Abstract
Spinocerebellar ataxia type 3 (SCA3) and type 1 (SCA1) are dominantly inherited neurodegenerative disorders that are currently incurable. Both diseases are caused by a CAG-repeat expansion in exon 10 of the Ataxin-3 and exon 8 of the Ataxin-1 gene, respectively, encoding an elongated polyglutamine tract that confers toxic properties to the resulting proteins. We have previously shown lowering of the pathogenic polyglutamine protein in Huntington’s disease mouse models using (CUG)7, a CAG repeat-targeting antisense oligonucleotide. Here we evaluated the therapeutic capacity of (CUG)7 for SCA3 and SCA1, in vitro in patient-derived cell lines and in vivo in representative mouse models. Repeated intracerebroventricular (CUG)7 administration resulted in a significant reduction of mutant Ataxin-3 and Ataxin-1 proteins throughout the brain of SCA3 and SCA1 mouse models, respectively. Furthermore, in both a SCA3 patient cell line and the MJD84.2 mouse model, (CUG)7 induced formation of a truncated Ataxin-3 protein species lacking the polyglutamine stretch, likely arising from (CUG)7-mediated exon 10 skipping. In contrast, skipping of exon 8 of Ataxin-1 did not significantly contribute to the Ataxin-1 protein reduction observed in (CUG)7-treated SCA1154Q/2Q mice. These findings support the therapeutic potential of a single CAG repeat-targeting AON for the treatment of multiple polyglutamine disorders.
Keywords: antisense oligonucleotide, SCA1, SCA3, polyglutamine disorders, exon skip, CAG repeat
Introduction
The spinocerebellar ataxias (SCAs) are a subgroup of hereditary ataxias, characterized by autosomal dominant mode of inheritance and progressive ataxia caused by degeneration of the cerebellum and often other connected brain regions. More than 30 types of SCAs have been identified to date (SCA1–SCA36), many of which are caused by point mutations, genomic rearrangements, or microsatellite repeat expansions in non-coding regions.1 The most common SCAs, however, are caused by translated CAG trinucleotide repeat expansions that encode elongated polyglutamine (polyQ) stretches in the respective disease proteins, including spinocerebellar ataxia type 1 (SCA1) and types 2, 3, 6, and 7.2 Presence of the elongated polyQ stretch confers pathogenic properties to the resulting protein, resulting in degeneration of specific neuronal subpopulations that differ between the different SCA types. Disease severity in patients with polyQ-associated SCAs seems to be greater compared with SCAs of different genetic cause, and survival is generally shorter.3 Worldwide, SCA3 is the most prevalent of all polyQ-associated SCAs, which collectively have an assumed prevalence of 3 cases per 100,000 people, followed by SCA1, SCA2, SCA6, and SCA7.4, 5
SCA3, also known as Machado-Joseph disease (MJD), is caused by a CAG repeat expansion in exon 10 of the Ataxin-3 (ATXN3 or MJD) gene, resulting in an abnormally long polyQ tract in the encoded protein Ataxin-3 (ATXN3). Affected individuals are usually heterozygous for the expansion and carry 52–86 CAG trinucleotide repeats in the expanded allele, whereas wild-type (WT) alleles have 12–44 CAG repeats.6 In SCA1, the expanded CAG repeat lies in exon 8 of the Ataxin-1 (ATXN1) gene, which encodes the polyQ-containing protein Ataxin-1 (ATXN1). Heterozygous SCA1 patients have 6–35 CAG repeats in the WT allele and 39–70 CAG repeats in the affected allele, often interrupted by CAT trinucleotides.7 In both diseases, age of onset and disease severity inversely correlate with the number of repeats.8
Disease manifestation is highly similar between SCA3 and SCA1, with both disorders presenting with ataxia of gait, stance, and limbs, dysarthria, and oculomotor abnormalities. Likewise, the neuropathology of both diseases overlaps to a large extent. Atrophy of the cerebellum is the pathological hallmark of both diseases, as well as associated atrophy of the brainstem and changes in the spinal cord.9 At the microscopic level, on the contrary, SCA3 and SCA1 exhibit loss of different cell types, reflecting divergent functions of the respective disease proteins. Whereas in the SCA1 brain there is an almost complete loss of cerebellar Purkinje neurons, these cells are relatively preserved in the brain of SCA3 patients, whereas selective degeneration of cerebellar dentate and fastigial nuclear cells occurs in the latter.9
To date, there are no disease-modifying therapies for SCA3 or SCA1. Because the polyQ proteins implicated in these diseases act through a dominant gain-of-function mechanism resulting in neurotoxicity, suppression of the mutant protein is an appealing and promising approach to slow or halt disease progression. Lowering levels of the mutant toxic protein is achievable by means of gene editing, RNAi, or the use of antisense oligonucleotides (AONs), as has already been demonstrated in the context of various SCAs through a number of studies (reviewed in Buijsen et al.10).
AONs are synthetic, single-stranded molecules, usually 13–30 nt long, that hybridize to a unique target RNA sequence, leading to RNA degradation or interference of RNA processing or translation.11 In contrast with gene editing and RNAi, AONs represent a non-viral gene suppression approach, and they are emerging as a promising means of treatment for neurodegenerative disorders.12 Recently, the first CNS-targeting AON was approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for the treatment of spinal muscular atrophy (SMA).13, 14 Many AONs have entered clinical trials for other neurodegenerative disorders, including the polyQ-associated disorder Huntington’s disease (HD; ClinicalTrials.gov identifier NCT: NCT02519036), where AON-mediated reduction of pathogenic polyQ protein levels was observed in the cerebrospinal fluid (CSF) of treated trial participants.15
Using a 2′-O-methyl-modified AON with a phosphorothioate backbone that specifically targets expanded CAG stretches and does not activate RNase H-dependent RNA degradation, we have previously demonstrated lowering of the mutant polyQ protein levels in patient cell lines and mouse models for HD.16, 17 In two different HD mouse models, the R6/2 N-terminal fragment model and the Q175 knock-in model, only six repeated weekly intracerebroventricular (ICV) administrations of (CUG)7 already reduced levels of the mutant Huntingtin (mutHTT) protein by approximately 15%–60% in three key brain regions affected in HD, i.e., striatum, cortex, and hippocampus. This level of mutHTT lowering resulted in correction of the motor phenotype in multiple motor tests, a brain volume increase, positive changes in striatal metabolite profile, and an increase of the striatal marker Darpp-32. The Q175 study further demonstrated that the mutHTT lowering was persistent and lasted for up to 18 weeks post-infusion, which suggests that a less frequent dose regimen is feasible.16 We hypothesized that in HD, (CUG)7 acts via binding to the extended CAG repeat in exon 1 of the mutHTT transcript close to the translation initiation site, thus sterically hindering translation initiation and/or elongation and resulting in lower mutHTT protein levels.16
The aim of this study was to investigate whether the same (CUG)7 AON could also display potential therapeutic benefit in SCA3 and SCA1. Initial in vitro results showed a significant reduction of mutant ATXN3 and ATXN1 proteins in SCA3 and SCA1 patient-derived fibroblasts, respectively, upon transfection with (CUG)7. In a next phase, in vivo studies were performed in the MJD84.2 mouse model for SCA3 and the SCA1154Q/2Q mouse model. Repeated ICV administration of (CUG)7 replicated the in vitro data, clearly demonstrating a significant reduction of levels of pathogenic polyQ proteins in multiple brain regions. These results underscore the therapeutic potential of (CUG)7 and support the development of a single CAG repeat-targeting AON for the treatment of multiple polyQ disorders, including HD, SCA3, and SCA1.
Results
(CUG)7 Induces Skipping of ATXN3 Exon 10 in Human SCA3 Cells
It has been previously demonstrated that targeting the expanded CAG repeat in ATXN3 pre-mRNA by means of single-stranded short interfering RNAs (ss-siRNAs), peptide nucleic acids (PNAs), or bridged nucleic acids (BNAs) induces skipping of exon 10.18 A similar finding was obtained with AONs targeting the regions immediately upstream or downstream of the ATXN3 CAG repeat.19 In order to assess whether our 2′-O-methyl phosphorothioate RNA AON (CUG)7 can also induce exon 10 skipping, SCA3 patient-derived fibroblast cells (GM06153) were transfected with increasing concentrations of (CUG)7, and 48 h post-transfection, total RNA was isolated. GM06153 cells are heterozygous for the mutant ATXN3 allele, carrying 23 repeats on the WT allele and 71 repeats on the mutant allele. Applying PCR primers spanning the CAG repeat (Figure 1A), we obtained a novel, shorter product of approximately 300 bp, which displayed a dose-dependent increase in intensity with increasing (CUG)7 concentration (Figure 1B). Sequence analysis confirmed exon 10 skipping and the precise splicing of exon 9 to exon 11 (Figure 1C). Despite the visible reduction of mutATXN3 levels (Figure 1B), no conclusions could be drawn on the origin of the skip product with regard to the WT or mutant transcript, or on the relative amount of the exon 10 skip product because of the inherent bias of the PCR method toward smaller products and its non-quantitative nature. In order to assess levels of the skipped ATXN3 transcript, we performed absolute quantification by digital droplet PCR (ddPCR) and calculated it to be approximately 10% of all ATXN3 transcripts amplified (mutATXN3 + wtATXN3 + skip product) (Figures 1A and 1D).
Figure 1.
(CUG)7 AON Induces Skipping of ATXN3 Exon 10 in Human SCA3 Fibroblasts
(A) Scheme of repeat region in ATXN3 mRNA and exon 10 skip mRNA product; location of PCR primers for skip assessment and ddPCR primers and probes (in red) for skip level quantification is indicated. (B) Agarose gel analysis of ATXN3 transcript of SCA3 fibroblasts treated with increasing AON concentrations (10-50-200 nM), using PCR primers flanking exon 10. (C) Results of Sanger sequencing of 301 bp amplicon showing skip of exon 10 and conjoining of exon 9 to exon 11. (D) Absolute quantification of ATXN3 exon 10 skip levels by ddPCR. Data are presented as mean ± SEM of three technical replicates. H2O, no template control; L, SmartLadder Small Fragment (Eurogentec); NT, non-treated; RT, reverse transcriptase negative control.
Reduction of ATXN3 Protein Levels in Human SCA3 Cells after (CUG)7 Treatment and Generation of Truncated Protein
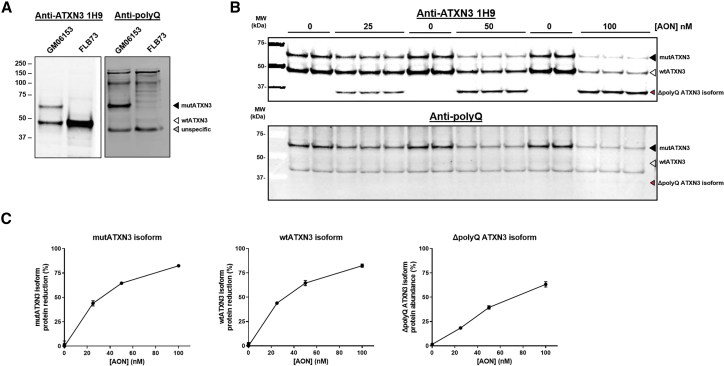
In a subsequent in vitro experiment, we tested to what extent (CUG)7 was able to reduce levels of the mutant ATXN3 protein. To enable assessment of ATXN3 protein levels, we validated antibodies for specific detection of the protein on western blot. Anti-ATXN3 antibody 1H9 was shown to detect WT ATXN3 (wtATXN3) in both the SCA3 (GM06153) and the healthy control cell line (FLB73), although it specifically recognized mutant ATXN3 (mutATXN3) in the SCA3 cell line only (Figure 2A, left panel). Although the 23Q-containing human ATXN3 is a 42-kDa protein, it migrated at a higher molecular weight (50 kDa), which probably reflects retardation caused by the polyQ motif. Similarly, mutATXN3 migrated at 65 kDa, as opposed to its theoretical molecular weight of 49 kDa. Stripping and reprobing the membrane with an antibody recognizing polyQ stretches (anti-polyQ) confirmed the presence of a polyQ repeat in the mutATXN3 band, whereas this antibody did not react with wtATXN3 (Figure 2A, right panel).
Figure 2.
(CUG)7 Reduces Levels of ATXN3 Protein and Results in Formation of the Truncated ΔpolyQ ATXN3 Isoform in Human SCA3 Fibroblasts
(A) Validation of anti-ATXN3 antibody 1H9 and anti-polyglutamine (polyQ) antibody for specific detection of mutATXN3 in GM06153 protein lysate (input = 10 μg). (B) Western blot analysis of protein samples derived from (CUG)7-treated GM06153 fibroblasts, using 1H9 or anti-polyQ antibody (input = 10 μg). (C) Quantification of western blot band intensity, plotted as mutATXN3 and wtATXN3 protein reduction percentage and ΔpolyQ ATXN3 isoform percentage (calculated as the level of ΔpolyQ ATXN3 isoform signal divided by the sum of wtATXN3, mutATXN3, and ΔpolyQ ATXN3 isoform signal). Revert Total Protein whole-lane signal was used for normalization of amount of loaded protein per sample. Data are presented as mean ± SD of three or six technical replicates.
SCA3 patient-derived fibroblast cells GM06153 were transfected with (CUG)7 at three increasing concentrations in three technical triplicates and harvested 96 h post-transfection for protein isolation. Western blot analysis showed a strong dose-dependent reduction of both wtATXN3 and mutATXN3, as well as the emergence of a truncated ATXN3 protein of around 34 kDa (Figure 2B). Stripping and reprobing the blot with the anti-polyQ antibody demonstrated that this shorter protein did not contain the toxic polyQ stretch and will hereafter be referred to as the ΔpolyQ ATXN3 isoform. Given the confirmed formation of the transcript lacking exon 10, it is highly likely that this truncated protein is derived from translation of that transcript; indeed, the skipping of exon 10 results in the formation of a premature stop codon at the start of exon 11 as previously described.19 Quantification of the western blot band intensity suggested a reduction of both mutATXN3 and wtATXN3 protein of over 80% at the highest AON concentration, relative to non-treated cells. The amount of the ΔpolyQ ATXN3 isoform was estimated to be approximately 60% of the total ATXN3 protein (wtATXN3 and mutATXN3 combined) (Figure 2C).
(CUG)7 Reduces Pathogenic mutATXN3 Protein Levels and Results in Generation of a Truncated Protein in SCA3 Mouse Brain
To obtain in vivo molecular proof-of-concept for (CUG)7-induced reduction of mutATXN3 protein levels, we used the SCA3 mouse model MJD84.2.20 MJD84.2 mice carry the full-length human ATXN3 gene with an expanded polyQ tract consisting of 84 CAG repeats on a yeast artificial chromosome (YAC), along with all the enhancers and long-range regulatory elements needed for cell-specific expression at physiological levels. We chose to use hemizygous MJD84.2 mice because of the better resemblance to the predominantly heterozygous SCA3 patient population, carrying the CAG repeat expansion on a single ATXN3 allele. Hemizygous MJD84.2 mice display a mild and late-onset motor phenotype, first detectable at 75 weeks of age.21
Mice at the age of 10–14 weeks (n = 14 per group) received weekly ICV infusions in the right hemisphere with 150 μg (CUG)7 or vehicle (artificial CSF [aCSF]) for 6 consecutive weeks and were sacrificed 1 week after last infusion (Figure 3A). Body weights were recorded on a weekly basis and just before sacrifice, and no significant differences in weight were observed between treatment groups (data not shown). Quantification of AON levels by sandwich hybridization ELISA revealed a widespread distribution of (CUG)7 throughout the brain of the MJD84.2 mice, with higher AON concentrations at brain regions closest to the site of injection, but limited lateralization between hemispheres (Figure 3B).
Figure 3.
(CUG)-Induced Reduction of mutATXN3 Levels in the MJD84.2 SCA3 Mouse Model
(A) Study design in the MJD84.2 mouse model, indicating treatment groups and sample size per treatment group. At first week of treatment, mice were 10–14 weeks old. (B) (CUG)7 distribution in right and left hemisphere regions of the MJD84.2 brain (n = 12–13 for right hemisphere groups and n = 5 for left hemisphere groups). Data are presented as mean ± SEM. The average difference in AON levels observed between left and right hemispheres of 19% was not significant, as assessed by two-tailed t test. (C) ddPCR analysis of exon 10 skipping levels in different brain regions. Data are presented as mean ± SEM. (D) Validation of anti-ATXN3 antibody in mouse brain, using 30 μg of cortex protein lysate as input (left panel), and representative example of western blot of some of the cerebellum samples of mice treated with VEH or (CUG)7, showing the emergence of the ΔpolyQ ATXN3 isoform after (CUG)7-treatment. (E) Mutant ATXN3 protein levels in various right hemisphere brain regions after treatment, as assessed by western blot analysis. Blot signals, normalized to Revert Total Protein whole-lane signal, are plotted on the right. Protein level data are presented as mean ± SEM. Percentages above bars indicate reduction in protein levels, compared with VEH set to 100%. Significance was assessed using a two-tailed t test comparing (CUG)7-treated mice with VEH-treated controls (*p < 0.05, **p < 0.01, ***p < 0.001).
Similar to the in vitro experiment in the SCA3 cells, the exon 10 skip product was detected in the brain of (CUG)7-treated MJD84.2 mice using ddPCR, and ranged between 0.8% in cortex and 2.2% in brainstem (Figure 3C). This was thus considerably lower than the 10% observed in the SCA3 cells.
To enable assessment of ATXN3 protein levels in mouse brain lysates, the anti-ATXN3 antibody 1H9 was validated and showed to specifically detect mutATXN3 in the MJD84.2 cortex (Figure 3D). Similar to observations in the patient cell line, mutATXN3 migrated at a higher molecular weight than expected (65 kDa, as opposed to the theoretical weight of 51.6 kDa), which may have been caused by retardation due to the repeat motif. The epitope of 1H9 is mapped at amino acids E214–L233 of human ATXN3, but the antibody also reacts with the mouse ATXN3, as well as with products truncated at the C terminus.
Analysis of ATXN3 protein levels in (CUG)7-treated MJD84.2 mice showed a significant reduction of mutATXN3 levels ranging from 20% to 30% in cerebellum, brainstem, and hippocampus 1 week after last infusion, but no significant reduction in cortex (Figure 3E). In addition to reduction of pathogenic mutATXN3 levels, a shorter ATXN3 protein of ∼34 kDa was detected in brain tissue of (CUG)7-treated mice. This truncated product was also observed, albeit to a much lesser extent, in vehicle-treated mice, indicating that it may be a naturally occurring isoform, possibly because of spontaneous skip of ATXN3 exon 10. To confirm that this shorter protein indeed was the ΔpolyQ ATXN3 isoform, one of the generated western blots was stripped and reprobed with anti-polyQ antibody. As expected, this failed to detect the shorter 34-kDa protein and detected only the larger mutATXN3 containing the stretch of 84 glutamines (Figure S1). Quantification of the ΔpolyQ ATXN3 isoform signal demonstrated levels of up to 20% of total human ATXN3 protein, which was slightly lower (4%–11% depending on the brain region) than the observed reduction of the mutant protein. No effect of (CUG)7 was observed on mouse ATXN3 levels; the short repeat stretch (five CAGs) in the mouse Atxn3 is an unlikely target for (CUG)7. Therefore, in the MJD84.2 mouse brain, the ΔpolyQ ATXN3 isoform arises predominantly from skipping exon 10 of the mutant Atxn3 transcript.
(CUG)7 Induces Low Levels of ATXN1 Exon 8 Skip in Patient Fibroblasts
We also assessed the effect of (CUG)7 in SCA1 fibroblasts and the SCA1 mouse model. SCA1 patient-derived fibroblasts GM06927 were transfected with increasing concentrations of (CUG)7, and total RNA was isolated 96 h after transfection. Heterozygous GM06927 cells carry 29 CAG repeats on the WT ATXN1 allele and 52 repeats on the mutant allele. Primers were designed to span the CAG repeat exon (Figure 4A), and a PCR product of approximately 190 bp was detected in both non-treated and (CUG)7-treated samples, corresponding in size to an exon 8 skip fragment (Figure 4B). Sequencing indeed confirmed the specific skipping of exon 8 and the precise splicing of exon 7 to exon 9 (Figure 4C). The presence of this skip product in non-treated samples indicates that it is a naturally occurring transcript and likely the product of alternative splicing. Absolute quantification of exon 8 skip levels by ddPCR analysis demonstrated very low copies in both (CUG)7-treated and non-treated samples, with a maximum skip percentage of 0.68% in samples treated with 100 nM AON (Figure 4D). ATXN1 protein levels were not assessed, because mutATXN1 protein cannot be detected in this cell line, possibly because of too low expression (data not shown).
Figure 4.
Very Low Levels of ATXN1 Exon 8 Skipping Observed in Non-treated and (CUG)7-Treated Human SCA1 Fibroblasts
(A) Scheme of repeat region on ATXN1 mRNA and exon 8 skip mRNA product; location of PCR primers for exon 8 skip detection on gel and of ddPCR primers and probes for quantification of skipping levels are indicated. (B) Agarose gel analysis of PCR products of ATXN1, using RNA extracted from SCA1 fibroblasts treated with 100 or 200 nM concentrations. (C) Results of Sanger sequencing of 183 bp amplicon showing skip of exon 8 and conjoining of exon 7 to exon 9. Non-coding strand, sequenced with reverse primer, is shown. (D) Absolute quantification of ATXN1 exon 8 skip levels by ddPCR. Data are presented as mean ± SEM of three technical replicates. One-way ANOVA followed by Dunnett’s multiple comparison post hoc test (*p < 0.05, **p < 0.01, compared with 0 nM AON). H2O, no template control; L, SmartLadder (Eurogentec); NT, non-treated, RT, reverse transcriptase negative control.
Reduction of Pathogenic ATXN1 Protein Levels in SCA1 Mouse Brain after (CUG)7 Treatment
For in vivo analysis of the (CUG)7 effect in a SCA1 mouse, SCA1154Q/2Q mice were used.22 SCA1154Q/2Q mice are heterozygous knock-in mice carrying 154 CAG repeats in the targeted mouse Atxn1 allele and only two repeats in the WT non-targeted mouse allele. The mice develop a progressive neurological phenotype that resembles human SCA1 and exhibit Purkinje cell loss, as well as age-related hippocampal synaptic dysfunction. We administered 75 or 150 μg of (CUG)7 or aCSF (vehicle [VEH]) by ICV infusion into the right lateral ventricle of 7-week-old mice, once weekly for a total of 8 weeks (Figure 5A). Mice were sacrificed 2 weeks after last infusion, and brain and spinal cord tissues were collected for pharmacokinetics (PK) and protein analysis. Body and brain weights were recorded during the study and at endpoint. No significant differences in weight were observed between treatment groups (data not shown). Quantification of AON levels by sandwich hybridization ELISA in several brain regions and spinal cord lysates revealed a widespread distribution of (CUG)7 with concentrations increasing with dose and no significant differences in AON levels between different brain regions (Figure 5B). Interestingly, AON levels in the spinal cord were similar to those in the brain despite the distance from the injection site.
Figure 5.
(CUG)7-Induced Reduction of mutATXN1 Levels in the SCA1154Q/2Q Mouse Model
(A) Study design in the SCA1154Q/2Q mouse model, indicating treatment groups and sample size per treatment group. (B) (CUG)7 distribution in right hemisphere brain regions and spinal cord (SC) sections of the SCA1 mouse (n = 10–11). Data are presented as mean ± SEM. No significant differences in AON levels observed between different regions of mice treated with the same dosing regimen (one-way ANOVA and Tukey’s multiple comparison post hoc test). Unpaired two-tailed t test revealed significant differences between the low and high (CUG)7 dose in brainstem (**p < 0.01), cerebellum (*p < 0.05), and lumbar spinal cord (**p < 0.01). (C) Validation of anti-ATXN1 antibody in vivo, using 90 μg of cerebellum protein lysate as input. Blot was initially probed with anti-polyQ antibody 1C2, then stripped and reprobed with an anti-ATXN1 antibody (Origene) to confirm band identity. Secondary antibodies that fluoresce in different wavelengths were used in order to avoid false interpretations due to leftover signal. (D) ATXN1 protein levels in various right hemisphere brain regions and spinal cord sections after treatment, as assessed by western blot analysis. Protein level data of each mouse are plotted normalized to Revert Total Protein whole-lane signal and presented as mean ± SEM. Percentages above bars indicate reduction in protein levels, compared with VEH set to 100%. Significance was assessed using one-way ANOVA followed by Dunnett’s multiple comparison post hoc test (*p < 0.05, **p < 0.01, ***p < 0.001, compared with VEH). H, high-dose AON, 150 μg; L, low-dose AON, 75 μg; VEH, vehicle.
Similar to observations in the SCA1 patient cells, the exon 8 skip product was detectable in the brain of (CUG)7-treated SCA1154Q/2Q mice using ddPCR, but only at very low levels (data not shown).
To enable western blot analysis of mutATXN1 expression in the SCA1154Q/2Q mouse brain, we tested various anti-ATXN1 antibodies on brain lysates, but failed to detect mutATXN1, although wtATXN1 was usually detectable (Figure S2). Only the generic anti-polyQ antibody 1C2 resulted in specific detection of mutATXN1 protein in SCA1154Q/2Q cerebellum lysate (Figure 5C). This antibody had previously been shown to detect mutATXN1 in various SCA1 models, including SCA1154Q/2Q.22, 23 The molecular weight of mutATXN1 protein was higher than expected (120 kDa as opposed to the theoretical molecular weight of 103 kDa). A potential explanation for this is altered protein mobility due to the expanded repeat motif, as has been observed before for mutant human ATXN1 and ATXN3.23, 24 We confirmed the identity of the detected band by stripping the membrane and reprobing with an anti-ATXN1 antibody (Origene; Figure S2), which detected a band at the exact same height as the 1C2 antibody (Figure 5C). However, because the ATXN1-specific antibody required a low dilution in combination with a high protein input in order to detect mutATXN1, the 1C2 antibody was used for protein level analysis in other brain regions.
In (CUG)7-treated SCA1154Q/2Q mice, a significant reduction of mutATXN1 levels was observed in all brain regions relevant to SCA1 pathology (i.e., cerebellum, brainstem, spinal cord) with both (CUG)7 doses, as well as in the hippocampus, striatum, and thalamus (Figure 5D). The reduction showed a clear dose-response in most brain and spinal cord regions, with the higher (CUG)7 dose yielding the strongest mutATXN1 reduction, reaching >50% in brainstem and in all three spinal cord regions and >40% in the hippocampus, striatum, and olfactory bulb (Figure 5D). In cortex, a trend toward reduction was observed, but statistical significance was not reached with both (CUG)7 doses (Figure 5D). WT mouse ATXN1 (wtATXN1) protein levels were not assessed, because the protein cannot be detected by the anti-polyQ antibody 1C2. However, we would not expect wtATXN1 levels to be affected by (CUG)7 treatment, because the murine wtAtxn1 allele carries only two CAG repeats and should theoretically not bind to the AON.
Discussion
CAG trinucleotide repeat expansions, which translate into an abnormally long polyQ stretch in the encoded proteins, are the root cause of various neurodegenerative disorders, collectively called polyQ diseases. Because the toxic polyQ proteins are proximal targets in the pathogenesis of these disorders, reducing their levels by RNA modulation is currently considered a promising therapeutic approach. These polyQ protein-lowering approaches make use of nucleic acids in the form of AONs or virally delivered RNAi molecules that target and bind to specific sequences in the (pre-)mRNA encoding the polyQ protein and trigger RNase H-mediated RNA degradation or disrupt RNA processing or translation, thus hindering pathogenic protein expression. To date, the clinical development of RNA modulating polyQ-lowering therapies is by far most advanced for HD, with an upcoming phase 3 clinical trial for the HTT-lowering AON IONIS-HTTRx (RG6042) and phase 1 clinical trials for several other HTT-lowering compounds in preparation.25 Among the HTT-lowering RNA therapeutics currently in development, different classes can be distinguished that target different regions within the HTT mRNA, ranging from areas far downstream of the repeat to specific SNPs to the CAG repeat itself (reviewed in Tabrizi et al.25). Because the CAG-repeat expansion is the shared disease-causing feature in polyQ disorders, targeting the CAG repeat with a single AON holds promise as a potential therapy for multiple of these neurodegenerative diseases, including HD, the SCAs caused by a CAG-repeat expansion (SCA types 1, 2, 3, 6, and 7), dentatorubral-pallidoluysian atrophy (DRPLA), and spinal bulbar muscular atrophy (SBMA).17, 26, 27, 28, 29, 30
We have previously demonstrated the therapeutic potential of a (CUG)7 2′-O-methyl phosphorothioate RNA AON, targeting the CAG repeat, for HD, hypothesized to act via steric hindrance of translation initiation and thus interference with protein translation.16 Repeated ICV administration of (CUG)7 resulted in a widespread and long-lasting (>18 weeks) reduction of mutHTT protein throughout the brain of the R6/2 and Q175 mouse models of HD.16 Here we set out to investigate whether we could demonstrate the applicability of (CUG)7 to SCA3 and SCA1, the most prevalent polyQ expansion disorders after HD.31 We successfully demonstrated that (CUG)7 was able to lower levels of the mutATXN3 and ATXN1 proteins carrying the elongated polyQ stretch in patient cell lines and mouse models for these diseases.
With its non-gapmer chemical structure, (CUG)7 is not a substrate for RNase H-mediated cleavage. Its proposed mechanism of action (MoA) is steric hindrance of translation of the polyQ-protein.16 Interestingly, we identified exon skipping as an additional and/or alternative MoA of (CUG)7, leading to lower levels of mutATXN3 in a SCA3 patient cell line and in the MJD84.2 mouse brain. In hindsight, perhaps this is not so unexpected, given that (CUG)7 has the same chemistry to splice-switching AONs used to induce exon skipping in Duchenne muscular dystrophy by sterically hindering binding of splicing factors to exonic splice enhancers.32 (CUG)7-induced skipping of exon 10, which contains the CAG-repeat expansion, and conjoining of exon 9 to exon 11 result in formation of a premature stop codon at the beginning of exon 11, and it has been previously documented that the resulting mRNA encodes a truncated ATXN3 protein consisting of 291 amino acids with a predicted mass of 34 kDa lacking the polyQ stretch and the C terminus, the ΔpolyQ ATXN3 isoform.19 We indeed detected a smaller ATXN3 protein of the expected molecular weight that was not recognized by an anti-polyQ antibody and therefore highly likely represents the translated ΔpolyQ ATXN3 isoform.
Skipping of ATXN3 exon 10 has been observed before with repeat-targeting ss-siRNAs, PNAs, and BNAs, or AONs targeting other sequences in ATXN3 exon 10, especially toward its 5′ end.18, 19 Although the mechanism behind these observations is not clear, various hypotheses have been suggested. It has been shown that expanded CAG repeat motifs bind the SRSF6 splicing factor, which regulates splicing.33 (CUG)7 binding may dislocate SRSF6 from the expanded repeat and therefore interfere with normal splicing of exon 10. It has also been proposed that CAG repeat tracts can function as exonic splicing enhancers (ESEs).34 In addition, ESEs located within 70 nt of the acceptor splice site have been reported to be more active than ESEs farther downstream.35 Furthermore, previous studies in Duchenne muscular dystrophy have shown that effective AONs were located significantly closer to splice acceptor sites.36 Taken together, these data suggest that the CAG repeat stretch in ATXN3, which is located only 13 nt downstream of the 5′ splice acceptor of exon 10, may constitute a potent ESE, whose “masking” by (CUG)7 AON leads to skipping of the exon.
ATXN3 functions mainly as a deubiquitinating enzyme and contains three ubiquitin-interacting motifs (UIMs). The third UIM is encoded by a sequence present in exon 11, which raises concerns regarding the functionality of the ΔpolyQ ATXN3 isoform. However, it has been shown that the interaction between ATXN3 and poly-ubiquitin chains is dependent on the first two UIMs, whereas the exon 11-encoded UIM appears to be dispensable.37, 38 Furthermore, the ΔpolyQ ATXN3 isoform has been shown to still bind and cleave ubiquitin chains, similar to WT protein.19 Besides loss of function due to lack of a UIM region, it is also critical that the ΔpolyQ ATXN3 isoform does not have a toxic gain of function. Despite widespread presence of the ΔpolyQ ATXN3 isoform in the brain of (CUG)7-treated MJD84.2 mice, repeated ICV injections with (CUG)7 were well tolerated. No toxicity of (CUG)7 treatment was apparent, judging by the fact that no adverse effects were noticed during daily inspection of the mice, nor was there any effect of treatment on body weight development or survival. Moreover, the ΔpolyQ ATXN3 isoform was also detected in VEH-treated mice, albeit to a lesser extent (Figure 3E, lower panel). The fact that this protein isoform occurs naturally further supports the notion that the protein is not a toxic variant. Similar conclusions were drawn in a previous study where this protein species was generated because of AON-mediated exon 10 skip.19
It is important to note that reduced levels of the pathogenic mutATXN3 protein still remain a more relevant molecular readout than formation of the ΔpolyQ ATXN3 isoform per se, and it therefore is essential to above all quantify levels of the disease-causing mutant protein. For example, Toonen et al.19 observed high levels of the ΔpolyQ ATXN3 isoform without any clear reduction of the full-length toxic protein. In addition, caution should be taken to not overinterpret RNA levels of transcripts either containing or lacking exon 10, because differences in translation efficacy and/or protein stability can give rise to different protein levels of the full-length and ΔpolyQ ATXN3 proteins. Indeed, although only very low levels (<2.5%) of the exon 10 skip RNA transcript were observed in the MJD84.2 brain after (CUG)7 treatment, ΔpolyQ ATXN3 isoform levels were only slightly lower than that of the full-length WT protein, suggesting that the ATXN3 RNA transcript lacking exon 10 and the CAG-repeat expansion may be more efficiently translated.
The extent of ATXN3 protein reduction observed in vivo upon (CUG)7 treatment was between 4% and 11% higher than the levels of the ΔpolyQ ATXN3 isoform in the cerebellum, brainstem, and hippocampus, suggesting that a secondary MoA may additionally play a role to reduce pathogenic ATXN3 protein levels. Similar to in HD, it is conceivable that binding of (CUG)7 to the repeat may also sterically hinder the translation machinery, likely via hampering translation elongation. Translation elongation arrest has previously been observed using PNAs, artificially synthesized polymers that resemble DNA and RNA and bind to nucleic acids.39 We therefore suggest that in SCA3, (CUG)7 has two complementary MoAs: induction of exon 10 skipping of the pre-mRNA molecule in the nucleus, resulting in a truncated ATXN3 protein lacking the toxic repeat, and inhibition of translation elongation of the full-length polyQ-containing protein in the cytoplasm, together leading to lower levels of the toxic protein (Figure 6A). These two MoAs not only occur in different cellular compartments but may also have different temporal kinetics. Indeed, in the cortex, levels of the ΔpolyQ ATXN3 isoform were comparable with in the cerebellum, brainstem, and hippocampus, whereas no reduction in mutATXN3 levels was observed (Figure 3E). A potential explanation for this could be that in the cortex the mutant protein has an overall higher stability or is more slowly degraded because of a different protein stoichiometry in cortex. Notably, we also observed a slower response in the reduction of mutant HTT protein levels in the cortex of Q175 HD mice compared with other brain regions.16
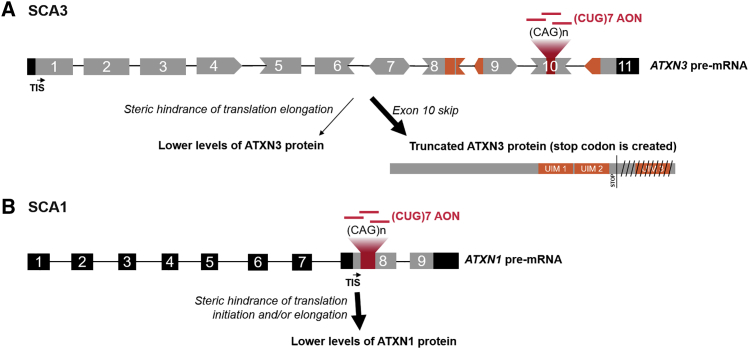
Figure 6.
Proposed Mechanism of Action of (CUG)7 in SCA3 and SCA1
(A) (CUG)7 AONs bind to the (expanded) CAG repeat on exon 10 of the ATXN3 pre-mRNA. During pre-mRNA processing this can result in incorrect splicing of exon 10, leading to exclusion of exon 10 in mature mRNA and conjoining of exon 9 to exon 11. This junction results in generation of a stop codon in the beginning of exon 11, and therefore ATXN3 mRNA is translated into a truncated product that lacks regions encoded by exons 10 (i.e., polyQ stretch) and 11 (i.e., UIM 3). In addition, it is proposed that binding of (CUG)7 to the repeat on mature mRNA sterically hinders translation elongation, thereby conferring lower levels of ATXN3 protein. (B) (CUG)7 binds to the expanded CAG repeat on exon 8 of the ATXN1 (pre-)mRNA, causing steric hindrance of translation initiation and/or elongation, thereby lowering levels of ATXN1 protein. 5′ and 3′ UTRs and introns are in black; coding regions are in gray; exonic regions encoding UIMs and UIMs on protein are in orange. TIS, translation initiation site.
A limitation of the current study is that, although we obtained clear molecular proof of concept (PoC), we did not assess any neuropathological hallmarks, such as nuclear accumulation of ATXN3 or neuronal loss in the MJD84.2 brain. The mice we used in the current study were relatively young at sacrifice (16–20 weeks old) at an age when there is no quantifiable loss of neurons of the deep cerebellar nuclei or of Purkinje cells, or any motor phenotype in hemizygous MJD84.2 mice.21 In order to assess whether the level of mutATXN3 reduction obtained by (CUG)7 treatment is sufficient to prevent neuronal loss and improve motor performance, a long-term study would need to be performed following the mice until they are at least 75 weeks, the age when a mild motor phenotype first becomes detectable.21 However, the levels of exon 10 skipping we observed are in a range that has previously been shown to resolve nuclear accumulation of ATXN3. Cerebellar levels of the ΔpolyQ ATXN3 isoform of 20% have previously been shown sufficient to reduce insoluble ATXN3 levels in the cerebellum and nuclear accumulation in the substantia nigra in this same mouse model.19
After obtaining promising results for (CUG)7 in MJD84.2 mice, we continued with assessing (CUG)7-mediated suppression of mutant protein in the SCA1 context, using the mouse model SCA1154Q/2Q. We demonstrated a significant reduction of mutATXN1 protein levels throughout the SCA1154Q/2Q brain and spinal cord after treatment with (CUG)7 using western blot analysis. In the cerebellum and brainstem, brain regions with the most neuronal loss in SCA1, the high dose of (CUG)7 reduced mutATXN1 levels by 45% and 56%, respectively.
Detection of mutATXN1 protein in the brain of SCA1154Q/2Q mice using western blot analysis proved to be challenging. The protein fraction generally assessed by western blot analysis corresponds to cellular proteins that are soluble in the buffers applied during lysis and isolation. It was previously shown that SCA1154Q/2Q show an age-dependent decrease in the detectability of soluble mutATXN1, with levels being barely detectable in whole-brain extracts from the age of 9 weeks onward.22 At the same time, a clear increase of aggregated mutATXN1 was observed, caused by progressive, age-dependent protein accumulation into nuclear aggregates, rendering the mutant protein less extractable.22 Our results are consistent with the findings by Watase et al.,22 with only 2 out of 14 tested antibodies able to detect mutATXN1 levels in the brain and spinal cord of SCA1154Q/2Q mice. We were not able to detect the aggregated form of mutATXN1 (data not shown), which likely requires a different protein extraction procedure. The mutATXN1 reduction we observed corresponds to the soluble fraction and may not be indicative of overall reduction or correlate with aggregated protein levels. However, the soluble form of the protein is most likely most relevant for SCA1 pathology. Watase et al.22 observed higher soluble mutATXN1 levels in neurons most vulnerable to degeneration, a finding consistent with earlier studies on SCA1 mice showing that mutATXN1 toxicity was much greater when it was not sequestered in nuclear inclusions.40 Although still a matter of debate, there are indications that nuclear inclusion may not be pathogenic per se, but instead their formation may constitute a protective mechanism against the toxicity of mutATXN141 and other expanded polyQ proteins (reviewed in Woulfe42).
In contrast with SCA3, in SCA1, skipping of exon 8 containing the CAG repeat does not seem to be a relevant MoA to reduce levels of the toxic polyQ protein. Because the repeat-containing exon of ATXN1 harbors the translation initiation site (TIS), skipping of the exon upon (CUG)7 treatment would result in reduction of protein levels because of loss of the initiation codon without formation of a truncated protein. However, because very low (<1%) exon skip levels were observed in SCA1 fibroblasts, it is unlikely that the mechanism behind this mutant protein reduction after (CUG)7 treatment is exclusion of the repeat-containing exon. Because the expanded CAG repeat in exon 8 of ATXN1 is located over 700 nt downstream of the splice acceptor site, it may comprise a much weaker ESE site, incapable of altering splicing upon AON binding. We previously reported that in HD the potential huntingtin-lowering MoA of (CUG)7 is steric hindrance of translation initiation,16 given that the CAG-repeat expansion is located in close proximity (51 nt upstream) of the TIS in exon 1 of huntingtin. However, in ATXN1, although located in the same exon, the TIS is located farther away (588 nt) from the CAG repeat, and thus unlikely to be able to interfere with translation initiation. We therefore hypothesize that in the context of SCA1, (CUG)7 leads to reduced pathogenic ATXN1 protein levels through steric hindrance of translation elongation rather than of translation initiation (Figure 6B).
Because the focus of this study was to obtain molecular PoC for a wider application of the (CUG)7 AON to SCA3 and SCA1, we did not perform histological or functional assessments. Recently another study was published on RNA modulation in the SCA1154Q/2Q model using an AON gapmer that operates via RNase H-mediated degradation of the ATXN1 RNA transcript.43 In this study they showed a significant reduction of ATXN1 RNA in different brain regions and mitigation of motor deficits combined with a prolonged survival, demonstrating the therapeutic value of RNA modulation in ATXN1.43 In this study they did not assess levels of the pathogenic mutATXN1 protein, most likely because of the difficulties described above in detecting ATXN1 protein. Therefore, it is not possible to conclude from this study how much ATXN1 protein reduction was required to obtain the improvement of motor performance and prolonged survival. Previous studies using genetic approaches in this same mouse model that indirectly resulted in lowering of ATXN1 levels have indicated that a reduction of ATXN1 protein levels around 20% already resulted in detectable improvement in motor performance and cerebellar pathologies.44, 45 Given that we observed a much higher ATXN1 reduction in the cerebellum of 46% and even higher reductions above 50% in the brainstem and spinal cord, the molecular effects obtained in our study are well above the range where improvements of pathology and motor performance are to be expected. This would need to be confirmed in a follow-up study.
A concern of CAG-targeting AON approaches is the potential for off-target effects on other non-expanded CAG repeat-containing genes. We and others have previously shown that CAG-targeting AONs display relative allelic preference for expanded repeat stretches with effects on shorter non-expanded repeats occurring with much lower efficacy.16, 27, 28 Using the same (CUG)7 AON in the Q175 HD mouse model, we found no evidence of off-target lowering of levels of a panel of five mouse genes with non-expanded CAG repeats ranging from 5 to over 20 CAGs, including Atxn3 and Atxn7.16 The reduction of off-targets by CAG-targeting AONs is therefore expected to be limited. Moreover, the harmfulness of lowering levels of non-expanded CAG proteins has not been demonstrated. A human mutation leading to a 50% reduction of HTT levels was not associated with any pathology.25 Consistent with this, a recent publication on the phase 1/2a IONIS-HTTRx clinical trial results reported the absence of any serious adverse events, despite that both WT and mutant HTT protein are equally targeted in this non-allele selective, RNase H-mediated therapeutic approach.25 This also appears to be the case for other CAG proteins, such as ATXN3, the complete functional knockout of which was tolerated in mice without affecting fertility or lifespan.46 Taking this together, we consider the risk associated with a potential mild reduction of non-expanded CAG proteins acceptable given the clear benefit of reducing levels of pathogenic CAG proteins with expanded repeats and the obvious advantages of being able to use a single AON approach for multiple diseases caused by CAG expansion.
The use of AONs with different MoAs for treatment of monogenic neurodegenerative disorders holds great promise for future therapies, as demonstrated by the groundbreaking results obtained with the recently approved splice-switching AON nusinersen for treatment of SMA, which alters splicing of the SMN2 pre-mRNA.47, 48 AONs are particularly suitable for application in the CNS, where they rapidly distribute throughout the brain via the CSF and have a half-life of several months, exerting long-lasting effects and requiring infrequent dosing.25, 48 CNS-administered AONs appear to be well tolerated because they are mostly retained in the CNS compartment, and therefore the well-known safety issues related to peripheral accumulation of AONs are absent.
In conclusion, we have demonstrated the therapeutic potential of the repeat-targeting AON (CUG)7 in HD16 and now also in SCA3 and SCA1, supporting the broader applicability of a single AON to multiple polyQ disorders. This holds obvious advantages with regard to development costs and speed. Considering the highly unmet medical need for these diseases, such an approach would clearly be advantageous in a clinical development process.
Materials and Methods
CAG Repeat-Targeting AON
The CAG repeat-targeting AON used in all experiments was a 2′-O-methyl phosphorothioate (2OmePS)-modified oligoribonucleotide (21-mer) with the sequence 5′-C*UGC*UGC*UGC*UGC*UGC*UGC*UG-3′ [abbreviated as (CUG)7] in which all cytosine residues were 5-methyl-C modified (C*, 5-methylcytosine; U, uracil; G, guanine). (CUG)7 was developed to selectively target a pure CAG repeat sequence. AONs were synthesized by BioMarin Nederland (Leiden, the Netherlands).
In Vivo Studies in Mouse Models of SCA3 and SCA1
For both mouse studies, (CUG)7 AON was formulated in aCSF (Harvard Apparatus, MA, USA).
Experimental Design of In Vivo Study in MJD84.2 Mice
MJD84.2 mice were obtained from the Jackson Laboratory, stock number 012705 (Bar Harbor, ME, USA) and were bred and genotyped at the Leiden University Medical Center (LUMC, the Netherlands). Animal experiments were approved by Leiden University Animal Ethical Committee and performed according to the European Communities Council Directive 2010/63/EU. Mice were singly housed after surgery in individually ventilated cages with a 12-h light-dark cycle and with food and water available ad libitum. Mixed gender hemizygous MJD84.2 mice were divided over two experimental groups: (1) VEH (aCSF), n = 14; and (2) 150 μg (CUG)7, n = 14. At 10–14 weeks of age, mice were implanted unilaterally with a cannula into the right lateral ventricle to allow for ICV infusion of AON or VEH (aCSF). Infusions (5 μL per infusion at a rate 0.3 μL/min) were started directly after cannula placement during the same operation procedure and continued once weekly for a total of 6 weeks until the mice were 16–20 weeks old. Prior to each ICV administration, body weight was monitored. One week after the last ICV infusion, mice were sacrificed and their brain dissected into the following regions: cortex, hippocampus, striatum, thalamus, cerebellum, brainstem, and olfactory bulb (left and right hemispheres were collected separately). Brain regions, liver, and kidney were snap frozen (in isopentane bain-marie in liquid nitrogen) and stored at −80°C.
Experimental Design of In Vivo Study in SCA1154Q/2Q Mice
Mixed gender SCA1154Q/2Q mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Animal experiments were carried out at Charles River Laboratories (Finland) according to the Guidelines for the Care and Use of Laboratory Animals (NIH) and were approved by the National Animal Experiment Board of Finland. A total of 30 SCA1154Q/2Q mice were divided into three experimental groups as follows: (1) VEH (aCSF), n = 10; (2) 75 μg (CUG)7, n = 10; and (3) 150 μg (CUG)7, n = 10.
At 7 weeks of age mice were implanted unilaterally with a cannula into the right lateral ventricle to allow for ICV infusion of AON or VEH (aCSF). Infusions (5 μL per infusion at a rate 0.3 μL/min) were started directly after cannula placement during the same operation procedure and continued once weekly for a total of 8 weeks until the mice were 14 weeks old. Prior to each ICV administration, body weight was monitored. Two weeks after the last ICV infusion, mice were sacrificed and their brain dissected into the following regions: cortex, hippocampus, striatum, thalamus, cerebellum, brainstem, and olfactory bulb (left and right hemispheres were collected separately). Brain regions, liver, and kidney were snap frozen and stored at −80°C. Finally, the spinal cord was excised from the spinal column and sectioned into three pieces: lumbar, thoracic, and cervical portions. Spinal cord samples were weighed and snap frozen in liquid nitrogen.
Cell Culture and Transfections
Patient-derived SCA1 (GM06927) and SCA3 (GM06153) fibroblasts (Coriell Cell Repositories, Camden, NJ, USA) and healthy control fibroblasts FLB7317 were cultured at 37°C, 5% CO2, in Minimal Essential Medium (MEM) (GIBCO, Invitrogen, Carlsbad, CA, USA), supplemented with 15% non-inactivated fetal bovine serum (FBS) (GIBCO, Invitrogen, Carlsbad, CA, USA), 1% GlutaMAX (GIBCO, Invitrogen, Carlsbad, CA, USA), and 100 U/mL penicillin-streptomycin (P/S) (GIBCO, Invitrogen, Carlsbad, CA, USA).
Fibroblasts were transfected with (CUG)7 using polyethylenimine (PEI; 3.3 μL/μg AON) or Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). AON and PEI were diluted in 150 mM NaCl to a total volume of 100 μL per well of a six-well plate, and mixtures were incubated for 15 min at room temperature. Transfection was performed in 900 μL total volume, in MEM supplemented with 5% FBS, 1% P/S, and 1% GlutaMAX, for 4 h at 37°C, 5% CO2. After this incubation time, transfection medium was added to a total volume of 2 mL, and the next day the medium was changed to proliferation medium. Transfections with RNAiMAX were performed according to the manufacturer’s instructions, except a different ratio of RNAiMAX to AON was used (2 μL RNAiMAX was added for every 125 pmol AON). RNA and protein were isolated 48 or 96 h post-transfection.
RNA Isolation, PCR, and ddPCR
At 48 or 96 h post-transfection, fibroblasts were harvested by trypsinization, washed, and total RNA was isolated using either Aurum Total RNA Mini-Kit (Bio-Rad Laboratories, Hercules, CA, USA) or NucleoZOL (Macherey-Nagel, Düren, Germany), according to the manufacturers’ protocols. RNA was reverse transcribed using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) or GoScript Reverse Transcriptase (Promega, WI, USA) and random hexamers.
PCR amplification of Ataxin-3 fragments was performed using 1 μL cDNA and Q5 High Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA), according to the manufacturer’s protocol. PCR was performed with primers hATXN3_ex8_Fw and hATXN3_ex11_Rv (for sequences, see Table S1). The PCR cycling program started with 30-s initial denaturation at 98°C, followed by 34 cycles of 7-s denaturation at 98°C, 15-s annealing at 64°C, and 15-s elongation at 72°C, after which a final elongation step was performed at 72°C for 2 min. PCR amplification of Ataxin-1 fragments was performed using 1 μL cDNA, 0.8 M Betaine per reaction, and Expand High Fidelity PCR System (Roche, Basel, Switzerland), according to the manufacturer’s protocol. PCR was performed with primers hATXN1_ex7_Fw and hATXN1_ex9_Rv (for sequences, see Table S1). The PCR cycling program started with 4 min initial denaturation at 95°C, followed by 34 cycles of 30-s denaturation at 94°C, 30-s annealing at 55°C, and 80-s elongation at 72°C (elongation time was extended 5 s in each cycle after cycle 10), after which a final elongation step was performed at 72°C for 7 min.
ddPCRs for quantification of ATXN3 exon 10 skip and ATXN1 exon 8 skip were performed as previously described49 using the primer sets or primer-probe sets for non-skipped and for exon-skipped ATXN3 or ATXN1 mRNA detailed in Table S1. Samples were loaded onto the QX200 droplet reader, and ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad Laboratories, Hercules, CA, USA). The percentage of exon skip was expressed as the total exon skip transcript copies to the percentage of total (skipped + non-skipped) transcript copies.
Protein Isolation and Western Blotting
Protein was isolated from fibroblasts 96 h after transfection and from mouse tissue using Cellytic MT Cell Lysis Reagent (Sigma-Aldrich, St. Louis, MO, USA), supplemented with Complete Protease Inhibitor Cocktail (one tablet per 10 mL; Roche, Basel, Switzerland) or radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher, Waltham, MA, USA), supplemented with 1× Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, Waltham, MA, USA). A different lysis buffer was used for better protein solubility and stability. Mouse tissue was homogenized in MagNA Lyzer (Roche, Basel, Switzerland). Protein concentrations were measured using Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA).
For western blot analysis, protein extracts were denatured and loaded on 10% acrylamide-bis midi-gels for ATXN3 detection, whereas for ATXN1 detection, proteins were separated by SDS-PAGE on 5% acrylamide-bis midi-gels. Proteins were transferred to Trans-Blot Turbo Midi Nitrocellulose Transfer Pack membranes (Bio-Rad Laboratories, Hercules, CA, USA), using the Trans-Blot Turbo Transfer System (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were stained with Revert Total Protein Staining (LI-COR Biosciences, Lincoln, NE, USA) according to the manufacturer’s protocol, scanned, and destained. Blocking was performed with 5% non-fat milk Elk (Campina, Zaltbommel, the Netherlands) in 1× TBST (0.1% Tween 20). Membranes were incubated overnight at 4°C with primary antibodies in blocking buffer and 3 h at room temperature with corresponding IRDye800CW or IRDye680LT secondary antibodies (LI-COR Biosciences, Lincoln, NE, USA). Fluorescent signal was detected in Odyssey CLx (LI-COR Biosciences, Lincoln, NE, USA). For membrane stripping, the NewBlot Nitrocellulose 5× Stripping Buffer (LI-COR Biosciences, Lincoln, NE, USA) was used according to the manufacturer’s protocol. For western blot signal quantification, Image Studio Lite version 5.2 software (LI-COR Biosciences, Lincoln, NE, USA) was used. Total Protein signal, as obtained by Revert Total Protein staining, was used as loading control for normalization of data.
Primary antibodies used were anti-ATXN3 1H9 (1:3,300; MAB5360; Millipore, Burlington, MA, USA), anti-polyQ (1:3,300; P1874; Sigma-Aldrich, St. Louis, MO, USA), anti-polyQ 1C2 (1:2,000; MAB1574; Millipore, Burlington, MA, USA), and anti-ATXN1 (1:500; TA349697; Origene, Rockville, MD, USA).
Sample Preparation for PK Analysis and ELISA
A sandwich hybridization ELISA method was used to quantify concentrations of (CUG)7 in tissue samples. The capture probe was a 12-mer DNA probe with two LNA modifications and a conjugated biotin label at the 3′ end. The detection probe was a 9-mer DNA probe with two LNA modifications and a conjugated digoxigenin (DIG) label at the 5′ end. Together, the probes comprised a sequence fully complementary to the analyte. The assay is specific for the parent compound: products of 3′ exonuclease degradation are not detected.
Samples were weighed and homogenized in 100 mM Tris-HCl (pH 8.5), 200 mM NaCl, 0.2% SDS, 5 mM EDTA, and 2 mg/mL Proteinase K using MagNa Lyzer green bead tubes in a MagNa Lyzer (Roche Diagnostics, the Netherlands), then diluted 1:60 in PBS. Brain tissue from VEH-treated mice was similarly homogenized and diluted to derive blank matrix.
For each 96-well plate run, eight calibrators (range 2.00 to 0.0156 nM) and three quality control (QC) samples (1, 0.25, and 0.05 nM) were prepared in duplicate by spiking the blank matrix with (CUG)7 stock solution, then serial dilution in the blank matrix. Study sample homogenates were diluted in the blank matrix to concentrations within the calibration curve range. With a minimum total dilution of 1,000× (16.7× in Proteinase K buffer, then a minimum of 60× in blank matrix), the tissue sample lower limit of quantification (LLOQ) was 0.0156 nmol/g (or 0.212 μg/g).
The capture probe solution (25 nM) was added to streptavidin-coated 96-well plates and incubated at 37°C for 30 min, followed by a wash step to remove the unbound probe. Calibrators, QCs, diluted tissue samples, and two blanks (blank matrix) were added and incubated at 37°C for 30 min, followed by a wash step to remove any unhybridized AON. Detection probe solution (2 nM) was added and incubated at 37°C for 30 min, followed by a wash step to remove unhybridized detection probe. To detect the DIG-label, anti-DIG peroxidase (POD; 1:5,000 in 1% milk powder blocking buffer) was added and incubated for 30 min in the dark at room temperature, followed by a wash step to remove any unbound anti-DIG POD. Following addition of 3,3′,5,5′-tetramethylbenzidine (TMB) and incubation for a maximum of 60 min, the reaction was stopped by addition of maleic acid (345 mM). Absorption was directly measured at 450 nm.
Each sample was analyzed in two dilutions (factor of 10 difference in dilution) in duplicate, allowing a maximum of 18 study samples per 96-well plate. A second order polynomial was fitted to the calibrator data and used to estimate tissue sample concentrations from observed optical densities.
Statistical Analysis
All values are presented as mean ± SEM, unless stated otherwise, and differences were considered to be statistically significant at the p < 0.05 level. Statistical analyses of all data were performed using t test or one-way ANOVA (when appropriate) followed by post hoc testing (GraphPad Prism Version 7.03), unless stated otherwise.
Author Contributions
Conceptualization, J.C.T.v.D. and N.A.D.; Formal Analysis, E.K., R.W., A.G.B., S.B., and N.A.D.; Investigation and Methodology, E.K., R.W., A.G.B., M.M., R.V., J.P., and J.T.; Resources, B.G.; Project Administration, N.A.D.; Supervision, N.A.D.; Validation, E.K., R.W., A.G.B., M.M., R.V., J.P., and J.T.; Visualization, E.K., A.G.B., and N.A.D.; Writing – Original Draft Preparation, E.K. and N.A.D.; Writing – Review & Editing, E.K., A.G.B., J.C.T.v.D., and N.A.D.
Conflicts of Interest
E.K., R.W., A.G.-B., M.M., R.V., S.B., B.G., J.C.T.v.D., and N.A.D. are (former) employees of BioMarin Nederland BV (formerly Prosensa Therapeutics BV) and performed the work with the company budget in the form of salaries, equipment, and facilities. J.P. and J.T. are employees of Charles River Research Discovery Services in Finland and have no financial conflict of interest related to the submitted manuscript.
Acknowledgments
We thank Dr. W. van Roon-Mom and Maurice Overzier (LUMC) for assistance with the SCA3 in vivo study. Funding was provided by BioMarin.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.004.
Supplemental Information
References
- 1.Bird T.D. Hereditary Ataxia Overview. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., editors. GeneReviews. University of Washington,; Seattle: 1993. pp. 1993–2019. [PubMed] [Google Scholar]
- 2.Paulson H.L., Shakkottai V.G., Clark H.B., Orr H.T. Polyglutamine spinocerebellar ataxias—from genes to potential treatments. Nat. Rev. Neurosci. 2017;18:613–626. doi: 10.1038/nrn.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monin M.L., Tezenas du Montcel S., Marelli C., Cazeneuve C., Charles P., Tallaksen C., Forlani S., Stevanin G., Brice A., Durr A. Survival and severity in dominant cerebellar ataxias. Ann. Clin. Transl. Neurol. 2015;2:202–207. doi: 10.1002/acn3.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krysa W., Sulek A., Rakowicz M., Szirkowiec W., Zaremba J. High relative frequency of SCA1 in Poland reflecting a potential founder effect. Neurol. Sci. 2016;37:1319–1325. doi: 10.1007/s10072-016-2594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Warrenburg B.P., Sinke R.J., Verschuuren-Bemelmans C.C., Scheffer H., Brunt E.R., Ippel P.F., Maat-Kievit J.A., Dooijes D., Notermans N.C., Lindhout D. Spinocerebellar ataxias in the Netherlands: prevalence and age at onset variance analysis. Neurology. 2002;58:702–708. doi: 10.1212/wnl.58.5.702. [DOI] [PubMed] [Google Scholar]
- 6.Bettencourt C., Santos C., Montiel R., Kay T., Vasconcelos J., Maciel P., Lima M. The (CAG)n tract of Machado-Joseph Disease gene (ATXN3): a comparison between DNA and mRNA in patients and controls. Eur. J. Hum. Genet. 2010;18:621–623. doi: 10.1038/ejhg.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menon R.P., Nethisinghe S., Faggiano S., Vannocci T., Rezaei H., Pemble S., Sweeney M.G., Wood N.W., Davis M.B., Pastore A., Giunti P. The role of interruptions in polyQ in the pathology of SCA1. PLoS Genet. 2013;9:e1003648. doi: 10.1371/journal.pgen.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 9.Fratkin J.D., Vig P.J. Neuropathology of degenerative ataxias. Handb. Clin. Neurol. 2012;103:111–125. doi: 10.1016/B978-0-444-51892-7.00005-X. [DOI] [PubMed] [Google Scholar]
- 10.Buijsen R.A.M., Toonen L.J.A., Gardiner S.L., van Roon-Mom W.M.C. Genetics, Mechanisms, and Therapeutic Progress in Polyglutamine Spinocerebellar Ataxias. Neurotherapeutics. 2019;16:263–286. doi: 10.1007/s13311-018-00696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 12.Wurster C.D., Ludolph A.C. Antisense oligonucleotides in neurological disorders. Ther. Adv. Neurol. Disorder. 2018;11 doi: 10.1177/1756286418776932. 1756286418776932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.US Food and Drug Administration. (2016). FDA News Release: FDA approves first drug for spinal muscular atrophy, December 23, 2016. https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-spinal-muscular-atrophy.
- 15.Ionis Pharmaceuticals, Inc. (2018). Press Release: new data from IONIS-HTT Rx phase 1/2 study demonstrates correlation between reduction of disease-causing protein and improvement in clinical measures of Huntington’s disease, April 24, 2018. https://ir.ionispharma.com/news-releases/news-release-details/new-data-ionis-htt-rx-phase-12-study-demonstrates-correlation.
- 16.Datson N.A., González-Barriga A., Kourkouta E., Weij R., van de Giessen J., Mulders S., Kontkanen O., Heikkinen T., Lehtimäki K., van Deutekom J.C. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS ONE. 2017;12:e0171127. doi: 10.1371/journal.pone.0171127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evers M.M., Pepers B.A., van Deutekom J.C., Mulders S.A., den Dunnen J.T., Aartsma-Rus A., van Ommen G.J., van Roon-Mom W.M. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS ONE. 2011;6:e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Yu D., Aiba Y., Pendergraff H., Swayze E.E., Lima W.F., Hu J., Prakash T.P., Corey D.R. ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res. 2013;41:9570–9583. doi: 10.1093/nar/gkt693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toonen L.J.A., Rigo F., van Attikum H., van Roon-Mom W.M.C. Antisense Oligonucleotide-Mediated Removal of the Polyglutamine Repeat in Spinocerebellar Ataxia Type 3 Mice. Mol. Ther. Nucleic Acids. 2017;8:232–242. doi: 10.1016/j.omtn.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cemal C.K., Carroll C.J., Lawrence L., Lowrie M.B., Ruddle P., Al-Mahdawi S., King R.H., Pook M.A., Huxley C., Chamberlain S. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum. Mol. Genet. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- 21.Costa Mdo.C., Luna-Cancalon K., Fischer S., Ashraf N.S., Ouyang M., Dharia R.M., Martin-Fishman L., Yang Y., Shakkottai V.G., Davidson B.L. Toward RNAi therapy for the polyglutamine disease Machado-Joseph disease. Mol. Ther. 2013;21:1898–1908. doi: 10.1038/mt.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watase K., Weeber E.J., Xu B., Antalffy B., Yuva-Paylor L., Hashimoto K., Kano M., Atkinson R., Sun Y., Armstrong D.L. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 23.Trottier Y., Lutz Y., Stevanin G., Imbert G., Devys D., Cancel G., Saudou F., Weber C., David G., Tora L. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 24.Servadio A., Koshy B., Armstrong D., Antalffy B., Orr H.T., Zoghbi H.Y. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat. Genet. 1995;10:94–98. doi: 10.1038/ng0595-94. [DOI] [PubMed] [Google Scholar]
- 25.Tabrizi S.J., Ghosh R., Leavitt B.R. Huntingtin Lowering Strategies for Disease Modification in Huntington’s Disease. Neuron. 2019;101:801–819. doi: 10.1016/j.neuron.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 26.Rué L., Bañez-Coronel M., Creus-Muncunill J., Giralt A., Alcalá-Vida R., Mentxaka G., Kagerbauer B., Zomeño-Abellán M.T., Aranda Z., Venturi V. Targeting CAG repeat RNAs reduces Huntington’s disease phenotype independently of huntingtin levels. J. Clin. Invest. 2016;126:4319–4330. doi: 10.1172/JCI83185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnon K.T., Pendergraff H.M., Deleavey G.F., Swayze E.E., Potier P., Randolph J., Roesch E.B., Chattopadhyaya J., Damha M.J., Bennett C.F. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiszer A., Mykowska A., Krzyzosiak W.J. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caplen N.J., Taylor J.P., Statham V.S., Tanaka F., Fire A., Morgan R.A. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum. Mol. Genet. 2002;11:175–184. doi: 10.1093/hmg/11.2.175. [DOI] [PubMed] [Google Scholar]
- 31.Bettencourt C., Hensman-Moss D., Flower M., Wiethoff S., Brice A., Goizet C., Stevanin G., Koutsis G., Karadima G., Panas M., SPATAX Network DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol. 2016;79:983–990. doi: 10.1002/ana.24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aartsma-Rus A., Straub V., Hemmings R., Haas M., Schlosser-Weber G., Stoyanova-Beninska V., Mercuri E., Muntoni F., Sepodes B., Vroom E., Balabanov P. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther. 2017;27:251–259. doi: 10.1089/nat.2017.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. USA. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorbunova V., Seluanov A., Dion V., Sandor Z., Meservy J.L., Wilson J.H. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol. Cell. Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J.Y., Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 36.Aartsma-Rus A., De Winter C.L., Janson A.A., Kaman W.E., Van Ommen G.J., Den Dunnen J.T., Van Deutekom J.C. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- 37.Mao Y., Senic-Matuglia F., Di Fiore P.P., Polo S., Hodsdon M.E., De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc. Natl. Acad. Sci. USA. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris G.M., Dodelzon K., Gong L., Gonzalez-Alegre P., Paulson H.L. Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS ONE. 2010;5:e13695. doi: 10.1371/journal.pone.0013695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanvey J.C., Peffer N.J., Bisi J.E., Thomson S.A., Cadilla R., Josey J.A., Ricca D.J., Hassman C.F., Bonham M.A., Au K.G. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 40.Cummings C.J., Mancini M.A., Antalffy B., DeFranco D.B., Orr H.T., Zoghbi H.Y. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 41.Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 42.Woulfe J.M. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol. 2007;33:2–42. doi: 10.1111/j.1365-2990.2006.00819.x. [DOI] [PubMed] [Google Scholar]
- 43.Friedrich J., Kordasiewicz H.B., O’Callaghan B., Handler H.P., Wagener C., Duvick L., Swayze E.E., Rainwater O., Hofstra B., Benneyworth M. Antisense oligonucleotide-mediated ataxin-1 reduction prolongs survival in SCA1 mice and reveals disease-associated transcriptome profiles. JCI Insight. 2018;3 doi: 10.1172/jci.insight.123193. e123193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J., Al-Ramahi I., Tan Q., Mollema N., Diaz-Garcia J.R., Gallego-Flores T., Lu H.C., Lagalwar S., Duvick L., Kang H. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature. 2013;498:325–331. doi: 10.1038/nature12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jafar-Nejad P., Ward C.S., Richman R., Orr H.T., Zoghbi H.Y. Regional rescue of spinocerebellar ataxia type 1 phenotypes by 14-3-3epsilon haploinsufficiency in mice underscores complex pathogenicity in neurodegeneration. Proc. Natl. Acad. Sci. USA. 2011;108:2142–2147. doi: 10.1073/pnas.1018748108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Switonski P.M., Fiszer A., Kazmierska K., Kurpisz M., Krzyzosiak W.J., Figiel M. Mouse ataxin-3 functional knock-out model. Neuromolecular Med. 2011;13:54–65. doi: 10.1007/s12017-010-8137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., ENDEAR Study Group Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 48.Mercuri E., Darras B.T., Chiriboga C.A., Day J.W., Campbell C., Connolly A.M., Iannaccone S.T., Kirschner J., Kuntz N.L., Saito K., CHERISH Study Group Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2018;378:625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 49.Verheul R.C., van Deutekom J.C., Datson N.A. Digital Droplet PCR for the Absolute Quantification of Exon Skipping Induced by Antisense Oligonucleotides in (Pre-)Clinical Development for Duchenne Muscular Dystrophy. PLoS ONE. 2016;11:e0162467. doi: 10.1371/journal.pone.0162467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.