There is limited information about the uncoating of enteroviruses under physiological conditions. Here, we focused on physiologically relevant factors that likely contribute to opening of echovirus 1 and other B-group enteroviruses. By combining biochemical and structural data, we show that, before entering cells, extracellular albumin is capable of priming the virus into a metastable yet infectious intermediate state. The ionic changes that are suggested to occur in endosomes can further contribute to uncoating and promote genome release, once the viral particle is endocytosed. Importantly, we provide a detailed high-resolution structure of a virion after treatment with albumin and a preset ion composition, showing pocket factor release, capsid expansion, and fenestration and the clearly visible genome still anchored to the capsid. This study provides valuable information about the physiological factors that contribute to the opening of B group enteroviruses.
KEYWORDS: cryoEM structure, albumin, biochemistry, enterovirus, fatty acids, ions, uncoating, virology
ABSTRACT
There is limited information about the molecular triggers leading to the uncoating of enteroviruses under physiological conditions. Using real-time spectroscopy and sucrose gradients with radioactively labeled virus, we show at 37°C, the formation of albumin-triggered, metastable uncoating intermediate of echovirus 1 without receptor engagement. This conversion was blocked by saturating the albumin with fatty acids. High potassium but low sodium and calcium concentrations, mimicking the endosomal environment, also induced the formation of a metastable uncoating intermediate of echovirus 1. Together, these factors boosted the formation of the uncoating intermediate, and the infectivity of this intermediate was retained, as judged by end-point titration. Cryo-electron microscopy reconstruction of the virions treated with albumin and high potassium, low sodium, and low calcium concentrations resulted in a 3.6-Å resolution model revealing a fenestrated capsid showing 4% expansion and loss of the pocket factor, similarly to altered (A) particles described for other enteroviruses. The dimer interface between VP2 molecules was opened, the VP1 N termini disordered and most likely externalized. The RNA was clearly visible, anchored to the capsid. The results presented here suggest that extracellular albumin, partially saturated with fatty acids, likely leads to the formation of the infectious uncoating intermediate prior to the engagement with the cellular receptor. In addition, changes in mono- and divalent cations, likely occurring in endosomes, promote capsid opening and genome release.
IMPORTANCE There is limited information about the uncoating of enteroviruses under physiological conditions. Here, we focused on physiologically relevant factors that likely contribute to opening of echovirus 1 and other B-group enteroviruses. By combining biochemical and structural data, we show that, before entering cells, extracellular albumin is capable of priming the virus into a metastable yet infectious intermediate state. The ionic changes that are suggested to occur in endosomes can further contribute to uncoating and promote genome release, once the viral particle is endocytosed. Importantly, we provide a detailed high-resolution structure of a virion after treatment with albumin and a preset ion composition, showing pocket factor release, capsid expansion, and fenestration and the clearly visible genome still anchored to the capsid. This study provides valuable information about the physiological factors that contribute to the opening of B group enteroviruses.
INTRODUCTION
The Enterovirus B species consists of tens of clinically relevant viruses, including over 30 serotypes of echoviruses and coxsackievirus B1 to B6 and A9. They can cause a wide variety of mild and severe infections, and many of them are also associated with the onset of type I diabetes and coeliac disease (1–4). These viruses infect primarily gut epithelial cells and are naturally stabile in an acidic environment. The latter important feature makes the enterovirus B (EV-B) group viruses different from, for example, rhinoviruses that are known to rely on acidification during virus uncoating (5, 6).
Earlier, we showed that echovirus 1 (E1) and coxsackievirus A9 trigger a clathrin-independent entry pathway (7–10). Within 15 min of entry, viruses are first localized in the endosomes that develop into pH-neutral multivesicular bodies. The genome uncoating continues for up to 2 h postinfection (8, 11, 12). A large number of host cell factors have been pinpointed as important regulators for the entry of enteroviruses (13). However, there is very little information on the possible extracellular and cointernalized soluble factors that potentially contribute to uncoating during infection.
Recently, we described a novel uncoating intermediate particle of E1 which formed during infection and could be isolated from the cells at early stages of infection (14). This particle proved to be stable, infectious, containing all of its capsid proteins, and still capable of receptor binding (14). Previously, several studies characterized uncoating intermediates for entero- and rhinoviruses that have been termed as A- or 135S-particles based on their altered conformation and lighter sedimentation in sucrose gradients (15). The formation of these particles has been suggested to be induced by receptor binding (16, 17), low pH (18), or by nonphysiological high temperatures (19). They have also been found to exist in purified virus preparations (20). One study showed that treatment with fatty acid-free bovine serum albumin (BSA) converts echovirus 12 into A-particles, but the mechanism of action was not investigated further (21). In addition, there are only few studies reporting what effects other physiological factors, such as changes in ionic conditions, may have on the virus particle (21–25). X-ray crystallography as well as cryo-electron microscopy (cryoEM) and single particle reconstruction have been used to gain structural information on picornavirus particles at different stages before genome release (20, 26–33). Due to methodological challenges, it has been difficult to exactly map the spatiotemporal events during the uncoating process and to link that to structural information. Furthermore, the physiological conditions in the tissues where virus infection takes place have not been carefully studied.
Here we show that albumin triggers the uncoating process of E1 at 37°C, in a manner dependent upon the balance between fatty acids and albumin present. We further show that changes in mono- and divalent cations, likely reflecting the endosomal concentrations, also trigger a slower uncoating process of E1, which is clearly boosted by albumin.
RESULTS
Serum at physiological temperature drives transformation from intact E1 virion to an uncoating intermediate.
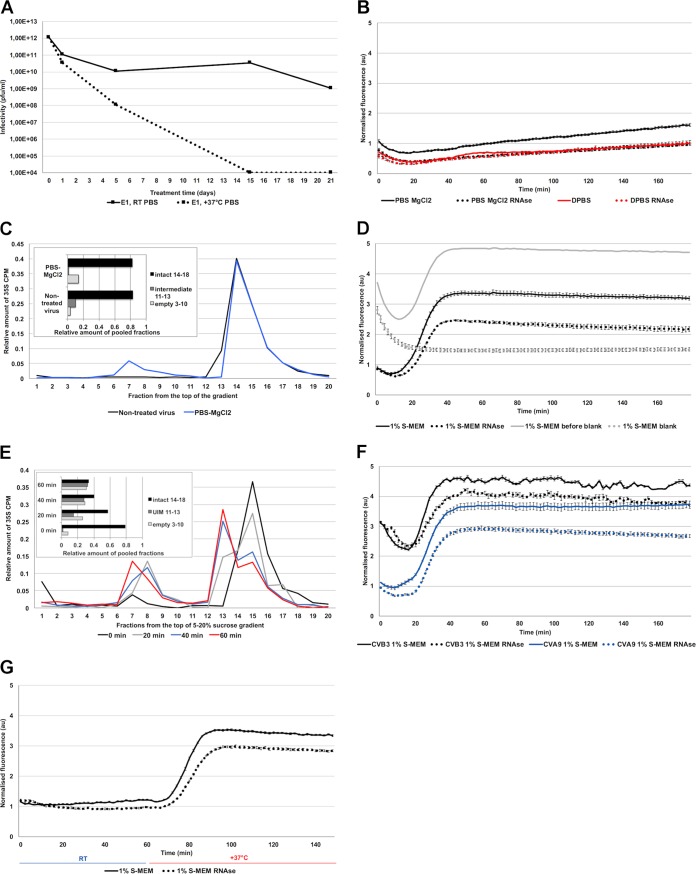
In this study, we investigated physiologically relevant factors that promote the uncoating process of E1. First, we noticed that in phosphate-buffered saline (PBS), E1 remained infectious up to 21 days at room temperature, and remarkably, at 37°C, still some virus stayed infectious under such conditions after 5 days (Fig. 1A). A 3-h incubation of E1 in PBS-MgCl2 at 37°C resulted in only a minor formation of empty virus particles and no formation of the uncoating intermediate, as detected by real-time fluorescence measurement in the presence or absence of RNase (Fig. 1B). This finding was further confirmed by gradient centrifugation of 35S-labeled E1 (Fig. 1C). The virus was even more stable in DPBS buffer (PBS containing also CaCl2) (for buffer compositions see Table 1) throughout the 3-h treatment (Fig. 1B). If enteroviruses are so stable, what molecules efficiently trigger uncoating in the right location, i.e., endosomes? We observed that conversion of E1 virions to intermediate particles was significantly enhanced by treating the virus with cell culture medium containing 1% serum (1% S-MEM): the SYBR green II fluorescence increased considerably after a 10- to 15-min incubation at 37°C, reaching maximal fluorescence around 40 to 50 min (Fig. 1D). At 45 min, approximately 68% of the intensity originated from intermediate particles and 32% from empty capsids (Fig. 1D). The increase of both forms was again confirmed by sucrose gradient centrifugation using 35S E1 (Fig. 1E). A similar effect was also observed by fluorescence measurements of coxsackievirus B3 and coxsackievirus A9 (Fig. 1F), suggesting that this phenomenon was not restricted to E1. Virus conversion to the intermediate form was strictly temperature dependent, as 1% S-MEM did not induce changes in the virus capsid at room temperature but did induce the formation quickly after raising the temperature to 37°C (Fig. 1G). Altogether, these results show that while E1 is stable in physiological buffers, 1% bovine serum effectively induces formation of the uncoating intermediate and empty capsids at 37°C.
FIG 1.
Echovirus 1 stability. (A) Infectivity of E1 after 1, 5, 15, or 21 days in PBS at 37°C or room temperature. (B) Fluorescence measurement of SYBR green II (SGII) dye in the presence of 1 μg E1 in PBS-MgCl2 and DPBS with or without RNase during 3 h at 37°C. (C) Sucrose gradient (5% to 20%) separation of 35S-labeled E1 when treated for 1 h at 37 °C in PBS-MgCl2 versus nontreated virus at 4°C in PBS-MgCl2. The amounts of different virus populations are shown in the inset. (D) Fluorescence measurement of SGII dye in the presence of 1 μg E1 in 1% S-MEM at 37°C. In gray, as an example, results of the same virus measurements before and subtracting the blank measurements. In the presented results of 1% S-MEM, and in all of the presented fluorescence results in this paper, the blank, which contains all other factors except the virus (gray dotted lines), is subtracted from the measurement with virus (gray solid lines). (E) Sucrose gradient (5% to 20%) separation of 35S-labeled E1 when treated for 0 to 60 min at 37°C in 1% S-MEM. The amounts of different virus populations are shown in the inset. (F) Fluorescence measurements of SGII dye in the presence of 1 μg of CVB3 or CVA9 in 1% MEM at 37°C during 3 h. (G) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in 1% S-MEM at room temperature (23°C) for 60 min after which the multilabel reader heater was turned to 37°C for 90 min. The transition from room temperature to 37°C lasted about 12 min. In all of the fluorescence measurements, in this and other figures, the y axes are normalized to the final fluorescence values of the control DPBS treatment (panel B, red solid line). Treatments in each buffer with and without RNase are marked with the same colors but with dotted and solid lines, respectively. All fluorescence measurements are averages from a minimum of five measurements and the presented error bars are ± standard errors of the means.
TABLE 1.
Exact compositions of the buffers used in the paper
| Buffer or compound | Concn (mM) |
faf-BSA (%) | pH | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NaCl | Na2HPO4 | Total Na | KCl | KH2PO4 | K2HPO4 | Total K | MgCl2 | CaCl2 | |||
| PBS | 137 | 8 | 145 | 3 | 2 | 5 | 7.34 | ||||
| PBS-MgCl2 | 137 | 8 | 145 | 3 | 2 | 5 | 2 | 7.22 | |||
| DPBS | 138 | 8.1 | 146.1 | 2.7 | 1.5 | 4.2 | 0.5 | 0.9 | 7.25 | ||
| NKMC | 20 | 20 | 6 | 12 | 30 | 0.5 | 0.2 | 7.18 | |||
| NK | 20 | 20 | 6 | 12 | 30 | 7.25 | |||||
| EM buffer | 29 | 29 | 5.6 | 11.2 | 28 | 0.145 | 0.0093 | 7.28 | |||
Ion composition mimicking endosomal conditions also triggers the E1 uncoating process.
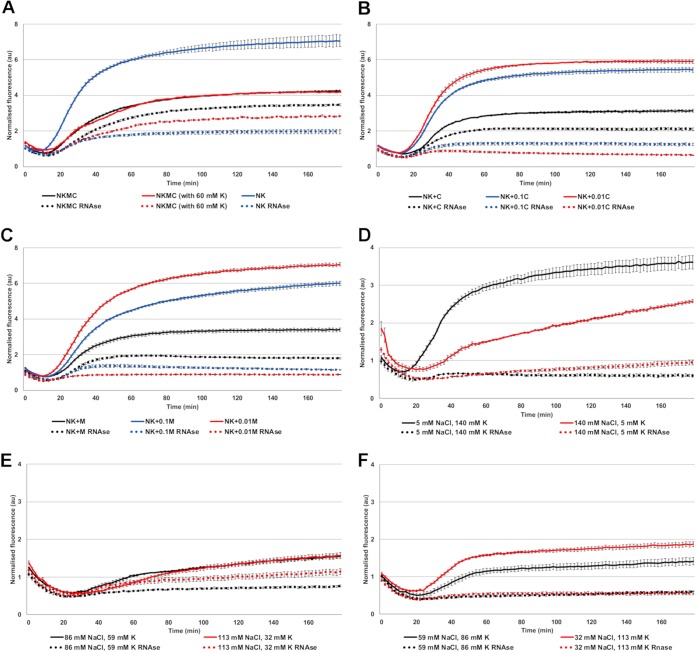
Uncoating of E1 and coxsackievirus A9 is known to be independent of endosome acidification (13), which suggests that ion concentrations other than H+, such as K+, Na+, Mg2+, and Ca2+, might be more important for E1 uncoating inside the endosomes. We thus chose a combination of ion concentrations based on the measurements made from endosomes (34–38) and the information provided by Scott and Gruenberg in their review on endosomal ionic conditions (39) and tested their effect on promoting E1 uncoating. The “endosomal ionic solution” used in this study contained 20 mM NaCl, 30 mM K+, 0.5 mM MgCl2, and 0.2 mM CaCl2 and is referred to as NKMC. The spectroscopy results showed that the hypotonic NKMC solution promoted a slow formation of the uncoating intermediate within a 3-h period at 37°C, as detected by the gradual increase of fluorescence to a roughly 4-times higher level when normalized to that of the DPBS treatment (Fig. 2A). Therefore, NKMC facilitates the formation of mainly porous particles, most of which still contain RNA. Increasing the concentration of K+ (60 mM) in relation to Na+ (20 mM) appeared to promote RNA release, suggesting that the increase of K+ found in the endosomes may be an important factor to promote the final RNA release (Fig. 2A). We also found a larger amount of RNA released when we omitted the divalent cations from the buffer (i.e., NK solution containing 20 mM NaCl and 30 mM K+ without Mg2+ or Ca2+). Without Mg2+ or Ca2+, the fluorescence signal mainly comprised released RNA, indicating the presence of mainly empty capsids (Fig. 2A). This is in line with the previously observed stabilizing effect of divalent cations on viral particles (24, 40–45). The stabilizing effect of divalent cations was further verified by complementing the NK solution with different concentrations of Ca2+ or Mg2+ ions. Similarly to the NK solution without divalent cations, concentrations of 0.002 and 0.02 mM Ca2+, 100-fold and 10-fold dilutions of Ca2+ compared to NKMC, respectively, as well as 0.005 and 0.05 mM Mg2+, 100-fold and 10-fold dilutions of Mg2+ compared to NKMC, respectively, mainly caused the formation of empty virions (Fig. 2B and C). In contrast, the presence of 0.2 mM Ca2+ or 0.5 mM Mg2+ in NK solution prevented RNA release from a significant number of the virions (Fig. 2B and C, black lines).
FIG 2.
Virus stability in NK solution with different amounts of K+, Mg2+ (M), and Ca2+ (C) at 37°C. (A) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in NKMC, containing 30 mM K phosphate (black lines), NKMC with higher, 60 mM, K phosphate (red lines), and NK (i.e., without Mg2+ and Ca2+, blue lines) solutions. (B) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in NK solution with different amounts of CaCl2 (C = 200 μM, 0.1C = 20 μM, 0.01C = 2 μM). (C) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in NK solutions with different amounts of MgCl2 (M = 500 μM, 0.1M = 50 μM, 0.01M = 5 μM). (D) Fluorescence measurements of SGII dye at 37°C in the presence of 1 μg E1 in either intra- and extracellular Na+ and K+ concentrations, combined with intracellular 800 μM MgCl2 and 0.2 μM CaCl2 concentrations. (E and F) Fluorescence measurements of SGII dye at 37°C in the presence of 1 μg E1 in various Na+ and K+ concentrations between intra- and extracellular values, but constant intracellular 800 μM MgCl2 and 0.2 μM CaCl2 concentrations.
We also tested different concentrations of sodium and potassium ranging between their extra- and intracellular values (extracellular concentrations roughly 140 mM Na+ and 5 mM K+ versus intracellular 5 mM Na+ and 140 mM K+). The concentrations of Mg2+ and Ca2+ were kept at their cytoplasmic values of 0.8 mM and 0.2 μM, respectively, while we changed Na+ and K+ concentrations step by step from 5 mM to 140 mM and 140 mM to 5 mM, respectively. The results showed that at 37°C, cytoplasmic ionic concentrations caused a clear increase in the number of empty virus particles: RNA release started already within 15 min (Fig. 2D). This suggests that the intracellular ion concentrations, 140 mM K+, 5 mM Na+, 0.5 mM MgCl2, and 0.2 μM Ca2+, are effective in promoting RNA release. Notably, the extracellular concentrations of Na+ (high) and K+ (low) resulted in the formation of empty particles in the presence of intracellular Mg2+ (0.5 mM) and Ca2+ (0.2 μM) concentrations (Fig. 2D). The concentrations of K+ and Na+ between the extremes were less effective at promoting RNA release (Fig. 2E and F).
In summary, the ionic conditions found in endosomes, with lowered Na+ and Ca2+ and higher K+ concentrations compared to that in the extracellular space, trigger a slow uncoating process. Even further reduction in the Ca2+ concentration and increase in K+ concentration, as may happen in the endosomes, facilitates also the RNA release from the virions.
Albumin triggers the E1 uncoating process.
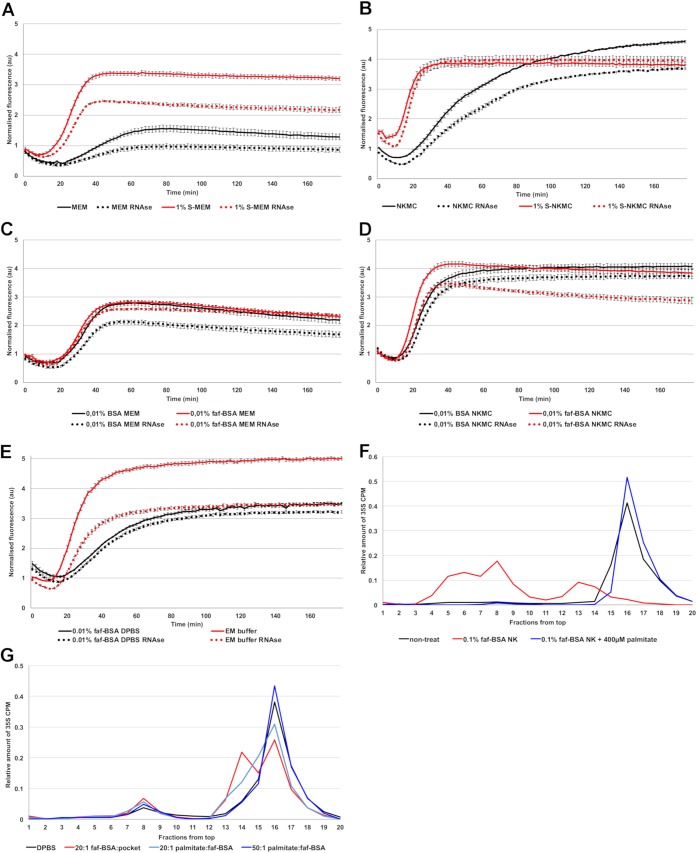
Comparison of 1% S-MEM and MEM without serum clearly showed that serum triggered the uncoating process, resulting in increases of both the uncoating intermediate and empty virus (Fig. 3A). We then tested if serum would have any additive effect when administered in the NKMC solution. We observed a clear increase in the rate of the fluorescence signal appearance: with NKMC, the maximal fluorescence signal was reached only at the end of the 3-h measurement, whereas in the presence of 1% serum, this time was shortened to approximately 30 min (Fig. 3B). Interestingly, with 1% serum, the amount of RNA released from the virions decreased and the intermediate particles remained stable throughout the measurement.
FIG 3.
Serum and albumin promote virus opening at 37°C. (A) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in MEM and 1% S-MEM. (B) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in NKMC and 1% S-NKMC. (C) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in MEM supplemented with 0.01% BSA or faf-BSA. (D) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in NKMC solution supplemented with 0.01% BSA or faf-BSA. (E) Fluorescence measurements of SGII dye in the presence of 1 μg E1 in DPBS supplemented with 0.01% faf-BSA and in EM buffer. (F) Effect of faf-BSA and fatty acids on virus uncoating and priming using 5% to 20% sucrose gradient analysis of metabolically labeled 35S E1. Nontreated virus in comparison to fully opened virus (red line; 1 h of incubation with 0.1% faf-BSA NK at 37°C) and fatty acid-rescued virus after addition of 400 μM palmitate (blue). (G) Virus treated for 1 h at 37°C in DPBS in comparison to DPBS supplemented with 1.5 μM faf-BSA (20:1 ratio of faf-BSA to virus hydrophobic pockets, red line), or, in addition to 1.5 μM faf-BSA, with increasing (20:1 and 50:1) molar ratio of palmitate to the faf-BSA (light blue and dark blue lines, respectively).
In order to narrow down the serum components responsible for the boosting effect, we focused on albumin, as it is the most abundant protein in serum (46). As 1% serum corresponds to approximately 0.04% albumin solution, we decided to use 0.01% to 0.1% BSA concentrations to see if albumin was responsible for the changes that we observed with 1% S-MEM. As it is known that albumin is a high-affinity fatty acid carrier in the blood (46) and the lipid moiety, typically palmitate (47), present in the hydrophobic pocket of many enteroviruses is important for the virion stability (15), we tested also fatty acid-free BSA (faf-BSA) in our experiments.
The spectroscopy measurements showed that both BSA and faf-BSA triggered the uncoating process in MEM in a similar manner to 1% S-MEM (Fig. 3A and C), suggesting that the major factor in serum initiating the uncoating process is indeed the serum albumin. Moreover, both albumin forms promoted serum-like effects in NKMC (Fig. 3D). After observing that BSA and faf-BSA resulted in similar results as serum in both isotonic MEM and hypotonic NKMC, we tested faf-BSA in DPBS and saw that it induced the uncoating process similarly to MEM (Fig. 3E). Spectroscopy analysis of the virus treated with EM buffer prepared for cryoEM imaging showed a large amount of the uncoating intermediate particles (Fig. 3E).
Since albumin appeared to be the major factor inducing and boosting the virus priming, we tested if the addition of fatty acids would prevent the observed effect. We started with conditions that most efficiently caused RNA release: we treated the 35S-labeled E1 with NK solution supplemented with 0.1% faf-BSA (Fig. 3F, red curve peak fractions 4 to 9). The presence of approximately 100-fold molar excess of palmitate (400 μM) with respect to the amount of albumin in the assay fully protected the E1 virions from the structural changes as detected by sucrose gradient analysis (Fig. 3F, blue curve).
We next explored in more detail the molar ratio between BSA and virus required for efficient triggering of the uncoating process. We found that an albumin-to-virion ratio of 1,200:1 (comparable to 0.01% faf-BSA solution with 1 μg of E1 used in spectroscopy assays), which corresponds to an albumin-to-hydrophobic pocket ratio of 20:1, efficiently triggered the formation of the uncoating intermediate (Fig. 3G). In contrast, an albumin-to-virion ratio of 120:1 showed only a mild effect, and a ratio of 17:1 induced no changes compared to the control condition (data not shown). The virus treatment with albumin did not significantly increase the amount of empty viruses.
Then we tested the concentration of free fatty acids needed to prevent the uncoating process. Adding increasing amounts of palmitate to the faf-BSA virus mixture reduced the formation of uncoating intermediate and concomitantly increased the amount of intact virions (Fig. 3G). Full protection against uncoating was gained by adding a 50-fold molar excess of palmitate with respect to the faf-BSA molecules, whereas a 20-fold excess showed an intermediate effect (Fig. 3G). The effect of a 10-fold excess was still notable, but an equimolar ratio showed no protection (data not shown).
These results show that albumin is the major component of the serum that triggers the uncoating process. It further stresses the fact that the net balance between fatty acids and albumin is important in enterovirus particle uncoating.
Structural details of the treated virion.
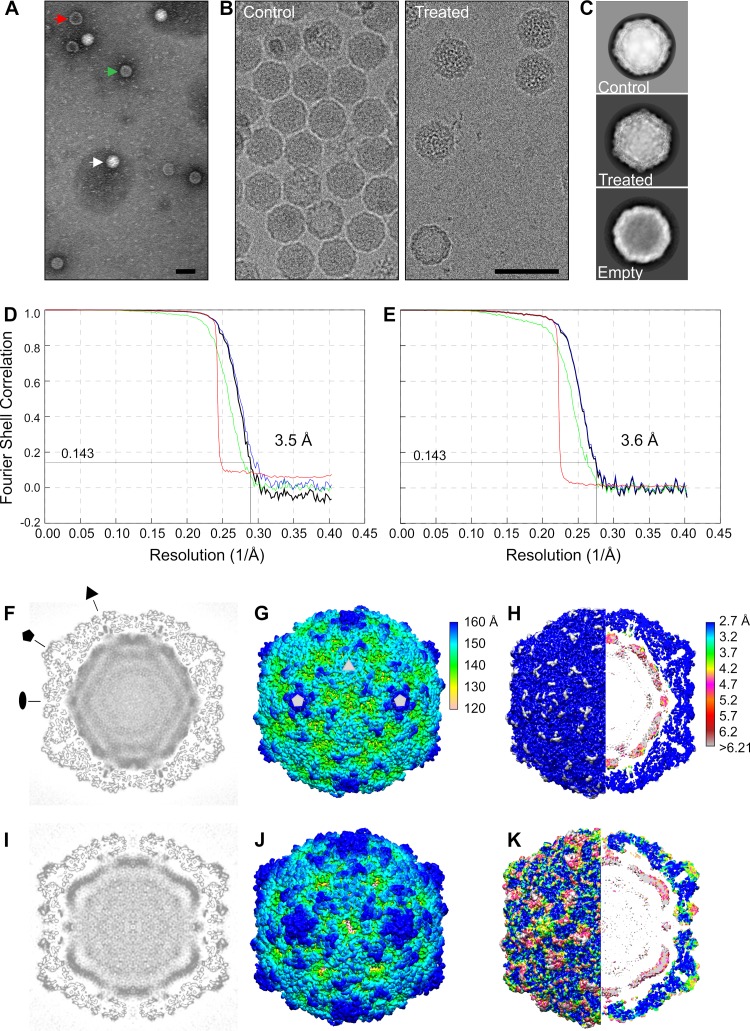
Having ascertained the physiological factors that could reproducibly start the uncoating process, we studied the treated virions by negative staining as well as by cryoEM and single particle reconstruction. Negatively stained preparations of cryoEM buffer-treated sample revealed three distinct forms of E1: intact, intermediate, and empty particles (Fig. 4A). In the EM buffer-treated virus sample, 29% of particles were intact, 65% were intermediate, and 6% were empty (of 2,108), whereas in nontreated sample, the percentages were 95%, 4%, and 1% (of 2,624), respectively. In cryoEM micrographs, we could not distinguish between intact and the intermediate particles by eye, but after image analysis and classification, 70% of all particles were intermediate (23,983 of 34,160) processed from the treated sample’s micrographs (Fig. 4B). Thus, the reconstruction of the treated particle described below represents the averaged structure for the most populated viral particle in the treated sample. Two-dimensional averages of the control and treated particles clearly show the density for both the capsid and RNA (Fig. 4C).
FIG 4.
Electron microscopy and cryoEM reconstructions of E1 intact and treated virions. (A) Negative stain of E1 after 1 h treatment at 37°C with 29 mM NaCl, 28 mM K, 0.145 mM MgCl2, 0.0093% faf-BSA. (B) Cryo-electron micrographs of nontreated (control) E1 and E1 after similar treatment to that for panel A. The white arrow indicates an intact virion, the red arrow an empty particle, and the green arrow an intermediate particle. Bars, 50 nm. (C) Example of 2D class averages showing intact (control, n = 16,298), treated (n = 2,015), and empty (n = 212) class averages, where n is the number of particles contributing to the averages shown. Box size is 496 Å (D) Fourier shell correlation curves for the control E1 reconstruction, giving a resolution of 3.5 Å with a cutoff of 0.143. (E) Fourier shell correlation curves for the treated particle reconstruction giving a resolution of 3.6 Å with a cutoff of 0.143. Corrected (black), unmasked (green), masked (blue), and phase randomized masked (red) maps (71). (F) Control E1 reconstruction central plane. Pentagon, triangle, and oval indicate 5-fold, 3-fold, and 2-fold symmetry axes, respectively. (G) Radially colored isosurface representation of the control E1 reconstruction viewed down a 2-fold axis of symmetry at 1.5 standard deviations (SDs) above the mean. Pentagons and triangle mark 5-fold and 3-fold axes, respectively. Two-fold axis is in the middle between the two marked 5-fold axes. (H) Control E1 colored by local resolution (79). (I) Treated particle reconstruction central plane. (J) Isosurface representation of treated particle viewed down a 2-fold axis of symmetry at 1.5 SDs above the mean. Radially colored according to the color key in panel G. (K) Treated particle colored by local resolution. Resolution colored according to the color key in panel H.
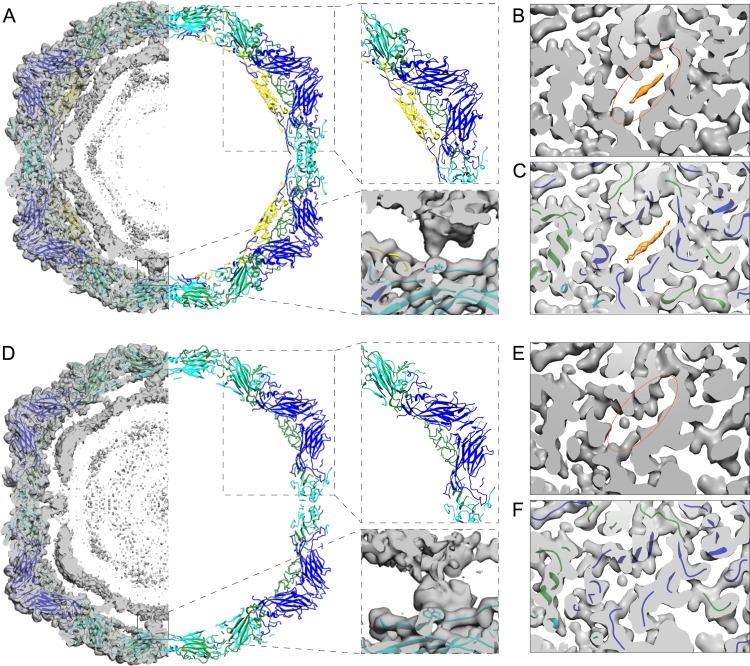
Icosahedral reconstruction of the intact E1 virus from the control sample’s cryoEM micrographs to 3.5-Å resolution was similar to the published X-ray structure PDB identifier (ID) 1EV1 (48). The atomic model fitted into the intact E1 reconstruction included the lipid factor in the lipid pocket and density for all four capsid proteins, VP1, VP2, VP3, and VP4 (Table 2, Fig. 4D, F, G, and H, and Fig. 5A to C) (48). The root mean square deviation (RMSD) compared to the X-ray structure was 0.77 Å. The intact E1 reconstruction shows icosahedrally averaged RNA density, which has not been reported earlier (Fig. 4F and H; Fig. 5A). The majority of the RNA follows the outline of the capsid and is distributed at a distance of 0 to 10 Å from the inner capsid surface, with the highest density between radii 94 and 113 Å (from the virion center). The RNA has clear connections around the 2-fold axes of symmetry to Trp 38 of VP2 (Fig. 5A). In addition, Arg 13 and Arg 27 of VP1 as well as a poorly resolved VP4 loop contact the RNA density. Similar RNA-capsid interactions involving VP2 Trp 38 and VP1 N terminus have been described in intact rhinovirus particles (30).
TABLE 2.
Summary of cryoEM data collection, refinement, and validation statistics
| Category | Value |
|
|---|---|---|
| Treated E1 | Control E1 | |
| Data collection | ||
| Voltage (kV) | 200 | 200 |
| Electron exposure (e−/Å2 × s) | 30 | 30 |
| Pixel size (Å) | 1.24 | 1.24 |
| No. of micrographs | 1,246 | 979 |
| Reconstruction | ||
| No. of particles | 14,615 | 45,309 |
| B factor (Å2) | −70 | −70 |
| FSCa threshold | 0.143 | 0.143 |
| Resolution (Å) | 3.6 | 3.5 |
| Model building (aa)b | ||
| VP1 (full protein, 281 aa) | 55–130 and 137–280 | 1–281 |
| VP2 (full protein, 261 aa) | 12–54 and 58–261 | 7–261 |
| VP3 (full protein, 239 aa) | 3–174 and 184–237 | 1–239 |
| VP4 (full protein, 68 aa) | none | 1–14 and 22–68 |
| Model validation | ||
| MolProbity score (percentile) | 1.25/100thc | 1.08/100thd |
| Ramachandran outliers (%) | 4.3 | 2.06 |
| Poor rotamers (%) | 1.7 | 1.24 |
| Clash score | 0 | 0 |
FSC, Fourier shell correlation.
aa, amino acid.
N = 342, 3.25 Å to 3.85 Å.
N = 27,675, 0 Å to 99 Å.
FIG 5.
Comparison of reconstructions and atomic models of control and treated E1 virions. (A) Slab of the virion atomic model (1EV1) (48) shown in ribbon, fitted into the control E1 density (left). Most of the capsid density is accounted for by the atomic model, but the inner density from the RNA is not. The upper right inset highlights the structure of one pentamer with VP4. The lower right inset highlights the interaction of the RNA and VP2 Trp 38 next to the 2-fold axis. (B) Section of control E1 reconstruction revealing the pocket and the lipid factor. (C) The same as in panel B with the atomic model fitted. (D) Slab of the treated atomic model (wwPDB deposition ID 6O06) shown in ribbon, fitted into the treated particle density (left). Most of the capsid density is accounted for by the atomic model, but the inner density from the RNA is not. The position of the RNA has moved radially outwards as the capsid expanded, maintaining the 2-fold connections. The upper right inset highlights the structure of one pentamer without VP4 modeled as there was no apparent density for it. The lower right inset highlights the interaction of the RNA and VP2 Trp 38 next to the 2-fold axis. (E) Section of treated particle reconstruction revealing the collapsed pocket and no evident density for the lipid factor. (F) The same as in panel E with the atomic model fitted. For all panels, EM density shown in transparent gray, lipid factor EM density in orange, VP1 (blue ribbon), VP2 (cyan ribbon), VP3 (green ribbon), VP4 (yellow ribbon), lipid factor (orange stick). In panels B and E, orange ovals indicate the corresponding positions of the pocket.
In comparison, the 3.6-Å resolution treated particle reconstruction has undergone a 4% expansion (Fig. 4E, I, J, and K). It was possible to model most of the VP1, VP2, and VP3 but none of VP4 in the reconstruction (Table 2 and Fig. 5D to F). The atomic modeling revealed that the capsid expansion occurred through rotation and outwards translation of the capsid proteins VP1, VP2, and VP3. This results in a fenestrated capsid, with prominent holes on the edges of the capsid close to the 2-fold axes of symmetry at the VP2 dimer interface, a hallmark of A-particles described for other enteroviruses (Fig. 4J and Fig. 6A and B). The atomic model emphasizes these holes, somewhat artificially, as not all the electron density has been accounted for (Fig. 6B and D). The pocket factor has been released (Fig. 5E and F). The GH loop on VP1, thought to be important in pocket factor release, has moved, collapsing the pocket. The RNA is clearly visible, its average conformation has changed, but it still maintains connections to the capsid via VP2 Trp 38, Glu 40, and Tyr 41 (Fig. 5D). The contacts mediated by the VP1 N termini and VP4 have been lost in the treated particle, but new connections appeared via the N termini of VP3 below the 5-fold vertices. In the control particle, these VP3 termini interact with VP4. Similar RNA-capsid interactions mediated by VP2 Trp 38 and VP3 N termini were seen in expanded rhinovirus particles (30). The averaged RNA still follows the profile of the inner surface of the capsid, but has moved outwards (Fig. 4 to 6), with the highest density between radii 102 and 122 Å. Thus, the RNA is less densely packed in the treated intermediate particle.
FIG 6.
Treated particle expansion and unmodeled cryoEM density. (A) Atomic model of control E1 seen along 2-fold axis. (B) Atomic model of the treated intermediate virion seen along 2-fold axis showing opening between two VP2 helices. (C) Unmodeled cryoEM density of control E1 (gray isosurface). Inset shows unmodeled density at the 3-fold axis. Red arrows indicate N termini of VP2 (Cys 7) in the vicinity of unmodeled density blob. Triangle indicates 3-fold symmetry axis. (D) Unmodeled cryoEM density of the treated virion (gray isosurface). Inset shows unmodeled density spanning the capsid near 2-fold axis. Red arrows indicate N termini of VP1 (Asn 55) in the vicinity of the unmodeled density spanning the capsid. Pentagon and oval indicate 5-fold and 2-fold symmetry axes, respectively. For panels A to D: VP1 (blue), VP2 (cyan), VP3 (green), VP4 (yellow), lipid factor (orange).
Besides the poorly ordered density inside the capsid assigned to viral RNA, the difference map between the control reconstruction and the atomic model of intact E1 showed only a small unassigned density in the capsid protein region at the 3-fold axes of symmetry (Fig. 6C). This unassigned density could be attributed to six unmodeled residues of the VP2 N terminus (Fig. 6C, red arrows in the inset). The difference map between the treated particle reconstruction and atomic model revealed a less-well-ordered lower resolution density near the 2-fold axes spanning the capsid from the interior to the exterior and at the 5-fold axes on the particle surface (Fig. 6D). The first modeled residue of the VP1 N terminus (Asn 55) lies in close proximity to the unassigned density near the 2-fold axis inside the particle; therefore, the unassigned density could be the VP1 N termini now traversing the capsid and exposed on the surface of the treated capsid similarly to the interpretation for expanded particles of other enteroviruses (Fig. 6D, red arrows in the inset) (20, 26, 49). In addition, part of this poorly ordered density seen on the exterior could be attributed to a flexible VP3 loop (residues Thr 175 to Asp 183). The poorly defined density seen at the 5-fold axes on the capsid surface is most likely attributed to disordered loops in VP1 (residues Thr 131 to Asn 136). Although VP4 could not be modeled in the density, its partial presence in the treated sample was confirmed by autoradiography (Fig. 7). Moreover, the treated virus sample showed approximately 2 logs lower infectivity (decrease from 8.23 × 1011 to 1.01 × 1010), confirming our previously published data for the E1 uncoating intermediate particle (14). Hence, VP4 could contribute to the poorly ordered density on the inside of the capsid close to the vertices, attributed primarily to RNA, as well as to the density spanning the capsid. In corroboration of this finding, the presence of a tiny amount of VP4 was recently reported in A-particles of enterovirus D68 induced by acid treatment (49).
FIG 7.

Autoradiography observation of 35S-labeled VP4 protein. Metabolically labeled E1, nontreated, heated to 50°C for 10 min, and EM buffer-treated sample.
DISCUSSION
We showed previously that during entry into cells, E1 undergoes structural changes that were first discovered as an increased permeability to the small molecule dye, SYBR green II, and Cs-ions (14). Here, we showed by cryoEM that temperature-dependent structural changes, under physiological conditions, involved expansion of the virus particle, loosening of the genome packing, loss of the lipid factor, and formation of larger openings at the VP2 dimer interface, explaining the increased permeability to the small molecular dye and the resistance to RNase treatment observed previously (14). We also showed by spectroscopy that serum priming of coxsackievirus B3 and coxsackievirus A9 caused similar changes in permeability to that of E1, suggesting that also other enteroviruses behave similarly (Fig. 1F).
Our in vitro experiments demonstrated two important factors in the serum and extracellular space affecting the integrity and the dynamics of E1 virion at 37°C, albumin and fatty acids, one of the most abundant components in the serum and interstitial fluids. Albumin concentration in serum has been reported as 640 μM (46) and the albumin-bound fatty acids concentration ranging from 200 to 700 μM in serum (50). As mentioned before, albumin serves as the major fatty acid carrier in the blood, with two to three high-affinity and four to five intermediate-affinity binding sites for fatty acids (46). Also, the fatty acid binding capacities of BSA and faf-BSA have been measured to be around 4.8 mol and 7 mol per mol albumin, respectively (51). The measurements further showed that BSA is approximately 50% occupied by serum fatty acids, meaning that roughly 50% of the fatty acid binding capacity is still left (51). Furthermore, both Penn et al. (51) and van der Vusse (46) showed that bovine and human albumin have very similar fatty acid binding capacities, making bovine albumin a good surrogate for human albumin. These data provide further support that under physiological conditions, BSA is not saturated with fatty acids. Thus, the most likely explanation for the increased intermediate particle formation at low fatty acid concentration is the partitioning of the fatty acid moiety from the capsid into a more hydrophobic environment, such as into the fatty acid binding sites on the albumin. Higher temperature will result in higher mobility of the fatty acid. Furthermore, the fatty acid saturation level of albumin has a great effect on capsid stability. This was demonstrated by the prevention of virus expansion and RNA release using increasing concentrations of palmitate in comparison to albumin (Fig. 3E and F). Considering the observed molar excess of fatty acids required to prevent the formation of an albumin-induced intermediate particle (Fig. 3F), and the approximately 1:1 ratio of albumin and fatty acids observed in serum, it seems probable that under physiological conditions, the albumin present outside the cells starts the uncoating process. The ability of faf-BSA to induce echovirus A-particle formation was previously shown using a radioactive gradient; however, the nature of this process was not studied further (21). Our results here explain the mechanism by which faf-BSA induces uncoating in enteroviruses, such as echoviruses and coxsackieviruses. Moreover, we suggest that the loss of the pocket factor likely precedes receptor binding, promoting the formation of an infectious intermediate particle already in the extracellular space.
The expansion of E1 capsid does not affect the receptor, α2I-domain, binding site (14, 52). However, the expansion does cause changes in the amino acid composition exposed on the capsid surface, such as the exposure of the VP1 N terminus, which may also give rise to new secondary receptor binding sites. If some lipid factors would still be bound to the capsid in the endosome initially, they could well be exchanged into the hydrophobic environment of the endosomal membrane that would extract the lipid factor from the capsid. In support of this hypothesis, albumin readily donates fatty acids to other fatty acid binding proteins in the vicinity of the plasma membrane. Furthermore, the binding and release of the lipid factor in enteroviruses is known to be a dynamic process (53, 54). A potential endosomal membrane protein that could help to sequester the lipid factor is the lipid-modifying enzyme PLA2G16 (55). It is thought to affect the insertion of VP4 into the endosomal membrane but could potentially help to sequester the lipid factor too (56).
The next and final step in picornavirus uncoating is the RNA release. Accumulated results on CVA9 and E1 uncoating show that it starts as early as 15 to 30 min postinfection, and the extent of uncoating increases strongly until 1 to 2 h postinfection (57, 58). However, the first signs of replication, including negative- and positive-strand RNA production, occur as late as 2 h postinfection (59). This suggests that the ambient endosomal ion conditions develop gradually to better promote the uncoating and the final RNA release into the cytoplasm. Current information on the intraendosomal ionic changes over time suggests that the sodium and calcium concentrations in endosomes decrease during their maturation and potassium concentration rises relative to the extracellular values (39). Our results demonstrated that a combination of low sodium and calcium and elevated potassium in the presence of a physiological magnesium concentration was able to trigger the uncoating process; however, it was more rapid in the presence of albumin (Fig. 3B and D). Furthermore, in addition to the formation of the uncoating intermediate, the right combination of ions led to more efficient RNA release (Fig. 2A) that could be further increased with faf-BSA (Fig. 3E, fluorescence data not shown). In the lack of thorough absolute ion concentration measurements inside the endosomes, the present study may underestimate the complexity and actual combination of the ions present. No ions have been resolved in the X-ray structure of E1, and so we cannot directly interpret the effects of the ions on the capsid or the RNA stability. However, both K+ and Mg2+ ions can stabilize RNA tertiary structures (60). Thus, we hypothesize that changes in these ions may contribute to the expansion of the RNA, providing an additional force to promote conformational change in the capsid and eventual release of the RNA in the endosome. Changes in the Ca2+ and Na+ ion concentrations probably affect the protein stability, through electrostatic interactions. Ca2+ has been shown to be important for the capsid stability of many viruses (24, 40–44).
The RNA-capsid interactions were resolved in structures of A-particles published for several enteroviruses, such as enterovirus 71, coxsackievirus A16, and rhinovirus 2, suggesting their importance in assisting RNA release (20, 30, 31). The genome is unique within the capsid; nevertheless, in some picornaviruses, the RNA is so well ordered that individual bases can be identified, ranging from one base to several (61–66). Here, the icosahedral reconstructions also showed for the first time details of the RNA, though not resolved to the atomic level, inside the intact E1 particle and in the uncoating intermediate structure. The RNA has a high density in proximity to the intact capsid, which suggests high occupancy, and the probable interaction of the RNA with both the ordered capsid proteins (notably with VP1, VP4, and VP2 shown in Fig. 5A) and their disordered termini (67). Trp 38 in VP2, a highly conserved residue in picornaviruses, points toward ordered RNA (61–66). This interaction between the RNA and capsid is directly adjacent to the major fenestrations that appear in the treated capsid, where the RNA has moved radially outwards, maintaining this specific interaction (Fig. 5D). Interestingly, this RNA-capsid interaction is in proximity to the N terminus of VP1 that appears to be extruded from the treated capsid, similarly to the VP1 in expanded poliovirus, coxsackievirus A16, and enterovirus D68 (20, 26, 49). VP4 could not be identified in the reconstruction, suggesting that it is no longer icosahedrally ordered or the occupancy is much reduced as suggested also by autoradiography. This implies that the RNA interaction with the capsid via VP4 under the vertices was significantly altered compared to the intact particle (Fig. 5). VP4 may have collapsed into the density attributed to RNA that is still prominent below the VP3 annulus at the vertices (Fig. 4 and 5). Changes in the RNA packaging density and its interaction with the capsid proteins, along with the expansion of the particle, are probably also responsible for the increase in permeability to dye.
Altogether, our results suggest that based on the dynamic nature of albumin-mediated fatty acid binding and the fact that both albumin and fatty acids are present outside cells, the majority of the enterovirus particles may reside in a triggered, intermediate, metastable state before entering cells. According to our results, the albumin-triggered intermediate state is likely to lead to more-efficient RNA release when it is further affected by the ambient concentrations of monovalent and divalent cations in endosomes.
MATERIALS AND METHODS
E1 production and purification.
E1 was produced and purified as described earlier (14). Confluent 5-layer bottles of green monkey kidney (GMK) cells, obtained from the American Type Culture Collection (ATCC), were infected with E1 (Farouk strain; ATCC) for 16 to 20 h at 37°C with 5% CO2. After infection, the cells and media were collected and lysed with three freeze-thaw cycles. The lysate was pelleted by centrifugation with a JA-10 rotor (6,080 rpm, 30 min, 4°C), after which, the supernatant was precipitated for 16 to 20 h at 4°C using polyethylene glycol 6000 (Sigma-Aldrich, St. Louis, MO, USA) (8% [wt/vol]), and NaCl (2.2%[wt/vol]). The precipitated supernatant was then centrifuged with a JA-10 rotor (8,000 rpm, 45 min, 4°C), and the resulting pellet was dissolved in R buffer (10 mM Tris-HCl [pH 7.5], 200 mM MgCl2, 10% [wt/vol] glycerol). For disrupting the remaining cellular membranes, 0.3% (wt/vol) sodium deoxycholate (Sigma-Aldrich) and 0.6% (vol/vol) Nonidet P-40 (Sigma-Aldrich) were mixed with the supernatant and incubated for 30 min on ice. The remaining debris was pelleted by centrifuging in a TX-200 rotor (4,700 rpm, 15 min, 4°C), and the resulting supernatant was loaded on top of 10-ml linear 10% to 40% (wt/vol) sucrose gradients in R buffer. The gradients were ultracentrifuged in an SW-41 rotor (30,000 rpm, 3 h, 4°C) and fractioned into 500-μl aliquots. The optical density at 260 nm of each fraction was measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) to identify fractions containing virus. These fractions were dialyzed against 2 mM MgCl2-phosphate-buffered saline (PBS-MgCl2) in a Spectra/Por Micro Float-A-Lyzer with Biotech cellulose ester membranes and 300-kDa cutoff (Spectrum Laboratories Inc., USA) at 4°C. Finally, the viruses were pelleted by ultracentrifugation using a 70-Ti rotor (35,000 rpm, 2 h, 4°C) and resuspended in PBS-MgCl2. Several purified virus batches were used, ranging between 0.5 and 1.3 mg/ml and 1 × 1011 and 1 × 1012 PFU/ml.
35S E1 production and purification.
35S-labeled E1 for gradient analysis was produced in GMK cells as described earlier (14). Semiconfluent cell culture bottles were washed at 37°C for 15 min with PBS and infected with E1 (Farouk strain; ATCC) using low-methionine-cysteine medium supplemented with 1% fetal bovine serum (FBS) for 3 h. After this, the medium was changed into the above-mentioned medium supplemented with 50 μCi/ml of [35S]methionine-cysteine, and the infection was allowed to proceed for 16 to 24 h until nearly all of the cells were detached. The cells were lysed via 3 freeze-thaw cycles, and the cell debris was pelleted using a TX-200 rotor (4,000 rpm for 15 min, 4°C). The supernatant was further treated with 0.3% (wt/vol) sodium deoxycholate and 0.6% (vol/vol) Nonidet P-40 for 30 min on ice, and the supernatant was further cleared by centrifugation (TX-200, 4,700 rpm, 15 min, 4°C). The supernatant was loaded onto the top of a 2-ml 40% sucrose cushion at 4°C. The cushions were ultracentrifuged at 30,000 rpm (SW-41) for 2.5 h at 4°C, after which the liquid above the cushion and one 500-μl fraction from the cushion were discarded and the three next 500-μl fractions were collected. These fractions were diluted in PBS-MgCl2; the virus was pelleted by centrifuging at 35,000 rpm (70-Ti) for 2 to 3 h at 4°C, after which the pellet was resuspended in 1.5 ml of PBS-MgCl2 and the different virus populations were further separated in a 5% to 20% sucrose gradient in R buffer via gradient centrifugation (35,000 rpm, SW-41, 2 h, 4°C). The virus-containing fractions were identified via liquid scintillation counting (Perkin Elmer, Waltham, MA, USA). The three fractions containing the largest amount of intact virus particles were again collected and pelleted as above. The resulting pellet was dissolved in 400 μl of PBS-MgCl2, divided into smaller aliquots, and stored at −80°C after determination of cpm/μl of the 35S E1 stock.
CVB3 and CVA9 production and purification.
CVB3 (ATCC Nancy strain) and CVA9 (ATCC Griggs strain) were purified similarly to E1, except they were separated in 5% to 20% sucrose gradients. From the gradient, fractions 1 to 10 were discarded and 11 to 20 collected. The collected fractions were diluted with PBS-MgCl2 to a final volume of 25 ml and concentrated as described for E1.
Endpoint dilution assay.
The endpoint dilution infection assay was performed as described earlier (68). Briefly, GMK cells were cultured in 10% serum-containing MEM and infected with E1 in a dilution series in the presence of 1% serum. Typically, after 3 days of infection at 37°C, the 96-well plate was stained with 50 μl of crystal violet stain (8.3 mM crystal violet, 45 mM CaCl2, 10% ethanol, 18.5% formalin, and 35 mM Tris base) for 10 min. The 50% tissue culture infective dose (TCID50) was determined by calculating the numbers of infected and uninfected wells for the eight replicates in one 96-well plate at each dilution. PFU/ml was calculated by multiplying the TCID50/ml value by 0.7 according to the Poisson distribution estimation.
Real-time fluorescence uncoating measurements.
The measurements were performed using a PerkinElmer 2030 Multilabel Reader Victor X4 with F485 lamp filter and F535 emission filter and 1 s counting time. In each measurement, 1 μg of nonlabeled virus (corresponding to 0.77 to 2 μl of PBS-MgCl2, depending on the stock concentration) was treated in 100 μl of the buffer described for each experiment in a single well of a 96-well plate (Sarstedt, Nümbrecht, Germany) in the presence of 10× SYBR green II fluorescent dye. The composition of the buffers is presented in Table 1. All of the buffers were neutral in pH ranging between 7.18 and 7.44. Where indicated, RNase A was added to the wells at a final concentration of 10 μg ml−1. By adding RNase to the assay, we distinguished between the fluorescence originating from the porous intermediate particles, i.e., from RNA inside the virus capsid (protected from the RNase activity), and the fluorescence of RNA released from the particles (sensitive to RNase treatment) indicating the presence of empty particles. Intact virion is inaccessible to the dye and thus gives a low fluorescence signal (14). For each virus treatment, a corresponding blank well with all other factors except virus was also measured, and the fluorescence was subtracted to eliminate fluorescence originating from other factors than the virus. Each well was measured either every minute or every other minute for a 3-h time period at 37°C if not otherwise stated. The plate was prepared on ice, from where it was placed into a preheated measurement chamber. The results were processed and plotted using Microsoft Excel. The error bars in the figures represent the standard errors of the means from a minimum of five technical replicates from at least two separate biological replicates. In all graphs, the amount of fluorescence is normalized to the end fluorescence value of the control, 3-h DPBS treatment.
Gradient analysis.
One microliter (approximately 10,000 to 80,000 cpm) of 35S E1 together with 1 μg of nonradioactive E1 was treated for 1 h at 37°C in 100 μl of the relevant buffer. The samples were then cooled on ice and loaded onto a cooled 10-ml linear 5% to 20% sucrose gradient. The gradients were centrifuged using an SW-41 rotor (35,000 rpm, 2 h, 4°C), and 500-μl fractions were collected and mixed with scintillation cocktail (Ultima Gold MW; Perkin Elmer). The samples were analyzed using Tri-Carb 2910 TR (Perkin Elmer) and plotted in Excel. Every treatment was repeated at least once, and similar effects were observed.
In the albumin-virus-fatty acid ratio experiment, the molarity of E1 was calculated using molecular weight estimation of 8 MDa for E1, which resulted in 1.25 nM E1 solution with 1 μg of virus in 100 μl of buffer. According to this, 1,200 times higher faf-BSA (Sigma-Aldrich, in double-distilled water [ddH2O]) concentration (1.5 μM, 1,200 albumins per virus) was used for a 20:1 ratio of albumin molecules to virus pocket factors, and 120 times higher concentration was used for a 2:1 ratio; 17 times higher concentration was used for 2:1 ratio between the albumin’s high- to medium-affinity fatty acid binding sites (7 per albumin) versus virus pocket factors (60 per virus). Palmitate (Sigma-Aldrich) from a 15.2 mM stock in 50% methanol (MeOH) was added at either a 20:1 or 50:1 ratio into the 1.5 μM faf-BSA solution.
EM sample preparation, imaging, particle processing, and model building.
Virus sample for cryoEM and negative staining was prepared as follows. Purified E1 in PBS-MgCl2 buffer at a concentration of 1.27 μg/μl was mixed with 20 mM NaCl, 30 mM K+, and 0.01% faf-BSA buffer giving a final concentration of 0.1 μg/μl virus, 29 mM NaCl, 28 mM K+, 0.145 mM MgCl2, and 0.0093% faf-BSA (EM buffer) and incubated for 1 h at 37°C. The uncoating intermediate formation was confirmed by spectroscopy measurement (Fig. 3E). Control samples were prepared from purified E1 in PBS-MgCl2.
Butvar-coated copper grids were glow discharged (EMS/SC7620 Mini sputter coater), after which the virus sample was added and incubated for 15 s. The excess virus was blotted away, and the grids were negatively stained using phosphotungstic acid (H2O, pH 7.4) for 60 s; the excess stain was blotted away. The samples were stored overnight at room temperature (RT) before imaging with a JEM-1400 (JEOL) transmission electron microscope (80 kV, Olympus SIS Quemesa bottom-mounted 11 Megapixel charge-couple-device [CCD] camera, ×12,000 to ×40,000 magnification).
Sample volumes of 3 μl of control or EM buffer-treated E1 particle were applied to glow-discharged Quantifoil holey carbon R2/2 grids and vitrified using a custom-made manual plunger. The vitrified samples were imaged on a 200-kV Talos Arctica microscope equipped with a Falcon III direct electron detector at a nominal ×120,000 magnification corresponding to a calibrated pixel size of 1.24 Å. Each exposure was 47.8 s long and collected as a movie containing 30 frames with an accumulated dose of 30 e/Å2 using Thermo Fisher Scientific’s automatic data acquisition software.
Dose-fractionated image stacks were aligned using MotionCor2 (69). The contrast transfer function parameters for each micrograph were estimated using Gctf (70). Images containing drift or astigmatism were discarded. Particle selection, two-dimensional (2D) classification, initial model generation, and three-dimensional (3D) classification were performed using RELION 2.1 (71). Final 3D refinement followed by sharpening (B-factor, −70 Å2) of 45,309 intact particles from control sample (∼45% of total particles) resulted in a 3.5-Å resolution map. After applying the same procedure to 14,615 uncoating intermediate particles from treated sample (∼59% of total particles), a 3.6-Å resolution map was generated.
Model building.
A crystal structure of the E1 virion (PDB ID 1EV1) served as an initial starting model for the intact E1 particle (48). An initial atomic model for the uncoating intermediate particle was generated using the I-TASSER server (72) based on the crystal structure of the CVA16 uncoating intermediate (PDB ID 4JGY) (20, 73). The atomic coordinates of the initial models were docked manually into the electron density maps using UCSF Chimera and further optimized using the “Fit in Map” command (74). In the case of the intact particle, one residue (Cys 7) was added to the VP2 N terminus of 1EV1 model using Coot 0.8.8, and the fit was further optimized using molecular dynamics flexible fitting (MDFF) software used along with NAMD and VMD (69–72, 75). A scale factor of 1 was used to weigh the contribution of the cryoEM map to the overall potential function used in MDFF. Simulations included 10,000 steps of minimization and 100,000 steps of molecular dynamics under implicit solvent conditions with secondary structure restraints in place. The atomic model of the uncoating intermediate particle was refined in Coot 0.8.8, and this served as an input for MDFF, NAMD, and VMD software (75–78). Simulations with scale factor of 1 included 20,000 steps of minimization and 100,000 steps of molecular dynamics under implicit solvent conditions with secondary structure restraints in place. To minimize atom clashes in the atomic model processed in MDFF, seven asymmetric units were simultaneously refined in MDFF using the same secondary structure restraints as above. To analyze unassigned density in intact or treated virion reconstruction, the atomic model was converted to 3.5-(intact) or 3.6-Å (treated) resolution electron density map using the “molmap” command in UCSF Chimera. The map generated for the accounted density was then subtracted from the reconstruction using the “vop subtract” command in UCSF Chimera. The treated particle expansion was estimated by measuring the particle diameter between the 5-fold vertices in intact and treated virions.
Gel separation of viral proteins and autoradiography.
One microgram of nonradioactive E1 with 1 μl of 35S E1 (approximately 10,000 to 80,000 cpm) was treated for 1 h at 37°C in EM buffer. Nontreated virus was used as a VP4 detachment negative control, and virus incubated for 10 min at 50°C was used as a positive control. Samples were dialyzed against 1 liter of PBS-MgCl2 for 40 min using a Slide-A-Lyzer mini dialysis device with 10-kDa cutoff (Thermo Fisher Scientific) to remove the possibly detached VP4 from the samples. The samples were boiled for 10 min with the sample buffer and ran in a 4% to 20% gradient gel (MINI-PROTEAN TGX precast gel; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Precision plus protein standard 10 to 250 kDa (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to distinguish the molecular weights. The gel was fixed in 30% methanol with 10% acetic acid and treated with autoradiography enhancer (Enlightning; Perkin Elmer). The gel was dried at 70°C for 2 h (Gel dryer 583; Bio-Rad Laboratories, Inc., Hercules, CA, USA) and subjected to autoradiography. Protein bands were analyzed using ImageJ gel analyzer tool.
Data availability.
The final density maps have been deposited in the Electron Microscopy Databank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/) with accession codes EMD-4903 (control E1) and EMD-0565 (expanded E1). The atomic models have been deposited in the Protein Data Bank (and Worldwide Protein Data Bank [wwPDB]) with accession codes 6RJF (control E1) and 6O06 (expanded E1). The raw data have been deposited in the EMPIAR database (https://www.ebi.ac.uk/pdbe/emdb/empiar/) with deposition EMPIAR-10284.
ACKNOWLEDGMENTS
We thank Benita Löflund and Pasi Laurinmäki (University of Helsinki), as well as Instruct-FI, the Biocenter Finland National cryo-electron microscopy unit, HILiFE-Institute of Biotechnology, and the CSC-IT Center for Science Ltd., for providing technical assistance and facilities to carry out the work. We also thank Tino Kantoluoto (University of Jyväskylä) for virus production and purification.
This work was supported by the Academy of Finland (275199 and 315950 to S.J.B.; 257125 to V.M.), the Sigrid Juselius Foundation (S.J.B.), and Jane and Aatos Erkko foundation (V.M.).
V.M., V.R., A.D., and S.J.B. conceptualized the study. M.P., A.D., V.R., S.J.B., and V.M. curated data. M.P., V.R., M.L., A.D., S.J.B., and V.M. developed the methodology. V.R. and A.D. validated the data. V.R. and A.D. visualized the data. V.R., M.L., A.D., S.J.B., and V.M. wrote the manuscript. V.M. and S.J.B. acquired funding. A.D., S.J.B., and V.M. supervised the study. S.J.B. and V.M. served as project administrators.
REFERENCES
- 1.Kahrs CR, Chuda K, Tapia G, Stene LC, Marild K, Rasmussen T, Ronningen KS, Lundin Kea Kramna L, Cinek O, Stordal K. 2019. Enterovirus as trigger of coeliac disease: nested case-control study within prospective birth cohort. BMJ 364:l231. doi: 10.1136/bmj.l231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roivainen M, Klingel K. 2009. Role of enteroviruses in the pathogenesis of type 1 diabetes. Diabetologia 52:995–996. doi: 10.1007/s00125-009-1332-9. [DOI] [PubMed] [Google Scholar]
- 3.Laitinen OH, Honkanen H, Pakkanen O, Oikarinen S, Hankaniemi MM, Huhtala H, Ruokoranta T, Lecouturier V, Andre P, Harju R, Virtanen SM, Lehtonen J, Almond JW, Simell T, Simell O, Ilonen J, Veijola R, Knip M, Hyoty H. 2014. Coxsackievirus B1 is associated with induction of beta-cell autoimmunity that portends type 1 diabetes. Diabetes 63:446–455. doi: 10.2337/db13-0619. [DOI] [PubMed] [Google Scholar]
- 4.Sioofy-Khojine A-B, Lehtonen J, Nurminen N, Laitinen OH, Oikarinen S, Huhtala H, Pakkanen O, Ruokoranta T, Hankaniemi MM, Toppari J, Vähä-Mäkilä M, Ilonen J, Veijola R, Knip M, Hyöty H. 2018. Coxsackievirus B1 infections are associated with the initiation of insulin-driven autoimmunity that progresses to type 1 diabetes. Diabetologia 61:1193–1202. doi: 10.1007/s00125-018-4561-y. [DOI] [PubMed] [Google Scholar]
- 5.Nurani G, Lindqvist B, Casasnovas JM. 2003. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J Virol 77:11985–11991. doi: 10.1128/jvi.77.22.11985-11991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brabec M, Baravalle G, Blaas D, Fuchs R. 2003. Conformational changes, plasma membrane penetration, and infection by human rhinovirus type 2: role of receptors and low pH. J Virol 77:5370–5377. doi: 10.1128/JVI.77.9.5370-5377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karjalainen M, Kakkonen E, Upla P, Paloranta H, Kankaanpaa P, Liberali P, Renkema GH, Hyypia T, Heino J, Marjomaki V. 2008. A Raft-derived, Pak1-regulated entry participates in alpha2beta1 integrin-dependent sorting to caveosomes. Mol Biol Cell 19:2857–2869. doi: 10.1091/mbc.e07-10-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karjalainen M, Rintanen N, Lehkonen M, Kallio K, Maki A, Hellstrom K, Siljamaki V, Upla P, Marjomaki V. 2011. Echovirus 1 infection depends on biogenesis of novel multivesicular bodies. Cell Microbiol 13:1975–1995. doi: 10.1111/j.1462-5822.2011.01685.x. [DOI] [PubMed] [Google Scholar]
- 9.Marjomaki V, Pietiainen V, Matilainen H, Upla P, Ivaska J, Nissinen L, Reunanen H, Huttunen P, Hyypia T, Heino J. 2002. Internalization of echovirus 1 in caveolae. J Virol 76:1856–1865. doi: 10.1128/jvi.76.4.1856-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heikkila O, Susi P, Tevaluoto T, Harma H, Marjomaki V, Hyypia T, Kiljunen S. 2010. Internalization of coxsackievirus A9 is mediated by β2-microglobulin, dynamin, and Arf6 but not by caveolin-1 or clathrin. J Virol 84:3666–3681. doi: 10.1128/JVI.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soonsawad P, Paavolainen L, Upla P, Weerachatyanukul W, Rintanen N, Espinoza J, McNerney G, Marjomaki V, Cheng RH. 2014. Permeability changes of integrin-containing multivesicular structures triggered by picornavirus entry. PLoS One 9:e108948. doi: 10.1371/journal.pone.0108948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huttunen M, Waris M, Kajander R, Hyypia T, Marjomaki V. 2014. Coxsackievirus A9 infects cells via nonacidic multivesicular bodies. J Virol 88:5138–5151. doi: 10.1128/JVI.03275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marjomaki V, Turkki P, Huttunen M. 2015. Infectious entry pathway of enterovirus B species. Viruses 7:6387–6399. doi: 10.3390/v7122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myllynen M, Kazmertsuk A, Marjomaki V. 2016. A novel open and infectious form of echovirus 1. J Virol 90:6759–6770. doi: 10.1128/JVI.00342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuthill TJ, Groppelli E, Hogle JM, Rowlands DJ. 2010. Picornaviruses. Curr Top Microbiol Immunol 343:43–89. doi: 10.1007/82_2010_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casasnovas JM, Springer TA. 1994. Pathway of rhinovirus disruption by soluble intercellular adhesion molecule 1 (ICAM-1): an intermediate in which ICAM-1 is bound and RNA is released. J Virol 68:5882–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan G, Freistadt MS, Racaniello VR. 1990. Neutralization of poliovirus by cell receptors expressed in insect cells. J Virol 64:4697–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prchla E, Kuechler E, Blaas D, Fuchs R. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J Virol 68:3713–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curry S, Chow M, Hogle JM. 1996. The poliovirus 135S particle is infectious. J Virol 70:7125–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward T, Powell RM, Chaudhry Y, Meredith J, Almond JW, Kraus W, Nelsen-Salz B, Eggers HJ, Evans DJ. 2000. Fatty acid-depleted albumin induces the formation of echovirus A particles. J Virol 74:3410–3412. doi: 10.1128/jvi.74.7.3410-3412.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonberg-Holm K, Gosser LB, Shimshick EJ. 1976. Interaction of liposomes with subviral particles of poliovirus type 2 and rhinovirus type 2. J Virol 19:746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cords CE, James CG, McLaren LC. 1975. Alteration of capsid proteins of coxsackievirus A13 by low ionic concentrations. J Virol 15:244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wetz K, Kucinski T. 1991. Influence of different ionic and pH environments on structural alterations of poliovirus and their possible relation to virus uncoating. J Gen Virol 72:2541–2544. doi: 10.1099/0022-1317-72-10-2541. [DOI] [PubMed] [Google Scholar]
- 25.Ward T, Powell RM, Evans DJ, Almond JW. 1999. Serum albumin inhibits echovirus 7 uncoating. J Gen Virol 80:283–290. doi: 10.1099/0022-1317-80-2-283. [DOI] [PubMed] [Google Scholar]
- 26.Butan C, Filman DJ, Hogle JM. 2014. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol 88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J, Cheng N, Hogle JM, Steven AC, Belnap DM. 2013. Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy. J Immunol 191:884–891. doi: 10.4049/jimmunol.1202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Shingler KL, Organtini LJ, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2016. The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci Adv 2:e1501929. doi: 10.1126/sciadv.1501929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. 2014. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol 88:5755–5765. doi: 10.1128/JVI.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickl-Herk A, Luque D, Vives-Adrian L, Querol-Audi J, Garriga D, Trus BL, Verdaguer N, Blaas D, Caston JR. 2013. Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle. Proc Natl Acad Sci U S A 110:20063–20068. doi: 10.1073/pnas.1312128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog 9:e1003240. doi: 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauss M, Filman DJ, Belnap DM, Cheng N, Noel RT, Hogle JM. 2015. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J Virol 89:4143–4157. doi: 10.1128/JVI.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Lee LY, Roivainen M, Filman DJ, Hogle JM, Belnap DM. 2012. Structure of the Fab-labeled “breathing” state of native poliovirus. J Virol 86:5959–5962. doi: 10.1128/JVI.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerasimenko JV, Tepikin AV, Petersen OH, Gerasimenko OV. 1998. Calcium uptake via endocytosis with rapid release from acidifying endosomes. Curr Biol 8:1335–1338. doi: 10.1016/S0960-9822(07)00565-9. [DOI] [PubMed] [Google Scholar]
- 35.Christensen KA, Myers JT, Swanson JA. 2002. pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci 115:599–607. [DOI] [PubMed] [Google Scholar]
- 36.Weinert S, Jabs S, Supanchart C, Schweizer M, Gimber N, Richter M, Rademann J, Stauber T, Kornak U, Jentsch TJ. 2010. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl− accumulation. Science 328:1401–1403. doi: 10.1126/science.1188072. [DOI] [PubMed] [Google Scholar]
- 37.Hara-Chikuma M, Yang B, Sonawane ND, Sasaki S, Uchida S, Verkman AS. 2005. ClC-3 chloride channels facilitate endosomal acidification and chloride accumulation. J Biol Chem 280:1241–1247. doi: 10.1074/jbc.M407030200. [DOI] [PubMed] [Google Scholar]
- 38.Steinberg BE, Huynh KK, Brodovitch A, Jabs S, Stauber T, Jentsch TJ, Grinstein S. 2010. A cation counterflux supports lysosomal acidification. J Cell Biol 189:1171–1186. doi: 10.1083/jcb.200911083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott CC, Gruenberg J. 2011. Ion flux and the function of endosomes and lysosomes: pH is just the start: the flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. Bioessays 33:103–110. doi: 10.1002/bies.201000108. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer P, Herzog M, Hirth L. 1976. RNA viruses: stabilization of brome mosaic virus. Philos Trans R Soc Lond B Biol Sci 276:99–107. doi: 10.1098/rstb.1976.0100. [DOI] [PubMed] [Google Scholar]
- 41.Hull R. 1978. The stabilization of the particles of turnip rosette virus. III. Divalent cations. Virology 89:418–422. doi: 10.1016/0042-6822(78)90184-8. [DOI] [PubMed] [Google Scholar]
- 42.Bishop NE, Anderson DA. 1997. Early interactions of hepatitis A virus with cultured cells: viral elution and the effect of pH and calcium ions. Arch Virol 142:2161–2178. doi: 10.1007/s007050050233. [DOI] [PubMed] [Google Scholar]
- 43.Kivela HM, Mannisto RH, Kalkkinen N, Bamford DH. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:364–374. doi: 10.1006/viro.1999.9838. [DOI] [PubMed] [Google Scholar]
- 44.Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA. 2006. Removal of divalent cations induces structural transitions in Red clover necrotic mosaic virus, revealing a potential mechanism for RNA release. J Virol 80:10395–10406. doi: 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3:63–78. doi: 10.1016/S0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Vusse GJ. 2009. Albumin as fatty acid transporter. Drug Metab Pharmacokinet 24:300–307. doi: 10.2133/dmpk.24.300. [DOI] [PubMed] [Google Scholar]
- 47.Smyth M, Pettitt T, Symonds A, Martin J. 2003. Identification of the pocket factors in a picornavirus. Arch Virol 148:1225–1233. doi: 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 48.Filman DJ, Wien MW, Cunningham JA, Bergelson JM, Hogle JM. 1998. Structure determination of echovirus 1. Acta Crystallogr D Biol Crystallogr 54:1261–1272. doi: 10.1107/s0907444998002790. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Sheng J, van Vliet ALW, Buda G, van Kuppeveld FJM, Rossmann MG. 2018. Molecular basis for the acid-initiated uncoating of human enterovirus D68. Proc Natl Acad Sci U S A 115:E12209–E12217. doi: 10.1073/pnas.1803347115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdelmagid SA, Clarke SE, Nielsen DE, Badawi A, El-Sohemy A, Mutch DM, Ma DW. 2015. Comprehensive profiling of plasma fatty acid concentrations in young healthy Canadian adults. PLoS One 10:e0116195. doi: 10.1371/journal.pone.0116195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penn AH, Dubick MA, Torres Filho IP. 2017. Fatty acid saturation of albumin used in resuscitation fluids modulates cell damage in shock: in vitro results using a novel technique to measure fatty acid binding capacity. Shock 48:449–458. doi: 10.1097/SHK.0000000000000865. [DOI] [PubMed] [Google Scholar]
- 52.Xing L, Huhtala M, Pietiainen V, Kapyla J, Vuorinen K, Marjomaki V, Heino J, Johnson MS, Hyypia T, Cheng RH. 2004. Structural and functional analysis of integrin alpha2I domain interaction with echovirus 1. J Biol Chem 279:11632–11638. doi: 10.1074/jbc.M312441200. [DOI] [PubMed] [Google Scholar]
- 53.Trigatti BL, Gerber GE. 1995. A direct role for serum albumin in the cellular uptake of long-chain fatty acids. Biochem J 308:155–159. doi: 10.1042/bj3080155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martikainen M, Salorinne K, Lahtinen T, Malola S, Permi P, Hakkinen H, Marjomaki V. 2015. Hydrophobic pocket targeting probes for enteroviruses. Nanoscale 7:17457–17467. doi: 10.1039/C5NR04139B. [DOI] [PubMed] [Google Scholar]
- 55.Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, Thibaut HJ, Nieuwenhuis J, Janssen H, van Kuppeveld FJ, Perrakis A, Carette JE, Brummelkamp TR. 2017. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 541:412–416. doi: 10.1038/nature21032. [DOI] [PubMed] [Google Scholar]
- 56.Baggen J, Thibaut HJ, Strating JRPM, van Kuppeveld FJM. 2018. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol 16:368–381. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 57.Siljamaki E, Rintanen N, Kirsi M, Upla P, Wang W, Karjalainen M, Ikonen E, Marjomaki V. 2013. Cholesterol dependence of collagen and echovirus 1 trafficking along the novel alpha2beta1 integrin internalization pathway. PLoS One 8:e55465. doi: 10.1371/journal.pone.0055465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huttunen M, Turkki P, Maki A, Paavolainen L, Ruusuvuori P, Marjomaki V. 2017. Echovirus 1 internalization negatively regulates epidermal growth factor receptor downregulation. Cell Microbiol 19:e12671. doi: 10.1111/cmi.12671. [DOI] [PubMed] [Google Scholar]
- 59.Upla P, Marjomaki V, Nissinen L, Nylund C, Waris M, Hyypia T, Heino J. 2008. Calpain 1 and 2 are required for RNA replication of echovirus 1. J Virol 82:1581–1590. doi: 10.1128/JVI.01375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Draper DE. 2004. A guide to ions and RNA structure. RNA 10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hendry E, Hatanaka H, Fry E, Smyth M, Tate J, Stanway G, Santti J, Maaronen M, Hyypia T, Stuart D. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527–1538. doi: 10.1016/S0969-2126(00)88343-4. [DOI] [PubMed] [Google Scholar]
- 62.Muckelbauer JK, Kremer M, Minor I, Diana G, Dutko FJ, Groarke J, Pevear DC, Rossmann MG. 1995. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure 3:653–667. doi: 10.1016/S0969-2126(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 63.Arnold E, Rossmann MG. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 Å. J Mol Biol 211:763–801. doi: 10.1016/0022-2836(90)90076-X. [DOI] [PubMed] [Google Scholar]
- 64.Hadfield AT, Lee W, Zhao R, Oliveira MA, Minor I, Rueckert RR, Rossmann MG. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427–441. doi: 10.1016/S0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 65.Kalynych S, Palkova L, Plevka P. 2016. The structure of human parechovirus 1 reveals an association of the RNA genome with the capsid. J Virol 90:1377–1386. doi: 10.1128/JVI.02346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Domanska A, Flatt JW, Jukonen JJJ, Geraets JA, Butcher SJ. 2019. A 2.8-angstrom-resolution cryo-electron microscopy structure of human parechovirus 3 in complex with Fab from a neutralizing antibody. J Virol 93:e01597-18. doi: 10.1128/JVI.01597-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shakeel S, Evans JD, Hazelbaker M, Kao CC, Vaughan RC, Butcher SJ. 2018. Intrinsically disordered N termini in human parechovirus 1 capsid proteins bind encapsidated RNA. Sci Rep 8:5820. doi: 10.1038/s41598-018-23552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidtke M, Schnittler U, Jahn B, Dahse H, Stelzner A. 2001. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J Virol Methods 95:133–143. doi: 10.1016/S0166-0934(01)00305-6. [DOI] [PubMed] [Google Scholar]
- 69.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang K. 2016. Gctf: Real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 75.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 77.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. 2008. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The final density maps have been deposited in the Electron Microscopy Databank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/) with accession codes EMD-4903 (control E1) and EMD-0565 (expanded E1). The atomic models have been deposited in the Protein Data Bank (and Worldwide Protein Data Bank [wwPDB]) with accession codes 6RJF (control E1) and 6O06 (expanded E1). The raw data have been deposited in the EMPIAR database (https://www.ebi.ac.uk/pdbe/emdb/empiar/) with deposition EMPIAR-10284.