Abstract
Objectives
This study aims to investigate the modulation of estrogen receptors by estrogen and the role of genistein in the transcriptional process that regulates genes involved in the proliferation, apoptosis, and telomere activity.
Methods
The research was conducted in silico, wherein docking, the most important method, was carried out using Hex 8.0 software and HADDOCK web server. Interaction analysis was subsequently done to observe the interactions between genistein and several related proteins and BCLX, Casp3, Ki-67, CyclinD1, hTERT, and POT1 genes using Discovery Studio, LigPlus, and NUCPLOT.
Results
The interaction between ERα with genistein was not found to form a single bond. Thus, the interaction that may occur will not be effective because it is not stable. Conversely, when interacting with ERβ, two hydrogen bonds and four hydrophobic bonds, MPP dihydrochloride interacted with ERα via two hydrogen bonds and three hydrophobic bonds. The ERβ/eNOS complex will be comparatively easier to induced by the transcriptional activation of BCLX, Casp3, Ki-67, CyclinD1, hTERT and POT1 genes.
Conclusions
Administration of genistein can increase the genomic activities of the estrogen-eNOS receptor complexes related to apoptosis, proliferation, and telomere activity.
Keywords: Apoptosis, Genistein, hTERT, Estrogen receptor, POT1
الملخص
أهداف البحث
تهدف هذه الدراسة للتحقيق في تشكيل مستقبلات هرمون الاستروجين بواسطة تركيب مضاد –الاستروجين ودور الجينيستين ضد تنظيم عملية النسخ من الجينات المشاركة في الانتشار، وموت الخلايا المبرمج ونشاط التيلومير.
طرق البحث
تم إجراء البحث باستخدام أسلوب سيليكو بحيث يكون الإرساء هو أهم طريقة تم تنفيذها بواسطة برمجيات الهيكس ٨.٠ وقاعدة البيانات هادوك. ثم تم عمل تحليل التفاعل لملاحظة التفاعلات بين الجينيستين وعدد من البروتينات والجينات ذات الصلة باستخدام برامج الاكتشاف.
النتائج
لم يظهر التفاعل بين مستقبلات هرمون الاستروجين – الفا مع الجينيستين تشكيل أي رابطة. وهكذا التفاعل، الذي ممكن حدوثه، لن يكون فاعلا لأنه ليس مستقرا. وعلى العكس، عندما يكون التفاعل مع مستقبلات هرمون الاستروجين- بيتا، اثنان من الروابط الهيدروجينية وأربع من الروابط الطاردة للماء، هيدروكلوريد تفاعل مع مستقبلات هرمون الاستروجين – الفا بواسطة اثنان من الروابط الهيدروجينية وثلاثة من الروابط الطاردة للماء. سيكون من السهل للمركب الحث على تنشيط النسخ للجينات المدروسة.
الاستنتاجات
إعطاء الجينيستين ممكن أن يزيد النشاط الجينومي لمركبات مستقبلات هرمون الاستروجين التي ترتبط بموت الخلايا المبرمج، والانتشار ونشاط التيلومير.
الكلمات المفتاحية: موت الخلايا المبرمج, مستقبل هرمون الاستروجين, جينيستين
Introduction
Steroid hormone nuclear receptors and their hormonal ligands are the key mediator groups in the endocrine signalling pathway, which play an important role in the regulation of differentiation, growth, and metabolic homeostasis. Estrogen regulates differentiation and maintains reproductive tissues, muscles, and other tissues by activating their receptors.1 Ligands of estrogen receptors (ER) induce ER bonding at specific DNA response elements to regulate target gene expression. Receptors that can bind to DNA can positively or negatively regulate the transcription of target genes in specific cells or tissues. The ER agonist bonding in breast cancer cells can recruit a transcriptional co activator. In contrast, when it is antagonistic, the ER will actively recruit the corepressors, thereby causing transcriptional suppression of the target genes.2 Structural studies indicate that estrogen and the antagonist estrogen compounds can induce different conformational changes in estrogen receptor-alpha (ERα), which may further determine the recruitment of co activators or corepressors, thus leading to diverse biological effects. Estrogen also acts as a mitogen to increase cell proliferation in both normal and cancerous breast cells, where ERα is a prognostic marker and a therapeutic target for breast cancer.3 One of the anti-estrogen compounds is methyl-piperidino-pyrazole (MPP) dihydrochloride (C29H31N3O3·2HCl), which is selective and has a high antagonist affinity to ERα compared with estrogen receptor-beta (ERβ) (200×).
The most potential estrogen produced by human body is 17β-estradiol. There are two estrogen metabolites, estrone and estriol. Although both are high affinity ligands, they have lower agonist properties against ER.4 Cellular signalling of estrogen is mediated through two subtypes of ER, namely ERα and ERβ, both of which belong to the nuclear receptor transcription factor family. The most conserved domain of ER is the DNA-binding domain (DBD) involved in the introduction and binding of DNA, while the ligand binding occurs in the ligand-binding domain (LBD) at the COOH terminal region. Transcription activation is facilitated by two different activation (AF) functions, i.e. AF-1 which is continuously active and located at the NH2 terminal of the receptor, and AF-2 located at the COOH terminal of LBD.5 ERα and ERβ have a high degree of homology based on sequence alignment, except at the terminal NH2 domain. They also have similar affinity levels towards estrogen and bind to the same DNA response elements.6 Estrogen signalling begins when estrogen binds to ER where the transcriptional response depends upon the composition of the corrugating protein and the characteristics of the target gene promoter. The ER complex binds to the estrogen-responsive element (ERE) of the target gene, which subsequently recruits various cofactors, inducing the activation or repression of the target genes.7
Nitric oxide (NO) is produced by eNOS, which is a free radical involved in various cellular processes. Several studies have reported that activated eNOS can translocate to the nucleus upon binding to ERβ.8 The eNOS/ERβ complex determines chromatin remodelling, thus inducing the transcriptional activation of some prognostic genes, including hTERT, MSH2, CylinD1, and ps2. These genes are quite sensitive to estrogen stimulus or intracellular NO levels.9
Epidemiological reports show that the consumption of food items that contain a lot of phytoestrogens is thought to be associated with a reduced risk of cancer due to hormonal induction.10 Studies have also reported the antiproliferative effect of genistein a phytoestrogen compound found most abundantly in soybean. Genistein has a higher affinity for ERβ compared with ERα (9×).11
Anti-estrogen designed to inhibit ERα signalling are widely and effectively being used clinically for the treatment of breast cancer.12 However, these drugs have undesired side effects on non-target tissues, and long-term treatments render the cancer resistant to the anti-estrogen therapy. The idea of using ERβ agonist offers new possibilities for pharmacological intervention in cancer therapy,13 and because of its specificity towards ERβ, genistein is expected to have no undesirable side-effects upon substitution with estrogen. This study aimed to investigate the modulation of estrogen receptors by anti-estrogen and roles of genistein against the transcription process regulation of genes involved in the proliferation, apoptosis, and telomere activities.
Materials and Methods
Search for nucleotide and protein sequences
The structures of the components of genistein (CID: 5280961), 17β-estradiol (CID: 5757), and MPP dihydrochloride (CID: 45073474) were obtained from PubChem Open Chemistry Database. The sequences of the proteins ERα (GI: 11907837), ERβ (GI: 2970564), eNOS (GI: 266648), and nucleotides BCLX gene promoter sequence (GI: 488120), Casp3 (GI: 27979118), CyclinD1 (GI: 483600), Ki-67 (GI: 1944550), hTERT (GI: 4239869), and POT-1 (GI: 649107630) were obtained from sequence database of the National Center for Biotechnology Information (NCBI), United States National Library of Medicine (NLM), and National Institute of Health (NIH) (http://www.ncbi.nlm.nih.gov).
3D structure modelling of DNA, proteins, and bioactive components
The 3D structure modelling of ERα, ERβ, and eNOS was performed to predict the 3D conformations using SWISS-MODELLER web server by homology modelling.14, 15 The 3D model of the respective protein was then validated using Ramachandran Plot analysis. The 3D structures of BCLX, Casp3, CyclinD1, Ki-67, hTERT, and POT-1 gene promoters were predicted by 3D DART web server.16 The conversion of *.sdf files into *.pdb files from genistein, 17β-estradiol and MPP dihydrochloride was performed using OpenBabel software.17
Docking computation
Simulation of docking between genistein compounds, 17β-estradiol and MPP dihydrochloride on ERα and ERβ, as well as the docking between the ERβ + genistein/17β-estradiol + eNOS complexes in the target gene promoters were performed using HEX 8.0 software.18 The docking protocol consists of three stages of visualization, namely rigid-body energy minimization, semi-flexible repair, and finishing refinement in explicit solvent. After the execution of each stage, the docking confirmation was scored and sorted based on the scoring function to facilitate the best conformation selection to be used at a later stage.
Inter-protein interaction analysis
The results of the next docking analysis will be visualized using Discovery Studio 4.1, LigPlot +, and Chimera 1.6.2.19 Analysis of the interaction between the protein complexes of ERβ + genistein/17β-estradiol + eNOS with the target gene promoter was done by using NUCPLOT software. Analyses of the protein–protein and protein-DNA interactions were performed to visualize the hydrogen bonding, hydrophobic bonding, and van der Waals bonding. Pharmacophore analysis was also performed to see the residues directly involved in the interaction process, minimization energy analysis was performed to improve the structure and shape of the molecules during interaction.
Results
Genistein exhibits a selective affinity for ERβ
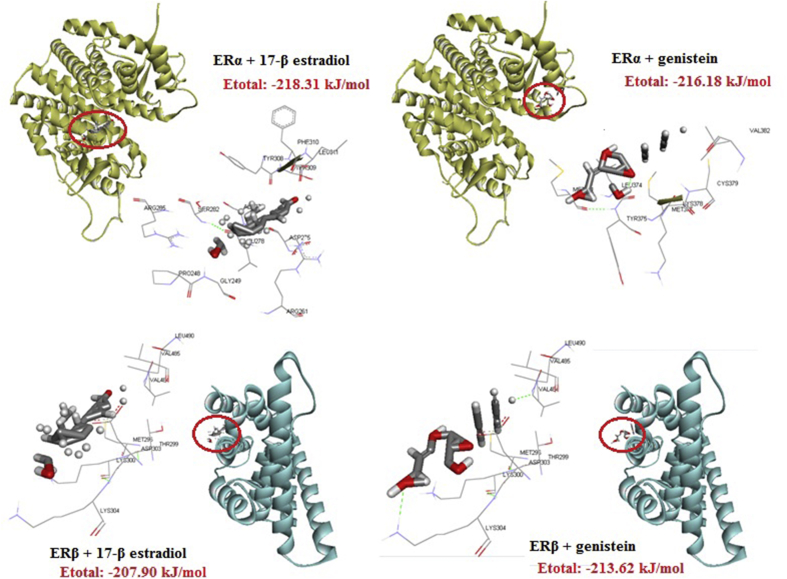
In this study, it was reported that genistein has a higher affinity for ERβ compared to ERα. The docking analysis showed that for the genistein-ERα interaction takes lesser energy (−216.18 kJ/mol) as compared to the genistein-ERβ interaction (−213.62 kJ/mol). However, the difference in energy is not significant enough. In addition, the docking analysis shows that the interaction between ERα with genistein does not involve formation of a chemical bond, because of which, the resultant unstable interaction may not be effective. Conversely, upon interacting with ERβ, two hydrogen bonds and four hydrophobic bonds between genistein and amino acid residues Lys304, Val485, Met296, Thr299, Val 485, and Leu490 of ERβ are formed, leading to a stronger interaction (Table 1 and Figure 1).
Table 1.
Possible interactions between 17 β-estradiol and genistein with ERα or ERβ.
| Molecule | Point interaction | Category | Donor atom | Acceptor atom | Binding energy |
|---|---|---|---|---|---|
| ERα – 17 β-estradiol | ARG261:H – 17 β-estradiol:C | Hydrophobic bond | H | C | −218.31 kJ/mol |
| PHE310:H – 17 β-estradiol:C | Hydrophobic bond | H | C | ||
| LEU311:H – 17 β-estradiol:C | Hydrophobic bond | H | C | ||
| ERα – genistein | – | – | – | – | −216.18 kJ/mol |
| ERβ – 17 β-estradiol | MET296:O – 17 β-estradiol:O | Hydrophobic bond | O | O | −207.90 kJ/mol |
| THR299:H – 17 β-estradiol:C | Hydrophobic bond | H | C | ||
| LYS300:O – 17 β-estradiol:C | Hydrophobic bond | O | C | ||
| ASP303:H – 17 β-estradiol:C | Hydrophobic bond | H | C | ||
| VAL485:O – 17 β-estradiol:O | Hydrophobic bond | O | O | ||
| ERβ – genistein | LYS304:HZ3 – genistein:O | Hydrogen bond | HZ3 | O | −213.62 kJ/mol |
| VAL485:H – genistein:O | Hydrogen bond | H | O | ||
| MET296:O – genistein:O | Hydrophobic bond | O | O | ||
| THR299:O – genistein:O | Hydrophobic bond | O | O | ||
| VAL485:O – genistein:O | Hydrophobic bond | O | O | ||
| LEU490:O – genistein:O | Hydrophobic bond | O | O |
Figure 1.
Analysis of docking and bond energy between estrogen receptor, 17β-estradiol, and genistein. Genistein more easily interacts with both estrogen receptors than the 17β-estradiol interactions with estrogen beta receptors.
Genistein has a higher affinity level in ERβ compared to 17β-estradiol
Table 1 and Figure 1 show the results of docking analysis. It can be clearly seen that the energy required by genistein for binding to ERβ is lower (−213.62 kJ/mol) than that required by 17β-estradiol (−207.90 kJ/mol). Docking analysis also showed that the interaction between genistein and ERβ was mediated by two hydrogen bonds and four hydrophobic bonds; whereas the interaction between 17β-estradiol and ERβ was mediated by five hydrophobic bonds only.
MPP dihydrochloride can inhibit the 17β-estradiol binding with ERα
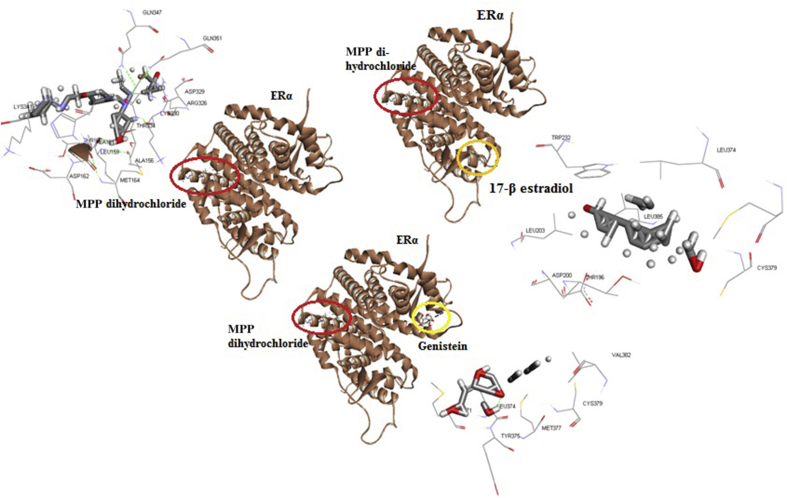
The analysis was conducted to see the counter mechanism of MPP dihydrochloride against ERα. It was seen that MPP dihydrochloride interacted with ERα via two hydrogen bonds and three hydrophobic bonds via ERα residues Asp333, Leu159, Arg326, Asp329, and Gln351. Interaction sites of MPP dihydrochloride and 17β-estradiol in ERα were differently located, but when MPP dihydrochloride binds to ERα, it brings about a change in 17β-estradiol interaction site in ERα (Figure 2 and Table 2). Interaction analysis also shows that the interaction site changes may cause an increase in the energy required to bind, but the bonds which mediate the interaction are not formed at all. It is suspected that when MPP dihydrochloride binds to ERα, the ERα conformation changes so as to inhibit the 17β-estradiol and ERα binding. Unlike with genistein, MPP dihydrochloride did not cause changes in the interaction site of genistein on ERα, but may cause the energy required for interaction become larger (−205.45 kJ/mol).
Figure 2.
Analysis of docking and bond energy between estrogen alpha receptor, 17β-estradiol, genistein, and MPP-dihydrochloride.
Table 2.
Possible interactions between MPP dihydrochloride and ERα.
| Molecule | Point interaction | Category | Donor atom | Acceptor atom | Binding energy |
|---|---|---|---|---|---|
| ERα – MPP dihydrochloride | MPP dihydrochloride:N – ASP333:OD2 | Hydrogen bond | N | OD2 | −341.22 kJ/mol |
| MPP dihydrochloride:H – LEU159:O | Hydrogen bond | H | O | ||
| MPP dihydrochloride:H – ARG326:O | Hydrophobic bond | H | O | ||
| ASP329:H - MPP dihydrochloride:C | Hydrophobic bond | H | C | ||
| GLN351:O - MPP dihydrochloride:C | Hydrophobic bond | O | C | ||
| ERα, MPP dihydrochloride + 17 β-estradiol | – | – | – | – | −216.66 kJ/mol |
| ERα, MPP dihydrochloride + genistein | – | – | – | – | −205.45 kJ/mol |
Genistein is more effective in inducing transcription of BCLX, Casp3, Ki-67, CyclinD1, hTERT, and POT1 genes
Out of the 6 genes analysed in this study, BCLX and Casp3 genes are involved in cell apoptosis, the Ki-67 and CyclinD1 genes are involved in cell proliferation, and hTERT and POT1 genes participate in the telomerase activity. The analysis showed that upon binding to genistein, the ERβ/eNOS complex/would be/was induced easily by the transcriptional activation of the above-mentioned genes. This is seen from the lower binding energy and higher number of the bonds formed (Figure 3, Figure 4, Table 3). In ERβ, the DNA binding domain extends from residues 149–214, the steroid binding domain is between residues 215–530, while the Zinc finger is positioned at residues 149–169 and 185–209.
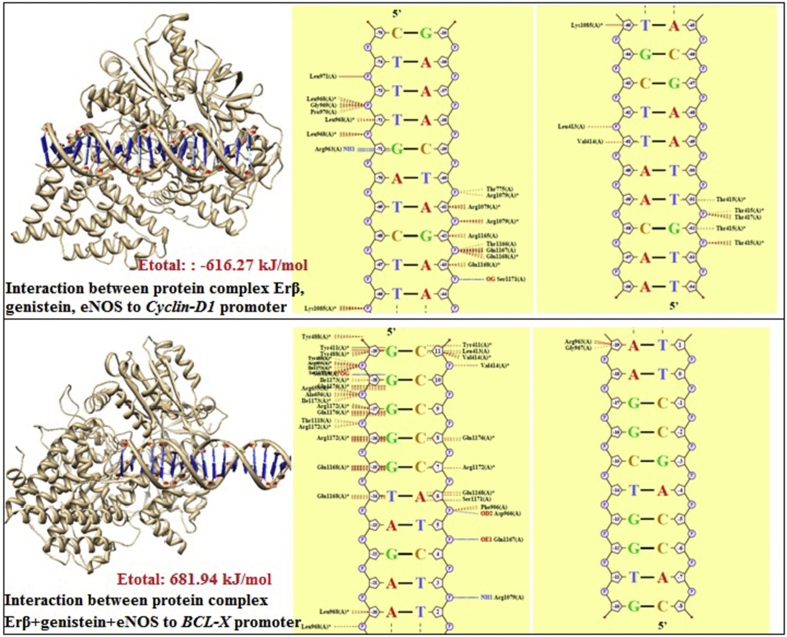
Figure 3.
Analysis of docking and bond energy between estrogen beta receptor, genistein, eNOS, with Cyclin D1 or Bcl-X.
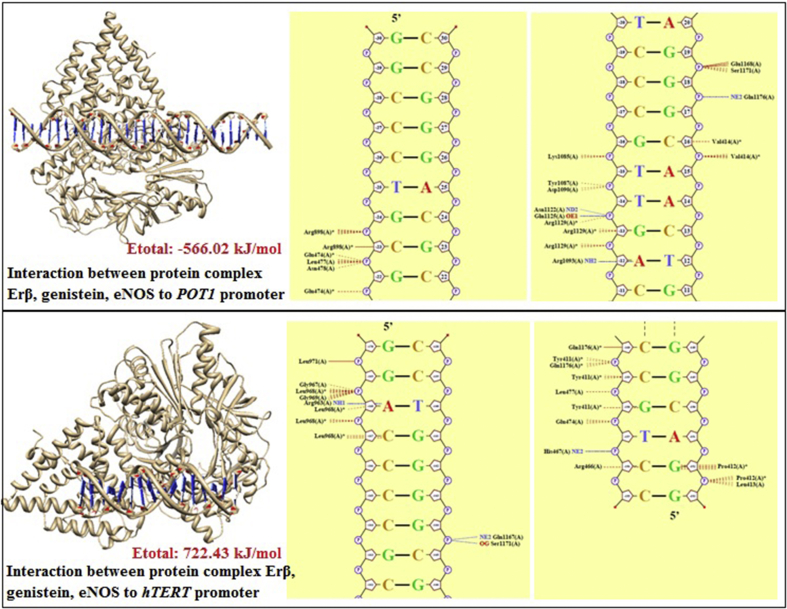
Figure 4.
Analysis of docking and bond energy between estrogen beta receptor, genistein, eNOS, with POT1 or hTERT.
Table 3.
Interactions of ERβ/eNOS/genistein-17β estradiol complexes on target gene promoters.
| Molecule | Number of interaction | Point interaction | Binding energy |
|---|---|---|---|
| ERβ, 17-β estradiol, eNOS + BCLX | 27 (4 hydrogen bonds, 23 van der waals interaction) | G(-29A) → Gln1167, Ser1171, Glu1168; G(-25A) → Ser 619; A(-23A) → Pro621, Ser617, Ser624; G(-22A) → Ser617, Ile616, Ser625; C(10B) → Glu1168; C(9B) → Arg1172, Asp620, Thr1118, Thr1121, Asn1122; C(8B) → Ser619, Gln1176, Ser619, Arg655, Ala656; C (7B) → Cys618, Ser619, Arg655, Ile1173, Ser1177; C(6B) → Gln1176. | −543.07 kJ/mol |
| ERβ, genistein, eNOS + BCLX | 36 (4 hydrogen bonds, 32 van der waals interaction) | G(-29A) → Tyr488, Tyr411, Arg655, Ile1173, Ser1177; G(-28A) → Ser619, Ile1173, Gln1176, Arg655, Ala656, Ile1173; G(-27A) → Arg1172, Gln1176, Thr1118, Arg1172; G(-26A) → Arg1172; G(-25A) → Glu1168; T(-24A) → Glu1168; A(-20A) → Leu968; A(-19A) → Arg963, Gly967; C(11B) → Tyr411, Leu413, Val414; C(8B) → Gln1176; C(7B) → Arg1172; A(6B) → Glu1168, Ser1171, Phe906, Asp966; T(5B) → Gln1167; T(3B) → Arg1079 | −681.94 kJ/mol |
| ERβ, 17-β estradiol, eNOS + Casp3 | 23 (4 hydrogen bond, 19 van der waals interaction) | T (-121A) → Ser1171; G(-115A) → Ser617, ys618, Pro621, Ser624; C(-114A) → Ser617; A (-81B) → Glu1168; A(-83B) → Arg1172, Asp620, Thr1118, Thr1121; G(-84B) → Ser619, Asp620, Arg1172, Arg655, Arg656; C(-85B) → Ile1173, Gln1176, Arg655, Gln1176, Ser1177. | −522.93 kJ/mol |
| ERβ, genistein, eNOS + Casp3 | 16 (one hydrogen bond, 15 van der waals interaction) | G(-115A) → Ala1098; C(-114A) → Thr1094, Glu1095; G(-107A) → Val414; A(-106A) → Ser619; G(-105A) → Glu1168, Arg1172; C(-104A) → Glu1168; (C(-91B) → Arg1093; G(-92B) → Arg1093, Gln1125, Thr1126, Arg1129; C(-93B) → Gln1089, Asp1090; C(-94B) → Lys1085. | −636.42 kJ/mol |
| ERβ, 17-β estradiol, eNOS + Ki-67 | 21 (4 hydrogen bond, 17 van der waals interaction) | G(-41A) → Gln1176, Ser1177; G(-40A) → Ser619, Arg655, Ile1173; G(-39A) → Ser619, Asp620, Thr1121; C(-38A) → Asp620, Arg1172, Asn1122; T(-37A) → Arg1172; C(-25B) → Ile616, Ser617, Ser624, Ser625; G(-26B) → Cys618, Ser624; C(-32B) → Glu1168. | −546.92 kJ/mol |
| ERβ, genistein, eNOS + Ki-67 | 29 (5 hydrogen bond, 24 van der waals interaction) | C(-44A) → Arg1049, Arg1079; C(-42A) → Gln1167; G(-41A) → Phe906, Asp966, Ser1171; G(-40A) → Glu1168, Ser1171; G(-39A) → Arg1172; G(-22B) → Arg963; G(-23B) → Arg963, Gly967, Leu968; C(-28B) → Glu1168; C(-29B) → Glu1168; G(-30B) → Arg1172, Thr1118; A(-31B) → Arg1172, Gln1176, Arg655, Ala656, Ile1173; C(-32B) → Ser619, Ile1173, Arg655, Ile1173, Ser1177. | −638.37 kJ/mol |
| ERβ, 17-β estradiol, eNOS + CyclinD1 | 26 (3 hydrogen bond, 23 van der waals interaction) | C(-75A) → Ser1171; T(-74A) → Arg1172; T(-69A) → Ser617, Cys618, Pro621, Ser617, Ser624; C(-68A) → Ser617, Ile616; A(-36B) → Glu1168; A(-37B) → Arg1172, Asp620, Thr1118, Thr1121; A(-38B) → Ser619, Asp620, Arg1172, Gln1176, Arg655, Ala656; C(-39B) → Ile1173, Gln1176, Arg655, Ile1173, Gln1176, Ser1177. | −542.26 kJ/mol |
| ERβ, genistein, eNOS + CyclinD1 | 26 (2 hydrogen bond, 24 van der waals interaction) | T(-74A) → Leu971; T(-73A) → Leu968, Gly969, Pro970; T(-72A) →Leu968; G(-71A) → Arg963; T(-66A) → Lys1085; T(-65A) → Lys1085; T(-62A) → Leu413; T(-61A) → Val414; T(-40B) → Thr775, Arg1079; A(-41B) → Arg1079; G(-42B) → Arg1165, Thr1166, Gln1167, Glu1168; A(-43B) → Glu1168, Ser1171; T(-51B) → Thr415, Thr417; G(-52B) → Thr415. | −616.27 kJ/mol |
| ERβ, 17-β estradiol, eNOS + hTERT | 20 (5 hydrogen bond, 15 van der waals interaction) | G(-163A) → Arg764, Ser765; C(-162A) → Ser765, Arg782; C(-161A) → His949; C(-160A) → Glu1033; C(-159A) → Thr948, Glu1033; G(-158A) → Ser947; C(-150B) → Ser947, Pro950, Gly787, Gly951; A(-151B) → Pro946, Pro950, Gln792, Tyr793, Pro946; G(-152B) → Ser448, Gln794. | −611.79 kJ/mol |
| ERβ, genistein eNOS + hTERT | 22 (4 hydrogen bond, 18 van der waals interaction) | G(-170A) → Leu971; G(-169A) → Gly967, Leu968, Gly969,; A(-168A) → Arg963, Leu968; C(-167A) → Leu968; C(-160A) →Gln1176, Tyr411, Gln1176; C(-159A) → Tyr411, Leu477; G(-158A) → Tyr411, Glu474; T(-157A) → His467; C(-156A) → Arg466; G(-144B) → Gln1167, Ser1171; G(-152B) → Pro412, Leu413. | −722.43 kJ/mol |
| ERβ, 17-β estradiol, eNOS + POT1 | 43 (5 hydrogen bond, 38 van der waals interaction) | C(-21A) →Trp345, Asp349; T(-20A) → Trp345, Asp349, Ile348, Phe387; C(-19A) → Asp349, Arg388, Glu389; C(-18A) → Asp349, Ala589, Glu592, Met593; C(-17A) → Ser638; G(-16A) → Asp637, Ser638; T(-15A) → Glu562, Asp637; G(-10A) → Ala515; T(-9A) → Ala515; A(20B) → Asn598; C(16B) → Lys353; A(15B) → Lys353, Glu562, Gly352, Lys353, Asp363; C(13B) → Glu562, His563, Glu564; T(12B) → Thr644. | −494.65 kJ/mol |
| ERβ, genistein eNOS + POT1 | 20 (4 hydrogen bond, 16 van der waals interaction) | G(-24A) → Arg898; C(-23A) → Arg898, Glu474, Leu477, Asn478; G(-22A) → Glu474; G(-16A) → Lys1085; T(-15A) → Tyr1087, Asp1090; T(-14A) → Asn1122, Gln1125, Arg1129; G(-13A) → Arg1129; A(-12A) → Arg1093; G(19B) → Glu1168, Ser1171; G(18B) → Gln1176; C(16B) → Val414; C(10B) → Thr1094, Glu1095; A(9B) → Ala1098. | −566.02 kJ/mol |
Discussion
Genistein is a phytoestrogen compound found mostly in soybean plant. In this study it was proven that the energy of the interaction between genistein and ERβ was slightly lower than that with ERα. This indicates that genistein can interact with both receptors, but it is relatively easier in case of ERβ. In addition, this type of bond with ERβ shows a more stable bonding compared to ERα. This finding is consistent with the previous reports stating that genistein can bind to both of the estrogen receptor isoforms.20, 21 In this study the interaction between genistein and Lys304, Val485, Met296, Thr299, Val 485, and Leu49 of ERβ has been elucidated. This investigation also proved the genistein-17β-estradiol interaction against ERβ. The energy of the interaction and the type of bonds formed indicate that genistein has a higher affinity for ERβ than 17β-estradiol. The hydrogen bonding is stronger as compared to the hydrophobic bonding. The stronger the bonds formed between the two molecules, the higher the stability of the interactions. Hence, it is suspected that genistein has a stronger affinity towards ERβ if the affinity levels between 17β-estradiol and ERβ are to be compared. The study supports the previous findings that genistein-ERβ interactions involve the —OH groups of His475, Glu305, and Arg306.22
Methyl-piperidino-pyrazole (MPP) is a very selective ERα antagonist. Its affinity for ERα is 200 times as compared to ERβ.23 Our study revealed that MPP dihydrochloride could inhibit the interaction between 17β-estradiol and ERα. The inhibitory mechanism is supposedly through the conformational changes in ERα, thus rendering 17β-estradiol non-functional for effective binding to ERα. MPP dihydrochloride also blocks the interaction between genistein and ERα by increasing the energy required for the interaction to occur.
Both ERα and ERβ can be activated by estrogen, wherein both their ligand-binding domains allow for the selection of binding ligands. DNA-binding domains of ERα and ERβ have high conservation rates and both share between 46% and 73% of chromatin binding sites.24 Although several clinical studies linked the ERβ expression to better clinical outcomes,25, 26 several other reports show an association between ERβ expression and the Ki-67 proliferation marker.27, 28 From our results of, it is speculated that ERβ can indeed induce the expression of Ki-67 as seen from the interaction between ERβ/eNOS/17β-estradiol-genistein with the Ki-67 gene promoter. In addition, it is suspected that genistein is more effective in inducing the occurrence of such interactions as compared to 17β-estradiol. This was seen from the lower energy required for interaction and the greater interaction of hydrogen and van der-Waals formed. Another proliferation marker analysed in this study is the CyclinD1 gene. This study reported that the ERβ/eNOS/17β-estradiol-genistein complex may also induce transcriptional activation of the CyclinD1 gene. The results from this study are not consistent with a previous study which reports that ERα is an activator of the CyclinD1 gene, whereas ERβ is its repressor.29
Different co activator/co repressor proteins determine the transcription complex to be recruited, thus causing differential gene transcriptions depending on the type of co regulator ER-complex dimer.30 We also evaluated the effect of formation of estrogen ligand complexes, and impact of eNOS on the promoter region of apoptotic and telomere activity genes. We revealed here that genistein ligand increases the ease and stability of interactions than the ligand estradiol indicating that genistein administration can increase the genomic activity of estrogen-eNOS receptor complexes related to apoptosis and telomere activity.
It was concluded that administration of genistein can increase the genomic activity of oestrogen-eNOS receptor complexes related to apoptosis, proliferation, and telomere activity.
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
This research has received ethical approval from the Research Ethics Committee, Faculty of Medicine, Brawijaya University, Malang, East Java, Indonesia.
Authors' contributions
HY, EH, TN, and HK conceived and designed the study, conducted research, provided research materials, and collected and organized data. HY analysed and interpreted data. HY, EH, and TN wrote the initial and final drafts of the article. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
Footnotes
Peer review under responsibility of Taibah University.
References
- 1.McKenna N.J., O'Malley B.W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108(4):465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 2.Smith C.L., O'Malley B.W. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 3.Thomas T., Gallo M.A., Thomas T.J. Estrogen receptors as targets for drug development for breast cancer, osteoporosis and cardiovascular disease. Curr Cancer Drug Targets. 2004;4(6):483–499. doi: 10.2174/1568009043332880. [DOI] [PubMed] [Google Scholar]
- 4.Kuiper G.G., Carlsson B., Grandien K., Enmark E., Haggblad J., Nilsson S., Gustafsson J.A. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138(3):863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson S., Makela S., Treuter E., Tujague M., Thomsen J., Andersson G., Enmark E., Pettersson K., Warner M., Gustafsson J.A. Mechanisms of estrogen action. Physiol Rev. 2001;81(4):1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 6.Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Strom A., Treuter E., Warner M., Gustafsson J.A. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87(3):905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 7.Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., Wang Q., Bekiranov S., Sementchenko V., Fox E.A., Silver P.A., Gingeras T.R., Liu X.S., Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 8.Gobeil F., Jr., Zhu T., Brault S., Geha A., Vazquez-Tello A., Fortier A., Barbaz D., Checchin D., Hou X., Nader M., Bkaily G., Gratton J.P., Heveker N., Ribeiro-da-Silva A., Peri K., Bard H., Chorvatova A., D'Orléans-Juste P., Goetzl E.J., Chemtob S. Nitric oxide signaling via nuclearized endothelial nitric-oxide synthase modulates expression of the immediate early genes iNOS and mPGES-1. J Biol Chem. 2006;281(23):16058–16067. doi: 10.1074/jbc.M602219200. [DOI] [PubMed] [Google Scholar]
- 9.Nanni S., Benvenuti V., Grasselli A., Priolo C., Aiello A., Mattiussi S., Colussi C., Lirangi V., Illi B., D'Eletto M., Cianciulli A.M., Gallucci M., De Carli P., Sentinelli S., Mottolese M., Carlini P., Strigari L., Finn S., Mueller E., Arcangeli G., Gaetano C., Capogrossi M.C., Donnorso R.P., Bacchetti S., Sacchi A., Pontecorvi A., Loda M., Farsetti A. Endothelial NOS, estrogen receptor β, and HIFs cooperate in the activation of a prognostic transcriptional pattern in aggressive human prostate cancer. J Clin Invest. 2009;119(5):1093–1108. doi: 10.1172/JCI35079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witorsch R.J. Endocrine disruptors: can biological effects and environmental risks be predicted? Regul Toxicol Pharmacol. 2002;36(1):118–130. doi: 10.1006/rtph.2002.1564. [DOI] [PubMed] [Google Scholar]
- 11.Barkhem T., Carlsson B., Nilsson Y., Enmark E., Gustafsson J.A., Nilsson S. Differential response of estrogen receptor a and estrogen receptor b to partial estrogen agonists/antagonists. Mol Pharmacol. 1998;54(1):105–112. doi: 10.1124/mol.54.1.105. [DOI] [PubMed] [Google Scholar]
- 12.Clarke M. Meta-analyses of adjuvant therapies for women with early breast cancer: the Early Breast Cancer Trialists' Collaborative Group overview. Ann Oncol. 2006;17(Suppl. 10):59–62. doi: 10.1093/annonc/mdl238. [DOI] [PubMed] [Google Scholar]
- 13.Montanaro D., Maggiolini M., Recchia A.G., Sirianni R., Aquila S., Barzon L., Fallo F., Ando S., Pezzi V. Antiestrogens upregulate estrogen receptor beta expression and inhibit adrenocortical H295R cell proliferation. J Mol Endocrinol. 2005;35(2):245–256. doi: 10.1677/jme.1.01806. [DOI] [PubMed] [Google Scholar]
- 14.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 15.Kiefer F., Arnold K., Kunzli M., Bordoli L., Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37:387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijk M., Bonvin A.M. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2008;37:235–239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Boyle N., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macindoe G., Mavridis L., Venkatraman V., Devignes M.D., Ritchie D.W. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;24(51):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson S., Kuiper G., Gustafsson J.A. ER β: a novel estrogen receptor offers the potential for new drug development. Trend Endocrinol Metab. 1998;9(10):387–395. doi: 10.1016/s1043-2760(98)00096-4. [DOI] [PubMed] [Google Scholar]
- 21.Mueller S.O., Simon S., Chae K., Metzler M., Korach K.S. Phytoestrogens and their human metabolites show distinct agonistic and antagonistic properties on estrogen receptor α (ERα) and ERβ in human cells. Toxicol Sci. 2004;80:14–25. doi: 10.1093/toxsci/kfh147. [DOI] [PubMed] [Google Scholar]
- 22.Mukund V., Mukund D., Sharma V., Mannarapu M., Alam A. Genistein: its role in metabolic diseases and cancer. Crit Rev in Oncol Hematol. 2017;119:13–22. doi: 10.1016/j.critrevonc.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Notch E.G., Mayer G.D. Efficacy of pharmacological estrogen receptor antagonists in blocking activation of zebrafish estrogen receptors. Gen Comp Endorcinol. 2011;173:183–189. doi: 10.1016/j.ygcen.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Matthews J., Gustafsson J.A. Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol Interv. 2003;3(5):281–292. doi: 10.1124/mi.3.5.281. [DOI] [PubMed] [Google Scholar]
- 25.Nakopoulou L., Lazaris A.C., Panayotopoulou E.G., Giannopoulou I., Givalos N., Markaki S., Keramopoulos A. The favourable prognostic value of oestrogen receptor beta immunohistochemical expression in breast cancer. J Clin Pathol. 2004;57(5):523–528. doi: 10.1136/jcp.2003.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiura H., Toyama T., Hara Y., Zhang Z., Kobayashi S., Fujii Y., Iwase H., Yamashita H. Expression of estrogen receptor beta wild-type and its variant ERbetacx/beta2 is correlated with better prognosis in breast cancer. Jpn J Clin Oncol. 2007;37(11):820–828. doi: 10.1093/jjco/hym114. [DOI] [PubMed] [Google Scholar]
- 27.Honma N., Horii R., Iwase T., Saji S., Younes M., Ito Y., Akiyama F. Ki-67 evaluation at the hottest spot predicts clinical outcome of patients with hormone receptor-positive/HER2-negative breast cancer treated with adjuvant tamoxifen monotherapy. Breast Cancer. 2015;22(1):71–78. doi: 10.1007/s12282-013-0455-5. [DOI] [PubMed] [Google Scholar]
- 28.O'Neill P.A., Davies M.P., Shaaban A.M., Innes H., Torevell A., Sibson D.R., Foster C.S. Wild-type oestrogen receptor beta (ERbeta1) mRNA and protein expression in Tamoxifen-treated postmenopausal breast cancers. Br J Cancer. 2004;91(9):1694–1702. doi: 10.1038/sj.bjc.6602183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M.M., Albanese C., Anderson C.M., Hilty K., Webb P., Uht R.M., Price R.H., Pestell R.G., Kushner P.J. Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem. 2002;277(27):24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- 30.Margeat E., Bourdoncle A., Margueron R., Poujol N., Cavailles V., Royer C. Ligands differentially modulate the protein interaction of the human estrogen receptors alpha and beta. J Mol Biol. 2003;326(1):77–92. doi: 10.1016/s0022-2836(02)01355-4. [DOI] [PubMed] [Google Scholar]