Anti-influenza drugs with broad-spectrum efficacy against antigenically diverse influenza viruses can be highly useful when no vaccines are available. To develop new anti-influenza drugs, we screened a number of small molecules and identified a strong candidate, IY7640. When added at the time of or after influenza virus infection, IY7640 was observed to successfully inhibit or reduce viral replication in cells. We subsequently discovered that IY7640 targets the stalk region of the influenza HA protein, which exhibits a relatively high degree of amino acid sequence conservation across various (sub)types of influenza viruses. Furthermore, IY7640 was observed to block HA-mediated membrane fusion of H1N1, H3N2, and influenza B viruses in cells. Although it appears less effective against strains other than H1N1 subtype viruses in a challenge study in mice, we suggest that the small molecule IY7640 has potential to be optimized as a new anti-influenza drug.
KEYWORDS: hemagglutinin stalk, membrane fusion, small molecule, antiviral agents, influenza virus
ABSTRACT
Combating influenza is one of the perennial global public health issues to be managed. Antiviral drugs are useful for the treatment of influenza in the absence of an appropriate vaccine. However, the appearance of resistant strains necessitates a constant search for new drugs. In this study, we investigated novel anti-influenza drug candidates using in vitro and in vivo assays. We identified anti-influenza hit compounds using a high-throughput screening method with a green fluorescent protein-tagged recombinant influenza virus. Through subsequent analyses of their cytotoxicity and pharmacokinetic properties, one candidate (IY7640) was selected for further evaluation. In a replication kinetics analysis, IY7640 showed greater inhibitory effects during the early phase of viral infection than the viral neuraminidase inhibitor oseltamivir. In addition, we observed that hemagglutinin (HA)-mediated membrane fusion was inhibited by IY7640 treatment, indicating that the HA stalk region, which is highly conserved across various (sub)types of influenza viruses, may be the molecular target of IY7640. In an escape mutant analysis in cells, amino acid mutations were identified at the HA stalk region of the 2009 pandemic H1N1 (pH1N1) virus. Even though the in vivo efficacy of IY7640 did not reach complete protection in a lethal challenge study in mice, these results suggest that IY7640 has potential to be developed as a new type of anti-influenza drug.
IMPORTANCE Anti-influenza drugs with broad-spectrum efficacy against antigenically diverse influenza viruses can be highly useful when no vaccines are available. To develop new anti-influenza drugs, we screened a number of small molecules and identified a strong candidate, IY7640. When added at the time of or after influenza virus infection, IY7640 was observed to successfully inhibit or reduce viral replication in cells. We subsequently discovered that IY7640 targets the stalk region of the influenza HA protein, which exhibits a relatively high degree of amino acid sequence conservation across various (sub)types of influenza viruses. Furthermore, IY7640 was observed to block HA-mediated membrane fusion of H1N1, H3N2, and influenza B viruses in cells. Although it appears less effective against strains other than H1N1 subtype viruses in a challenge study in mice, we suggest that the small molecule IY7640 has potential to be optimized as a new anti-influenza drug.
INTRODUCTION
Influenza virus is a negative-sense, single-stranded RNA virus of the family Orthomyxoviridae (1). As a human respiratory pathogen, influenza virus causes recurrent seasonal epidemics, with global circulation of two subtypes (H1N1 and H3N2) of influenza A virus (IAV) and the two antigenically distinct lineages (so-called Victoria and Yamagata) of influenza B virus (IBV), which leads to an estimated half million deaths each year (2). Sometimes, the virus also provokes a pandemic that affects global communities with incalculable socioeconomic damage (3, 4). These perpetuating events of influenza may result from antigenic drift or shift that cause strain and subtype changes of influenza viruses, respectively (5, 6). To deal with antigenic variation issues, universal intervention methods against various (sub)types of influenza viruses have been investigated (7–10), with vaccine designs and platforms being the primary considerations to develop universal vaccines inducing cross-reactive immunity in vaccinated individuals (11). While anti-neuraminidase (anti-NA) antibodies and other immune repertoires have been also investigated for the development of broadly effective therapeutics (12–16), several broadly neutralizing anti-hemagglutinin (anti-HA) antibodies have been discovered: C05, which recognizes the receptor binding site (RBS) of HA (17); CR6261 and CR8020, which inhibit the fusion activity of the groups 1 and 2 IAV HAs, respectively (17); FI6, which neutralizes IAVs of both HA groups (18); and CR9114, which has efficacy against both IAVs and IBVs (19). These antibodies can be used as universally effective therapeutics (20), although there are still issues to be solved in terms of their delivery routes and high manufacturing costs (21).
Antiviral drugs can be another option as universal intervention methods against various (sub)types of influenza viruses (22, 23). Most antivirals approved by the Food and Drug Administration are small molecules (24), and several types of antiviral drugs have been used to treat influenza (25). However, due to the possibility of the emergence and dissemination of antiviral-resistant strains (26, 27), new types of antivirals are always needed. Nine antiviral drug candidates (baloxavir marboxil, a cap-dependent endonuclease inhibitor [28], which has been licensed recently in the United States; FluDase, a recombinant sialidase fusion protein [29]; JNJ-5806, a polymerase inhibitor; laninamivir octanoate, a neuraminidase inhibitor [30]; MEDI8852, an HA stem-binding monoclonal antibody [31]; NT-300, an HA intracellular trafficking inhibitor; pimodivir, a PB2 cap-snatching inhibitor [32]; radavirsen, a genetic transcription inhibitor of antisense oligonucleotide [33]; and VIS-410, an HA stem-binding monoclonal antibody [34]) are in clinical trials (24), five of which are small molecules that have advantages of standardization and manufacturing processes.
Given the binding and fusion functions of the influenza virus HA protein, small molecules targeting the HA may be of great importance in the prevention and treatment of influenza in humans. Several HA-targeted candidates are currently under investigation (35), of which umifenovir (Arbidol) is an orally active antiviral agent that inhibits HA fusion activity (36) and has been used to treat influenza in China and Russia. However, this inhibitor is not approved for use in other countries. In addition, the HA-targeting small-molecule inhibitors RO-5487624 and RO-5464466 and natural molecules SQ-02-S5 and pentacyclic triterpenoids are in preclinical studies (35). Recently, a small molecule (JNJ4796) was reported to inhibit HA-mediated membrane fusion by binding the residues that CR6261 interacts with in the HA stalk region (37). Despite these efforts, no licensed HA-targeted inhibitors have been introduced against influenza.
To identify new antiviral drugs against various influenza viruses, in this study, we used the recombinant A/Puerto Rico/8/34 virus expressing green fluorescent protein (GFP; rPR8/GFP) for a rapid initial screen (38, 39). From several candidate compounds, the final candidate IY7640 was identified, a de novo synthesizable, anti-influenza virus small molecule. IY7640 targets the stalk region of the HA protein, which is highly conserved across different (sub)types of influenza viruses. In a series of in vitro experiments, IY7640 exhibited antiviral effects against various influenza viruses, including pH1N1 and oseltamivir-resistant strains, by inhibiting acid-dependent membrane fusion of the HA protein. We subsequently demonstrated the anti-influenza virus effects of this small molecule against lethal viral challenge in mice.
RESULTS
Efficacy of IY7640 against influenza viruses.
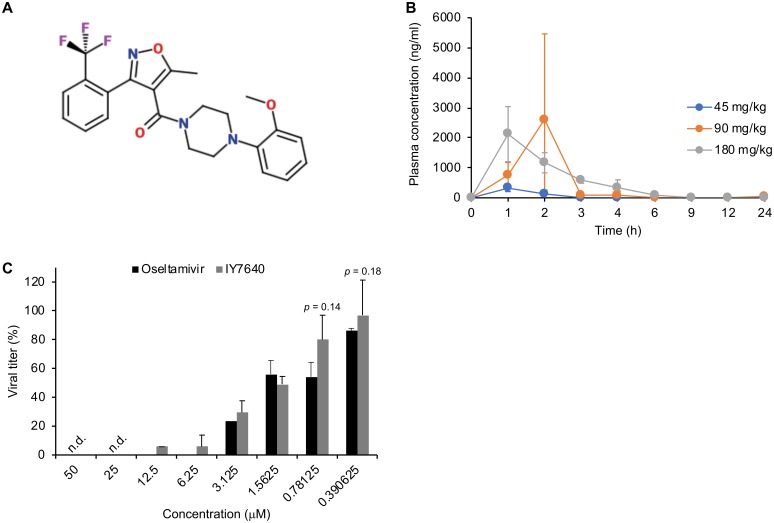
Among 9,687 chemicals tested, 8 compounds reduced GFP expression in rPR8/GFP-infected cells (Fig. 1). Of the eight hit compounds, IY7640 was selected based on the preliminary in vivo pharmacokinetic (PK) analyses in mice (Fig. 2A and B). Among the PK parameters, the area under the concentration-time curve (AUC) of IY7640 was determined in a dose-dependent manner, whereas the maximum plasma concentration (Cmax) was the highest with administration of 90 mg/kg of body weight of IY7640 (Table 1). IY7640 was then tested against H1N1, H3N2, H5N1, H7N9, and H9N2 subtypes of IAV, including the 2009 pandemic H1N1 (pH1N1) virus (A/Korea/01/2009, rK09) (40), and Victoria and Yamagata lineage strains of IBV in viral cytopathic effect (CPE) and plaque reduction assays. As oseltamivir carboxylate (oseltamivir) was used as a control chemical, oseltamivir-resistant pH1N1 strains were also included. Given the observed 50% effective concentration (EC50) values (EC50 values in the CPE assay, 0.62 to 221 μM for IY7640 and 0.09 to 1991 μM for oseltamivir; EC50 values in the plaque reduction assay, 0.76 to 832 μM for IY7640) (Table 2), IY7640 may primarily be effective against H1N1 subtype and oseltamivir-resistant strains, with relatively higher EC50 values against H5N1, H7N9, and H9N2 subtypes and IBVs than those of oseltamivir. IY7640 efficacy cannot be determined against H3N2 subtype strains because, compared with oseltamivir results in the CPE assay (0.09 to 518.4 μM), the EC50 values of IY7640 in the same assay were determined within a much smaller range (83.05 to 213.9 μM), whereas the plaque reduction assay resulted in slightly different EC50 values (111.1 to 1108 μM) (Table 2).
FIG 1.
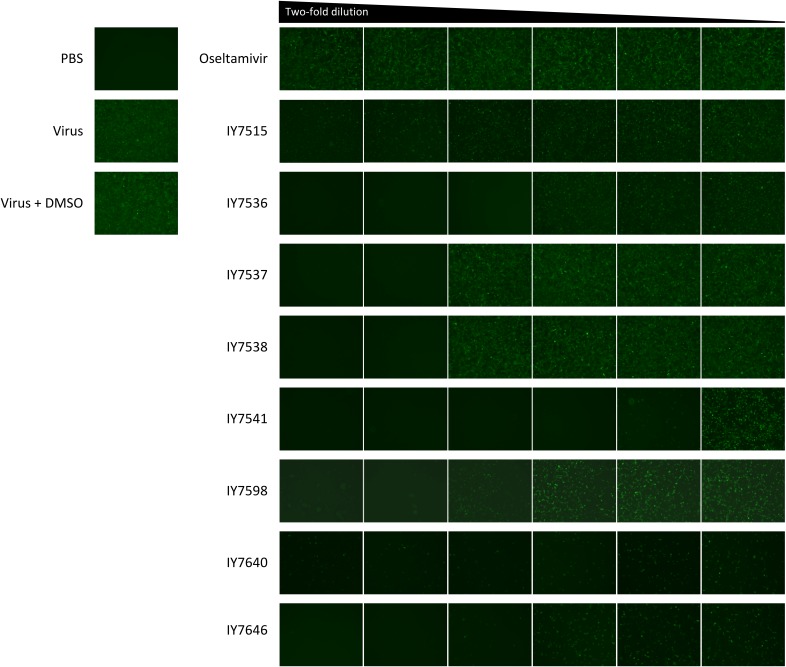
Candidate chemicals selected using the rPR8-GFP virus. By a plaque reduction assay in MDCK cells (96-well plate; 4 × 104 cells/well), eight candidate chemicals were selected based on the GFP expression of oseltamivir-treated wells. A total of 100 μg of each chemical dissolved in DMSO was serially 2-fold diluted in PBS and added to overlaying agar. The multiplicity of infection of rPR8-GFP was 0.5, and GFP expression was observed at 24 h postinfection. PBS was used for mock infection.
FIG 2.
PK evaluation and anti-influenza effects of IY7640. (A) Molecular structure of IY7640. (B) Graphic representation of IY7640 pharmacokinetics in mice. Mean values are plotted. (C) The anti-influenza effect of IY7640 in cultured cells. In MDCK cells, the inhibitory effect of IY7640 was determined against the rK09 virus compared with that of oseltamivir using a plaque reduction assay. n.d., below detection limit (10 PFU/ml).
TABLE 1.
Pharmacokinetic study of IY7640 in BALC/c mice
| Dose (mg/kg) | Value for indicated PK parameter |
|||
|---|---|---|---|---|
| AUC (ng · h/ml) | Cmax (ng/ml) | Tmax (h) | t1/2 (h) | |
| 45 | 552.78 | 351.41 | 1.20 | 3.67 |
| 90 | 4,062.27 | 2,699.16 | 1.60 | 5.43 |
| 180 | 4,769.26 | 2,141.14 | 1.20 | 3.04 |
TABLE 2.
Comparison of IY7640 and oseltamivir efficacies against group 1 and 2 influenza A viruses as well as influenza B viruses using the CPE assay
|
Virus |
EC50 (μM) |
||||
|---|---|---|---|---|---|
| CPE assay |
Plaque reduction assay, IY7640 | ||||
| Group | Subtype | Strain | IY7640 | Oseltamivir carboxylate | |
| 1 | H1N1 | A/Solomon Island/03/2006 | 0.7 | 206.5 | 0.76 |
| A/Brisbane/59/2007 | 0.62 | 7.11 | 1.53 | ||
| Seasonal/2008a | 3.81 | 964.3 | 1.95 | ||
| pH1N1 | A/Korea/01/2009 (rK09) | 1.95 | 2.17 | 1.58 | |
| A/Netherlands/602/2009 | 2.13 | 2.26 | 2.3 | ||
| A/Korea/2785/2009a | 7.1 | 1323 | 5.11 | ||
| H5N1 | A/chicken/IS/06/2006 (6:2)b | 59.6 | 0.19 | 16.16 | |
| H9N2 | A/chicken/Korea/01310/2001 | 33.39 | 0.63 | 52.23 | |
| 2 | H3N2 | A/Wisconsin/67/2005 | 83.05 | 0.09 | 111.1 |
| A/Brisbane/10/2007 | 110.7 | 493.7 | 1,108 | ||
| A/Perth/16/2009 | 213.9 | 518.4 | 832.4 | ||
| H7N9 | A/Anhui/1/2013 (6:2)b | 221 | 0.81 | 277.1 | |
| B_Victoria | B/Brisbane/60/2008 | 39.68 | 3.25 | 213.7 | |
| B_Yamagata | B/Wisconsin/01/2010 | 17.13 | 0.56 | 47.19 | |
| rg-virusc | pH1N1 | rK09/NA:Y275a | 1.92 | 1,302 | 2.6 |
| rK09/NA:Y275+V222a | 4.79 | 1,991 | 3.73 | ||
The NA proteins of these viruses retained the Y275 signature, which promotes resistance to oseltamivir.
6:2 viruses were generated on the PR8 backbone.
rg-viruses were generated on the K09 backbone.
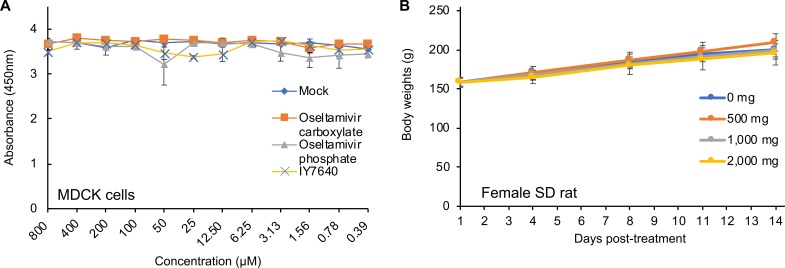
In agreement with the EC50 values toward H1N1 subtype strains, IY7640 successfully inhibited rK09 replication in cells at a level comparable to that of oseltamivir (Fig. 2C). Given the 50% cytotoxic concentration (CC50) of IY7649 (greater than 800 μM in Madin-Darby canine kidney [MDCK] cells) (Fig. 3A) and body weight increases of female rats administered once daily with 500-, 1,000-, and 2,000-mg/kg doses for 2 weeks (Fig. 3B), IY7640 appeared to have no significant toxicity.
FIG 3.
Toxicity evaluation of IY7640. (A) Cytotoxicity test of IY7640 in MDCK cells. The number of viable cells was measured fluorometrically using the CCK-8 reagent after incubating the cells with DMSO (mock) or the designated concentrations of IY7640 (dissolved in DMSO), oseltamivir-carboxylate, or oseltamivir phosphate. The experiments were performed in duplicate. (B) Two-week repeated-oral-dose toxicity test of IY7640. Female SD rats (5 rats per group) were administered once daily with 0, 500, 1,000, or 2,000 mg of IY7640 by gastric intubation for 2 weeks, and their body weights were recorded at 1, 4, 8, 11, and 14 days posttreatment.
Escape mutations and viral target of IY7640.
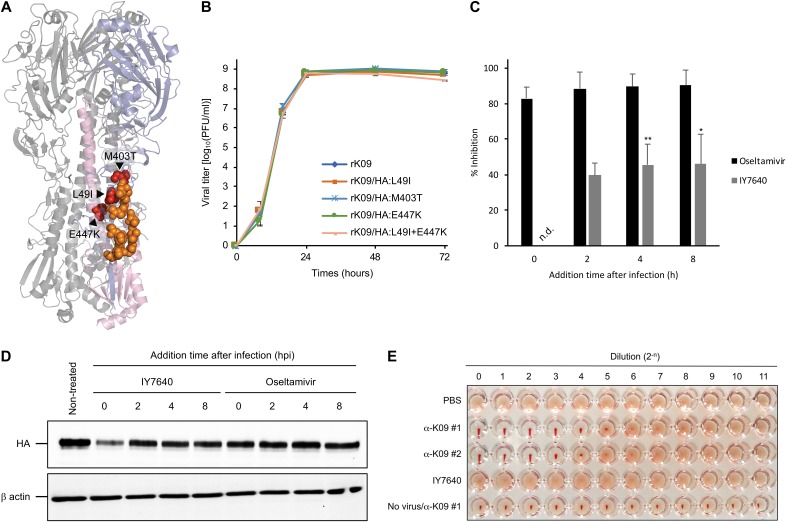
Elucidating the molecular target and mechanism of action of IY7640 is important for its development as an anti-influenza drug, either as a single therapy or in combination with other drugs (41). To determine whether IY7640 targets the virus and which viral proteins it targets, we induced the generation of escape mutants by sequentially passaging the rK09 virus in 2-fold increasing concentrations of IY7640. Escape mutants first appeared during the 5th passage (10 days with the chemical) in one set of experiments and during the 6th passage (12 days with the chemical) in the other set, which was approximately the same time frame as that in which amantadine and oseltamivir escape mutant viruses appeared in a previous study (42). We sequenced 3 plaque-purified viral clones from each set of experiments. All three of the escape mutants from the first experiment shared two HA mutations, L49I and E447K (numbered according to the HA amino acid sequence of K09, GenBank accession number ACQ84451), and those from the second experiment shared an M403T mutation (Table 3). In the HA protein structure, the L49I, M403T, and E447K mutations are located in the HA stalk region (Fig. 4A), which is crucial for HA fusion activity (43). To determine whether these HA mutations promoted viral resistance against IY7640, we generated recombinant viruses (rK09/HA:L49I, rK09/HA:M403T, rK09/HA:E447K, and rK09/HA:L49I+E447K) using reverse genetics, with each strain harboring an escape HA mutation(s) in the K09 virus backbone. In the replication kinetics in MDCK cells, the escape mutant viruses exhibited growth patterns and rates similar to those of rK09 (Fig. 4B). For the L49I and E447K mutations, E447K appeared to be a major contributor to the resistant phenotype, based on the EC50 values of rK09/HA:L49I (15.4 and 16.3 μM), rK09/HA:E447K (207.1 and 971.8 μM), and rK09/HA:L49I+E447K (69.46 and 767 μM) determined using the CPE and plaque reduction assays, respectively (Table 4). In the second set, the M403T mutation either was the only mutation or was present together with a P229S mutation in the HA head region, but the M403T mutation alone was sufficient to confer resistance to IY7640 (EC50 of rK09/HA:M403T, 116 and 445.8 μM as determined by the CPE and plaque reduction assays, respectively) (Table 4). These results indicate that the IY7640-induced escape mutations may cause resistance to IY7640. In addition to the HA escape mutant analysis, the time-to-addition experiment also suggested that the earliest functioning HA protein is the major, if not the only, target of IY7640, effectively inhibiting rK09 replication (Fig. 4C) and HA protein expression (Fig. 4D). However, IY7640 did not inhibit the hemagglutination activity of rK09 with turkey red blood cells (tRBCs) (Fig. 4E). These results suggested that IY7640 may target HA in a step after the virus binds to the cell, most likely during membrane-to-membrane fusion.
TABLE 3.
Amino acid mutations of K09 after the IY7640 treatment
| Expt | Clone | Escape mutation(s) identified in K09 proteina
|
|||
|---|---|---|---|---|---|
| PB2 | HA | NP | NA | ||
| 1 | 1 | − | L49I, E447K | N492K, E494G | − |
| 2 | − | L49I, E447K | − | − | |
| 3 | − | L49I, E447K | − | Q45L | |
| 2 | 1 | E208A | M403T | − | − |
| 2 | − | P229S, M403T | − | K347R | |
| 3 | − | M403T | − | − | |
−, no genetic mutations from two independent experiments.
FIG 4.
Molecular target of IY7640. (A) Escape mutations (L49I, M403T, and E447K) identified in the HA protein of rK09 after IY7640 treatment are adjacently located with the CR6261 epitopes in the HA structure. The colors represent the following: slate, HA1; light pink, HA2; red, escape mutations against IY7640; and orange, CR6261 epitopes. (B) Replication kinetics of escape mutant viruses were evaluated in MDCK cells. (C and D) Viral infection and replication efficiency were measured by the number of the newly generated virus particles via plaque assays (C), and the amounts of newly expressed HA were compared by Western blotting (D). (E) A hemagglutination assay using the K09 virus and 0.5% (vol/vol) tRBCs was performed in the presence of PBS (negative control), 2-fold serially diluted anti-K09 guinea pig sera (α-K09 #1 and #2; positive controls), or IY7640 (starting at 90 μM).
TABLE 4.
EC50 values of IY7640 escape mutant viruses
| Virus | EC50 (μM) of IY7640 |
|
|---|---|---|
| CPE assay | Plaque reduction assay | |
| rK09 | 1.95 | 1.58 |
| rK09/HA:L49I | 15.4 | 16.3 |
| rK09/HA:M403T | 116 | 445.8 |
| rK09/HA:E447K | 207.1 | 971.8 |
| rK09/HA:L49I+E447K | 69.46 | 767 |
Molecular docking simulation of IY7640 with the HA crystal structure.
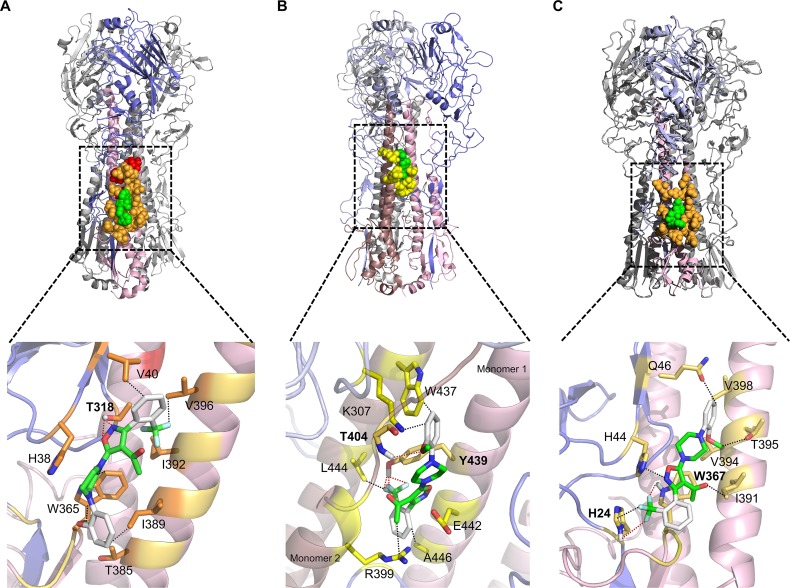
In further support of the molecular target of IY7640, the escape mutations were positioned around the CR6261 epitope (Fig. 4A), which is known to play a role in HA fusion (44). We then attempted a molecular docking simulation of IY7640 on the HA structure from a pH1N1 virus, A/California/04/2009 (Ca04; PDB ID 3UBQ) (45), using SYBYL-X (v2.1.1) Surflex Dock and the AutoDock Vina (46). In the simulation, IY7640 was observed to bind around the HA stalk region defined by the CR6261 epitope residues (Fig. 5A). However, it did not dock at the CR6261 epitope-equivalent region in the group 2 HA. Instead, it docked near the binding site of the fusion inhibitor tert-butyl hydroquinone (TBHQ) present in H3 HA (Fig. 5B), possibly due to the tryptophan at HA residue 365 (W365) (47, 48). In H5 HA of the group 1 HAs, IY7640 also docked at a similar stalk region (Fig. 5C).
FIG 5.
Structural interaction of IY7640 with H1, H3, and H5 HAs. The interaction simulation of IY7640 (green) with the CR6261 (orange) or the TBHQ (yellow) epitopes is depicted using the crystal structures of trimeric HAs (HA1 is slate-colored and HA2 is light pink) in one monomer—pH1N1 Ca04 HA of PDB ID 3UBQ in panel A, H3N2 A/Aichi/2/1968 HA of PDB ID 3ZTJ in panel B, and H5N1 A/Vietnam/1194/2004 HA of PDB ID 2IBX in panel C—with a respective enlarged image of the HA2 stalk region. The escape mutations against IY7640 are colored red. The residues are labeled according to H1, H3, or H5 numbering. The hydrogen bond interactions between IY7640 and HAs (connected by red dots) are given in angstroms: in the enlarged H1 HA (A), 2.109 and 2.468 to T318; in the enlarged H3 HA (B), 2.923 to T404 and 1.860, 1.470, and 2.016 to Y439; and in the enlarged H5 HA (C), 3.175 to H24, and 3.138 to W367. The non-hydrogen bond interactions (connected by black dots) are also shown: 3.334 to V40, 3.132 and 3.193 to W365, 3.368 to I389, and 2.767 to V396 in H1 HA (A); 2.617 to K307, 3.250 to R399, 3.104 to W437, 2.547 to L444, and 3.168 to A446 in H3 HA (B); and 3.013 to H24, 2.844 to H44, 3.003 to Q46, 2.817 to I391, 3.212 to V394, and 2.986 to T395 in H5 HA (C).
Inhibition of HA fusion activity and in vivo efficacy of IY7640.
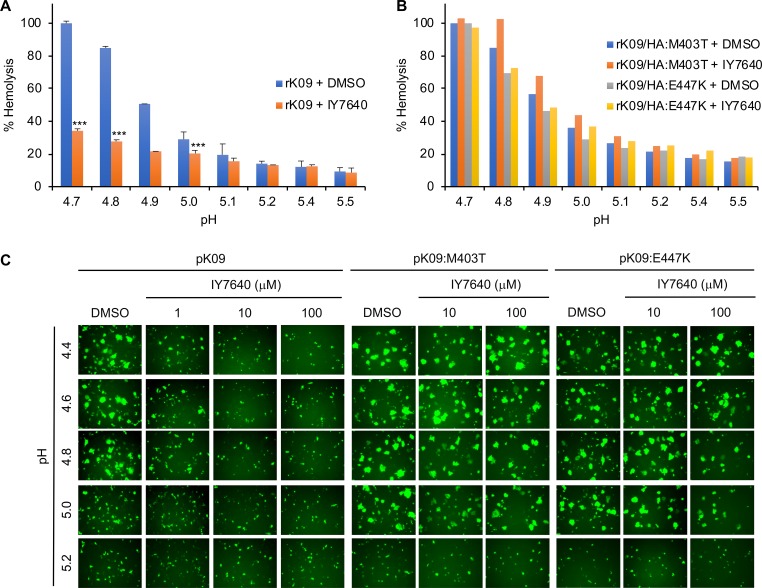
IY7640 binding to the CR6261 epitope residues strongly suggests that IY7640 may inhibit HA fusion activity (44, 49). To test this possibility, we applied a standard hemolysis assay, in which viral HA-mediated membrane fusion is indirectly demonstrated by the hemolysis of RBCs. In this assay, IY7640 surely inhibited acid-induced RBC hemolysis by the rK09 virus (Fig. 6A), which was not observed for the escape mutant viruses (rK09/HA:M403T and rK09/HA:E447K virus) (Fig. 6B). When using a GFP and HA cotransfection method in which the fusion activity of HA results in the spread of GFP to neighboring cells (49, 50), the pH required for the fusion of the escape mutant HAs (fusion pH for both pH 1/K09:M403T and pH 1/K09:E447K was approximately 5.0) increased by approximately 0.2 pH unit compared with that of the K09 (fusion pH for the pH 1/K09:M403T was approximately 4.8) (Fig. 6C). Compared with the escape mutant HAs, K09 HA fusion was inhibited only by 1 μM IY7640 (Fig. 6C).
FIG 6.
Inhibition of HA fusion activity by IY7640. (A and B) Inhibition of RBC hemolysis by IY7640. rK09 and the escape mutant (rK09/HA:M403T and rK09/HA:E447K) mixtures were preincubated with 50 μM of IY7640 in dimethyl sulfoxide (DMSO) (rK09 + IY7640) or DMSO alone (rK09 + DMSO). Then the mixtures were acidified to the designated pH and measured for the release of hemoglobin from lysed erythrocytes. (C) Inhibition of HA-mediated cell-to-cell fusion by IY7640. In Vero cells, the inhibition of HA-mediated cell-to-cell fusion was determined with IY7640 treatment (for pK09 HA, 1, 10, or 100 μM, and for pK09:M403T and pK09:E447K HAs, 10 or 100 μM) according to the pH ranges (pH 4.4 to 5.2). DMSO was used as a control.
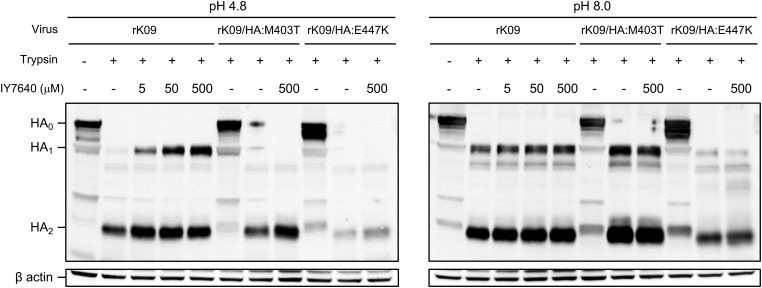
Influenza virus HA is synthesized as the inactive form HA0, and the active HA1 and HA2 dimer generated by trypsin-like proteases can adopt a fusion conformation under acidic conditions similar to those in endosomes (1). When the HA1 and HA2 dimer is in a fusion conformation, it is susceptible to cleavage by proteases, such as trypsin (51). However, IY7640 protected the HA in the fusion conformation from trypsin digestion. The rK09 HA maintained an intact HA1 in the presence of IY7640 at concentrations as low as 5 μM, whereas the M403T and E447K HAs did not maintain an intact HA1, even at IY7640 concentrations as high as 500 μM (Fig. 7), which was much higher than the EC50 values for the viruses with escape mutant HAs (Tables 2 and 4).
FIG 7.
HA susceptibility to protease and its inhibition by IY7640. MDCK cell lysates infected with the viruses were trypsinized followed by IY7640 treatment (5, 50, or 500 mM). After pH adjustment (pH 4.8 or 8.0), the cell supernatants were examined by the Western blot analysis.
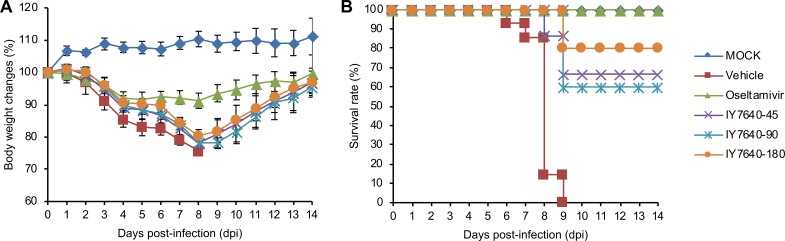
The anti-influenza efficacy of IY7640 determined in the CPE and plaque reduction assays appeared to be more focused against H1N1 subtypes (Table 2). Consistent with these results, IY7640 did not exhibit in vivo efficacy against H3N2 and H5N1 subtype challenges in mice (data not shown). Only for the pH1N1 rK09 viral challenge did IY7640 reduce body weight losses (vehicle group, 26.76% at 8 days postinfection [dpi]; oseltamivir, 8.77% at 8 dpi; IY7640 at 45 mg/kg/day, 22.56% at 8 dpi; IY7640 at 90 mg/kg/day, 23.65% at 9 dpi; and IY7640 at 180 mg/kg/day, 19.88% at 9 dpi) in the infected mice (Fig. 8A) and protect them with 60 to 80% survival rates in a dose-dependent manner (Fig. 8B).
FIG 8.
Anti-influenza effects of IY7640 against pH1N1 challenge in mice. The in vivo efficacy of IY7640 was evaluated in BALB/c mice (n = 10 per group). The mice were treated twice daily (half daily dose per treatment) with IY7640 (45 mg/kg/day [IY7640-45], 90 mg/kg/day [IY7640-90] or 180 mg/kg/day [IY7640-180]) for 8 days after pH1N1 rK09 infection. Body weight changes (A) and survival rates (B) were recorded for 14 dpi. Mice treated with oseltamivir (45 mg/kg/day) were used as a therapeutic control. Mice infected with PBS (mock) or treated only with a vehicle were also used as negative controls.
DISCUSSION
Through in vitro and in vivo studies, we identified an anti-influenza drug candidate, IY7640 (Fig. 1 and 2), which targets the HA stalk of influenza viruses without exhibiting significant toxicity in MDCK cells and rats (Fig. 3). As demonstrated for the antibodies CR6261 (44), FI6 (18), and CR9114 (19), a molecule targeting the HA stalk region may have broad-spectrum efficacy against IAVs and IBVs by inhibiting membrane-to-membrane fusion. Given the results of the CPE and plaque reduction assays (Table 2), escape mutant analysis (Fig. 4, 6, and 7 and Tables 3 and 4), docking simulation (Fig. 5), and GFP fusion assays (Fig. 6), the mechanism of action of IY7640 may also be fusion inhibition, highlighting the potential of IY7640 as a small-molecule counterpart of broadly neutralizing antibodies. However, the EC50 values of IY7640 were determined to be relatively higher than those of oseltamivir. Even against the same subtype strains, a wide range of concentrations were required for IY7640 efficacy (Table 1). Oseltamivir also exhibited fluctuating EC50 values against the H3N2 strains, but higher concentrations of IY7640 were needed to inhibit H5N1, H7N9, H9N2, and IBV strains (Table 2). Against the H1N1 and oseltamivir-resistant strains, IY7640 exhibited superb efficacy. As shown in Table 1, even less than 1 μM IY7640 could successfully inhibit various H1N1 strains and protect the mice from lethal challenge (Fig. 8). Given the global circulation of oseltamivir-resistant strains (52, 53), IY7640 may be useful under certain circumstances.
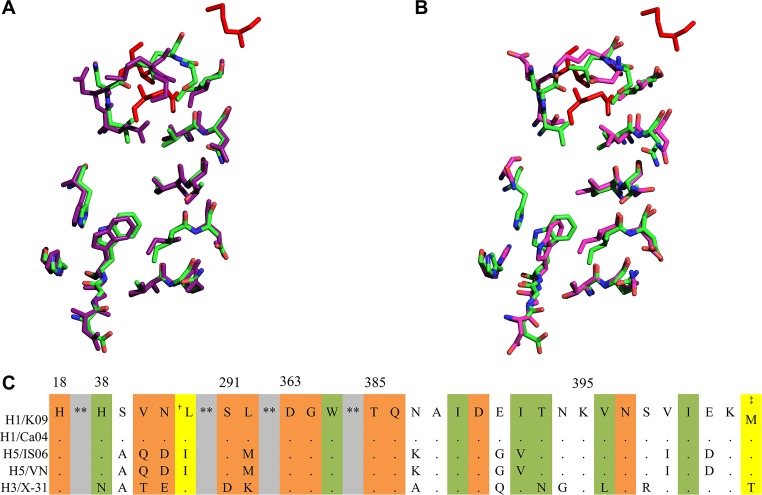
The observed EC50 values (Table 2) and docking simulations (Fig. 5) indicate that the binding of IY7640 to the CR6261 epitope region appeared to be associated with its efficacy. The CR6261 epitope residues in Ca04 HA that interact with IY7640 are well aligned with those of H5 HA (Fig. 9A) (54), with only one amino acid difference (isoleucine and valine in H1 and H5 HA residue 392, respectively) (Fig. 9C). However, differences of the EC50 values between the H1N1 and H5N1 viruses (Table 2) suggest how a single amino acid change can render the virus less sensitive to IY7640, as observed for the escape mutants. For H3 HA, IY7640 was not structurally accommodated in the CR6261 epitope-equivalent region, due to HA W365 residing in a protruding conformation and differences at three of the IY7640-interacting residues (Fig. 5B and 9B and C), as suggested in a recent study by Kadam et al. (48). The W365 residue is well conserved in the fusion peptide; mutating this residue abolishes its fusion activity (55), and it participates in the binding interaction of IY7640 with the Ca04 CR6261 epitope residues (Fig. 5A), which may be relevant to the anti-influenza activity of IY7640. Given a recent report on the group 1 IAV-specified HA fusion inhibitor candidate JNJ4796 (37), structural optimization may be a strongly recommended step to enhance the efficacy of IY7640. As demonstrated by variations in amino acids in the potential IY7640 binding residues (Fig. 5 and 9), more defined modifications of IY7640 derivatives or further investigation of active metabolites of IY7640 may lead to successful translation of IY7640 efficacy to in vivo and clinical studies. The discrepancy between the in vitro and in vivo efficacies of IY7640, which may be comparable to that of oseltamivir in vitro but is lower than that of oseltamivir in vivo (Table 2 and Fig. 8), also suggests an issue concerning IY7640 bioavailability in mice.
FIG 9.
Structural alignments of the CR6261 epitope (or equivalent) residues from H1, H3, and H5 HAs. (A) Alignment of the CR6261 epitope residues from H1 HA (PDB code 3UBQ [green]) and H5 HA (PDB code 2IBX [purple]). (B) The CR6261 epitope residues of H1 HA (PDB code 3UBQ [green]) and the equivalent H3 HA residues (PDB code 3ZTJ [magenta]). The alignments were performed using PyMOL. (C) Sequence alignment of the HA region comprising the CR6261 epitope or equivalent residues. Residues conserved compared with K09 HA are represented by dots. The HA residue numbers are given with H1 numbering. The residues highlighted with colors are the CR6261 epitope (orange) or equivalent residues. Shaded in green are the CR6261 epitope residues from H1 and H5 HA that interact with IY7640. Shaded in yellow are residues mutated in the K09 IY7640 escape mutants (†L49I and ‡M403T). **, the intervening sequences have been omitted. Abbreviations of the virus HAs are as follows: H1/Ca04, A/California/04/2009 (pH1N1); H5/IS06, A/chicken/IS/2006 (H5N1); H5/VN, A/Viet Nam/1203/2004 (H5N1); and H3/X-31, A/Aichi/2/1968 (H3N2).
As resistance to M2 ion channel blockers is widespread among influenza virus strains, the sole commonly used anti-influenza compound is oseltamivir. However, single-drug therapy runs the risk of rapidly selecting for mutations conferring resistance. Combination therapy, where two or more compounds with different targets are given at the same time, greatly decreases the chance of mutants arising (41, 42, 56, 57), as the virus must acquire multiple mutations at once. However, such a strategy requires the availability of multiple compounds. In this regard, IY7640 can be used in monotherapy as well as in a combination therapy, which enhances our ability to combat drug resistance problems of influenza viruses. Although IY7640 induced the generation of escape mutant viruses within a similar time frame as amantadine or oseltamivir (41, 42), a combination strategy for the anti-influenza compounds could increase the useful life span of the compound. Furthermore, because the IY7640 binding stalk region of HA is highly conserved, structurally degenerative mutations leading to IY7640 insensitivity may be self-limited, which is demonstrated by the mutation of residues of the structurally stable escape mutant HAs merely resulting in conversion to the counterpart residues in another subtype of HAs (the escape mutations L49I and M403T were merely a conversion to an H5 and H3 HA signature at those positions, respectively, as shown in Fig. 9C). Even though IY7640 appeared to be less effective against avian influenza viruses, the use of combination of IY7640 with other anti-influenza agents, such as NA inhibitors, will enhance the general preparedness against a potential influenza pandemic.
MATERIALS AND METHODS
Ethics statement.
To minimize animal suffering, all animal procedures performed in this study were conducted in accordance with the recommendations in the guidelines for the care and use of laboratory animals of the Animal and Plant Quarantine Agency of Korea (59). The protocols were approved by the Institutional Animal Care and Use Committee of the Korea University (approval no. KUIACUC-2014-225), Il-Yang Pharmaceutical Co. (approval no.: IYA201316), and Biotoxtech Co. (approval no. 130325).
Compounds.
Condensation of 2-(trifluoromethyl)benzaldehyde with hydroxylamine hydrochloride produced oxime at a 90% yield. Oxime was reacted with N-chlorosuccinimide (NCS) in N,N-dimethylformamide (DMF), followed by the addition of alkyl acetoacetate to generate isoxazole at an 80% yield, which was hydrolyzed to acid in a quantitative yield. Finally, the acid was reacted with 1-(2-methoxyphenyl)piperazine and 1-ethyl-3-(3′-dimethylaminopropyl)carbodiimide hydrochloride under basic conditions to produce IY7640 at an 80% yield. The purity was measured by analytical high-performance liquid chromatography (HPLC), and the spectra were recorded at 254 nm. The purity of IY7640 was greater than 99%.
Pharmacokinetic study of IY7640 in mice.
Five 7-week-old BALB/c mice (Orient Bio) were used for this study. Forty-five, 90, or 180 mg/kg of IY7640 was administered in 10% Tween 80 and 0.5% carboxymethyl cellulose (CMC) as a single dose via oral gavage, and blood samples were collected at 0, 1, 2, 3, 4, 6, 9, 12, and 24 h postadministration. The AUC, the Cmax, the time to peak concentration (Tmax), and the half-life (t1/2) were calculated using BA Calc 2007. The parameters were determined for each individual animal, and the sample population averages were calculated.
Cells and viruses.
MDCK and Vero cells were cultured in Eagle’s minimum essential medium (EMEM; Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Fisher Scientific, Waltham, MA), 100 U/ml of penicillin, and 100 μg/ml streptomycin (Gibco, Thermo Fisher Scientific) at 37°C in a 5% CO2 incubator. A/Solomon Island/03/2006 (H1N1), A/Brisbane/59/2007 (H1N1) (a seasonal H1N1 virus isolated in 2008 and retaining Y275 in the NA), A/Korea/01/2009 (pH1N1), A/Korea/2785/2009 (a pH1N1 isolate that retained Y275 in the NA), A/Wisconsin/67/2005 (H3N2), A/Brisbane/10/2007 (H3N2), A/Perth/16/2009 (H3N2), A/chicken/IS/06/2006 (H5N1 6:2 vaccine virus), B/Brisbane/60/2008 (Victoria lineage), and B/Wisconsin/01/2010 (Yamagata lineage) were obtained from the Korea Centers for Disease Control and Prevention (Osong, Republic of Korea). A/Puerto Rico/8/1934 virus expressing GFP (rPR8/GFP) (38) and A/Netherlands/602/2009 (pH1N1) were provided by Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai, New York, NY). X-31 (H3N2 6:2 vaccine virus) was provided by Peter Palese (Icahn School of Medicine at Mount Sinai). A/chicken/Korea/01310/2001 (H9N2) was provided by Young Ki Choi at Chungbuk National University (Cheongju, Republic of Korea). A/Shandong/09/1993 (H3N2) was provided by Chang-Seon Song at Konkuk University (Seoul, Republic of Korea). All viruses were propagated in embryonated chicken eggs.
Plasmids.
Eight genes of the K09 virus were cloned into a bidirectional pDZ vector (58) and used to generate rK09 and its mutant viruses by reverse genetics. The pDZ plasmids of PR8 gene segments were also used to generate 7:1 and 6:2 viruses. The HA genes of the above-named viruses were cloned into the pDZ plasmid, except for A/Anhui/01/2013 (H7N9, commercially synthesized). All plasmid sequences were confirmed before use in the experiments.
Screening system using an influenza virus expressing GFP.
The screening system using a GFP-expressing virus was previously described (38, 39). Briefly, MDCK cells in 96-well plates were infected with rPR8/GFP. After adsorption for 1 h, the unbound virus particles were removed and medium with 2-fold serially diluted chemicals was added to the cells. The cells were further incubated for 24 h, and the GFP signals were observed under a fluorescence microscope.
Plaque assay.
The plaque assay was performed to determine infectious viral titers. Briefly, MDCK cells in a monolayer were inoculated with 10-fold serially diluted viral allantoic fluids or cultured cell supernatants for 1 h. After adsorption for 1 h, the unbound virus particles were removed, and the cells were overlaid with a medium containing 0.2% agar and 1 μg/ml of TPCK-trypsin. After incubation for 72 h, the cells were stained with 1% crystal violet, and viral titers were determined by counting the plaques.
Plaque reduction assay.
MDCK cells were cultured in a 6-well cell culture plate and infected with approximately 102 PFU/ml of influenza virus. After 1 h, the unbound virus particles were washed and the medium was replaced by an overlay medium (containing N-tosyl-l-phenylalanine chloromethyl ketone [TPCK]-trypsin at 1 μg/ml, 2% agarose, and IY7640 at different concentrations). After incubation for 72 h, the cultures were stained with 1% crystal violet solution to detect the plaques. The EC50 was calculated using GraphPad Prism 5.0d (GraphPad software, Inc., La Jolla, CA).
Toxicity evaluation.
The cytotoxicity of IY7640 was evaluated using the CCK-8 reagent. MDCK cells were cultured in 96-well plates for 24 h. Subsequently, the culture medium was replaced with medium containing 2-fold serially diluted IY7640, and the cells were incubated for an additional 24 h. Next, the medium was replaced with medium containing 10 μl/ml of CCK-8 solution (Dojindo Molecular Technologies, Inc., Rockville, MD). Thereafter, the mixture was incubated at 37°C for 4 h, and the absorbance of the supernatant at 495 nm was measured by an enzyme-linked immunosorbent assay (ELISA) reader (Spectra Max 250; Molecular Devices, San Jose, CA). The CC50 value (concentration of the drug that leads to 50% cytotoxicity) was calculated as the concentration that reduced the number of surviving cells to 50% of the untreated controls.
The cumulative toxicity of IY7640 was evaluated in Sprague-Dawley rats. The 5-week-old rats (five male and five female rats per group) were administered 500, 1,000, and 2,000 mg/kg of IY7640. The dosing volume was 5 ml/kg of body weight. Individual doses were calculated for each animal based on the most recently recorded body weights. The rats were dosed once daily for two consecutive weeks via gastric intubation with 3-ml disposable syringes fitted with intubation tubes. Control animals received a vehicle only, corn oil, at 5 ml/kg. Body weight changes and clinical symptoms of the rats were recorded during the observation period.
Viral CPE assay.
MDCK cells were seeded into 96-well plates at 30,000 cells/well. The next day, the cells were inoculated with 300 PFU of influenza virus. After the viral adsorption using a standard infection procedure, 100 μl of medium with 2-fold serially diluted IY7640 (starting from 400 or 100 μM) and 1 μg/ml of TPCK-trypsin were added to each well. The infection culture was incubated at 37°C for 72 h. Next, the medium was replaced with fresh medium without IY7640, followed by the addition of 10 μl of a CCK-8 solution (Dojindo Molecular Technologies, Inc.) to the medium and incubation for 4 h at 37°C. The status of the viable cells in the infection culture was determined by measuring the absorbance at 450 and 650 nm, which was detected using an ELISA reader. The EC50 value was calculated using GraphPad Prism 5.0d (GraphPad software, Inc.).
Generation of escape mutant viruses.
The confluent MDCK cell monolayers in 6-well tissue culture plates were inoculated with 103 PFU of K09 virus, followed by a routine tissue culture infection procedure. The first 48 h of incubation with the infected culture included 0.5 μM IY7640 (below the EC50, as determined using the plaque reduction assay) and was considered passage 0. The viral titer was measured using a plaque assay after each passage, and 103 PFU of virus from the previous passage was used to inoculate cells for the next passage in medium with a 2-fold-higher IY7640 concentration. The virus yielded after each passage was also subjected to the plaque reduction assay in the presence of the chemical. When plaques appeared in the wells treated with 100 μM IY7640, 3 plaques were isolated and amplified in the MDCK cells without IY7640 addition. The amplified viruses were again tested for IY7640 resistance using the plaque reduction assay. The escape mutant virus generation procedure was performed twice, and the genome of the escape mutant virus was cloned into the pDZ vector using a standard molecular biology protocol and sequenced. A recombinant virus composed of 7 WT K09 segments and an escape mutation-containing segment (7:1 reassortant virus; K09 virus genome sequence gi:237651253/5/7, 22978366/68/70/73, 229892681) was generated using the 8-plasmid reverse genetics system and the 293T and MDCK coculture transfection method (5). The IY7640 resistance of the 7:1 reassortant virus was confirmed using the plaque reduction assay.
Time-to-addition assay.
Confluent MDCK cell monolayers in 6-well tissue culture plates were inoculated with 103 (for plaque assays) or 106 PFU (for Western blotting) of the rK09 virus. After 1 h of adsorption and the removal of unbound viruses, EMEM containing 0.3% bovine serum albumin (BSA) was added (0 h postinfection). IY7640 was added to the medium at 5 μM (for the plaque assay) or 40 μM (for Western blotting), at 0, 2, 4, or 8 h postinfection (hpi). The cells and supernatants (for the plaque assay) or cells only (for Western blotting) were collected 10 hpi. The plaque assay was performed following a standard procedure after subjecting the supernatant and cells to three freeze-thaw cycles. However, because the inoculated virus was in the IY7640-containing supernatant, washing after inoculation did not fully remove the IY7640; thus, the overall number of plaques for these samples was lower than the number for the untreated samples. To complement the plaque assay results, a Western blot analysis was performed in which newly expressed HA in the infected cells was detected by a primary antibody, a polyclonal sheep anti-pH1N1 HA antibody (anti-A/California/7/2009 H1N1 HA serum, NIBSC code 11/110; NIBSC, Hertfordshire, United Kingdom) and a secondary antibody, a horseradish peroxidase (HRP)-conjugated rabbit anti-sheep IgG (KPL, SeraCare Life Sciences, Milford, MA).
Hemagglutination inhibition assay.
In 96-well plates, 8 HA units of rK09 virus in a 25-μl volume was incubated with the same volume of guinea pig antisera raised against rK09 (positive control) or designated concentrations of IY7640 at 37°C. After 1 h of incubation, the mixtures were agglutinated with 50 μl of 0.5% (vol/vol) turkey red blood cells (tRBCs) for 30 min.
In silico docking studies.
The IY7640 molecular structure was converted to PDB format using PyMOL (The PyMOL Molecular Graphics System, v1.3, Schrödinger, LLC, New York, NY). A molecular docking experiment was performed using the SYBYL-X (v2.1.1) Surflex Dock and AutoDockVina software (46) in accordance with the author’s online tutorial (https://vina.scripps.edu/tutorial.html). To verify the validity of the docking method, TBHQ docking was performed on the HA of A/Aichi/2/1968 (H3N2) (PDB ID 3ZTJ), which docked at the exact position shown in the crystal structure of TBHQ and a group 2 HA (47) with a calculated affinity of −6.7 kcal/mol. The PDB accession codes for the IY7640 docking counterparts are given in the relevant figure legends. Interaction distances and graphics were prepared using SYBYL-X (v2.1.1) Surflex Dock and PyMOL.
Hemolysis assay.
IY7640 in dimethyl sulfoxide (DMSO) or DMSO alone was dissolved to a 5× final concentration in 100 μl, and the viruses (27 hemagglutination units [HAU]) were prepared in 100-μl volumes. The mixtures with IY7640 and the viruses were incubated for 1 h at 37°C. Thereafter, 2% (vol/vol) of tRBCs preincubated at 37°C in a 200-μl volume were added to the IY7640-virus mixture. The IY7640-virus-RBC mixture was incubated for 20 min at 37°C. Next, sodium acetate (0.5 M) at various pH values in a 100-μl volume was added to the IY7640-virus-RBC mixture. The pH values for this final mixture were predetermined, and after gentle resuspension, the mixture was incubated for 30 min at 37°C. Thereafter, the reaction was centrifuged for 6 min at approximately 1,000 × g and 4°C. Three hundred microliters of the supernatant was added to a flat-bottom 96-well plate, and the hemoglobin concentration in the supernatant was determined by measuring the absorbance at 540 nm.
Cell fusion assay.
Vero cells were grown for 24 h to approximately 70% confluency in 12-well plates. The cells were transfected with pCAGGSII-GFP and pDZ-WT K09 HA or pDZ-mutant K09 HA using transIT2020 (Mirus Bio, LLC, Madison, WI). After 48 h, upon confirmation of GFP expression, the cells were washed with phosphate-buffered saline (PBS) containing Mg2+ and Ca2+ and treated with 5 μg/ml of TPCK-trypsin for 10 min. The cells were then treated with a culture medium (Dulbecco modified Eagle medium [DMEM]) containing 10% FBS to inactivate the TPCK-trypsin and either DMSO or IY7640 for 4 h. Thereafter, the cells were treated for 3 min with acidic PBS adjusted to various pH values using citric acid. After the acidic PBS was removed, culture medium (DMEM) supplemented with 10% FBS and DMSO or IY7640 was added to the cells. For consistency, the cells were incubated for 24 h before an image was taken under a fluorescence microscope to allow GFP to diffuse through the pores generated by membrane fusion.
Protease susceptibility assay.
MDCK cell monolayers in 6-well plates were infected with 106 PFU of rK09 or escape mutant viruses, with the cells collected at 16 hpi. After centrifugation, each cell pellet was incubated with 100 μl of immunoprecipitation lysis buffer without protease inhibitor (Thermo Fisher Scientific) at 4°C for 10 min. After microcentrifugation at 14,000 rpm and 4°C for 10 min, the supernatant was transferred to a new tube and 30 μg/ml of TPCK-trypsin was added, followed by incubation for 30 min at 37°C. Next, IY7640 was added to the supernatant to yield the designated concentration, followed by incubation for 4 h at 37°C. The supernatant was then acidified by adding 30 μl of sodium acetate (the pH was regulated using a predetermined amount of HCl). After incubation for 30 min at 37°C, the reaction was neutralized by adding 20 μl of 0.5 M Tris and incubation for 30 min at room temperature. The reaction was stopped by adding 6× SDS loading dye (under reducing conditions). The reactions were loaded on a 10% SDS-PAGE gel followed by a standard Western blotting procedure. HA was detected using a primary polyclonal sheep anti-pH1N1 HA antibody (anti-A/California/7/2009 H1N1 HA serum, NIBSC code 11/110; NIBSC, Hertfordshire, United Kingdom) and an HRP-conjugated rabbit anti-sheep IgG secondary antibody (KPL). For the H5 HA protease susceptibility inhibition by IY7640, MDCK cells were infected with a 7:1 reassortant virus (rVN, H5N1). Mouse serum raised against the rVN virus was used for H5 HA detection in the Western blot.
Animal experiment.
For each experiment, 10 7- to 8-week-old female BALB/c mice (Nara Biotech, Seoul, Republic of Korea) were infected intranasally with 4 × 50% of the mouse lethal dose (MLD50) of influenza virus rK09m (an engineered recombinant K09 virus that encodes an NA containing N58S and N59S mutations to render the virus lethal to mice; MLD50 analysis data are available upon request), and IY7640 was administered orally at 45, 90, or 180 mg/kg/day twice daily for 8 days using a Sonde feeding apparatus beginning 1 h before infection. The designated amount of IY7640 or oseltamivir phosphate (Toronto Research Chemicals, Inc., North York, ON, Canada) was formulated with Tween 80 (Daejung Chemicals and Metals, Siheung, Republic of Korea) and carboxymethyl cellulose sodium salt (CMC; Sigma-Aldrich, St. Louis, MO). The control mice were orally administered the formulation without IY7640 or oseltamivir. The survival rates and body weight changes of the infected mice were monitored daily for 14 days. When the body weight of a test animal decreased to 75% of its initial body weight, the animal was anesthetized and humanely euthanized.
Statistical analysis.
The significance of the differences in efficacy between IY7640 and oseltamivir carboxylate and for the hemolysis assay was assessed using a Student’s t test. In figures, significant differences are indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Error bars in figures indicate standard deviations (SDs).
ACKNOWLEDGMENTS
This study was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea (grant no. NRF-2017R1A2B2003773).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no conflicts of interest to declare.
M.-S.P. designed the study; J.I.K., S.L., G.Y.L., S.P., J.-Y.B., J.H., H.-Y.K., S.-H.W., H.U.L., C.A.A., H.J.B., H.S.J., K.O., Y.B., and M.S.P. performed the experiments; J.I.K., S.L., G.Y.L., S.P., D.-J.C., J.S.S., D.-Y.K., and M.-S.P. analyzed the data; and J.I.K., S.L., G.Y.L., M.S.P., and M.-S.P. wrote and revised the manuscript.
REFERENCES
- 1.Palese P, Shaw M. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Johnson NP, Mueller J. 2002. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull Hist Med 76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 3.Palese P. 2004. Influenza: old and new threats. Nat Med 10:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 4.Neumann G, Kawaoka Y. 2011. The first influenza pandemic of the new millennium. Influenza Other Respir Viruses 5:157–166. doi: 10.1111/j.1750-2659.2011.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JI, Lee I, Park S, Hwang M-W, Bae J-Y, Lee S, Heo J, Park MS, García-Sastre A, Park M-S. 2013. Genetic requirement for hemagglutinin glycosylation and its implications for influenza A H1N1 virus evolution. J Virol 87:7539–7549. doi: 10.1128/JVI.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright PF, Neumann G, Kawaoka Y. 2013. Orthomyxoviruses, p 1186–1243. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Krammer F, Palese P. 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 9.Krammer F, Palese P, Steel J. 2015. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol 386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 10.Coughlan L, Palese P. 2018. Overcoming barriers in the path to a universal influenza virus vaccine. Cell Host Microbe 24:18–24. doi: 10.1016/j.chom.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Elbahesh H, Saletti G, Gerlach T, Rimmelzwaan GF. 2019. Broadly protective influenza vaccines: design and production platforms. Curr Opin Virol 34:1–9. doi: 10.1016/j.coviro.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9:e02332-17. doi: 10.1128/mBio.02332-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichelberger MC, Morens DM, Taubenberger JK. 2018. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 53:38–44. doi: 10.1016/j.coi.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angeletti D, Yewdell JW. 2018. Understanding and manipulating viral immunity: antibody immunodominance enters center stage. Trends Immunol 39:549–561. doi: 10.1016/j.it.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Thulin NK, Wang TT. 2018. The role of Fc gamma receptors in broad protection against influenza viruses. Vaccines (Basel) 6:E36. doi: 10.3390/vaccines6030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewnard JA, Cobey S. 2018. Immune history and influenza vaccine effectiveness. Vaccines (Basel) 6:28. doi: 10.3390/vaccines6020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachbagauer R, Krammer F. 2017. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect 23:222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill DS, Damle NK. 2006. Biopharmaceutical drug discovery using novel protein scaffolds. Curr Opin Biotechnol 17:653–658. doi: 10.1016/j.copbio.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Wu X, Sun Q, Zhang C, Yang S, Li L, Jia Z. 2017. Progress of small molecular inhibitors in the development of anti-influenza virus agents. Theranostics 7:826–845. doi: 10.7150/thno.17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naesens L, Stevaert A, Vanderlinden E. 2016. Antiviral therapies on the horizon for influenza. Curr Opin Pharmacol 30:106–115. doi: 10.1016/j.coph.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhuri S, Symons JA, Deval J. 2018. Innovation and trends in the development and approval of antiviral medicines: 1987-2017 and beyond. Antiviral Res 155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muthuri SG, Venkatesan S, Myles PR, Leonardi-Bee J, Al Khuwaitir TSA, Al Mamun A, Anovadiya AP, Azziz-Baumgartner E, Báez C, Bassetti M, Beovic B, Bertisch B, Bonmarin I, Booy R, Borja-Aburto VH, Burgmann H, Cao B, Carratala J, Denholm JT, Dominguez SR, Duarte PAD, Dubnov-Raz G, Echavarria M, Fanella S, Gao Z, Gérardin P, Giannella M, Gubbels S, Herberg J, Iglesias ALH, Hoger PH, Hu X, Islam QT, Jiménez MF, Kandeel A, Keijzers G, Khalili H, Knight M, Kudo K, Kusznierz G, Kuzman I, Kwan AMC, Amine IL, Langenegger E, Lankarani KB, Leo Y-S, Linko R, Liu P, Madanat F, Mayo-Montero E, McGeer A, Memish Z, Metan G, Mickiene A, Mikić D, Mohn KGI, Moradi A, Nymadawa P, Oliva ME, Ozkan M, Parekh D, Paul M, Polack FP, Rath BA, Rodríguez AH, Sarrouf EB, Seale AC, Sertogullarindan B, Siqueira MM, Skręt-Magierło J, Stephan F, Talarek E, Tang JW, To KKW, Torres A, Törün SH, Tran D, Uyeki TM, Van Zwol A, Vaudry W, Vidmar T, Yokota RTC, Zarogoulidis P, Nguyen-Van-Tam JS. 2014. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir Med 2:395–404. doi: 10.1016/S2213-2600(14)70041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayden FG, de Jong MD. 2011. Emerging influenza antiviral resistance threats. J Infect Dis 203:6–10. doi: 10.1093/infdis/jiq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvier NM, Rahmat S, Pica N. 2012. Enhanced mammalian transmissibility of seasonal influenza A/H1N1 viruses encoding an oseltamivir-resistant neuraminidase. J Virol 86:7268–7279. doi: 10.1128/JVI.07242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden FG, Sugaya N, Hirotsu N, Lee N, de Jong MD, Hurt AC, Ishida T, Sekino H, Yamada K, Portsmouth S, Kawaguchi K, Shishido T, Arai M, Tsuchiya K, Uehara T, Watanabe A, Baloxavir Marboxil Investigators Group. 2018. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med 379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 29.Colombo RE, Fiorentino C, Dodd LE, Hunsberger S, Haney C, Barrett K, Nabha L, Davey RT Jr, Olivier KN. 2016. A phase 1 randomized, double-blind, placebo-controlled, crossover trial of DAS181 (Fludase(R)) in adult subjects with well-controlled asthma. BMC Infect Dis 16:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe A, Chang SC, Kim MJ, Chu DW, Ohashi Y, Marvel Study Group. 2010. Long-acting neuraminidase inhibitor laninamivir octanoate versus oseltamivir for treatment of influenza: a double-blind, randomized, noninferiority clinical trial. Clin Infect Dis 51:1167–1175. doi: 10.1086/656802. [DOI] [PubMed] [Google Scholar]
- 31.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, Vorlaender MK, Bianchi S, Guarino B, De Marco A, Vanzetta F, Agatic G, Foglierini M, Pinna D, Fernandez-Rodriguez B, Fruehwirth A, Silacci C, Ogrodowicz RW, Martin SR, Sallusto F, Suzich JA, Lanzavecchia A, Zhu Q, Gamblin SJ, Skehel JJ. 2016. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleu S, Kakuda TN, Spittaels K, Vercauteren JJ, Hillewaert V, Lwin A, Leopold L, Hoetelmans R. 2018. Single- and multiple-dose pharmacokinetics and safety of pimodivir, a novel, non-nucleoside polymerase basic protein 2 subunit inhibitor of the influenza A virus polymerase complex, and interaction with oseltamivir: a phase 1 open-label study in healthy volunteers. Br J Clin Pharmacol 84:2663–2672. doi: 10.1111/bcp.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beigel JH, Voell J, Munoz P, Kumar P, Brooks KM, Zhang J, Iversen P, Heald A, Wong M, Davey RT. 2018. Safety, tolerability, and pharmacokinetics of radavirsen (AVI-7100), an antisense oligonucleotide targeting influenza A M1/M2 translation. Br J Clin Pharmacol 84:25–34. doi: 10.1111/bcp.13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tharakaraman K, Subramanian V, Viswanathan K, Sloan S, Yen HL, Barnard DL, Leung YH, Szretter KJ, Koch TJ, Delaney JC, Babcock GJ, Wogan GN, Sasisekharan R, Shriver Z. 2015. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc Natl Acad Sci U S A 112:10890–10895. doi: 10.1073/pnas.1502374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng LY, Yang J, Liu S. 2017. Investigational hemagglutinin-targeted influenza virus inhibitors. Expert Opin Investig Drugs 26:63–73. doi: 10.1080/13543784.2017.1269170. [DOI] [PubMed] [Google Scholar]
- 36.Kadam RU, Wilson IA. 2017. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci U S A 114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dongen MJP, Kadam RU, Juraszek J, Lawson E, Brandenburg B, Schmitz F, Schepens WBG, Stoops B, van Diepen HA, Jongeneelen M, Tang C, Vermond J, van Eijgen-Obregoso Real A, Blokland S, Garg D, Yu W, Goutier W, Lanckacker E, Klap JM, Peeters DCG, Wu J, Buyck C, Jonckers THM, Roymans D, Roevens P, Vogels R, Koudstaal W, Friesen RHE, Raboisson P, Dhanak D, Goudsmit J, Wilson IA. 2019. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 363:eaar6221. doi: 10.1126/science.aar6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JI, Park S, Lee I, Lee S, Shin S, Won Y, Hwang MW, Bae JY, Heo J, Hyun HE, Jun H, Lim SS, Park MS. 2012. GFP-expressing influenza A virus for evaluation of the efficacy of antiviral agents. J Microbiol 50:359–362. doi: 10.1007/s12275-012-2163-9. [DOI] [PubMed] [Google Scholar]
- 40.Kim JI, Lee I, Park S, Park MS. 2012. Surface glycoproteins determine the feature of the 2009 pandemic H1N1 virus. BMB Rep 45:653–658. doi: 10.5483/BMBRep.2012.45.11.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Govorkova EA, Webster RG. 2010. Combination chemotherapy for influenza. Viruses 2:1510–1529. doi: 10.3390/v2081510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilyushina NA, Bovin NV, Webster RG, Govorkova EA. 2006. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antiviral Res 70:121–131. doi: 10.1016/j.antiviral.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 43.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 44.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. 2012. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol 86:982–990. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell RJ, Kerry PS, Stevens DJ, Steinhauer DA, Martin SR, Gamblin SJ, Skehel JJ. 2008. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadam RU, Juraszek J, Brandenburg B, Buyck C, Schepens WBG, Kesteleyn B, Stoops B, Vreeken R, Vermond J, Goutier W, Tang C, Vogels R, Friesen RHE, Goudsmit J, van Dongen MJP, Wilson IA. 2017. Potent peptidic fusion inhibitors of influenza virus. Science 358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo G, Torri A, Harte WE, Danetz S, Cianci C, Tiley L, Day S, Mullaney D, Yu KL, Ouellet C, Dextraze P, Meanwell N, Colonno R, Krystal M. 1997. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J Virol 71:4062–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanderlinden E, Goktas F, Cesur Z, Froeyen M, Reed ML, Russell CJ, Cesur N, Naesens L. 2010. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J Virol 84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skehel JJ, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci U S A 79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. 2009. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007–08. Emerg Infect Dis 15:155–162. doi: 10.3201/eid1502.081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonges M, van der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. 2009. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005–2008. Antiviral Res 83:290–297. doi: 10.1016/j.antiviral.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, Shinya K, Sawada T, Kiso M, Usui T, Murata T, Lin Y, Hay A, Haire LF, Stevens DJ, Russell RJ, Gamblin SJ, Skehel JJ, Kawaoka Y. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 55.Cross KJ, Langley WA, Russell RJ, Skehel JJ, Steinhauer DA. 2009. Composition and functions of the influenza fusion peptide. Protein Pept Lett 16:766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- 56.Park S, Kim JI, Park M-S. 2012. Antiviral agents against influenza viruses. J Bacteriol Virol 42:284–293. doi: 10.4167/jbv.2012.42.4.284. [DOI] [Google Scholar]
- 57.Park S, Kim JI, Lee I, Lee S, Hwang MW, Bae JY, Heo J, Kim D, Jang SI, Kim H, Cheong HJ, Song JW, Song KJ, Baek LJ, Park MS. 2014. Combination effects of peramivir and favipiravir against oseltamivir-resistant 2009 pandemic influenza A(H1N1) infection in mice. PLoS One 9:e101325. doi: 10.1371/journal.pone.0101325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol 79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Animal and Plant Quarantine Agency of Korea. The guidelines for the care and use of laboratory animals. http://www.qia.go.kr/animal/protect/ani_prot_ani_testcommission.jsp.