold and 4-fold at day 14 and day 28 postchallenge, respectively (Fig. 5G). Taken together, these results demonstrated that WNV-ΔNS1 virus could protect mice from a highly lethal challenge with WT WNV.
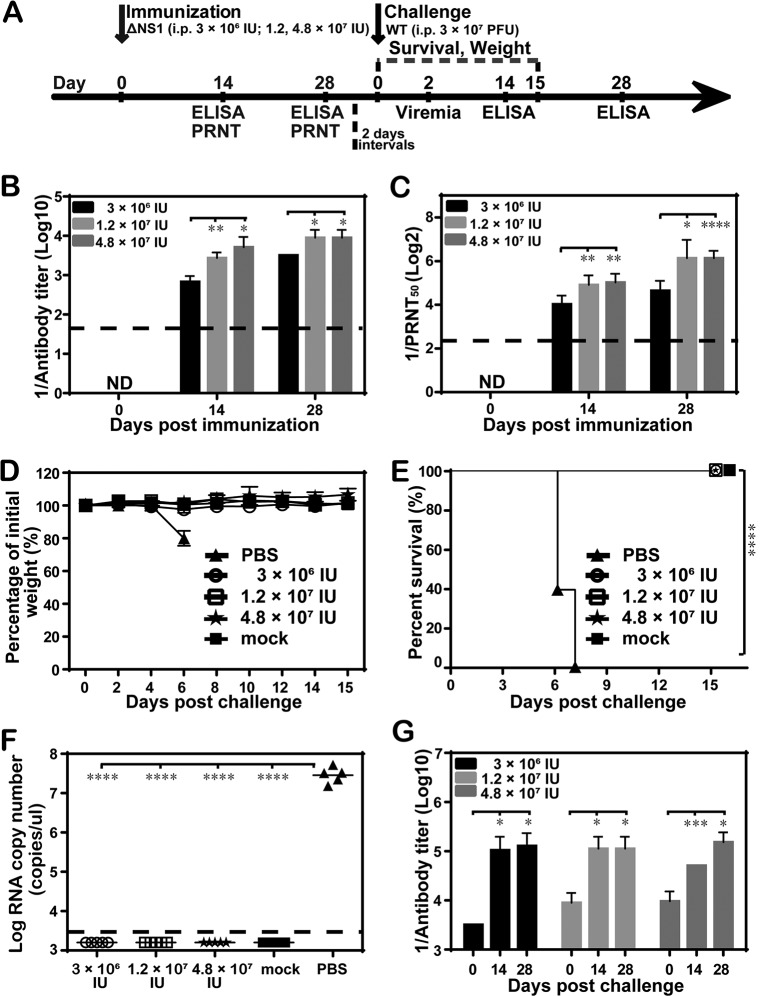
Protective efficacy of single immunization with WNV-ΔNS1 in C57BL/6 mice.
To further determine whether one-shot vaccination with WNV-ΔNS1 was sufficient to elicit protective immunity against WNV, C57BL/6 mice were immunized with WNV-ΔNS1 only once before challenge. Three dosages (3 × 106 IU, 1.2 × 107 IU, and 4.8 × 107 IU) of WNV-ΔNS1 were evaluated. Sera collected on days 14 and 28 were subjected to ELISA (Fig. 6A). The three dosages induced antibody responses in a dose- and time-dependent manner (Fig. 6B and C). On day 14, the average IgG titers were 1:800, 1:3,200, and 1:3,200 for the groups treated with 3 × 106 IU, 1.2 × 107 IU, and 4.8 × 107 IU, respectively; on day 28, the antibody titers rose to 1:3,200, 1:9,600, and 1:9,600, respectively. Corroborating the ELISA IgG titers, neutralizing antibody titers reached 1:20, 1:26, and 1:33 on day 14 and 1:26, 1:80, and 1:66 on day 28 for the lowest to the highest dosages, respectively (Fig. 6C). At 30 days postimmunization, all mice, including PBS-vaccinated ones, were i.p. challenged with 3 × 107 PFU of WT WNV. Mice were then monitored daily for weight loss and visible signs of disease. All mice vaccinated with WNV-ΔNS1, even at the lowest vaccine dosage, were fully protected and survived without any significant weight loss or detectable viremia, whereas the mice immunized with PBS exhibited severe disease or death, weight loss, and high levels of viremia (Fig. 6D to F). Additionally, the IgG antibody titers were measured at 0, 14, and 28 days postchallenge. The titers increased significantly in mice immunized with different dosages of WNV-ΔNS1, relative to their counterparts prior to challenge (Fig. 6B and G). Overall, our results illustrated that WNV-ΔNS1 could protect mice from a highly lethal challenge with only one dose of immunization.
FIG 6.
Protective efficacy of one-shot immunization with WNV-ΔNS1 in C57BL/6 mice. (A) Animal experimental schedule. (B and C) Antibody titer measurements. Four-week-old C57BL/6 female mice (n = 5 for each group) were i.p. immunized once with different dosages of WNV-ΔNS1 (3 × 106 IU, 1.2 × 107 IU, or 4.8 × 107 IU), and sera were collected on day 14 and day 28 for IgG antibody (B) and neutralizing antibody (C) titer measurements. ND, not detected. (D to G) Analyses of morbidity, survival, and virus and antibody levels. All mice were i.p. injected with 3 × 107 PFU of WT WNV at 30 days postimmunization and monitored for weight loss (D), survival (E), viremia (F), and postchallenge IgG antibody titers (G). The dashed lines in panels B, C, and F represent the limit of detection. Data represent the mean ± standard deviation of 5 mice at each time point in each group. The asterisks denote statistical differences between the indicated groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
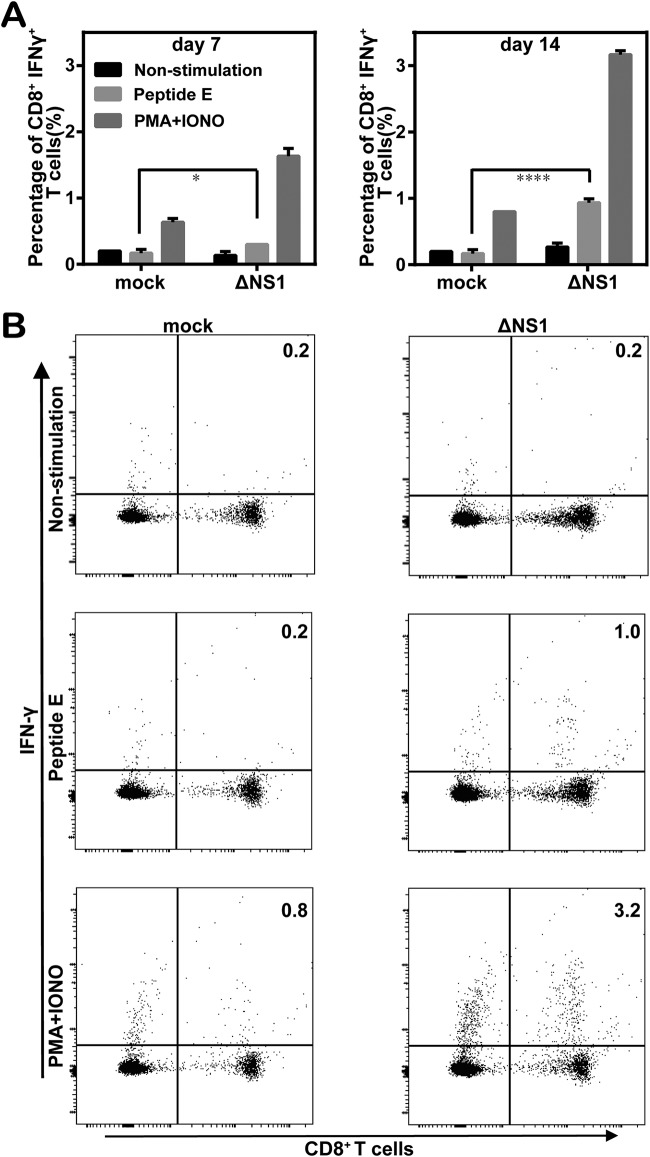
CD8+ T cell response in vaccinated mice.
To assess T cell responses in immunized mice, the mice were immunized with 2 × 108 IU WNV-ΔNS1, with a booster on day 28 after primary immunization. Splenocytes were isolated from mice on day 7 and day 14 after booster immunization, and the levels of CD8+ T cell activation were analyzed by flow cytometry after stimulation with the peptide of CD8+ T cell epitopes corresponding to the WNV envelope. Nonspecific stimulation with phorbol myristate acetate (PMA) and ionomycin (IONO) was used as a positive control. Time-dependent enhancement of IFN-γ production by CD8+ T cells (IFN-γ+ CD8+ cells) was observed in the WNV-ΔNS1-vaccinated mice in response to either specific (peptide E) or nonspecific (PMA plus IONO) stimulation. In contrast, antigen-specific stimulation did not cause any increase in the percentage of IFN-γ+ CD8+ cells in the mock-vaccinated group. There were significant differences in the percentages of antigen-specific IFN-γ+ CD8+ cells between the mock- and WNV-ΔNS1-vaccinated mice (0.17% versus 0.3% on day 7 [P < 0.05] and 0.17% versus 0.93% on day 14 [P < 0.0001]) (Fig. 7A). Taken together, these data suggested that WNV-ΔNS1 could induce T cell responses in mice.
FIG 7.
CD8+ T cell responses in WNV-ΔNS1-vaccinated C57BL/6 mice. (A) Summary of the percentages of IFN-γ+ CD8+ T cells in the total number of CD8+ T cells without stimulation (non-stimulation) or with stimulation with WNV peptide (peptide E) or PMA and IONO at 7 and 14 days after booster immunization. Mock-vaccinated mice were used as a negative control. Data represent the mean ± standard deviation of 5 mice at each time point in each group. The asterisks denote statistical differences between the indicated groups. *, P < 0.05; ****, P < 0.0001. (B) Representative flow cytometry contour plots of IFN-γ+ CD8+ T cells on day 14 after booster immunization. The CD8+ T cells were gated, and the percentages of IFN-γ+ CD8+ cells are indicated in the top right corners.
DISCUSSION
In this study, we assessed the potential of the replication-defective virus WNV-ΔNS1 as a vaccine candidate. WNV-ΔNS1 provides complete protection against WT WNV infection in mice, with strong neutralizing antibody responses. Viremia was not observed in the vaccinated mice, compared with the mock-vaccinated mice. No specific safety-associated adverse effects related to the vaccine were observed. Although effective CD8+ T cell responses were also observed in mice receiving the high dose (2 × 108 IU), the role of the cellular response in preventing WNV infection with WNV-ΔNS1 needs further study. CD8+ T cell depletion experiments following vaccination and prior to challenge (20, 34) will help elucidate how cellular responses contribute to survival and viral clearance.
WNV is a biosafety level 3 (BSL3) pathogen. We tried to obtain a balance between safety and immunogenicity by using WNV-ΔNS1 as a vaccine through trans complementation of NS1. WNV-ΔNS1 was safe in IFNAR−/− mice, without causing any symptoms of disease (Fig. 4C). The safety profile of WNV-ΔNS1 can be explained by the fact that WNV-ΔNS1 undergoes only viral entry and translation, without viral replication, in naive cells (Fig. 4B). This safety property is stably maintained after serial passaging in the VeroNS1 cell line (Fig. 4A). The replication-defective nature of WNV-ΔNS1 in normal cells, despite its ability to replicate to exceptionally high titers in VeroNS1 cells, highlights the safety advantage of WNV-ΔNS1 as a vaccine.
Owing to the defective replication of WNV-ΔNS1 in normal cells, a chemical or physical inactivation step is not required for the production of this vaccine candidate. Thus, native and functional antigens could be maximally preserved to provide an antigen presentation capacity as native virus particles (35). Without the need for inactivation, WNV-ΔNS1 combines the ease of production of live-attenuated vaccines and the safety of inactivated vaccines with immunogenicity and can be manufactured at relatively low cost. It should be noted that the viral titers were much higher in the complementary VeroNS1 cells than in the previously reported 293TNS1 cells (Fig. 1B). This may result from the inability of Vero cells to generate type I IFN (36). Additionally, different cell lines for selection may have different NS1 expression levels, which may lead to different complementing efficiencies.
In summary, our results showed that WNV-ΔNS1 produced from an NS1 complementary cell line provides a new flavivirus vaccine platform. Due to its nonreplicative nature, this vaccine platform is safe and immunogenic. We think that the NS1 complementation approach could be applied to vaccine development for other highly pathogenic flaviviruses, for which safety is one of the major concerns.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The VeroNS1 cell line was established following the same method as described previously for the generation of 293TNS1 (11). VeroNS1 and 293TNS1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) containing 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 5 μg/ml or 2 μg/ml of puromycin (Invitrogen), respectively, in 5% CO2 at 37°C. BHK-21 and Vero cells were grown in DMEM as described above without puromycin. Recombinant WT WNV was generated by electroporation of BHK-21 cells with in vitro-transcribed viral genomic RNA from the linearized infectious cDNA clone of strain 3356 from New York City (37, 38). The backbone of the WT WNV cDNA clone was used to construct WNV-ΔNS1 and WNV-ΔNS1-Gluc vectors, which were originally produced in the 293TNS1 cell line from a previous study (11), and the resultant replication-deficient WNV-ΔNS1 and WNV-ΔNS1-Gluc viruses were amplified in the VeroNS1 cell line in DMEM as described above but supplemented with 2% FBS. The supernatants of transfected cells were harvested at 72 h posttransfection and frozen at −80°C.
Monoclonal antibody 4G2 against the flavivirus envelope protein was kindly provided by Cheng-Feng Qin (Beijing Institute of Microbiology and Epidemiology, Beijing, China). Monoclonal neutralization antibodies FD011-1 and FD011-2 against WNV envelope were from the State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology (38). Anti-WNV capsid polyclonal antibody was reclaimed from the sera of BALB/c mice immunized with purified capsid proteins. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse/rabbit IgG and horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit IgG secondary antibodies were purchased from Proteintech (China).
Mice.
Four-week-old C57BL/6 mice were used for virus immunization and challenge studies, and 6-week-old type I IFN receptor-deficient IFNAR−/− mice, which are sensitive to WNV infection, were used to test the biosafety of WNV-ΔNS1. All mouse infection experiments were performed in an animal BSL3 facility at Wuhan Institute of Virology under a protocol approved by the Laboratory Animal Ethics Committee of Wuhan Institute of Virology (permit WIVA26201801).
Viral titer quantification with immunofluorescence assay.
VeroNS1 cells seeded on coverslips in 12-well plates (2 × 105 VeroNS1 cells per well) were infected with 10-fold dilutions of WNV or WNV-ΔNS1 for 1 h at 37°C and then overlaid with 2% methylcellulose. At 36 hpi, the infected cells were washed three times with PBS and then fixed with cold 5% acetone in methanol for 15 min at room temperature. The fixed cells were stained with 4G2 antibody for 1 h at room temperature. After three washes with PBS, the cells were incubated with FITC-conjugated goat anti-mouse IgG polyclonal secondary antibody. The coverslips were mounted on glass slides with 90% glycerol after three washes with PBS. Images were captured under a fluorescence microscope (Nikon Eclipse TE2000). The infectious titer of viruses was calculated as IU per milliliter.
Immunostaining-based focus-forming assay.
To compare the plaque sizes of WT WNV and WNV-ΔNS1, an immunostaining-based focus-forming assay was performed as described previously (39), in 12-well plates seeded with 2 × 105 VeroNS1 cells per well. The 4G2 antibody and goat anti-mouse IgG conjugated with HRP were used as primary and secondary antibodies, respectively. An enhanced HRP-3,3'-diaminobenzidine (DAB) chromogenic substrate kit (Tiangen) was used for color development.
Virus growth kinetics.
The virus growth kinetics of both WNV and WNV-ΔNS1 were assessed in VeroNS1 cells. The cells (in 12-well plates, at 2 × 105 cells per well) were infected with viruses at an MOI of 0.1. At the indicated times postinfection, the culture medium was collected and subjected to viral titer quantification as described above.
Concentration and purification of viral particles.
To obtain highly purified and concentrated viral stocks, four 175-cm2 flasks of VeroNS1 cells were infected with WT or WNV-ΔNS1 at an MOI of 0.1. The supernatants were reclaimed at 72 hpi and subjected to sequential centrifugation at 4°C for 10 min at 400 × g and for 20 min at 5,000 rpm, to remove cells and cell debris, respectively. The clarified supernatants were then incubated overnight at 4°C with PBS containing 8% (wt/vol) PEG8000 (Sigma), followed by centrifugation at 4°C for 50 min at 10,500 × g. The pellets were then gently resuspended in PBS and subjected to ultracentrifugation at 4°C for 1.5 h at 105,000 × g with a 24% sucrose cushion, using a SW41 rotor in an Optima MAX-XP ultracentrifuge (Beckman). The pellets were finally resuspended in 50 μl of PBS.
Gel electrophoresis.
Purified virions were denatured by heating at 95°C for 10 min in SDS sample buffer containing β-mercaptoethanol and were resolved on a 15% SDS-PAGE gel. The gel was then stained with Coomassie brilliant blue or subjected to Western blotting analysis, as described previously (11).
Luciferase assay.
The infected cells were lysed with 200 μl of lysis buffer (Thermo Scientific) and subjected to the luciferase assay according to the manufacturer’s instructions. Briefly, 20 μl of cell lysate was mixed with 50 μl of substrate (Thermo Scientific), followed by immediate measurement on a multimode microplate reader (Varioskan Flash; Thermo Fisher, Finland). The relative light units (RLUs) were recorded for each sample. All luciferase assays were performed in triplicate.
Animal safety test.
For safety evaluations, 6-week-old female IFNAR−/− mice were infected i.p. with 102 IU of WT WNV or 2 × 108 IU of WNV-ΔNS1. The animals were monitored for survival and body weight loss for 28 days.
Immunization and challenge studies.
Four-week-old female C57BL/6 mice were i.p. immunized with 4.8 × 107 IU of WNV-ΔNS1, followed by the same dose of booster 28 days later; unvaccinated mice were immunized with PBS as a negative control. At 30 days after booster vaccination, all mice were challenged i.p. with 3 × 107 PFU of WT WNV and monitored for survival and weight loss for 21 days. For one-shot immunization, mice were i.p. immunized once with 3 × 106, 1.2 × 107, or 4.8 × 107 IU of WNV-ΔNS1, challenged i.p. with 3 × 107 PFU of WT WNV at 30 days postimmunization, and monitored for weight loss and survival for 15 days. Viremia was measured, with a real-time reverse transcription (RT)-PCR assay, in the serum of challenged and unchallenged (mock-treated) mice at 2 days after challenge, as described previously (39); the limit of detection was 103.3 viral RNA copies/μl. Total RNA was extracted from 100 μl blood of mice using TRIzol reagent (TaKaRa, China), according to the manufacturer’s protocols. Postchallenge IgG antibody titers were also quantified on day 14 and day 28 after challenge.
Antibody titers.
WNV-specific IgG antibody levels of vaccinated mice were examined with an ELISA. Briefly, 96-well plates were coated with WNV-ΔNS1 and blocked with PBS containing 5% skim milk. The coated wells were incubated with 4-fold serial dilutions of heat-inactivated (56°C for 30 min) sera from vaccinated mice, starting with a 1:50 dilution, and the HRP-conjugated goat anti-mouse IgG secondary antibody. A two-component 3,3',5,5'-tetramethylbenzidine (TMB) color development kit (Beyotime Biotechnology) was used for color development for detection of bound antibodies. The optical density at 450 nm was measured after the addition of 1 M H2SO4 stop solution, using a multimode microplate reader (Varioskan Flash; Thermo Fisher), according to the manufacturer’s instructions. The IgG antibody titers were defined as the highest dilution of serum giving an optical density twice that of the nonimmune serum (20, 26, 40, 41).
The neutralizing antibody titers were determined with the standard 50% plaque reduction/neutralization titer (PRNT50) assay, as described previously (38, 39). Briefly, serial 2-fold dilutions of serum samples were preincubated with WT WNV for 1 h at 37°C and added to a monolayer of BHK-21 cells seeded into 12-well plates. The mixture was then replaced with a layer of 2% methylcellulose. After further incubation at 37°C in 5% CO2 for 3 days, the overlay was removed. The cells were then fixed with 3.7% formaldehyde containing 1% crystal violet for plaque formation, and the plaques were counted. The neutralizing antibody titers were calculated as the highest dilution of serum yielding a 50% reduction in virus control plaque numbers (42, 43).
Intracellular staining and flow cytometry.
C57BL/6 mice were immunized with 2 × 108 IU of WNV-ΔNS1, with a booster 28 days after the first immunization. On day 7 and day 14 after booster immunization, splenocytes were isolated from the spleens of immunized mice for cytokine analysis. Briefly, the isolated splenocytes were first stimulated with 1 μg/ml of WNV E peptide (IALTFLAV) (no. 3, Kb-restricted) (26, 34, 44, 45) or 50 ng/ml of PMA plus 1 μg/ml of IONO at 37°C for 5 h in the presence of GolgiPlug (1 μl/ml; BD Biosciences). The treated cells were then washed with staining buffer (2% FBS in PBS) and stained at 4°C for 30 min with anti-CD8 antibody conjugated with allophycocyanin (APC)-Cy7 (BD Pharmingen). After an additional wash, the cells were fixed and permeabilized with BD Fix/Perm buffer at 4°C for 20 min and stained with anti-IFN-γ antibody conjugated with phycoerythrin (PE)-Cy7 or an isotype control (BD Pharmingen). Finally, the cells were washed and analyzed by flow cytometry. The percentage of IFN-γ-positive CD8+ T cells in the total number of CD8+ T cells was calculated.
Statistical analysis.
The unpaired t test was used to determine whether there were significant (P < 0.05) differences for all experiments. Kaplan-Meier survival curves were analyzed by the log-rank test. The statistical analyses were performed using the nonparametric test in GraphPad Prism 5.0.
ACKNOWLEDGMENTS
We are grateful to the Core Facility and Technical Support staff (Pei Zhang, An-na Du, and Juan Min), the Center for Animal Experiments staff (Xue-fang An, Fan Zhang, He Zhao, and Li Li), and the BSL3 laboratory (Hao Tang) at the Wuhan Institute of Virology and the Wuhan Key Laboratory of Special Pathogens and Biosafety for helpful support during the course of the work.
This work was supported by the National Key Research and Development Program of China (grant 2016YFD0500400) and the National Natural Science Foundation of China (grant 81572003). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Tyler KL. 2014. Current developments in understanding of West Nile virus central nervous system disease. Curr Opin Neurol 27:342–348. doi: 10.1097/WCO.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Z, Fu SH, Cao L, Tang CJ, Zhang S, Li ZX, Tusong M, Yao XH, Zhang HL, Wang PY, Wumaier M, Yuan XY, Li MH, Zhu CZ, Fu LP, Liang GD. 2014. Human infection with West Nile virus, Xinjiang, China, 2011. Emerg Infect Dis 20:1421–1423. doi: 10.3201/eid2008.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anukumar B, Sapkal GN, Tandale BV, Balasubramanian R, Gangale D. 2014. West Nile encephalitis outbreak in Kerala, India, 2011. J Clin Virol 61:152–155. doi: 10.1016/j.jcv.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Suthar MS, Aguirre S, Fernandez-Sesma A. 2013. Innate immune sensing of flaviviruses. PLoS Pathog 9:e1003541. doi: 10.1371/journal.ppat.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Dong H, Zhou Y, Shi PY. 2008. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5' stem-loop of genomic RNA. J Virol 82:7047–7058. doi: 10.1128/JVI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Dong H, Ye H, Tilgner M, Shi PY. 2010. Genetic analysis of West Nile virus containing a complete 3'CSI RNA deletion. Virology 408:138–145. doi: 10.1016/j.virol.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Shi PY. 2014. Flavivirus NS5 prevents the inSTATement of IFN. Cell Host Microbe 16:269–271. doi: 10.1016/j.chom.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Rastogi M, Sharma N, Singh SK. 2016. Flavivirus NS1: a multifaceted enigmatic viral protein. Virol J 13:131. doi: 10.1186/s12985-016-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M Jr. 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crook KR, Miller-Kittrell M, Morrison CR, Scholle F. 2014. Modulation of innate immune signaling by the secreted form of the West Nile virus NS1 glycoprotein. Virology 458-459:172–182. doi: 10.1016/j.virol.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang HL, Ye HQ, Deng CL, Liu SQ, Shi PY, Qin CF, Yuan ZM, Zhang B. 2017. Generation and characterization of West Nile pseudo-infectious reporter virus for antiviral screening. Antiviral Res 141:38–47. doi: 10.1016/j.antiviral.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Lindenbach BD, Rice CM. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol 73:4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akey DL, Brown WC, Dutta S, Konwerski J, Jose J, Jurkiw TJ, DelProposto J, Ogata CM, Skiniotis G, Kuhn RJ, Smith JL. 2014. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343:881–885. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youn S, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, Diamond MS. 2012. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol 86:7360–7371. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol 73:10272–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenbach BD, Rice CM. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol 71:9608–9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muylaert IR, Chambers TJ, Galler R, Rice CM. 1996. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology 222:159–168. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- 18.Muylaert IR, Galler R, Rice CM. 1997. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol 71:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Throsby M, Ter Meulen J, Geuijen C, Goudsmit J, de Kruif J. 2007. Mapping and analysis of West Nile virus-specific monoclonal antibodies: prospects for vaccine development. Expert Rev Vaccines 6:183–191. doi: 10.1586/14760584.6.2.183. [DOI] [PubMed] [Google Scholar]
- 20.Pinto AK, Richner JM, Poore EA, Patil PP, Amanna IJ, Slifka MK, Diamond MS. 2013. A hydrogen peroxide-inactivated virus vaccine elicits humoral and cellular immunity and protects against lethal West Nile virus infection in aged mice. J Virol 87:1926–1936. doi: 10.1128/JVI.02903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer AV, Kousoulas KG. 2013. A review of vaccine approaches for West Nile virus. Int J Environ Res Public Health 10:4200–4223. doi: 10.3390/ijerph10094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amanna IJ, Slifka MK. 2014. Current trends in West Nile virus vaccine development. Expert Rev Vaccines 13:589–608. doi: 10.1586/14760584.2014.906309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zlatkovic J, Stiasny K, Heinz FX. 2011. Immunodominance and functional activities of antibody responses to inactivated West Nile virus and recombinant subunit vaccines in mice. J Virol 85:1994–2003. doi: 10.1128/JVI.01886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arroyo J, Miller C, Catalan J, Myers GA, Ratterree MS, Trent DW, Monath TP. 2004. ChimeriVax-West Nile virus live-attenuated vaccine: preclinical evaluation of safety, immunogenicity, and efficacy. J Virol 78:12497–12507. doi: 10.1128/JVI.78.22.12497-12507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteman MC, Li L, Wicker JA, Kinney RM, Huang C, Beasley DW, Chung KM, Diamond MS, Solomon T, Barrett AD. 2010. Development and characterization of non-glycosylated E and NS1 mutant viruses as a potential candidate vaccine for West Nile virus. Vaccine 28:1075–1083. doi: 10.1016/j.vaccine.2009.10.112. [DOI] [PubMed] [Google Scholar]
- 26.Welte T, Xie G, Wicker JA, Whiteman MC, Li L, Rachamallu A, Barrett A, Wang T. 2011. Immune responses to an attenuated West Nile virus NS4B-P38G mutant strain. Vaccine 29:4853–4861. doi: 10.1016/j.vaccine.2011.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XF, Zhao W, Lin F, Ye Q, Wang HJ, Yang D, Li SH, Zhao H, Xu YP, Ma J, Deng YQ, Zhang Y, Qin ED, Qin CF. 2013. Development of chimaeric West Nile virus attenuated vaccine candidate based on the Japanese encephalitis vaccine strain SA14-14-2. J Gen Virol 94:2700–2709. doi: 10.1099/vir.0.059436-0. [DOI] [PubMed] [Google Scholar]
- 28.Yamshchikov V, Manuvakhova M, Rodriguez E. 2016. Development of a human live attenuated West Nile infectious DNA vaccine: suitability of attenuating mutations found in SA14-14-2 for WN vaccine design. Virology 487:198–206. doi: 10.1016/j.virol.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Yamshchikov V, Manuvakhova M, Rodriguez E, Hebert C. 2017. Development of a human live attenuated West Nile infectious DNA vaccine: identification of a minimal mutation set conferring the attenuation level acceptable for a human vaccine. Virology 500:122–129. doi: 10.1016/j.virol.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Martin JE, Pierson TC, Hubka S, Rucker S, Gordon IJ, Enama ME, Andrews CA, Xu Q, Davis BS, Nason M, Fay M, Koup RA, Roederer M, Bailer RT, Gomez PL, Mascola JR, Chang GJ, Nabel GJ, Graham BS. 2007. A West Nile virus DNA vaccine induces neutralizing antibody in healthy adults during a phase 1 clinical trial. J Infect Dis 196:1732–1740. doi: 10.1086/523650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao F, Li XF, Yu XD, Deng YQ, Jiang T, Zhu QY, Qin ED, Qin CF. 2011. A DNA-based West Nile virus replicon elicits humoral and cellular immune responses in mice. J Virol Methods 178:87–93. doi: 10.1016/j.jviromet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Chang DC, Liu WJ, Anraku I, Clark DC, Pollitt CC, Suhrbier A, Hall RA, Khromykh AA. 2008. Single-round infectious particles enhance immunogenicity of a DNA vaccine against West Nile virus. Nat Biotechnol 26:571–577. doi: 10.1038/nbt1400. [DOI] [PubMed] [Google Scholar]
- 33.Ohtaki N, Takahashi H, Kaneko K, Gomi Y, Ishikawa T, Higashi Y, Todokoro M, Kurata T, Sata T, Kojima A. 2011. Purification and concentration of non-infectious West Nile virus-like particles and infectious virions using a pseudo-affinity Cellufine Sulfate column. J Virol Methods 174:131–135. doi: 10.1016/j.jviromet.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 34.Shrestha B, Ng T, Chu HJ, Noll M, Diamond MS. 2008. The relative contribution of antibody and CD8+ T cells to vaccine immunity against West Nile encephalitis virus. Vaccine 26:2020–2033. doi: 10.1016/j.vaccine.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erasmus JH, Auguste AJ, Kaelber JT, Luo H, Rossi SL, Fenton K, Leal G, Kim DY, Chiu W, Wang T, Frolov I, Nasar F, Weaver SC. 2017. A chikungunya fever vaccine utilizing an insect-specific virus platform. Nat Med 23:192–199. doi: 10.1038/nm.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chew T, Noyce R, Collins SE, Hancock MH, Mossman KL. 2009. Characterization of the interferon regulatory factor 3-mediated antiviral response in a cell line deficient for IFN production. Mol Immunol 46:393–399. doi: 10.1016/j.molimm.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol 76:5847–5856. doi: 10.1128/jvi.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang PT, Shan C, Li XD, Liu SQ, Deng CL, Ye HQ, Shang BD, Shi PY, Lv M, Shen BF, Qin CF, Zhang B. 2016. Generation of a recombinant West Nile virus stably expressing the Gaussia luciferase for neutralization assay. Virus Res 211:17–24. doi: 10.1016/j.virusres.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Zou G, Zhang B, Lim PY, Yuan Z, Bernard KA, Shi PY. 2009. Exclusion of West Nile virus superinfection through RNA replication. J Virol 83:11765–11776. doi: 10.1128/JVI.01205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shrestha B, Wang T, Samuel MA, Whitby K, Craft J, Fikrig E, Diamond MS. 2006. Gamma interferon plays a crucial early antiviral role in protection against West Nile virus infection. J Virol 80:5338–5348. doi: 10.1128/JVI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Chen H, Wu N, Fan D, Liang G, Gao N, An J. 2013. Characterization of immune responses induced by inactivated, live attenuated and DNA vaccines against Japanese encephalitis virus in mice. Vaccine 31:4136–4142. doi: 10.1016/j.vaccine.2013.06.099. [DOI] [PubMed] [Google Scholar]
- 42.Li SH, Dong H, Li XF, Xie X, Zhao H, Deng YQ, Wang XY, Ye Q, Zhu SY, Wang HJ, Zhang B, Leng QB, Zuest R, Qin ED, Qin CF, Shi PY. 2013. Rational design of a flavivirus vaccine by abolishing viral RNA 2'-O methylation. J Virol 87:5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrovsky N, Larena M, Siddharthan V, Prow NA, Hall RA, Lobigs M, Morrey J. 2013. An inactivated cell culture Japanese encephalitis vaccine (JE-ADVAX) formulated with delta inulin adjuvant provides robust heterologous protection against West Nile encephalitis via cross-protective memory B cells and neutralizing antibody. J Virol 87:10324–10333. doi: 10.1128/JVI.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purtha WE, Myers N, Mitaksov V, Sitati E, Connolly J, Fremont DH, Hansen TH, Diamond MS. 2007. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur J Immunol 37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- 45.Brien JD, Uhrlaub JL, Nikolich-Zugich J. 2007. Protective capacity and epitope specificity of CD8+ T cells responding to lethal West Nile virus infection. Eur J Immunol 37:1855–1863. doi: 10.1002/eji.200737196. [DOI] [PubMed] [Google Scholar]