Highlights
-
•
In stored honeys, bacterial composition consists mainly of Bacillus spp.
-
•
MK-7 and MK-8 were predominant menaquinones produced by the residing bacteria.
-
•
MK-7 and MK-8 were predominant menaquinones extracted from honey.
-
•
MKs identification was based on UHPLC–ESI-MS/MS and quadrupole orbitrap.
-
•
Maximal accumulation of soluble MK-7 coincides with a massive cell death of Bacillus spp.
-
•
Bacterial cell death and lysis is a likely mechanism of the MK-7 release to honey.
Keywords: Menaquinones, MK-7, MK-8, UPLC–ESI-MS/MS, Exact mass measurements, Bacillus spp., Honey bacterial composition, B. amyloliquefaciens, B. methylotrophicus, B. cereus, B. thuringiensis, B. pumilus, Vitamin K2
Abstract
Long-chain menaquinones (MK) are of bacterial origin. We investigated the possibility that MKs observed in honey are also the products of bacteria present in honey. The bacterial composition of honey was analyzed using culture-dependent methods. 16S rRNA gene sequencing and MALDI-TOF showed prevalence of the members of Bacillus subtilis and Bacillus cereus groups. The dominant menaquinones in both bacteria and honey were menaquinones MK-7 and MK-8 as indicated by UHPLC–ESI-MS/MS coupled to quadrupole orbitrap. The EICs showed alignment of mass ions of MK-7 and MK-8 from culture supernatants with that of honey. The unique MS/MS fragmentation pattern indicated that fragment ions were arising from the same menaquinone present in both samples. During Bacillus growth, the accumulation of MK-7 in supernatants occurred in a stationary phase and coincided with cell death. These novel findings suggest that the soluble MKs in honey originate from shedding of cell membranes of dead vegetative cells.
1. Introduction
Menaquinones (MK), homologues of vitamin K2, are low-molecular-weight compounds bound to the inner cytoplasmic membranes of bacteria (Collins & Jones, 1981). They share a common structure consisting of hydrophilic head group of 2-methyl-1, 4-naphtoquinone and hydrophobic isoprenyl side-chain attached to the C3 of naphtoquinone. The amphiphilic character of menaquinones allows binding of hydrophobic, polyisoprene side-chains to fatty acyl groups of phospholipids, while hydrophilic groups of MK, protruding from the membrane, are involved in oxidation-reduction reactions typical for quinones. The redox process involves passing one-or two electrons from the electron donors (NADH dehydrogenases) via menaquinones and via several lipoprotein carriers (such as cytochrome bc1 complex) to the terminal electron acceptor, oxygen. The energy formed through the electron transport is used to move the H+ or protons out of the cell, thereby generating an electrochemical gradient known as proton motive force (PMF). The energy of the PMF provides energy to the F0F1ATPase complex to convert ADP into ATP (Kurosu & Begari, 2010).
The menaquinone-based system of energy production operates in all Gram-positive bacteria and anaerobic Gram-negative bacteria (Nowicka and Kruk, 2010, Sherman et al., 1989). In Gram-positive bacteria, gene encoding enzymes involved in menaquinones synthesis belong to a class of essential genes (Kobayashi et al., 2003). The interruption of the electron transport chain by blocking the synthesis of menaquinones or by blocking their functions is lethal for bacteria (Bald et al., 2017, Kurosu and Begari, 2010).
Menaquinones are synthesized by eukaryotes and prokaryotes, but the long chain menaquinones with side-chains consisting of more than six isoprene units are typically produced by bacteria (Collins & Jones, 1981). Gram-positive, facultative anaerobic lactic acid bacteria and Bacillus spp. are major dietary sources of long-chain menaquinones (MK-7 to MK-10) in fermented foods. Lactobacillus spp., such as Lactobacillus subtilis, Lactococcus lactis ssp. cremoris and lactis, are used to produce fermented milk, cheese and yogurt (Booth, 2012, Walther and Chollet, 2017). High levels of menaquinone MK-7 are found in natto, fermented soybean that is produced by Bacillus subtilis (B. subtilis subsp. subtilis BEST195) (Berenjian et al., 2011, Ikeda and Doi, 1990, Kubo et al., 2011, Sato et al., 2001a, Sato et al., 2001b).
The newly reported presence of long-chain menaquinones in honey suggests that they also might be of bacterial origin (Kim & Brudzynski, 2018). Raw honeys may initially contain a number of environmental microorganisms originating from soil, pollen and nectar of flowers that are introduced by bees during honey harvesting (Iurlina and Fritz, 2005, Snowdon and Cliver, 1996). However, their growth in raw honeys is strongly inhibited, primarily by the high sugar concentration, low water activity and antibacterial action of intrinsic honey factors (hydrogen peroxide or non-peroxide type compounds). Vegetative and non-sporulating bacteria cannot survive in honey longer than 30 days (Olaitan, Adeleke, & Ola, 2007). In contrast, the spores of spore-forming Bacillus spp. are well adapted to adverse conditions and can survive in honey over long periods of time (Olaitan et al., 2007, Snowdon and Cliver, 1996). When conditions become more favourable (e.g. increased water content, decreased osmolarity), the dormant spores can return to active growth by germination.
To develop a better understanding of the origin of long-chain menaquinones in honey we aimed to investigate a possible link between the production of specific types of menaquinones by bacteria present in honey and honey’s menaquinone pool.
2. Materials and methods
2.1. Honey
Five raw buckwheat honeys were directly obtained from the Ontario apiaries during the 2015–2017 harvest seasons and were stored at −20 °C. The floral origin was specified by a beekeeper based on the location of the beehives. The honeys were assigned a number after arriving to the laboratory as buckwheat honey H177, H220, H229, H230 and H234.
2.2. Primary honey plates and total bacterial counts
Honey (1.35 g) was added to 1 ml of filter-sterilized water to prepare 50% w/v solution. The completely fluid solution was filtered through a glass fiber disposable microfilter unit (S&S CENTREX, Schleicher & Schuell, Keene, New Hampshire, NE, USA) to remove solid particles. Aliquots (100 μl) of honey solution were inoculated in triplicate, onto Mueller-Hinton Agar (MHA) plates (primary plates). Plates were incubated at 37 °C. The number of colonies forming units (CFU) was counted after 48 h.
2.3. Isolation and preparation of pure cultures
Colonies of distinct morphological features were selected from a mixed culture of the honey primary plates. They were picked with a wire loop and transferred to the separate trypticase soy broth (TSB) tubes and incubated with shaking for 24 h at 37 °C. The small amount of inoculum was streaked out onto MHA plates using the streak plate method to obtain pure cultures. Plates were incubated overnight at 37 °C. An isolated, discrete colony from the pure culture plate was subcultured again by re-streaking into a new MHA plate. The single, isolated colony from the subcultured plate was incubated in TSB overnight at 37 °C with shaking. The portion of the pure culture was used for bacterial identification while the rest was stocked as glycerol stock and stored at – 80 °C until required for use.
2.4. Colony imaging
Morphological differences of bacterial colonies on agar plates, such as size, shape and color, were visually observed. Images of the colonies were taken using a Leica L2 stereoscope (Leica Microsystems Ltd Business Unit SM CH-9435 Heerbrugg (Switzerland).
2.5. Bacterial identification by 16S rRNA sequencing
Bacterial cells from pure liquid cultures were centrifuged at 10,000×g for 5 min. A cell pellet of each pure culture, #1A, 1B, 2, 3, 4, and 5 was used for DNA isolation, PCR amplification and 16S rRNA gene sequencing. Primers design, detection and documentation were conducted by Laragene Sequencing and Genotyping (Culver City, CA, USA). Bacteria were identified to genus and species levels.
2.6. Bacterial identification by MALDI-TOF
Bacteria from pure cultures #2 and #4 were identified by Matrix Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF). The protein mass spectra were acquired with the use of an automated mass spectrometer Bruker MALDI-TOF BioTyper (Bruker Daltonics, Germany) within the range of m/z 3 000 to 15 000. The Bruker MALDI-TOF BioTyper target plate was inoculated with bacterial test standard (Bruker #8255343) and calibration was performed according to the recommendations of the manufacturer. Using the direct colony transfer method, the tested bacteria were applied to the target spot, allowed to dry and covered with 1 µl of 70% formic acid, followed by 1 µl of matrix (Bruker, HCCA #8255344). The bacterial identification was obtained using a Microflex LT instrument (Bruker Daltonics, Germany) with Flex Control (version 3.0) software (Bruker Daltonics, Germany). The mass spectra were matched with the spectral database carried out by automated software, the MALDI BioTyper automation (version 2.0) software (Bruker Daltonics, Germany). The identification was conducted by Clinical Microbiology, London Health Science Centre, London, Ontario.
2.7. Menaquinone extraction
Menaquinone extraction was carried out according to the method described by Sato et al., 2001a, Sato et al., 2001b, using n-hexane:2-propanol (2:1, v/v) as the extracting solution. Honey solution (50% w/v) or bacterial culture supernatants were mixed with extracting solution (1:4, v/v) by vigorous vortexing for 10 min and centrifuged at 3000 rpm for 3 min, (IEC Centra-8R centrifuge). The resultant organic solvent layer was transferred to microtubes and centrifuged at 10,000×g for 5 min. The organic phase was filter-sterilized through a 0.45 nm filter, transferred to clean tubes and evaporated to dryness at 55 °C. The residue was dissolved in 100 µl HPLC-grade methanol.
2.8. Solid-phase extraction (SPE)
The filtrate was purified using an Oasis® HBL 3 cc extraction cartridges following the manufacture’s recommendations (Waters Corp. Milford, MA, USA). The extracted material was eluted with 1 ml methanol. Five µl of the eluted fraction was subjected to ultra- high-performance liquid chromatography (UHPLC).
2.9. Analysis of MK by UHPLC
Menaquinones were analyzed using UHPLC-ESI-MS operated on the positive ion mode. The sample was resolved on an Acquity BEH C18 column (150 mm × 2.1 mm, 1.8 μm) supplied by Waters, Milford, MA, USA. Flowrate was 0.5 ml/min at 30 °C. The mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Chromatographic separation was 6-min. The gradient was 0 min, 8% B; 1 min, 8% B; 4.3 min, 25% B; 5 min, 8% B; 6 min, 8% B. The detection was by means of diode-array which was set up at 248 nm. The method of ionization was electrospray ionization (ESI). The cone voltage was set to 30 V, the source temperature at 147 °C; the desolvation gas flow was 483 L/h at a temperature of 300 °C.
The data acquisition and processing was obtained using Thermo Xcalibur 2.2 software (Thermo Fisher Scientific, U.S.A).
2.10. The exact mass measurements using Q-Exactive mass spectrometer
The LC–MS platform consisted of a Dionex Ultimate 3000 UHPLC system and a Q-Exactive mass spectrometer equipped with a HESI II source (Thermo Scientific). Control of the system and data handling was performed using Thermo XCalibur 2.2 software and Chromeleon 7.2 software. Separation by liquid chromatography was conducted on an Acquity BEH C18 column (50 mm × 2.1 mm, 1.7 μm particle size). The pump was run at a flow rate of 200 μl/min. Solvent A was water containing 0.1% formic acid; solvent B was acetonitrile containing 0.1% formic acid. The gradient was 0 min, 8% B; 6 min, 95% B; 8 min, 95% B; 9 min, 8% B; 15 min, 8% B. Autosampler temperature was maintained at 10 °C and injection volume was 15 μl. Data collection was done in positive ionization mode with MS1 scan range m/z 100–1000, resolution 70 000, GC target of 3e6 and a maximum injection time of 200 ms, MS2 data was collected using a TOP5 method, 1 m/z isolation window, 20, 30 stepped NCE, 17,500 resolution, AGC target 1e5 and a maximum injection time of 50 ms.
2.11. Statistical analysis
Analyses were performed using GraphPad statistical package (GraphPad Software Inc. La Jolla, CA, USA). Results are expressed as means ± SEM. For correlation, non-parametric Spearman r coefficient was used. Critical p-value for significance was p < 0.05.
3. Results
3.1. Morphology of bacterial colonies in honey
The bacterial composition and diversity in five buckwheat honey samples (H177, H220, H229, H230 and H234) was examined using conventional culturing methods. These methods were preferred over culture-independent methods for two reasons: (a) the levels of bacteria in honey were very low, ranging from 2 × 101 (H230), 2.6 × 102 (H234), 3 × 102 (H177 and H229) to 6 × 102 (H220) CFU/g of honey as indicated by the standard plate count, and (b) bacterial cultures allowed assessing the levels of culturable bacteria, including spore-formers as relevant contributors to honey menaquinones.
The microbial composition, the presence or absence and relative abundance of bacterial colonies in different honey samples, was investigated in primary MHA plates, followed by preparation of pure cultures from a single, discrete bacterial colony isolated, one by one, from the bacterial mixture from MHA plates. Visually, several colonies in primary MHA plates showed similar colony morphology, suggesting that the composition of honey microbiota did not differ between these honeys. Only six colonies out of 76 were morphologically distinct. They were isolated and subcultured until a pure culture was obtained. Streak plates of pure cultures of colonies #1A, 1B, 2, 3, 4, and 5 were visually examined and at microscopic levels (Fig. 1). Their morphological features were assessed (Table 1).
Fig. 1.
Microscopic comparison of morphologically different bacterial colonies from pure cultures isolated from honey primary plates. (I). Streak plates of colony 1, 2, 3 and 5, scale bar = 10 mm. (II). Higher magnification of colonies that were identified as follow: 1A – B. amyloliquefaciens, 1B – B. methylotrophicus, 2 – B. cereus, 3 – B. methylotrophicus, 4 – B. cereus/thuringiensis, 5 – B. pumilus. (Leica L2 stereoscope, magnification ×16).
Table1.
Colony morphology and bacterial identification.
| Colony | Color | Size | Shape | Bacterial ID | ID method | Similarity (%) | Genetic Distance |
|---|---|---|---|---|---|---|---|
| 1A | Cream | Medium | Irreg | Bacillus amyloliquefaciens | 16S rRNA | 99 | 0 |
| opaque | raised | Bacillus methylotrophicus | 99 | 0.001 | |||
| 1B | Cream | Medium | Lobate | Bacillus methylotrophicus | 16S rRNA | 99 | 0.003 |
| opaque | mucoid | Bacillus amyloliquefaciens | 99 | 0.004 | |||
| 2 | White | Large | Irreg, | Bacillus cereus | MALDI-TOF* | 2.04 | |
| 3 | Cream | Medium | Lobate, | Bacillus methylotrophicus | 16S rRNA | 99 | 0.002 |
| opaque | mucoid | Bacillus amyloliquefaciens | 99 | 0.003 | |||
| 4 | White | Large | Irreg | Bacillus cereus/ | MALDI-TOF | 2.94 | |
| Bacillus thuringiensis | |||||||
| 5 | Cream | Small | Round | Bacillus stratosphericus | 16S rRNA | 99 | 0 |
| opaque | raised | Bacillus pumilus | 99 | 0.005 | |||
MALDI Biotyper IDs scores: Scores of 2.3 indicate the reliable identification to species and genus (highly probable), score values between 2.0 and 2.3 represent probable identification to species level, score values between 1.7 and 2.0 represent a reliable genus level, and scores of 1.7 are regarded as unreliable (Szabados et al., 2012).
Although the colony composition among honeys was similar, the abundance of a specific phenotype in a given sample varied. Phenotypes #2 and #4 were prevalent in samples H229, H230 and H177, while honeys H220 and H234 were enriched in phenotypes #1A, 1B and 3. All samples contained colony #5.
3.2. Bacterial identification
To obtain a definite assessment of bacterial composition and diversity, bacteria in colonies #1A, 1B, 2, 3, 4 and 5 were subjected to genotyping or proteotyping using 16S rRNA gene sequencing or MALDI-TOF. The identification of bacterial isolates is presented in Table 1. The reliability of bacterial identification is reflected by the sequence similarity values of 99%.
3.3. Colony composition and diversity at genetic level
Regardless of the fact that five different honeys were used, the core bacteria that dominated all honey samples belonged to the genus Bacillus. Among the six isolates, three clustered into B. subtilis group; Bacillus amyloliquefaciens, Bacillus methylotrophicus and Bacillus stratosphericus/B. pumilus and two isolates clustered into Bacillus cereus group; Bacillus cereus and Bacillus thuringiensis (Table 1). Both groups consist of closely related species of bacteria as indicated by a close genetic distance obtained from 16S rRNA gene sequences (Table 1).
Both Bacillus subtilis and Bacillus cereus groups are ubiquitous “environmental” microbes associated with plant surfaces and soil. The main respiratory menaquinone produced by Bacillus spp. is menaquinones MK-7 (Berenjian et al., 2011, Sato et al., 2001a, Sato et al., 2001b).
3.4. Isolation and identification of menaquinones
Menaquinones in honey occur in very low levels as indicated by the work of Kim and Brudzynski (2018). To increase the efficiency of menaquinone extraction, a protocol described by Sato et al., 2001a, Sato et al., 2001b was adopted. Both the filtered culture supernatants and honey solutions were extracted using a mixture of n-hexane:2-propanol (2:1), followed by solid-phase extraction using Oasis C18 cartridges. The extracts were subjected to ESI-UHPLC-tandem mass spectrometry coupled to high-resolution quadrupole orbitrap mass spectrometry. The strategy was to use an extracted-ion chromatogram (EIC) to specifically analyze menaquinones containing long, isoprenyl side-chains. The selected ions for the EIC corresponded to [M+H]+: MK-6, 581.3; MK-7, 649.5; MK-8, 717.4; MK-10, 853.8 and MK-11, 921.7. The chromatographic peaks, extracted within a mass tolerance below 5 ppm, allowed the identification of MK-7, MK-8 and MK-11 at m/z 649.5264, 717.4589 and 921.7893, respectively, in both bacterial culture supernatants and in honey samples (Table 2). The most abundant menaquinones in honey and in bacterial culture supernatants appeared to be MK-7 followed by MK-8, based on ion intensity signals (Table 2).
Table 2.
Mass spectra information of menaquinones found in the culture supernatants and in honey samples.
| MK | RT | [M+H]+ | Peak intensity |
||||
|---|---|---|---|---|---|---|---|
| Bacterial colonies |
Honey |
||||||
| #1A | #1B | #2 | H220 | H229 | |||
| MK-6 | 6.85 | 581.3594 | 2.47E+07 | 3.78E+07 | |||
| MK-7 | 6.76 | 649.5264 | 1.60E+09 | 1.82E+09 | 5.69E+08 | 2.55E+07 | 1.89E+07 |
| MK-8 | 8.51 | 717.4589 | 1.88E+09 | 8.08E+08 | 9.83E+08 | 1.80E+08 | 3.02E+07 |
| MK-11 | 10.4 | 921.7893 | 3.02E+07 | ||||
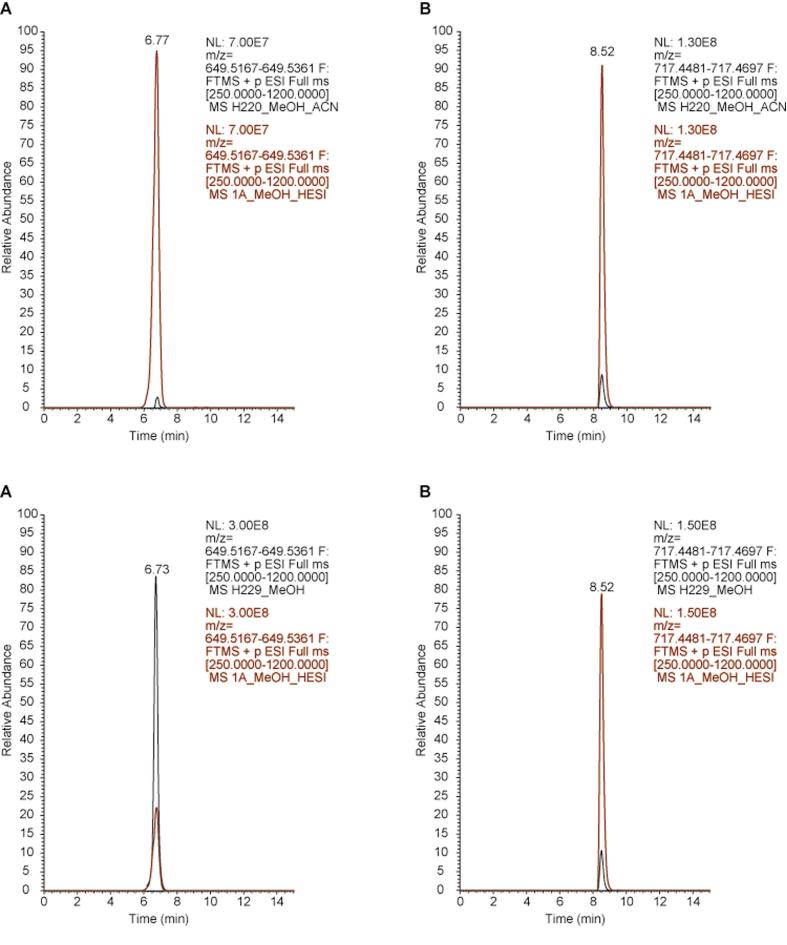
Fig. 2 shows that the EIC peaks are of high quality as indicated by their sharpness, Gaussian symmetry and no background noise. The overlay of the EIC peaks of MK-7 and MK-8 showed a perfect match of aligned mass ions between honey and culture supernatants (colony #1A) (Fig. 2). There were also no differences in the MS/MS fragments generated from menaquiones isolated from either honey or bacterial culture supernatants (Fig. 3). Each of the parent ions, m/z 649.5248 and m/z 717.4580 generated a unique, abundant fragment of m/z 274.2738 and m/z 191.1428, respectively (Fig. S1). These product ions were identical regardless of whether menaquinone originated from honey or from the supernatant, reaffirming that each unique fragment ion was arising from the same menaquinone present in both samples (Fig. 3).
Fig. 2.
Alignment of the extracted ion chromatogram peaks for m/z 649.5167 (MK-7) and 717.4481 (MK-8) from supernatant of bacterial cultures (red line) and honey (black line). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Full-scan mass spectrum of menaquinones MK-7 and a tentative assignment of fragment ions.
Although the peak intensities for both menaquinones were higher in the culture supernatants than in honey samples these differences were expected. The low numbers of bacteria found in honey (up to 6 × 102 CFU/ml) contributed less to menaquinone concentration compared to the bacterial numbers in pure cultures (∼109 CFU/ml) from which menaquinones were isolated. Thus, honey menaquinones MK-7 and MK-8 were identified as the major menaquinones produced by bacteria present in honey.
3.5. Levels of soluble menaquinones during bacterial growth
To investigate further likelihood that the soluble menaquinones derived from Bacillus spp. present in honey, the levels of menaquinone-7 (MK-7) released from B. amyloliquefaciens, B. methylotrophicus and B. cereus (colony #1A, 1B and 2, respectively) to the culture supernatants during their growth were analyzed. Bacterial growth was monitored by turbidity measurements (OD600) and by the CFU count in order to differentiate between culturable/viable and dead cells. After a lag phase, the exponential growth of all three bacterial cultures occurred between 6 and 18 h, reaching the peak at 18 h. Between 18 and 24 h, cells cease to divide and entered the stationary phase (Fig. 4A). During the stationary phase, the rapid, steep reduction of culturable/viable cells was observed. The 1–2 log reduction in CFU counts indicated substantial bacterial death. The CFU counts reached the lowest point at 24 h incubation in all three bacterial cultures and persisted to post-stationary phase (24–27 h). The growth resumed when a sporulation, induced by cell death, gave rise to a new population of cells (Fig. 4A).
Fig. 4.
Changes in the levels of MK-7 in the supernatants during the growth stages of B. amyloliquefaciens (colony 1A), B. methylotrophicus (colony 1B) and B. cereus (colony # 2). (A) A growth curve of colonies 1A, 1B and 2 measured spectrophotometrically at OD 600 nm (dotted line) and by CFU counts (solid line), (B) Comparison of the levels of MK-7 as relative intensity of UPLC peak (columns) with the changes in culturable/viable cell counts (CFU/ml, dotted line) during bacterial growth phases: late lag phase/early exponential phase (6 h), exponential growth phase (6–18 h), stationary/cell death phase (24 h) and the early exponential growth of new generation after sporulation (27 h).
The pattern of accumulation of soluble MK-7 in culture supernatants did not follow the Bacillus growth curve. The abundance of MK-7 in supernatants reached its maximum at the 6 h incubation time, that is, at the end of lag phase when cells started to divide by binary fission. The MK-7 levels were one order of magnitude higher at the 6 h than at 18 h incubation. At the peak of the exponential growth, at 18 h, the levels of soluble MK-7 were the lowest (Fig. 4B). Subsequently, the MK-7 abundance in the supernatants increased again at the stationary phase at 24 h and coincided with the height of the cell death and lysis, as indicated by CFU counts (Fig. 4B). The MK-7 levels in the culture supernatants remained relatively high throughout the post-stationary phase, up to the lag phase and beginning of growth of the new bacterial generations from the spores released by dying cells (at 27 h and longer).
These results indicate the inverse relationship between the bacterial growth and accumulation of soluble MK-7 in supernatants. At low bacterial growth, at the beginning and at the end of the growth curve, the levels of MK-7 were very high (at 6 h and 24 h, Fig. 4B), while at the peak of bacterial growth, the levels of MK-7 were very low (18 h, Fig. 4B). Despite some variability in the MK-7 measurements for each point of growth (shown as relative intensity of the peak), a significant negative correlation was found between stages of bacterial growth and the levels of soluble MK-7 in supernatants (Spearman’s r = −0.53, n = 15, p = 0.04 on log-transformed data obtained from all colonies). Thus, the increase of soluble MK-7 in supernatants was triggered by the death of Bacillus cells.
Morphological changes were also observed during the course of growth of pure cultures #1A, 1B, representing members of Bacillus subtilis group, specifically B. amyloliquefaciens (colony #1A). The changes included transition from the mucoidal morphology to the dry, crusted appearance with elaborate biofilm structure. The reduction in colony numbers from exponential growth (at 18 h) to the cell death phase (24–27 h) is clearly observable. The growth of new generation of bacteria at 30 h of incubation formed colonies that resemble colony structure of mother cells at lag to the early exponential phase of growth (3–6 h) (Fig. S2).
4. Conclusions
In this study, we evaluated the concept that the long-chain manequinones in honey derive from bacteria present in honey. Firstly, we assessed microbial composition in six buckwheat honeys, using both culture-dependent and molecular approaches. Bacterial identifications by 16S rRNA gene sequencing and MALDI-TOF showed that the predominant bacterial species in honey belonged to the genus Bacillus and consisted of two groups: B. subtilis and B. cereus. Each group contained several closely related species, including B. amyloliquefaciens, B. methylotrophicus, B. stratosphericus/B. pumilus in the Bacillus subtilis group and two isolates; B. cereus and B. thuringiensis in the Bacillus cereus group.
The presence of Bacillus spp. in honey was not surprising considering their common encounter in natural environment. They are introduced into beehives by bees together with nectars, pollen, soil and dust. The prominent presence of Bacillus spp. in honey is likely related to the bacterium ability to survive unfavourable environmental conditions by the process of sporulation. Highly resistant spores are metabolically dormant and can remain in this state in honey for long periods of time until growth conditions became favourable (Olaitan et al., 2007). This is the reason why the microbial diversity in honey was found to be low and comparable between honeys from distant geographic areas (Pajor, Worobo, Milewski, & Szweda, 2018).
Although the presence of Bacillus spp. in honey was expected, our finding that they are the source of the long-chain menaquinones in honey is novel.
The genus Bacillus, like many Gram-positive bacteria, uses only MK in their electron transport systems, specifically MK-7. With this in mind, the presence of limited types of menaquinones, namely MK-7 and MK-8, in the supernatants of the residing Bacillus spp. and in honey was investigated. The method included extracting from the UHPLC–ESI-MS scan the mass chromatogram (EIC) of selected mass ions representing [M+H]+ m/z 649.5 for MK-7 and m/z 717.4 for MK-8. The high-resolution EIC in conjugation with quadrupole orbitrap led to the identification of MK-7 (m/z 649. 5264) and MK-8 (717.4589) in both, the culture supernatants and in honey. The exact mass measurements of these mass ions were within 5 ppm error of expected theoretical formulas. The high quality of the EIC, characterized by the symmetry, sharpness and no background noise, reaffirmed that each mass peak was generated by an ion arising from menaquinone present in these samples. The precise alignment of MK-7 and MK-8 mass ion peaks between two experimental groups (honey and culture supernatants) observed in the overlay of EIC suggested that these menaquinones originated from the same source. This observation was further supported by the generation of the identical, unique MS/MS fragments from the parent mass ions of MK-7 and MK-8.
The results of this study confirmed that the honey menaquinones MK-7 and MK-8 are the main forms of menaquinones produced by the genus Bacillus found in honey.
Strain-specific differences in the production of these two menaquinones were also found. Honey H220 that showed higher prevalence of colonies of B. amyloliquefaciens (B. subtilis group) showed higher peak intensity of MK-7 over MK-8 (2.55E+07 to 1.89E+07). In contrast, honey H229 that contained mostly colonies of B. cereus/B. thuringiensis showed higher peak intensity of MK-8 over MK-7 (3.02E+07 to 1.80E+07). Similar strain-specific production of menaquinones were previously observed by others; the production of MK-7 was reported as a characteristic property of B. subtilis (Berenjian et al., 2011, Sato et al., 2001a, Sato et al., 2001b) while the production of MK-8 in addition to MK-7 was a property of B. thuringiensis (Hess, Holländer, & Mannheim, 1979). Thus, our data lend further support for the causal relationship between production of MK-7 and MK-8 by Bacillus spp. residing in honey and the predominant presence of MK-7 and MK-8 in honey.
Since both Bacillus strains were able to produce soluble menaquinones, the abundance of these compounds in the culture supernatants was investigated in relation to bacterial growth phases. The most abundant soluble MK-7 was present in the stationary (24 h incubation) and post-stationary (27 h) phases of growth, irrespective of the Bacillus strain responsible for its production. The maximum of MK-7 peak intensity in the cultures supernatants coincided in time with the marked decline in number of culturable/viable cells. On the other hand, the minimum MK-7 peak intensity in the supernatants corresponded to the peak of exponential growth. The rapid appearance of MK-7 in the supernatants was then in the inverse relationship with the growth (Spearman’s r = −0.53, n = 15, p = 0.04). Results presented in this paper are in contrast with those of Berenjian et al. (2011) who reported that the majority of MK-7 production was found during the logarithmic growth phase of B. subtilis natto, but in agreement with those of Ikeda and Doi (1990). The latter work indicated that while menaquinone biosynthesis inside of Bacillus subtilis increased in parallel with exponential growth, the appearance of external, soluble MK-7 took place in the stationary phase of growth. Data presented here expand this observation further by indicating that the increase in soluble MK-7 in supernatants in the stationary phase was triggered by cell death.
Bacillus amyloliquefaciens was the predominant producer of soluble MK-7. The rate of dying cells of B. amyloliquefaciens at the post-stationary stage exceeded that of B. cereus. It was reflected on MHA plates by the higher net loss of B. amyloliquefaciens culturable cells than that of B. cereus. Thus, the abundance of soluble MK-7 in the cultured supernatants seemed to be related to the number of dead cells. Dying cells undergo lysis and subsequently can release membrane fragments, together with the embedded menaquinones, to the external environments.
In conclusion, the major advance in this study is the understanding of the link between the death of vegetative forms of Bacillus spp., due to a hostile environment, and the accumulation of soluble menaquinones in honey. Cell death and membrane degradation, via cell lysis, appeared to be a prerequisite for the release of MK-7. Data can now explain the process by which intrinsically insoluble, membrane-bound menaquinones can be shed into aqueous solutions, such as honey. The release of soluble forms of MK-7 from dead bacteria ultimately enriched honey in this nutritionally important vitamin K2 homologue.
Acknowledgments
Acknowledgments
We thank Dr. Johan Delport, Clinical Microbiology/Molecular Microbiology, London Health Science Centre, London, Ontario for bacterial identification using MALDI-TOF, and Dr. A. Starostine, University of Toronto, BioZone.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing financial interest
The authors declare no competing financial interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2019.100008.
Contributor Information
Katrina Brudzynski, Email: kbrudzynski@brocku.ca.
Robert Flick, Email: robert.flick@utoronto.ca.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S2.
References
- Bald D., Villellas C., Lu P., Koul A. Targeting energy metabolism in Mycobacterium tuberculosis, a new paradigm in antimycobacterial drug discovery. mBio. 2017;8 doi: 10.1128/mBio.00272-17. e00272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenjian A., Mahanama R., Talbot A., Biffin R., Regtop H., Valtchev P. Efficient media for high menaquinone-7 production: Response surface methodology approach. New Biotechnology. 2011;28(6):665–672. doi: 10.1016/j.nbt.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Booth S.L. Vitamin K: Food composition and dietary intakes. Food & Nutrition Research. 2012;56(5505):1–5. doi: 10.3402/fnr.v56i0.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M., Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiology Reviews. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess A., Holländer R., Mannheim W. Lipoquinones of some spore-forming rods, lactic-acid bacteria and actinomycetes. Journal of General Microbiology. 1979;115(1):247–252. doi: 10.1099/00221287-115-1-247. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Doi Y. A vitamin-K2-binding factor secreted from Bacillus subtilis. European Journal of Biochemistry. 1990;192:219–224. doi: 10.1111/j.1432-1033.1990.tb19218.x. [DOI] [PubMed] [Google Scholar]
- Iurlina M.O., Fritz R. Characterization of microorganisms in Argentinean honeys from different sources. International Journal of Food Microbiology. 2005;105(3):297–304. doi: 10.1016/j.ijfoodmicro.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Kim L., Brudzynski K. Identification of menaquinones (Vitamin K2 homologues) as novel constituents of honey. Food Chemistry. 2018;249:184–192. doi: 10.1016/j.foodchem.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M. Essential Bacillus subtilis genes. Proceeding of National Academy of Science of the United States of America. 2003;100(8):4578–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y., Alejandro P., Rooney A.P., Tsukakoshi Y., Nakagawa R., Hasegawa H. Phylogenetic analysis of Bacillus subtilis strains applicable to natto (fermented soybean) production. Applied and Environmental Microbiology. 2011;77(18):6463–6469. doi: 10.1128/AEM.00448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu M., Begari E. Vitamin K2 in electron transport system: Are enzymes involved in vitamin K2 biosynthesis promising drug targets? Molecules. 2010;15:1531–1553. doi: 10.3390/molecules15031531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka B., Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochimica et Biophysica Acta. 2010;1797:1587–1605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Olaitan P.B., Adeleke O.E., Ola I.O. Honey: A reservoir for microorganisms and an inhibitory agent for microbes. African Health Science. 2007;7(3):159–165. doi: 10.5555/afhs.2007.7.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajor M., Worobo R., Milewski W., Szweda P. The antimicrobial potential of bacteria isolated from honey samples produced in the apiaries located in Pomeranian Voivodeship in Northern Poland. International Journal of Environmental Research and Public Health. 2018;15(9):E2002. doi: 10.3390/ijerph15092002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Yamada Y., Ohtani Y., Mitsui N., Murasawa H., Araki S. Efficient production of menaquinone (vitamin K2) by a menadione-resistant mutant of Bacillus subtilis. Journal of Industrial Microbiology and Biotechnology. 2001;26(3):115–120. doi: 10.1038/sj.jim.7000089. [DOI] [PubMed] [Google Scholar]
- Sato T., Yamada Y., Ohtani Y., Mitsui N., Murasawa H., Araki S. Production of menaquinone (vitamin K2)-7 by Bacillus subtilis. Journal of Bioscience and Bioengineering. 2001;91(1):16–20. doi: 10.1263/jbb.91.16. [DOI] [PubMed] [Google Scholar]
- Sherman M.M., Petersen L.A., Poulter C.D. Isolation and characterization of isoprene mutants of Escherichia coli. Journal of Bacteriology. 1989;171:3619–3628. doi: 10.1128/jb.171.7.3619-3628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowdon J.A., Cliver D.O. Microorganisms in honey. International Journal of Food Microbiology. 1996;31(1–3):1–26. doi: 10.1016/0168-1605(96)00970-1. [DOI] [PubMed] [Google Scholar]
- Szabados F., Tix H., Anders A., Kaase M., Gatermann S.G., Geis G. Evaluation of species-specific score cutoff values of routinely isolated clinically relevant bacteria using a direct smear preparation for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based bacterial identification. European Journal of Clinical Microbiology & Infectious Diseases. 2012;31(6):1109–1119. doi: 10.1007/s10096-011-1415-7. [DOI] [PubMed] [Google Scholar]
- Walther B., Chollet M. Menaquinones, Bacteria and foods: Vitamin K2 in the diet. 2017. pp. 63–82.https://cdn.intechopen.com/pdfs/50921 [Chapter 4] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.