Abstract
Regenerating the periodontal ligament (PDL) is a crucial factor for periodontal tissue regeneration in the presence of traumatized and periodontally damaged teeth. Various methods have been applied for periodontal regeneration, including tissue substitutes, bioactive materials, and synthetic scaffolds. However, all of these treatments have had limited success in structural and functional periodontal tissue regeneration. To achieve the goal of complete periodontal regeneration, many studies have evaluated the effectiveness of decellularized scaffolds fabricated via tissue engineering. The aim of this study was to fabricate a decellularized periodontal scaffold of human tooth slices and determine its regeneration potential. We evaluated two different protocols applied to tooth slices obtained from human healthy third molars. The extracellular matrix scaffold decellularized using sodium dodecyl sulfate and Triton X-100, which are effective in removing nuclear components, was demonstrated to preserve an intact structure and composition. Furthermore, the decellularized scaffold could support repopulation of PDL stem cells near the cementum and expressed cementum and periodontal-ligament-related genes. These results show that decellularized PDL scaffolds of human teeth are capable of inducing the proliferation and differentiation of mesenchymal stem cells, thus having regeneration potential for use in future periodontal regenerative tissue engineering.
Introduction
The regeneration of periodontal tissue is the aim of periodontal therapy to ensure the healing of traumatized teeth. Several kinds of stem cells such as dental pulp stem cells (DPSCs), stem cells from the apical papilla, dental follicle precursor cells, periodontal ligament stem cells (PDLSCs), and stem cells from human exfoliated deciduous teeth (SHED) have been evaluated for their usefulness in the regeneration of dental tissue[1–4]. Periodontal ligament (PDL) tissue contains multipotent stem cells that exhibit a self-renewing capacity to differentiate into various types of cells to form PDL, cementum, and alveolar bone [5, 6]. Stem cells derived from PDL cells have been shown to exhibit superior regenerative properties compared with other cells derived from gingival connective tissue and alveolar bone cells[7].
There have been many attempts to regenerate periodontium, such as using cell- and scaffold-based engineering, but no effective method for regenerating periodontal tissue has yet been reported [8–10]. The various attempts made to achieve PDL regeneration have had limitations, such as gene therapy possibly inducing (human) host responses and tumorigenesis, growth factors being unstable, and biomaterial being likely to fail.
The selection of an appropriate scaffold material is crucial in tissue engineering. The principal functions of the scaffold are to maintain mechanical integrity, supply growth factors, control cell growth, and induce cell differentiation. Commonly used scaffolds such as ceramics, synthetic polymers, and natural polymers have highly adjustable mechanical properties and good production repeatability, but they show low bioactivity [11]. Since extracellular matrix (ECM) scaffolds prepared by the decellularization of mammalian tissues show no immune responses and inherently contain the tissue-specific factors involved in cell growth and differentiation [12, 13], they have been used in studies of regenerative medicine and dentistry [14]. Clinical products derived from the decellularization of tissues are currently used for the replacement and reconstruction of organs and tissues, including human dermis, porcine urinary bladder, human pericardium, and porcine heart valves [15].
Various studies have attempted to apply scaffolds in the field of dentistry. Human dental pulp ECM scaffold that was successfully decellularized promotes the proliferation and differentiation of autologous mesenchymal stem cells [16]. The use of decellularized PDL cell-sheet constructs for periodontal regeneration has also been reported [17, 18]. Decellularized human PDL (dHPDL) cell sheets were shown to maintain the structural integrity of the ECM, retain growth factors, and support allogenic cell repopulation in vitro [17]. Decellularized PDL cell-sheet constructs that promote the differentiation of PDL and mesenchymal stem cells were also shown to support periodontal attachment in a rat periodontal defect model [18]. However, although the application of decellularized PDL tissue to cell sheets has been investigated, the use of human teeth including hard tissue has not been.
A decellularization protocol can be applied to the delayed replantation of an avulsed tooth, autologous transplantation, and periodontal therapy, via the decellularization of PDL tissue following recellularization with PDL stem cells (PDLSCs) derived from orthodontically extracted teeth, primary teeth, and supernumerary teeth. The aim of this study was to apply and evaluate a decellularization protocol that can retain structural integrity and remove cell and DNA effectively, and to test the periodontal recellularization potential of the decellularized human tooth model in vitro.
Materials and methods
Tooth-slice sample preparation and cell culture
Third molars that were free of caries and restorations were collected randomly from patients aged 17 to 25 years under approved guidelines set by the Institutional Review Board of the Dental Hospital, Yonsei University (approval no. #IRB 2-2016-0030). Teeth were washed with phosphate-buffered saline (PBS; Invitrogen, Carlsbad, CA, USA) and frozen at –80°C until use. After thawing at room temperature, the teeth were submerged in 0.5% chloramine-T (Sigma-Aldrich, St. Louis, MO, USA) for 2 hours at 4°C, followed by washing in cold running water. The pulp tissues were removed with a barbed broach, and tooth slices were prepared using an precision saw (IsoMet 1000, Buehler, Lake Buff, IL, USA) as described previously (Cordeiro et al. 2008). Samples were collected in cold PBS and immediately subjected to the decellularization procedures.
Previously characterized PDLSCs at passages 3–6 were used in repopulation experiments in a basal cell culture medium comprising alpha minimum essential medium (Invitrogen) containing 10% fetal bovine serum (Invitrogen), 1% L-glutamine/penicillin/streptomycin solution (Invitrogen), and 0.2% amphotericin B solution (Invitrogen) at 37°C in 5% CO2.
Decellularization and repopulation
Based on a review of the literature and a previous study[16], two decellularization protocols were evaluated in this study, as described in Table 1: Protocol I (PI) and Protocol II (PII). All treatment steps were applied to tooth slices at room temperature with constant gentle agitation of the samples in an orbital shaker (SH30, Fine PCR, Gunpo, Gyeonggi, Korea) in the presence of protease inhibitor cocktail (EMD, Millipore, Darmstadt, Germany). At the end of each protocol, samples were rinsed with 10% ethylenediaminetetraacetic acid (EDTA, Fisher Scientific, Houston, TX, USA) at pH 7.4 for 5 min, followed by three rinses of 10 min each with PBS (Invitrogen).
Table 1. The tested decellularization protocols.
| Protocol | Treatment |
|---|---|
| Protocol I | 2% Triton X-100 and 0.1% NH4OH for 72 hours |
| Protocol II | Three cycles of 1% SDS for 24 hours and 1% Triton X-100 for 24 hours |
SDS, sodium dodecyl sulfate.
For repopulation of the decellularized tooth slices (dHPDL), PDL at a density of 1×107 cells/mL in rat tail collagen I (Col I; Corning, Corning, NY, USA) was pipetted directly onto dHPDL using PII or into empty wells (control) in 12-well culture plates (Corning). After 1 hour, 1 ml of basal culture media was applied to each well, and this was changed every 3 days. Cells were cultured at 37°C in 5% CO2 for either 2 or 5 weeks.
Residual DNA quantification
Immediately after each decellularization protocol, samples (n = 10~15) were homogenized and residual DNA was isolated using the QIAamp DNA Investigator Kit (Qiagen, Valencia, CA, USA) and quantified using a spectrophotometer (NanoDrop NE-2000, Thermo Scientific, Waltham, MA, USA).
Scanning electron microscopy
Samples from the control, PI, and PII groups were incubated in fixative solution (2% paraformaldehyde [PFA], 2% glutaraldehyde, and 0.5% calcium chloride) and then postfixed in 1% osmium tetroxide. Next, the samples were dehydrated sequentially in a graded series of ethanol solutions, coated with platinum, and visualized using a scanning electron microscope (SEM; Hitachi S-3000N, Hitachi Science Systems, Tokyo, Japan) at an accelerating voltage of 20.0 kV.
Cell viability analysis
The survival of PDL seeded onto dHPDL from PII (n = 10) was compared after 2 weeks of culture. dHPDL without cell seeding served as a negative control. Samples were incubated with the Cell Counting Kit-8 assay (Dojindo Laboratories, Kumamoto, Japan) for 1 hour, and the quantity of water-soluble colored formazan formed by the activity of dehydrogenases in living cells was measured using a spectrophotometer (Benchmark Plus microplate spectrophotometer, Bio-Rad Laboratories, Hercules, CA, USA) at 450 nm. All samples were run in triplicate.
Histological and immunohistochemistry staining
For histological staining, tooth slices with intact control PDL, dHPDL derived from PI and PII, or repopulated decellularized PDL were fixed with 4% PFA for 1 h, decalcified with EDTA (pH 7.4; Fisher Scientific) for 6 weeks at room temperature, embedded in paraffin, sectioned at a thickness of 4 μm, and stained with hematoxylin and eosin (H&E).
The expression levels of Col I, collagen XII (Col XII), fibronectin, osteocalcin (OC), and cementum protein 23 (CP23) were evaluated by immunohistochemistry (IHC). Protease K (Dako, Carpinteria, CA, USA) was used to retrieve the antigen for the OC and CP23 staining, while no such treatment was performed for Col I, Col XII, and fibronectin. In brief, samples from the control group and dHPDL derived from PI and PII were immersed in 3% hydrogen peroxide for 10 min to inactivate endogenous peroxidase activity, and then incubated with the primary antibody overnight. The primary antibodies were a 1:10000 dilution of antihuman Col I rabbit monoclonal antibody (ab138492, Abcam, Cambridge, UK), a 1:1000 dilution of antihuman Col XII rabbit polyclonal antibody (sc-68862, Santa Cruz Biotechnology, Santa Cruz, CA, USA), a 1:4000 dilution of antihuman fibronectin rabbit polyclonal antibody (F3648, Sigma-Aldrich), a 1:8000 dilution of antihuman OC rabbit polyclonal antibody (#AB10911, Millipore, Temecula, CA, USA), and a 1:1000 dilution of antihuman CP23 goat polyclonal antibody (sc-164031, Santa Cruz Biotechnology). The sections were subsequently incubated for 20 min with horseradish-peroxidase-labeled polymer conjugated with the secondary rabbit antibody in the EnVision+ system kit (Dako) or for 30 min with the Vectastain Elite ABC Kit (PK-6105, Vector Laboratories, Burlingame, CA, USA; goat IgG, diluted 1:200). The color was developed using 3,3'-diaminobenzidine substrate (Dako) and counterstained with Gill’s hematoxylin solution (Merck, Darmstadt, Germany). Negative control sections were stained in the same manner but without the primary antibody reaction procedure.
Fluorescence staining
PDL specimens were fixed, permeabilized, and blocked following the Image-iT Fix-Perm kit (Life Technologies, Carlsbad, CA, USA) recommendations. NucBlue Fixed Cell ReadyProbes (Life Technologies) were used for staining the nuclei. Immunoreactivity was visualized with a confocal microscope (Zeiss LSM 700) combined with the Zen software (Carl Zeiss, New York, NY, USA).
Real-time reverse-transcription polymerase chain reaction assay
After 5 weeks of cell seeding and culture, total RNA was extracted from the repopulated tooth slices and from PDL cultured onto cultured dishes (control group) using the RNeasy Mini Kit (Qiagen) (n = 4 for the control group and n = 12 for each experimental group). RNA (100 ng) was reverse transcribed to synthesize cDNA using the Maxime RT premix kit [oligo d(T)15 primer; Intron Biotechnology, Seongnam, Gyeonggi, Korea] according to the manufacturer’s instructions. A quantitative real-time polymerase chain reaction (qPCR) assay was performed with SYBR Premix Ex Taq (Takara Bio, Otsu, Japan) and a real-time polymerase chain reaction (PCR) system (ABI 7300, Applied Biosystems, Carlsbad, CA, USA) as per the manufacturer’s instructions. The qPCR conditions were 95°C for 10 sec followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, with a final 5-min extension at 72°C. The sequence and size of the primers used are listed in Table 2. The expressions level of each gene was normalized to that of GAPDH (the gene encoding glyceraldehyde-3-phosphate dehydrogenase), and the relative expression levels of the studied genes were calculated using the 2–ΔΔCt method[19].
Table 2. Primers used for the real-time polymerase chain reaction assay.
| Gene | Primer sequence (5’–3’) | Size (bp) |
|---|---|---|
| ALP |
F: GGACCATTCCCACGTCTCAC R: CCTTGTAGCCAGGCCCATTG |
137 |
| Col XII |
F: CGGACAGAGCCTTACGTGCC R: CTGCCCGGGTCCGTGG |
180 |
| CP23 |
F: AACACATCGGCTGAGAACCTCAC R: GGATACCCACCTCTGCCTTGAC |
142 |
| OC |
F: CAAAGGTGCAGCCTTTGTGTC R: TCACAGTCCGGATTGAGCTCA |
150 |
| GAPDH |
F: TCCTGCACCACCAACTGCTT R: TGGCAGTGATGGCATGGAC |
100 |
F, forward; R, reverse; ALP, alkaline phosphatase; Col XII, collagen XII; CP23, cementum protein 23; OC, osteocalcin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Statistical analysis
All experiments were performed at least in triplicate. Statistical analysis was performed with SPSS (version 23.0, Chicago, IL, USA). The normality of the data was evaluated using the Shapiro-Wilk test, with a significance criterion of p<0.05. The Kruskal-Wallis test followed by the post-hoc Bonferroni test (Bonferroni correction; p<0.017) was used for comparing residual DNA contents, with a significance criterion of p<0.05. The Mann-Whitney U test was performed for cell viability after repopulation and real-time reverse-transcription PCR, also with a significance criterion of p<0.05.
Results
Evaluation of different decellularization protocols of human PDL
Residual nuclei and DNA contents after decellularizing PDL were evident compared to the control group not subjected to decellularization (Fig 1A–1C). Compared to the control group, nuclei and DNA were eliminated in the PI and PII groups, and there was a notable DNA reduction for PII of up to approximately 62.32% (Fig 1A and 1B). As seen in H&E and DAPI (4',6-diamidino-2-phenylindole) staining, disperse nuclei were evident for PI (Fig 1A-b and 1A-e), while almost no nuclei were observed for PII (Fig 1A-c and 1A-f). SEM images revealed that some Sharpey’s fibers had been removed, but they had retained their morphology in both the PI and PII groups compared to controls. The density of Sharpey’s fibers was preserved more for PII than for PI (Fig 1C-a and 1C-b).
Fig 1. Comparison of different decellularization protocols for periodontal tissue in human tooth slices.
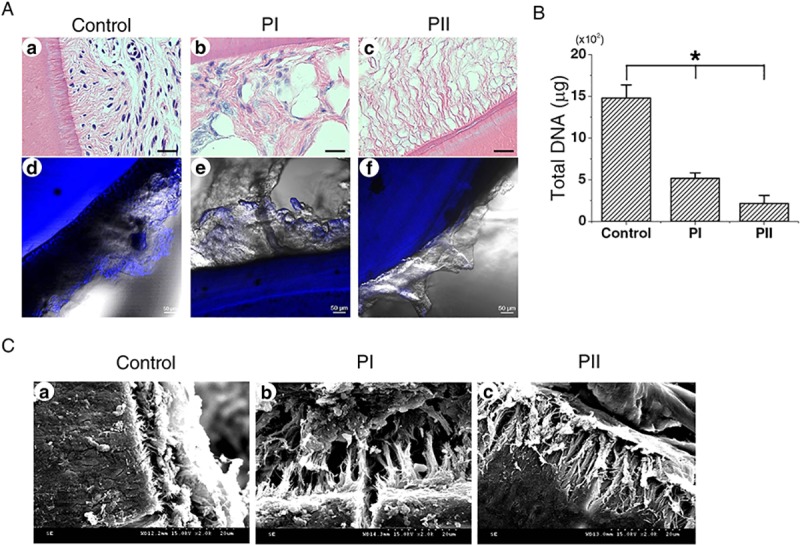
(A) Residual nuclei (arrowheads) are indicated by hematoxylin and eosin (H&E) staining (a–c) and fluorescence staining with 4',6-diamidino-2-phenylindole (DAPI) (d–f). More cell nuclei were removed in Protocol II (PII) than Protocol I (PI) after decellularization in H&E and DAPI staining. (B) Residual total DNA contents after decellularization of the periodontal ligament (PDL). The normality of the data was evaluated using the Shapiro-Wilk test (p<0.05). *p<0.05 in Kruskal-Wallis test followed by the post-hoc Bonferroni test (Bonferroni correction; p<0.017). (C) Scanning electron microscopy images of the different protocol groups. Arrowheads indicate remaining Sharpey’s fibers in the PDL. D, dentin. Scale bars: 20 μm in A(a–c), 50 μm in A(d–f), and 20 μm in C.
ECM characterization of decellularized PDL
IHC staining of samples for PI and PII indicated that Col I was almost intact compared with the control group (Fig 2A–2C). Fibronectin was notably diminished for PI and PII (Fig 2G–2I). Col XII was decreased for PI but increased for PII compared to controls (Fig 2D–2F).
Fig 2. Immunohistochemistry (IHC) staining of collagen I (Col I), collagen XII (Col XII), and fibronectin in the decellularized PDL of tooth slices.
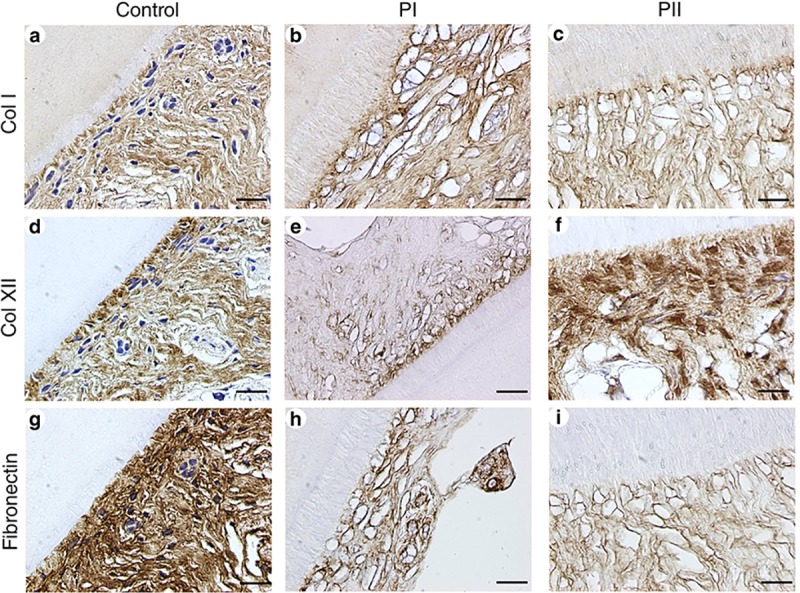
Scale bars: 20 μm in a–i.
Recellularization potential of human PDLSCs in decellularized PDL
PDLSCs were viable after 2 weeks of seeding PDLSCs onto decellularized scaffold for PII followed by culturing (Fig 3). PDL cells were repopulated on decellularized scaffold in H&E and Masson’s trichrome staining. Immunostaining of histological sections for OC showed positive staining in PDL cells adjacent to cementum, while significant CP23 staining was also found (Fig 3B).
Fig 3. Characterization of decellularized human PDL periodontal ligament (dHPDL) recellularized with PDL stem cells (PDLSCs).
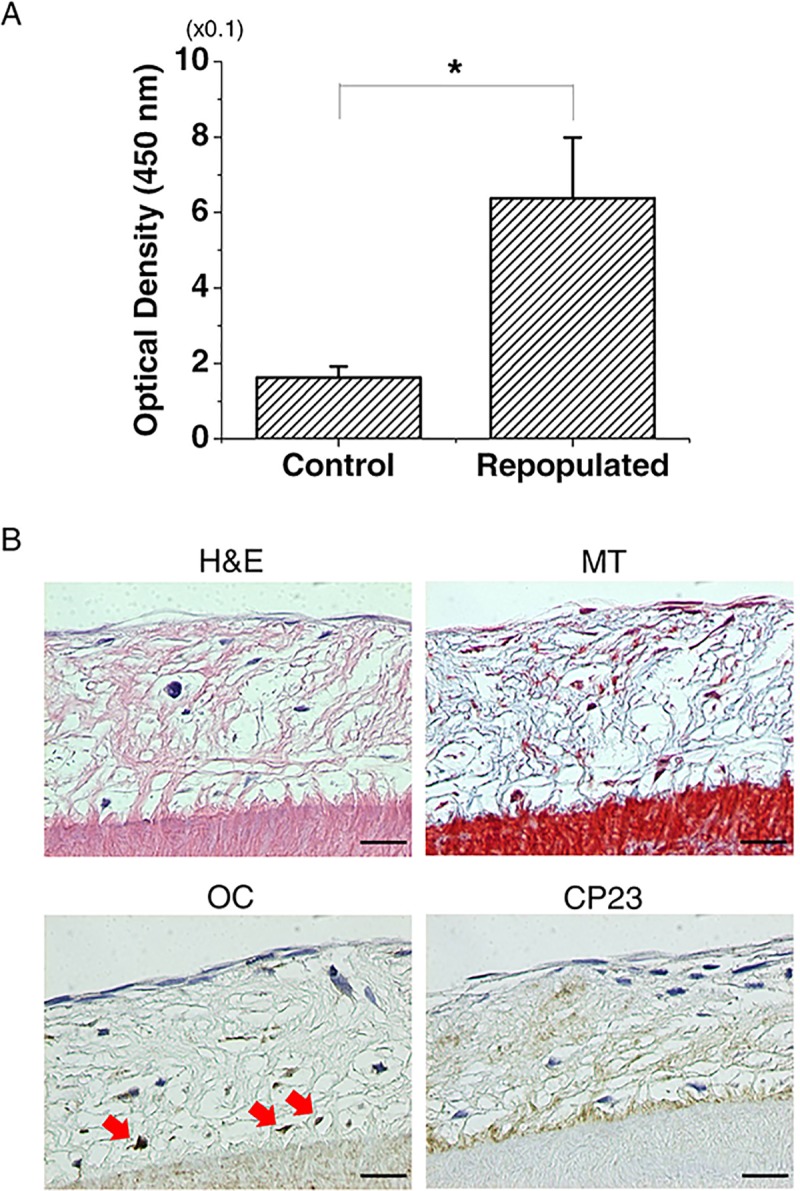
(A) Cell viability for PII after recellularization. PDLSCs repopulated onto dHPDL when applying PII were viable compared with the control group. The Mann-Whitney U test (*p<0.05) was used to assess cell viability. (B) PDLSCs seeded onto decellularized tooth slices in PII repopulated and infiltrated toward the cementum (black arrows) in H&E and Masson’s trichrome (MT) staining. PDLSCs expressed osteocalcin (OC) and cementum protein 23 (CP23) near the cementum (red arrows) in recellularized human tooth slices in IHC staining. C, cementum. Scale bars: 20 μm.
Differentiation of human PDLSCs in decellularized PDL
Real-time PCR was performed to assess the gene expression of cementum/PDL markers (Table 2) by PDLSCs repopulated on decellularized PDL scaffolds of the tooth slices. The expression levels of alkaline phosphatase (ALP), CP23, and OC were significantly up-regulated after recellularization, by 5.61±0.70-, 1.96±0.30-, and 275.87±201.47-fold (mean±SD), respectively. Col XII was also up-regulated (by 1.46±0.24-fold), but the change was not statistically significant (Fig 4).
Fig 4. Gene expression in the recellularized human PDL group analyzed using the quantitative real-time reverse-transcription polymerase chain reaction.
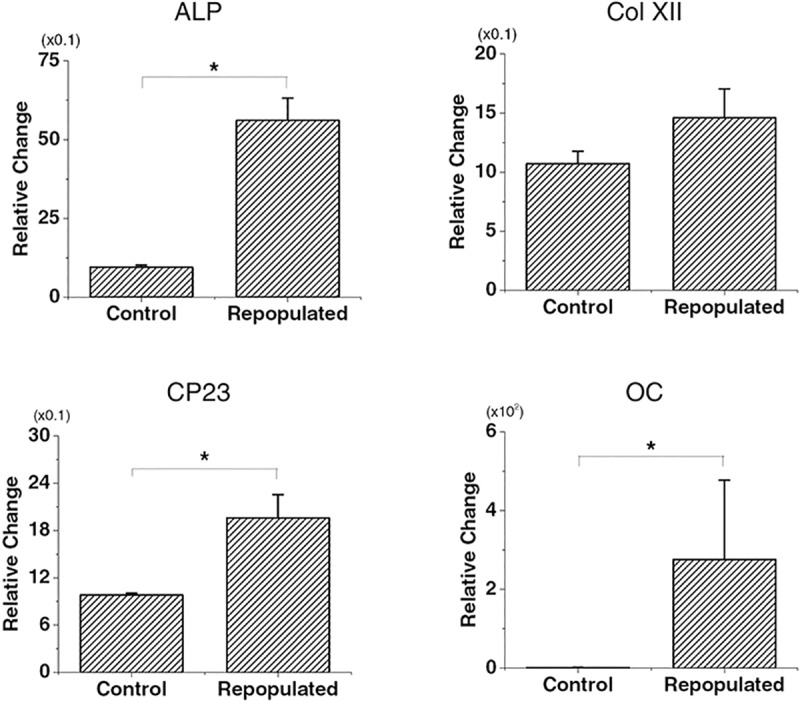
PDLSCs were seeded in the control group or in the decellularized group using PII. *p<0.05 for alkaline phosphatase (ALP) in Mann-Whitney U test; *p<0.05 for CP23 and OC in Student’s t-test.
Discussion
Decellularization protocols have been applied to various tissues and organs to fabricate bioactive ECM scaffold that could eliminate cells and cellular components completely without affecting the ECM [20–22]. Incompletely decellularized tissue might induce immune responses [23, 24], while a suitably tissue-engineered scaffold was found to not elicit immune responses in vivo [25]. The present study decellularized PDL in human tooth slices to assess the efficacy of nuclei and DNA removal while maintaining their native structure and composition. Furthermore, the recellularization potential of decellularized PDL scaffold was assessed by real-time PCR with the aid of periodontal tissue markers.
The successful application decellularization techniques might be dependent on many factors, including the types of tissue and organs, and the thickness, density, and cellularity of the tissue [26]. Previous studies found that using NH4OH and Triton X-100 could successfully remove cellular components [17, 26]. Decellularization with 1% Triton X-100 and 1% sodium dodecyl sulfate (SDS)—as used in PII in the present study—was effective in porcine anterior cruciate ligament and monkey kidney [27, 28]. Different methods using NH4OH, Triton X-100, hypertonic buffer, and SDS have recently been applied to human dental pulp tissue [16], but there had been no clinical investigation of decellularizing PDL obtained from human tooth slices. We applied a combination of NH4OH, Triton X-100, and SDS, which demonstrated effective decellularization compared to the other combinations of agents used in the previous studies.
In the present study, PII removed up to 62.32% of the DNA content, which was more than for PI. It seems that the SDS used in PII provided more complete removal of nuclear remnants than Triton X-100 [27, 28]. Ionic detergents such as SDS are more disruptive to tissue structure than is Triton X-100 [29]. However, the removal effectiveness was lower in the present study than in previous studies: at least 90% of host DNA was eliminated by SDS treatment in tissues and organs including rat forearm [30], porcine cornea [31], porcine heart valve [32], human vein [33], and human heart [34]. It appears that the specific characteristics of the tooth structure including its constituent hard tissue result in less elimination of hard tissue, and so additional agents for the complete removal of DNA (e.g., DNAase) need to be investigated in the future.
Sharpey’s fibers, which are the principal PDL fibers functioning in both tooth attachment and PDL regeneration, were shown to have a denser arrangement in PII. For evaluating the integrity of decellularized ECM, structural proteins such as collagen, fibronectin, and laminin as well as glycosaminoglycans should be evaluated [35]. Although fibronectin was notably decreased for PI and PII, Col I (which is the predominant type of collagen in PDL) was preserved in IHC staining, whereas Col XII was enhanced for PII. These findings could be attributed to teeth being involved in the mature/functional stage of PDL, as compared with developing stages, while the expression of Col I decreases with maturity [10, 36]. Furthermore, Col XII is responsible for the organization of collagenous fibers in response to mechanical loading in mature PDL [36]. The findings from both the present SEM and IHC analyses indicated that utilizing SDS and Triton X-100 preserved collagen fibrous network without any marked loss of structural proteins. This is consistent with [13] demonstrating the efficacy of 1% SDS and 1% Triton X-100 for the decellularization of heart-valve leaflets while preserving the integrity of the ECM scaffold.
Few studies have evaluated the influence of a decellularized scaffold on gene expression and cell differentiation. The gene expression markers selected in the present study are relevant to wound healing and the regeneration of bone, cementum, and PDL tissue (Table 2). The expression levels of CP23, ALP, and OC were higher than that of Col XII at 5 weeks after cell seeding. This could be explained by the induction of a later differentiation response of the repopulating PDL cells after the initial proliferative phase [18].
The human PDLSCs in this study were viable and able to be recellularized onto decellularized periodontal scaffold, as demonstrated by the cell viability analysis and H&E staining. OC expression was more prominent than CP23 expression in IHC staining. The expression of OC surrounding the cementum in the recellularized constructs suggests that decellularized scaffolds have the potential of inducing biomineralization and bone remodeling [7]. It is noteworthy that PDLSCs could be infiltrated and repopulated close to the cementum area from outside of the ECM.
Tooth avulsion accounts for 16% of all traumatic injuries in the permanent dentition, and it is accompanied by severe damage to the PDL [37]. PDL cells are damaged by inadequate storage of the avulsed teeth. Following the replantation of avulsed teeth, inflammatory root resorption and further tooth loss could occur. In this study, decellularized PDL scaffolds of tooth sections preserved their structure, and collagen remained in the ECM. They also have potential for recellularization and to induce the expression of genes related to PDL and bone regeneration. The application of autologous mesenchymal stem cells to the decellularized periodontal scaffold of avulsed tooth that potentially promotes the proliferation and differentiation of PDL cells could be a novel approach for PDL regeneration.
Data Availability
All relevant data are within the manuscript.
Funding Statement
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (https://ernd.nrf.re.kr/index.do) grant NRF-2018R1D1A1B07041657 to JSS and grant NRF-2018R1D1A1B07046163 to HJC. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13625–30. Epub 2000/11/23. 10.1073/pnas.240309797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao S, Pan F, Prpic V, Wise GE. Differentiation of stem cells in the dental follicle. Journal of dental research. 2008;87(8):767–71. Epub 2008/07/25. 10.1177/154405910808700801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet (London, England). 2004;364(9429):149–55. Epub 2004/07/13. 10.1016/s0140-6736(04)16627-0 . [DOI] [PubMed] [Google Scholar]
- 4.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5807–12. Epub 2003/04/30. 10.1073/pnas.0937635100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCulloch CA, Bordin S. Role of fibroblast subpopulations in periodontal physiology and pathology. J Periodontal Res. 1991;26(3 Pt 1):144–54. Epub 1991/05/01. . [DOI] [PubMed] [Google Scholar]
- 6.Isaka J, Ohazama A, Kobayashi M, Nagashima C, Takiguchi T, Kawasaki H, et al. Participation of periodontal ligament cells with regeneration of alveolar bone. J Periodontol. 2001;72(3):314–23. Epub 2001/05/01. 10.1902/jop.2001.72.3.314 . [DOI] [PubMed] [Google Scholar]
- 7.Dan H, Vaquette C, Fisher AG, Hamlet SM, Xiao Y, Hutmacher DW, et al. The influence of cellular source on periodontal regeneration using calcium phosphate coated polycaprolactone scaffold supported cell sheets. Biomaterials. 2014;35(1):113–22. 10.1016/j.biomaterials.2013.09.074 [DOI] [PubMed] [Google Scholar]
- 8.Bratthall G, Söderholm G, Neiderud AM, Kullendorff B, Edwardsson S, Attström R. Guided tissue regeneration in the treatment of human infrabony defects Clinical, radiographical and microbiological results: a pilot study. Journal of Clinical Periodontology. 1998;25(11):908–14. 10.1111/j.1600-051X.1998.tb02389.x [DOI] [PubMed] [Google Scholar]
- 9.Han J, Menicanin D, Gronthos S, Bartold PM. Stem cells, tissue engineering and periodontal regeneration. Australian Dental Journal. 2014;59(s1):117–30. 10.1111/adj.12100 [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Florit M, Monjo M, Ramis JM. Quercitrin for periodontal regeneration: effects on human gingival fibroblasts and mesenchymal stem cells. Sci Rep. 2015;5:16593 Epub 2015/11/13. 10.1038/srep16593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter SD, Costa PF, Vaquette C, Ivanovski S, Hutmacher DW, Malda J. Additive Biomanufacturing: An Advanced Approach for Periodontal Tissue Regeneration. Ann Biomed Eng. 2017;45(1):12–22. Epub 2016/07/31. 10.1007/s10439-016-1687-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quint C, Kondo Y, Manson RJ, Lawson JH, Dardik A, Niklason LE. Decellularized tissue-engineered blood vessel as an arterial conduit. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):9214–9. Epub 2011/05/17. 10.1073/pnas.1019506108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syedain ZH, Bradee AR, Kren S, Taylor DA, Tranquillo RT. Decellularized tissue-engineered heart valve leaflets with recellularization potential. Tissue engineering Part A. 2013;19(5–6):759–69. Epub 2012/10/24. 10.1089/ten.TEA.2012.0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials. 2006;27(19):3675–83. 10.1016/j.biomaterials.2006.02.014 [DOI] [PubMed] [Google Scholar]
- 15.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32(12):3233–43. Epub 2011/02/08. 10.1016/j.biomaterials.2011.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song JS, Takimoto K, Jeon M, Vadakekalam J, Ruparel NB, Diogenes A. Decellularized Human Dental Pulp as a Scaffold for Regenerative Endodontics. Journal of dental research. 2017;96(6):640–6. Epub 2017/02/15. 10.1177/0022034517693606 . [DOI] [PubMed] [Google Scholar]
- 17.Farag A, Farag A, Vaquette C, Theodoropoulos C, Hamlet SM. Decellularized Periodontal Ligament Cell Sheets with Recellularization Potential. Journal of dental research. 2014;93(12):1313–9. 10.1177/0022034514547762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farag A, Hashimi SM, Vaquette C, Bartold PM, Hutmacher DW, Ivanovski S. The effect of decellularized tissue engineered constructs on periodontal regeneration. J Clin Periodontol. 2018;45(5):586–96. Epub 2018/03/04. 10.1111/jcpe.12886 . [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 20.Caralt M, Uzarski JS, Iacob S, Obergfell KP, Berg N, Bijonowski BM, et al. Optimization and Critical Evaluation of Decellularization Strategies to Develop Renal Extracellular Matrix Scaffolds as Biological Templates for Organ Engineering and Transplantation. American Journal of Transplantation. 2015;15(1):64–75. 10.1111/ajt.12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chun SY, Oh SH, Yoo JJ, Kwon TG. Fabrication and characterization techniques for decellularized organ scaffolds. Tissue Engineering and Regenerative Medicine. 2015;12(1):1–10. 10.1007/s13770-014-0421-0 [DOI] [Google Scholar]
- 22.Badylak SF, Taylor D, Uygun K. Whole-Organ Tissue Engineering: Decellularization and Recellularization of Three-Dimensional Matrix Scaffolds. Annual Review of Biomedical Engineering. 2011;13(1):27–53. 10.1146/annurev-bioeng-071910-124743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasimir MT, Rieder E, Seebacher G, Nigisch A, Dekan B, Wolner E, et al. Decellularization does not eliminate thrombogenicity and inflammatory stimulation in tissue-engineered porcine heart valves. Journal of Heart Valve Disease. 2006;15(2):278–86. WOS:000236097000026. [PubMed] [Google Scholar]
- 24.Simon P, Kasimir MT, Seebacher G, Weigel G, Ullrich R, Salzer-Muhar U, et al. Early failure of the tissue engineered porcine heart valve SYNERGRAFT in pediatric patients. Eur J Cardiothorac Surg. 2003;23(6):1002–6; discussion 6. Epub 2003/06/28. 10.1016/s1010-7940(03)00094-0 . [DOI] [PubMed] [Google Scholar]
- 25.Bloch O, Golde P, Dohmen PM, Posner S, Konertz W, Erdbrugger W. Immune response in patients receiving a bioprosthetic heart valve: lack of response with decellularized valves. Tissue engineering Part A. 2011;17(19–20):2399–405. Epub 2011/05/12. 10.1089/ten.TEA.2011.0046 . [DOI] [PubMed] [Google Scholar]
- 26.Lu H, Hoshiba T, Kawazoe N, Chen G. Comparison of decellularization techniques for preparation of extracellular matrix scaffolds derived from three-dimensional cell culture. J Biomed Mater Res A. 2012;100(9):2507–16. Epub 2012/05/25. 10.1002/jbm.a.34150 . [DOI] [PubMed] [Google Scholar]
- 27.Nakayama KH, Batchelder CA, Lee CCI, Tarantal A. Decellularized Rhesus Monkey Kidney as a Three-Dimensional Scaffold for Renal Tissue Engineering. Tissue Engineering Part A. 2010:100216005013075 10.1089/ten.TEA.2009.0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods T, Gratzer PF. Effectiveness of three extraction techniques in the development of a decellularized bone–anterior cruciate ligament–bone graft. Biomaterials. 2005;26(35):7339–49. 10.1016/j.biomaterials.2005.05.066 [DOI] [PubMed] [Google Scholar]
- 29.Nair R, Ngangan AV, McDevitt TC. Efficacy of solvent extraction methods for acellularization of embryoid bodies. J Biomater Sci Polym Ed. 2008;19(6):801–19. Epub 2008/06/07. 10.1163/156856208784522056 . [DOI] [PubMed] [Google Scholar]
- 30.Jank BJ, Xiong L, Moser PT, Guyette JP, Ren X, Cetrulo CL, et al. Engineered composite tissue as a bioartificial limb graft. Biomaterials. 2015;61:246–56. Epub 2015/05/26. 10.1016/j.biomaterials.2015.04.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang K, Du L, Wu X. A rabbit anterior cornea replacement derived from acellular porcine cornea matrix, epithelial cells and keratocytes. Biomaterials. 2010;31(28):7257–65. Epub 2010/07/06. 10.1016/j.biomaterials.2010.05.066 . [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Fritze O, Schleicher M, Wendel HP, Schenke-Layland K, Harasztosi C, et al. Impact of heart valve decellularization on 3-D ultrastructure, immunogenicity and thrombogenicity. Biomaterials. 2010;31(9):2549–54. Epub 2010/01/12. 10.1016/j.biomaterials.2009.11.088 . [DOI] [PubMed] [Google Scholar]
- 33.Schaner PJ, Martin ND, Tulenko TN, Shapiro IM, Tarola NA, Leichter RF, et al. Decellularized vein as a potential scaffold for vascular tissue engineering. J Vasc Surg. 2004;40(1):146–53. Epub 2004/06/26. 10.1016/j.jvs.2004.03.033 . [DOI] [PubMed] [Google Scholar]
- 34.Guyette JP, Charest JM, Mills RW, Jank BJ, Moser PT, Gilpin SE, et al. Bioengineering Human Myocardium on Native Extracellular Matrix. Circ Res. 2016;118(1):56–72. Epub 2015/10/28. 10.1161/CIRCRESAHA.115.306874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilpin A, Yang Y. Decellularization Strategies for Regenerative Medicine: From Processing Techniques to Applications. Biomed Res Int. 2017;2017:9831534 Epub 2017/05/26. 10.1155/2017/9831534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaku M, Yamauchi M. Mechano-regulation of collagen biosynthesis in periodontal ligament. J Prosthodont Res. 2014;58(4):193–207. Epub 2014/10/15. 10.1016/j.jpor.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreasen JO. Etiology and pathogenesis of traumatic dental injuries. A clinical study of 1,298 cases. Scand J Dent Res. 1970;78(4):329–42. Epub 1970/01/01. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.


