Abstract
Pluripotent stem cells (PSC) can be used as a model to study cardiomyogenic differentiation. In vitro modeling can reproduce cardiac development through modulation of some key signaling pathways. Therefore, many studies make use of this strategy to better understand cardiomyogenesis complexity and to determine possible ways to modulate cell fate. However, challenges remain regarding efficiency of differentiation protocols, cardiomyocyte (CM) maturation and therapeutic applications. Considering that the extracellular milieu is crucial for cellular behavior control, cardiac niche studies, such as those identifying secreted molecules from adult or neonatal tissues, allow the identification of extracellular factors that may contribute to CM differentiation and maturation. This review will focus on cardiomyogenesis modeling using PSC and the elements involved in cardiac microenvironmental signaling (the secretome – extracellular vesicles, extracellular matrix and soluble factors) that may contribute to CM specification and maturation.
Keywords: cardiomyocytes, pluripotent stem cell, secretome, cell differentiation, maturation
Introduction
Pluripotent stem cells (PSC), both embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC), show strong potential to proliferate and differentiate. PSC have already been successfully differentiated into a number of cell types, including cardiomyocytes (CM) (Murry and Keller, 2008). Key events that regulate lineage commitment can be reproduced in vitro and used as a model to study cardiomyogenesis, to generate CMs and produce clinically relevant cell populations, and to evaluate cardiac toxicity or model congenital abnormalities (Kehat et al., 2001; Xu et al., 2002; Laflamme et al., 2007; Kattman et al., 2011; Burridge et al., 2012). Despite the advances in this field, new challenges are emerging, mainly related to cardiac differentiation efficiency and the functional maturation of human PSC-derived cardiomyocytes (hPSC-CM).
This review discusses cardiac differentiation and hPSC-CM maturation approaches that use extracellular components of the cardiac microenvironment. Initially, an overview of hPSC cardiomyogenic differentiation protocols was described, indicating some of the essential signaling pathways that control CM commitment. However, the main focus is to explore the cardiac niche, its components and the strategies developed to mimic its complexity in vitro. After a brief description of important signals and interactions available in a tissue niche, we emphasize aspects related to cardiac extracellular matrix (ECM; composition or structure), soluble factors and extracellular vesicles (EVs) that could influence in vitro CM differentiation and maturation.
Overview of Heart Development
The heart is a complex muscular organ composed of several cell types, including CM, smooth muscle cells (SMC), endothelial cells (EC), cardiac fibroblasts (cFB), and cardiac progenitor cells (CPC). Although CM occupy most of the heart volume, they comprise only ∼40% of the total cells. The other 60% largely comprises EC and cFB, however, the percentage of each of them is still not certain (Banerjee et al., 2007; Bergmann et al., 2015; Pinto et al., 2016).
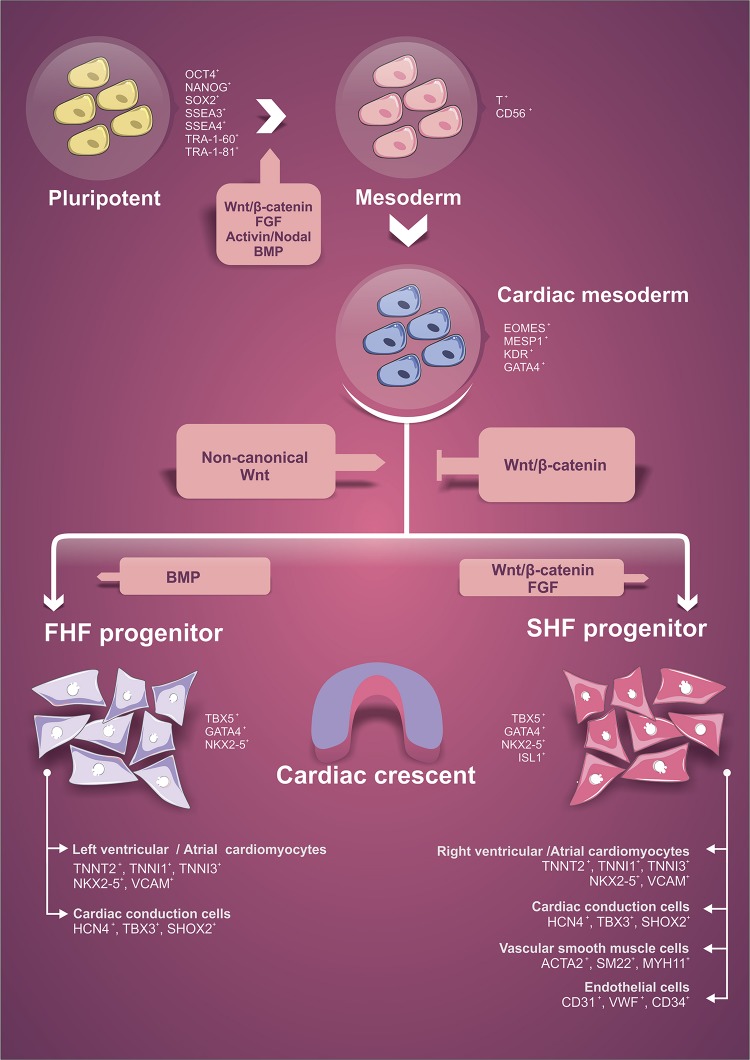
The heart is the first organ to become functional in the vertebrate embryo (Brand, 2003). Although the heart develops early, cardiogenesis is a highly regulated process involving differentiation and cellular specialization, spatial integration and coordination of several signaling pathways. Cardiac tissue is mostly derived from the mesodermal layer and the induction to the cardiomyogenic phenotype depends on signals derived from adjacent layers, such as endodermal and ectodermal cells (Wagner and Siddiqui, 2007; Sun and Kontaridis, 2018). The signaling factors modulated over heart development include members of bone morphogenetic proteins (BMPs), Activin and NODAL, fibroblast growth factor (FGF), and Wingless (Wnt) families (Brand, 2003; Wagner and Siddiqui, 2007; Liu and Foley, 2011; Brade et al., 2013; Sun and Kontaridis, 2018). In Figure 1, we briefly highlight some aspects of embryonic cardiac commitment that will be important to understand and support the in vitro differentiation protocols using PSC. The signaling pathways influencing the stages of cell differentiation and the cell markers expressed in these different stages are indicated (Figure 1). For more details about the morphogenesis, signaling pathways and factors involved in this process, see Wagner and Siddiqui (2007), Brade et al. (2013), Sylva et al. (2014), Paige et al. (2015), Sun and Kontaridis (2018).
FIGURE 1.
Schematic representation of the initial steps of cardiac lineage commitment. Indication of signaling pathways that influence each differentiation stage and the specific cellular markers expressed during lineage differentiation. FHF, first heart field. SHF, second heart field.
In vitro Differentiation of hPSC
Cardiac cell fate specification occurs through progressive steps that we are currently able to reproduce at the laboratory. There are three major strategies to derive CM from hPSC: (1) inductive coculture, (2) embryoid bodies, and (3) monolayer cultures. Table 1 summarizes these strategies and their main references [complete reviews can be found in Burridge et al. (2012), Mummery et al. (2012), Denning et al. (2016)].
TABLE 1.
Three major in vitro cardiac differentiation protocols.
|
Cardiac differentiation protocols | ||
| Strategy | Induction method | Main References |
| Inductive coculture with END-2 visceral endoderm-like cells | direct coculture or END-2 conditioned medium | Mummery et al., 2003, 2007 |
| Embryoid Body (EB) | spontaneous differentiation | Itskovitz-Eldor et al., 2000; Kehat et al., 2001; Zhang et al., 2009 |
| growth factors in defined media (GFs) | Yang et al., 2008; Kattman et al., 2011; Ren et al., 2011 | |
| Monolayer | growth factors in defined media (GFs) | Laflamme et al., 2007 |
| small molecules in defined media (SM) | Lian et al., 2012; Burridge et al., 2014 | |
Their main references are indicated.
The inductive coculture protocol uses visceral endoderm-like cells (END-2), which play an important role in signaling the adjacent mesoderm in developing embryos, to induce the differentiation of cardiogenic precursor cells. This protocol was developed by Mummery et al. (2003) and improved by them in Mummery et al. (2007). The convenience of the coculture is that it requires few cells and is less time-consuming. On the other hand, the efficiency of the protocol is very low, and it is not commonly used.
Three-dimensional aggregates, known as embryoid bodies (EBs), represent the first structure in which CM could be produced in vitro. Using a protocol to derive spontaneous differentiation, contracting structures containing CM and other germ layer derivatives were generated (Itskovitz-Eldor et al., 2000). These EBs could also be transferred to attachment culture plates and give rise to beating areas (Kehat et al., 2001; Zhang et al., 2009). Studies in animal models have helped with valuable information for the optimization of in vitro hPSC differentiation protocols (Marvin et al., 2001; Ueno et al., 2007). Cardiomyocyte derivation from hPSC can be manipulated and directed to cardiac lineage by specific elements, such as growth factors known to be involved in in vivo heart development (Vidarsson et al., 2010). The same signaling pathways mentioned above as essential for heart development are also used to modulate hPSC differentiation in vitro, such as BMP, Nodal, FGF, and Wnt (Filipczyka et al., 2007). Concentration and time of addition or removal of specific factors, such as BMP4, activin A and Wnt modulators, are critical for cardiac lineage specification and were adapted in the protocols to improve efficiency (Yang et al., 2008; Kattman et al., 2011; Ren et al., 2011). A similar idea was applied to the monolayer-based protocol, in which the use of growth factors and other molecules could improve efficiency and would not be interfered by the diffusional barrier present in EBs (Mummery et al., 2012). Using a defined serum-free medium supplemented with BMP4 and activin A, Laflamme et al. (2007) reported better efficiency in CM differentiation in the monolayer system compared to the EB method. Other adaptations were made (Lian et al., 2012; Burridge et al., 2014), including the use of more specific and low cost signaling small molecules, such as CHIR99021 (an inhibitor of glycogen synthase kinase 3), leading to the monolayer protocol becoming the most popular and routinely employed cardiogenic differentiation method (Denning et al., 2016). In addition, the ECM influence was also tested to improve differentiation efficiency. Combining ECM with growth factor signaling in a protocol that uses a double Matrigel layer, so-called matrix sandwich, CM could be generated with high purity (Zhang J. et al., 2012).
Advances in methodologies to direct cardiac differentiation helped to improve efficiency in the protocols, increasing the final percentage of CM (usually cardiac troponin T positive – cTnT+). However, the purity of these populations is still a limitation in the field. Some approaches were developed to enrich CM, rather than other cell types. For example, genetic selection strategies were based on the expression of a drug resistance gene or reporter protein gene under the control of a cardiac-specific promoter (Xu et al., 2008; Kita-Matsuo et al., 2009; Elliott et al., 2011). Reporter protein genes could also be applied in flow cytometry sorting, as well as selection by markers from distinct stages of differentiation (Yang et al., 2008; Lin et al., 2012; Den Hartogh et al., 2015). Purification of CM using a Percoll gradient (Xu et al., 2002) and energy metabolism differences (Tohyama et al., 2013) were also established. In addition to the purity of final populations, another limitation of in vitro cardiogenic differentiation is that the currently available methods generate a heterogeneous CM population that includes a mix of subtypes, such as ventricular, atrial, pacemaker, and non-contractile cells (Kolanowski et al., 2017; Friedman et al., 2018). Strategies to derive specific cardiac cell subtypes are being developed and could help the demand for therapeutic applications of these cells (Zhang et al., 2011; Karakikes et al., 2014; Devalla et al., 2015; Protze et al., 2016; Lee J.H. et al., 2017).
Another challenge in the field of hPSC cardiac differentiation is related to the maturity of hPSC-CM: most of the protocols generate immature CM. In recent years, a great number of studies have focused on investigating strategies to improve the maturation of hPSC-CM and make them more similar to adult CM (Sartiani et al., 2007; Lundy et al., 2013; Robertson et al., 2013), which are multinucleated (25–30%) with highly organized sarcomeres (I, A, and Z bands, M lines and intercalated disks), T-tubules, high expression of sarcomeric and ion channel genes, fatty acid β-oxidation metabolism, higher contractile force and upstroke and conduction velocities (Yang et al., 2014; Dunn and Palecek, 2018; Machiraju and Greenway, 2019). Among the approaches to achieve this purpose are the prolonged time for culture (e.g., 120–360 days), addition of hormones, metabolites or other soluble factors, mechanical or electrical stimulus, microtissue development, coculture with other cell types (such as SMC, cFB, and EC), stiffness regulation, and tridimensional (3D) cultures with biomaterials or ECM components (reviewed by Yang et al., 2014; Besser et al., 2018; Dunn and Palecek, 2018; Machiraju and Greenway, 2019).
Hence, cardiac differentiation efficiency and, consequently, the purity of the cell population and the immature phenotype of hPSC-CM are among the challenges regarding the use of these cells for drug discovery models, development studies, tissue engineering or cellular therapies. Considering the importance of the microenvironment for cellular behavior, we believe that knowledge about the cardiac niche and the use of strategies to mimic its specificities could improve (or create new) hPSC cardiac differentiation and maturation approaches. Since the coordination of all cellular processes including how the cells participate in the microenvironment is governed, inter alia, by gene expression, transcriptomic and proteomic analyses can contribute to the elucidation of cardiac niche specificities and also help to overcome the challenges of using hPSC-CM.
High-throughput studies have allowed the investigation of the global changes in gene expression at the transcriptional, post-transcriptional and protein levels, contributing with considerable insights about gene regulatory programs that are crucial to control cardiac tissue formation (Table 2). Distinct gene expression patterns drive PSC into specific cell types, consequently, the investigation of sequential stages during differentiation might help to discover essential molecules that determines the final destination of a cell. For instance, Paige et al. (2012) and, more recently, Liu et al. (2017) showed a temporal alteration in chromatin structure when analyzing distinct time-points of in vitro cardiomyogenesis. Changes in DNA methylation and post-transcriptional regulation were also described during cardiac differentiation and potentially participate in the modulation of gene regulatory programs (Tompkins et al., 2016; Fu et al., 2018; Pereira et al., 2019). Key regulators of cardiovascular development identified in those studies could be used to improve differentiation protocols, e.g., activating or inhibiting a specific signaling pathway. In addition, the identification of new cell surface proteins, such as the studies of den Hartogh et al. (2016) and Van Hoof et al. (2010), could provide new markers for development stages, cell lineage, cell subtype or maturation. Proteomic approaches also have highlighted important aspects regarding the cardiac commitment process, such as the metabolic and mitochondrial maturation described by Poon et al. (2015), the identification of metabolic and cytoskeletal proteins by Konze et al. (2017b) and the identification of proteins related to specific metabolic process, e.g., ketogenesis, described by Kim et al. (2019). Genes related to the ECM were also shown as modulated throughout the cardiac commitment process (den Hartogh et al., 2016; Pereira et al., 2019), and it will be discussed later in this review.
TABLE 2.
Summary of transcriptomic and proteomic studies based on hPSC cardiac differentiation.
| Differentiation method | Time-points | High throughput method | Highlights | References |
| Monolayer in END-2 coculture | Days 0, 1, 3, 6, 9, and 12 | Agilent Microarray | Identification and validation of time-dependent gene expression patterns | Beqqali et al., 2006 |
| EB spontaneous differentiation | hESC, CM, and hF heart | Agilent Microarray | hESC-CM promoting recovery from cardiac ischemia reperfusion injury | Cao et al., 2008 |
| EB in END-2 conditioned medium | hESC, EBs, CM-Day 21, fetal heart and adult heart | Illumina microarray | Evaluation of the biological relevance of uncharacterized genes | Xu et al., 2009 |
| EB (GFs) | Days 0, 2, 5, 9, and 14 | ChIP-seq and Affymetrix array | Temporal alterations in chromatin structure identify key regulators of cardiovascular development | Paige et al., 2012 |
| EB (GFs) KDRlow/CD166 + | hESC or iPS, M-Day 6, Day 20: CM, SMC, EC | Illumina RNA-seq | Lineage-enriched genes and lncRNAs, RNA splicing isoforms | Li et al., 2015 |
| Monolayer (SM/C) | hESC, D3, D4 and CM-D31 | Illumina RNA-seq and MDB-seq | TFs, miRNAs, lncRNAs and methylome | Tompkins et al., 2016 |
| Monolayer (GFs, SM/C) * Day 3 MESP1 + sorting | Days 0, 3, 5, 7, 10, and 14 | Illumina Microarray | Regulation of ECM components and new cell surface markers | den Hartogh et al., 2016 |
| Monolayer cardiomyocyte differentiation kit (Thermo Fisher Scientific) | Days 0, 2, 4, and 30 | RNA-seq and ATAC-seq | Mapping open chromatin patterns | Liu et al., 2017 |
| Monolayer (GFs) | Days 0, 12 and 20 | Illumina Microarray and ChIP-seq | Genetic and epigenetic changes and a role for NR2F2 | Pursani et al., 2017 |
| Monolayer (SM/C) | Days 0, 2, 5, 15, and 30 | Illumina single-cell RNA-seq | Cardiomyocyte hypertrophy and maturation | Friedman et al., 2018 |
| Monolayer (SM/C) | Days 0, 5, 14, and 45 | Single-cell RNA-seq | Single-cell heterogeneity | Churko et al., 2018 |
| EB (SM/C) | Days 0, 2, 5, 15 and 30 | Illumina RNA-seq and RRB-seq | TFs, lincRNAs and DNA methylation changes | Fu et al., 2018 |
| EB (GFs) | Days 0, 1, 4, 9, and 15 | Illumina RNA-seq polysome | Polysomal RNAs and post-transcriptional regulation | Pereira et al., 2018 |
| Monolayer (SM/C) | Days 0, 2, 5, and 14 | RNA-seq and ATAC-seq | Interplay of local and global chromatin structure on gene regulation | Bertero et al., 2019 |
| Monolayer in END-2 coculture | differentiated hESC, enriched populations of hESC-derived CM and primary hF CM | SILAC-based quantitative MS | Identification of cell surface proteins for antibody-based selection | Van Hoof et al., 2010 |
| EB (GFs) | hESC, hESC-VCMs, hF-VCMs, and hA-VCMs. | 2D-Differential-In-Gel Electrophoresis followed by MS | Metabolic and mitochondrial maturation | Poon et al., 2015 |
| Monolayer (GFs, SM/C) | Days 0, 5, and 14 | label-free quantitative | Identification of known and unknown regulatory proteins | Hofsteen et al., 2016 |
| Monolayer (SM/C) | Days 0, 20, and 35 | SILAC-labeled | Metabolic and cytoskeletal proteins | Konze et al., 2017b |
| Monolayer (SM/C) | Days 0, 7, and 15 | SILAC upon PAL-based capture of sialylated glycoproteins (glyco-proteomic) | Global proteomic, sialo-glycoproteomic, and glycomic characterization | Konze et al., 2017a |
| Monolayer (SM/C) | Days 0, 5, and 15 | Three-plex tandem mass tag labeling | Identification of proteins associated with branched chain amino acid degradation and ketogenesis | Kim et al., 2019 |
CM, cardiomyocytes; END-2, visceral endoderm-like cells; EB, embryoid body; EC, endothelial cells; GFs, growth factors; hF, human fetal; hA, human adult; iPSC, induced pluripotent cell; lncRNA, long non-coding RNA; M, multipotential cardiovascular progenitor cells; miRNA, microRNA; MS, mass spectrometry SMC, smooth muscle cells; SM/C, small molecules and chemical compounds; TFs, transcriptional factors; VCM, ventricular cardiomyocytes.
Tissue Microenvironment: Composition, Interactions, and Importance
Stem cells are modulated by microenvironmental cues in which they are embedded. This surrounding microenvironment is called the stem cell niche and is able to support cell maintenance and regulate the expansion or differentiation of stem cell populations. Thus, the stem cell niche was defined as an anatomical structure that includes cellular and acellular components, which integrates local and systemic factors that regulate stem cell behavior (Jones and Wagers, 2008). Since first being described by Schofield (1978), the niche concept has been expanded; it currently includes many components: the stem cell itself and tissue stromal cells; the ECM and related molecules; the secreted factors and extracellular vesicles (EVs); the blood vessels that carry systemic signals and cells (as immune regulators); the neural stimuli; oxygen concentration; and physical cues, such as shear stress (Jones and Wagers, 2008; Ferraro et al., 2010).
Cell signaling in the niche should be pictured as a result of multiple interactions and stimuli involving cell-to-cell connections, adhesive and de-adhesive ECM proteins, soluble trophic factors, and EVs. Figure 2 depicts some of the interactions that might occur along with niche networking. The complex netting of fibrillar proteins, proteoglycans and glycoproteins that compose the ECM acts in support and structure cells in the niches. In addition, ECM proteins and bioactive fragments released from the ECM by enzymatic degradation (the so-called matrikines) trigger cellular responses through interaction with cell receptors (Rozario and DeSimone, 2010; Ricard-Blum and Salza, 2014). Additionally, the ECM provides signals to the cells through its mechanical features (e.g., stiffness and elasticity) and its ability to function as a reservoir of growth factors and other bioactive elements. These secreted soluble factors may act locally or may diffuse throughout the niche, generating a concentration gradient (Jones and Wagers, 2008; Brizzi et al., 2012) that activates/inhibits signal pathways in the cells through autocrine/paracrine routes. Furthermore, another important player to consider on the niche site is the EVs that load different types of cargo (e.g., proteins, small RNAs, lipids) and function in intercellular communication (Stik et al., 2017; Durand et al., 2018).
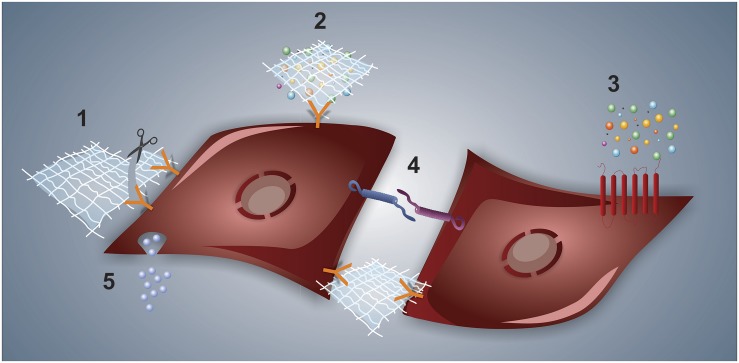
FIGURE 2.
Types of interactions at the cell–niche interface. In (1), a representative interaction between the cell and ECM is shown, whereas the matrikine-receptor interaction is pictured alongside the scissor, which represents matrix proteases. The ECM can also bind to and present a given trophic factor to the cell, as depicted in (2). The typical cytokine–receptor interaction is shown in (3). Cell-to-cell interactions are illustrated in (4), both by interacting membrane adhesive proteins and through ECM-integrin-mediated interaction. Finally, extracellular vesicle-driven signaling is represented in (5).
Distinct stem cell populations are arranged in unique and specific tissue microenvironments (Xin et al., 2016), within which the way stem cells behave depends on the interactions between specific cells and niche elements. Regarding the heart, cardiogenic niches are highly dynamic, presenting different functionalities and characteristics according to the stage of development of the heart and its physiological state. During the early stages of development, the elements of the cardiogenic niche play roles related primarily to cell expansion and specification, controlling the size and shape of cardiac structures. Subsequently, the niche elements stimulate the completion of differentiation and maturation of cardiac cells (revised by Christalla et al., 2012). In the adult heart, however, the current understanding of the cardiac niches is limited. Fatih Kocabas et al. (2012) identified putative cardiac niches in the heart epicardium and sub-epicardium with low oxygen tensions and housing a metabolically distinct population of glycolytic progenitor cells. Future attempts to characterize the stem cell niches in the heart atria and apex, however, are controversial. Nevertheless, cardiac niches may contain quiescent stem cells and progenitors in addition to the influence of all other heart cells (e.g., EC, CM, and SMC) that interacts and communicate together secreting a wide range of ECM proteins and soluble factors (reviewed by Aguilar-Sanchez et al., 2018).
Besides cell-to-cell interactions, ECM and soluble trophic factors present in the tissue microenvironment, other important modulators to consider are the biomechanical and electrical stimuli that influence the differentiation and maturation of cardiomyocytes, and the correct formation of the heart. Mechanical forces present during cardiac development include shear and strain stress, flow forces (i.e., blood flow), pressure and stretch. These mechanical signals are sensed by cells and converted into intracellular signals that activated pathways that induce gene expression and cellular commitment. The perception and transmission of signals involve both cell–cell and cell–ECM interactions (reviewed by Lindsey et al., 2014; Andrés-Delgado and Mercader, 2016; Stoppel et al., 2016). The mechanical force dynamics in the development of the heart involves, e.g., cFB that secrete a great amount of ECM proteins influencing the stiffness and topography of the organ while cardiomyocytes improves expression of sarcomeric proteins increasing the contractile stress and tissue strength. As a result, the heart pumps more blood, which increases the flow forces (reviewed by Majkut et al., 2014). All these biomechanical forces are interconnected and influence different processes during heart formation, from the proliferation and differentiation of cardiomyocytes to the correct formation of valves and cardiac chambers. Considering the importance of mechanical and electrical signals in cardiac development, many approaches have been developed in an attempt to understand and mimic these signals in vitro, with the aim of improving cardiomyocyte differentiation or maturation (reviewed by Zhu et al., 2014; Stoppel et al., 2016; Besser et al., 2018).
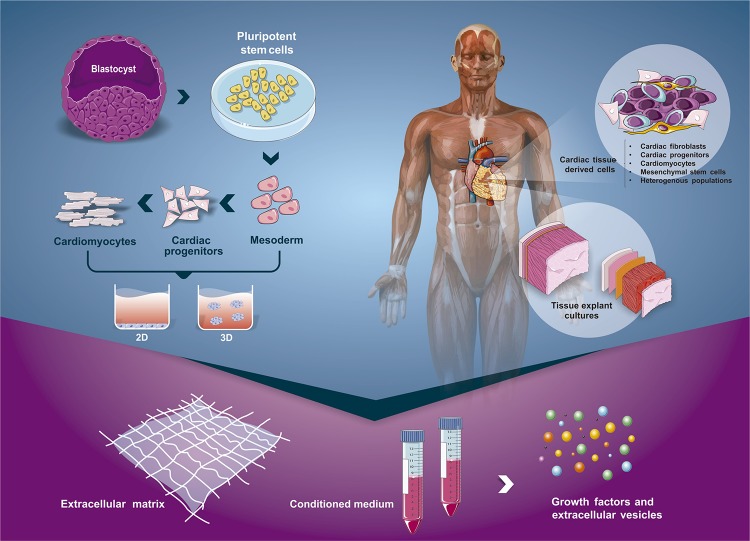
Some possibilities to approach cell-niche signaling relies on the use of PSC cardiomyogenic modeling, culture of cardiac tissue explant/cardiospheres and isolated cell populations (e.g., cardiac progenitor cells). Figure 3 summarizes the current strategies used to model cardiomyogenesis or cardiac cell/tissue behavior in vitro and ways to analyze the cardiac secretome (CS). The secretome comprises the complex set of secreted molecules/vesicles from cells to the extracellular space. At the cell–environment interface, the biological activity of the secretome is exerted by the modulation of signal transduction pathways that direct fundamental biological processes on cells, including cell fate and proliferation (Reus et al., 2016). Through the analysis of the ECM composition, soluble factors and EVs derived from cell and tissue cultures, we can search for essential signals and understand regulatory networks underlying human heart development. Regarding this issue, genomic and proteomic studies have provided important contributions (Reus et al., 2016; Wolling et al., 2018; Leitolis et al., 2019; Pereira et al., 2019). In the next sections, we will describe approaches to study the composition of cardiac secretome and discuss strategies that use the extracellular signals to induce PSC cardiac differentiation and maturation of PSC-CM.
FIGURE 3.
Scheme of different approaches to isolate the cardiac secretome. Extracellular matrix, soluble factors and extracellular vesicles can be obtained from the cardiomyogenic developmental process (using hPSC) or from adult tissues through the use of different strategies. The process of hPSC cardiac differentiation can occur using 2D (monolayer) or 3D (embryoid bodies) cultures. On the other hand, with fragments of neonate or adult heart, we could isolate specific cell populations from the tissue (e.g., fibroblasts, cardiac progenitors) or culture the entire or parts of the cardiac tissue.
Exploring the Cardiac ECM: Composition, Tissue Engineering Strategies and in vitro Modeling
Considering the actual challenges related to the use of PSC, the development of three-dimensional (3D) heart tissues constructs represents a strategy to improve in vitro models, both for studies concerning heart physiology and drug screening and to advance in vivo (translational) applications (Hirt et al., 2014; Ogle et al., 2016). However, recapitulating the complexity of cardiac tissue in vitro – structure, composition, and mechanical properties – is not an easy task. In this context, knowing the ECM, an important niche component, may have a relevant role in the development of new approaches to mimic heart tissue. Some methodologies use synthetic or natural biomaterials, singular ECM proteins or their combinations, and decellularized cardiac tissue (Smith et al., 2017; Zhang et al., 2017).
Cardiac ECM Composition
The ECM is not an inert scaffold restricted to supporting and structuring cells in tissues or organs but is also an important modulator of cellular responses either through its direct interaction with cell receptors, through the control of growth factor activities, controlling their diffusion or release, and transmitting mechanical signals (Rozario and DeSimone, 2010). ECM proteins comprise approximately 1–1.5% of the mammalian proteome called the “core matrisome,” which is composed of approximately 300 proteins shared among glycoproteins, proteoglycans and many collagen types. In addition, there are a great number of ECM-associated proteins, including the group of mucins and lectins, enzymes, matrix metallopeptidases, and secreted factors, such as chemokines, interleukins, and growth factors (Hynes and Naba, 2012; Naba et al., 2012, 2016). Considering the diversity of ECM components, the content of ECM varies among tissues, phase of development and according to the pathophysiological state of an organ (Christalla et al., 2012; Bonnans et al., 2014; Kular et al., 2014).
The cardiac ECM composition and function have been studied over the last few years. Several collagen types, fibronectin, laminin, fibrillins, proteoglycans, such as perlecan, agrin and glypicans, and glycosaminoglycans, mainly hyaluronan, are among the constituents of heart ECM (Lockhart et al., 2011; Rienks et al., 2014). As a dynamic milieu, the expression of the heart ECM varies during cardiac development and, accordingly, development of the heart region. Furthermore, it was previously described that the deletion of some ECM proteins causes serious heart defects, many of them causing embryonic lethality, as reviewed by Lockhart et al. (2011). Another component present in the ECM, which is important in the development of the heart and in its response to injury, is matricellular proteins. Although these proteins do not have structural function, they interact with surface receptors, growth factors, and ECM, among others, aiming to integrate the signals of the microenvironment. Among the members of this group are thrombospondin, tenascin, SPARC (acid secreted protein and cysteine rich), osteopontin, periostin and members of the CCN family (Frangogiannis, 2012, 2017; Rienks et al., 2014; Rienks and Papageorgiou, 2016).
Among heart cells, cFB are the main producers of cardiac ECM proteins. They are responsible for the ECM homeostasis, maintaining the balance between production and degradation of ECM proteins that are essential for the correct heart function. Despite its importance, there is still a lack of knowledge about the surface markers that could reveal their real molecular identity and, even, about functionality during the development of the heart. The cFBs are mainly derived from cells of the epicardium (with some contribution of the endothelium) that undergo epithelial-to-mesenchymal transition. In addition to ECM proteins, they secrete soluble factors (as will be discussed later in this review) and are also important in transducing and responding to electromechanical signals (Furtado et al., 2016; Civitarese et al., 2017; Tallquist and Molkentin, 2017). Interestingly, it was described that cFB expressed many specific cardiogenic genes, different of fibroblasts from other sources (Furtado et al., 2016). In adult heart, several factors can affect cFB activation, including response to injury. In this case, cFB can differentiate into myofibroblasts, which have increased proliferative and secretory capacity. The raise in the number of cFB and/or the deposition of ECM proteins causes fibrosis and, consequently, changes the rigidity of the tissue, the interaction between the cells, impairs myocyte contractility, oxygenation and metabolism (Tirziu et al., 2010; Martin and Blaxall, 2012; Fountoulaki et al., 2015), completely changing the cardiac microenvironment.
Originally, to better understand the role of cardiac ECM in embryonic development and elucidate its composition, the majority of studies used murine and other animal models (chicken, zebrafish) or were based on data related to congenital human cardiac diseases (Lockhart et al., 2011). Recently, one alternative is the use of hPSC cardiomyogenic differentiation to investigate ECM proteins secreted by cells. High-throughput analysis during hPSC differentiation showed that genes related to the ECM are modulated throughout the cardiac commitment process (den Hartogh et al., 2016; Pereira et al., 2019). Additionally, it was verified that during CM differentiation, total proteoglycan and hyaluronan decreased (Chan et al., 2010); meanwhile, using an endogenous optical signal, it was showed that elastin increased until day 9 of the murine EB differentiation protocol, and type I collagen (Col I) increased over 12 days (Thimm et al., 2015). Using a spontaneous differentiation protocol and isolating the beating areas, it was demonstrated that hESC-CM were surrounded by types I, IV, and XVIII collagens, laminin isoforms and fibronectin (FN) (van Laake et al., 2010).
In an attempt to characterize the CPC niche, Schenke-Layland et al. (2011) isolated fetal mouse and human hearts and identified a population of CPC that expressed Isl1+Flk1+. This population was delineated by a basement membrane with, primarily, collagen type IV (Col IV) and laminin, while FN and Col I were also present but more distant from it. Considering these results, it was shown that murine ESC (mESC) monolayer cultures with Col IV and laminin increase the number of CPC (Flk1+), a number that was even higher in 3D Col IV cultures (Schenke-Layland et al., 2011).
Myocardial Tissue Scaffolds
More recently, the understanding of the native ECM composition and its role in cell behavior has advanced due to decellularization approaches (Crapo et al., 2011). Decellularization techniques aimed to remove all cellular components of an organ or tissue, maintaining the ECM proteins and structure (Crapo et al., 2011; Tang-Quan et al., 2018). This approach leads to distinct possibilities, including the development of new cardiac tissue engineering strategies, through the use of decellularized/recellularized organs in transplants, as well as allowing the characterization of the cardiac ECM in normal or pathological conditions. Over the last 10 years, different decellularization and recellularization strategies were developed and performed with murine (Ott et al., 2008; Carvalho et al., 2012; Lu et al., 2013; Wang et al., 2019), porcine (Ott et al., 2008; Weymann et al., 2014; Ferng et al., 2017; Lee P.-F. et al., 2017), bovine (Arslan et al., 2018) and even human (Sánchez et al., 2015; Garreta et al., 2016; Guyette et al., 2016) heart tissue (revised by Scarrit et al., 2015; Tang-Quan et al., 2018).
Regarding ECM characterization, histochemical and immunofluorescent analyses were performed to visualize the ECM structure and some of its components. Large-scale proteomic analysis through mass spectrometry of matrix proteins could be difficult, since they are poorly soluble as a result of their macromolecular nature, extensive posttranslational modifications and the tendency to form protein complexes (Chang et al., 2016; Lindsey et al., 2018). Nevertheless, many approaches have sought to improve the protocols for mass spectrometry ECM characterization, allowing the identification of heart ECM proteins and collaborating with advances in the area (Lindsey et al., 2018). The number of extracellular proteins identified varies according to the form of ECM solubilization, analysis and equipment used.
Different extracellular protein combinations were identified after decellularization processes in the infarcted area of the mouse left ventricle (De Castro Brás et al., 2013), in normal porcine myocardium (Perea-Gil et al., 2018), after ischemia/reperfusion porcine heart injury (Barallobre-Barreiro et al., 2012), from rat hearts (Nguyen et al., 2018) and others. Human cardiac tissues were also characterized under normal conditions (Guyette et al., 2016; Johnson et al., 2016; Robert et al., 2017). For example, Johnson et al. (2016) used ECM-target and non-target methodologies to quantify 43 proteins in decellularized samples and identified more than 200 proteins in the global approach; these researchers also verified some variation in ECM composition between different heart donors. Despite the differences, in general, the matrisome components found in greater quantity in decellularized cardiac ECM are collagen types, glycoproteins, such as fibronectin, and members of the basal membrane, such as laminins and perlecan.
Rat neonatal CM, EC (rat or human origin), hCPC, mesenchymal stem cells (MSC), hPSC and hPSC-CM or hPSC-CPC were some of the cells used for tissue recellularization (Scarrit et al., 2015; Tang-Quan et al., 2018). Using undifferentiated hESC or hESC-derived mesendodermal cells, Ng et al. (2011) recellularized mouse hearts and, after 14 days under static culture conditions, cells began to express cardiac markers, such as cTnT, Nkx2.5, Myh6, and others. However, the cells were unable to contract, even in vivo (Ng et al., 2011). Lu et al. (2013) demonstrated that hiPSC-derived cardiac multipotent progenitors perfused into a decellularized mouse heart were able to migrate, proliferate and differentiate in situ into CM, SMC and EC, showing spontaneous contractions after 20 days, but they did not reach complete organ recellularization (Lu et al., 2013). Indeed, whole organ recellularization is an important barrier in cardiac tissue engineering. As a consequence, recent approaches include the use of cardiac patches with fragments/slices of decellularized matrices or its soluble form/hydrogel.
When undifferentiated mESC were cultured in slices of decellularized mouse hearts, they were able to express higher levels of cardiac markers in comparison with mESC cultured in slices of decellularized liver (Higuchi et al., 2013). Through the use of laser-cut sections of decellularized porcine myocardium, it was developed a system capable of maintaining hPSC-CM with organized sarcomeres and gap junctions that allowed the characterization of the biomechanical function of these EHT (Schwan et al., 2016). Pieces of decellularized rat heart were cultured with MSCs and hPSC-derived mature ventricular CM, which attached and formed a tissue-like structure with not only CM but also SMC and EC (Li et al., 2017). Other types of decellularized ECM were also tested both to support and differentiate PSC and to maintain PSC-CM. Hong et al. (2018) verified that decellularized mouse skeletal muscle was able to induce mESC to differentiate to a cardiac phenotype and supported mESC-CM (Hong et al., 2018). In addition, human placenta-derived hydrogel is another decellularized ECM that proved to be sufficient to maintain and generate more synchronized and electrically coupled hiPSC-CM (Francis et al., 2017).
Using human decellularized cardiac tissue, Oberwallner et al. (2015) showed that this ECM could support the mESC and iPSC and favored cardiac lineage differentiation (Oberwallner et al., 2015). More recently, slices of decellularized human heart cultured with PSC-CM presented spontaneous beating after 7–10 days, confirming cell–matrix interaction, demonstrated better electrophysiological response than in Matrigel and showed increased expression of cardiac ion channels in CM (Garreta et al., 2016). Additionally, Guyette et al. (2016), using two different tissue thicknesses and PSC-CM, showed spontaneous tissue contraction after 4–10 days, maintenance in culture for 60 or 120 days and the formation of a mechanical and electrical active myocardial tissue (Guyette et al., 2016).
Despite 3D approaches, an alternative is the use of hydrogels derived from decellularized ECM. Hydrogel prepared from decellularized porcine hearts combined with Col I, in proportion 75% ECM and 25% collagen, increased the number of cells expressing cTnT in comparison with those hydrogels with low ECM content (25%) or 100% collagen. This strategy also improved the maturation of hESC EBs, as demonstrated by the upregulation of connexin 43 (Cx43), the cardiac troponin I (cTnI) striation patterns, the improvement in the number of contracting cells and in the contraction amplitude (Duan et al., 2011). Fong et al. (2016) used hydrogels derived from decellularized bovine adult and fetal hearts in culture with hiPSC-CM in 2D (coating) and 3D (cardiac ECM + fibrinogen) approaches. These researchers demonstrated that 3D cultures with adult cardiac heart showed better improvement in CM maturation, mainly related to higher expression of mature cardiac genes (MYL2, Cx43, SERCA2a, and HCN4) and increased calcium signaling and kinetics compared with CM in 2D cultures (Fong et al., 2016). Together, these results indicate that native ECM provides a microenvironment capable of differentiating cells and providing a scaffold for the culture of CM, improving, at least in part, the maturation of these cells. These and other strategies used to induce differentiation and maturation of PSC-CM are summarized in Table 3.
TABLE 3.
Summary of ECM approaches to improve PSC cardiomyogenic differentiation and/or PSC-CM maturation.
| Decellularized heart ECM | Method | Cells | Highlights | References |
| Whole organ | Mouse ECM. Injected cells. | hESC or hESC-derived mesendodermal cells | After 14 days, both cell types expressed cardiac markers genes (cTnT, NKX2.5). No spontaneous contraction. | Ng et al., 2011 |
| Mouse ECM. Cells perfunded with growth factors. | hPSC-derived cardiac multipotent progenitors | Cells differentiate in situ in CM, SMC, and EC. Spontaneous contraction after 20 days. No complete recellularization. | Lu et al., 2013 | |
| Human ECM. Injected cells. Human heart bioreactor. | hiPSC-CM | After 14 days, cells remained viable, integrate with matrix and showed a range of maturity. No complete recellularization. | Guyette et al., 2016 | |
| Slices | Mouse ECM. 60-μm thick slices. | mESC | Higher levels of cardiac markers in comparison with liver decellularized ECM. | Higuchi et al., 2013 |
| Porcine ECM. 150-μm thick slices. Laser-cut sheets. | hPSC-CM | Developed of an EHT for biomechanical characterization of PSC-CM. Cells presented organized sarcomeres and formed gap junctions. | Schwan et al., 2016 | |
| Rat ECM. | MSCs and hPSC-derived ventricular CM | EHT with 75% CM and 25% MSC. After 2 weeks, cells formed a tissue-like structure with spontaneous contraction and CM, SMC and EC. | Li et al., 2017 | |
| Human ECM from patients with end-stage non-ischemic dilated cardiopathy. 300-μm thick slices. | mESC, miPSC | Supported PSC proliferation. Increase expression of cardiac markers. | Oberwallner et al., 2015 | |
| Human ECM. 400-μm thick slices. | PSC-CM | Spontaneous beating after 7–10 days. Better electrophysiological response. Uniform contraction, functional gap junctions. Increase expression of cardiac ion channels in CM. | Garreta et al., 2016 | |
| Human ECM. 200-μm thick slices. Cardiac fiber bundles with 15 mm length, 2.5 mm diameter. Injected cells. | PSC-CM | Cells adhered, remained viable and functional. Spontaneous beating after 4–10 days. Maintenance in culture for 60–120 days. Formation of mechanical and electrical tissue. | Guyette et al., 2016 | |
| Hydrogel | Different hydrogels composition: 75%, 25% or 0% of porcine ECM. | hESC EBs | The hydrogel composed of 75% porcine ECM:25% collagen increase the number of cells cTnT + Improve expression of Cx43, number of contracting cells and contraction amplitude. | Duan et al., 2011 |
| 2D (coating) or adult (cardiac patch) bovine adult and fetal ECM | hiPSC-CM | 3D cultures with adult tissue showed higher expression of mature cardiac genes. Increase calcium signaling. | Fong et al., 2016 | |
| 3D bioprinting | Porcine ECM. Different bioinks composition. | hCPC and/or MSC | Improve maturation of CPC. MSC + VEGF promoted vascular formation. | Jang et al., 2017 |
| Decellularized porcine ECM bioink. Custom digital light processing (DLP)-based scanningless and continuous 3D bioprinter. | hiPSC-CM | Improve expression of mature cardiac genes. | Yu et al., 2019 | |
| ECM preparations | ||||
| ECM proteins | Different proportions of fibronectin and laminin. | hESC | Ratio Fibronectin and Laminin (70:30) improve the number of differentiated cells (higher than 60% cTnI +). | Sa et al., 2014 |
| Systematic optimization of different ratios of type I collagen, laminin and fibronectin. | miPSC | Hydrogel composed of 61% type I collagen, 24% laminin-111 and 15% fibronectin increase number of cells (cTnT+). | Jung et al., 2015 | |
| Different combinations of laminin, fibronectin, types I, III and IV collagens. | hESC- derived CPC | Combinations of ECM proteins improve CPC attachment and survival. Fibronectin, types I, III and IV collagens showed better results. | Lu et al., 2017 | |
| Development of a Laminin-221 based cardiac differentiation protocol. | hESC | Combination of LN-521 + 221 matrix generated more CM (∼80%). High reproducibility confirmed by bulk and single-cell RNA-seq. | Yap et al., 2019 | |
| Biowire platform: Cells culture in type I collagen gel around a suture in a PDMS channel. Associate with electrical stimulation. | hPSC-CM and non-CM cells (e.g., FB, SMC, EC). | CM increase size, rod-like shape, organized sarcomeric banding, lower proliferative rate, improved Ca(+2) handling. | Nunes et al., 2013 | |
| Matrigel | Cells encapsulated in 3D cardiac strips composed of matrigel and type I collagen. Associated with mechanical cyclic stretch. | hESC-CM associated or not with non-CM cells | Conditions with non-CM cells improve more CM maturation. Cyclic stretch improved sarcomere size and expression of mature cardiac genes. | Zhang et al., 2017 |
| Cardiopatch hydrogel composed of matrigel, fibrinogen and cardiac media. | hPSC-CM | Improve expression of mature cardiac genes, sarcomeric banding, lower proliferative rate, more mature electrophysiology. | Shadrin et al., 2017 | |
| Coating with fibronectin or matrigel in glass coverslips or PDMS membranes. | hPSC-CM | Matrigel in PDMS improve CM electrophysiology, number of binucleated cells, expression of sarcomere and myofilament markers. | Herron et al., 2016 | |
| Matrigel Matress: 0.4–0.8 mm-thick of undiluted matrigel. | hiPSC-CM | CM developed rod-shape morphology, increase sarcomere size, upstroke velocity and expression of cardiac markers. | Feaster et al., 2015 | |
| Cells culture in matrigel or hyaluronan-based hydrogel associated or not with pro-survival factors. | hiPSC-CM | In vivo, CM in matrigel presented a more mature phenotype. | Ogasawara et al., 2017 | |
| Fibrin | 3D fibrin cardiac patch. | hESC-CM (SIRPA cells) | Improve sarcomere size, conduction velocity and expression of cardiac genes. | Zhang et al., 2013 |
| 3D fibrin matrix. | hiPSC-CM | Increase sodium current density and upstroke velocity. | Lemoine et al., 2017 | |
| Fibrin hydrogels associated with stretch and electrical stimulation | hiPSC-CM | Early-stage iPSC-CM associated with physical conditioning at an increasing intensity accelerated maturation, showed superior electrophysiological properties, CMs with increase cell size and sarcomere length. | Ronaldson-Bouchard et al., 2018 |
EBs, embryoid bodies; CM, cardiomyocytes; SMC, smooth muscle cells; EC, endothelial cells; VEGF, vascular endothelial growth factor; MSC, mesenchymal stem cells; CPC, cardiac progenitor cells.
Isolated ECM Components
In addition to the use of complex matrices and based on previous knowledge regarding cardiac ECM components, studies were performed with isolated ECM components. Many studies have used different combinations of ECM proteins to improve cardiac differentiation. hESC differentiated in a specific ratio of fibronectin (FN) and laminin (70:30) showed more differentiated cells than gelatin cultures, and its effects may be related to the integrin-mediated MEK/ERK signaling pathway (Sa et al., 2014). Furthermore, to optimize miPSC cardiac differentiation, Jung et al. (2015) investigated various combinations of ECM proteins. A systematic optimization indicated that a solution containing 61% Col I, 24% laminin-111 and 15% FN increased the number of cells expressing cTnT, MHCa, and α-actinin compared with suboptimal solutions (Jung et al., 2015). Additionally, it was demonstrated that the combination of ECM components, including FN, types I, III, and IV collagens, was important to allow hESC-derived cardiac progenitors to attach and survive (Lu et al., 2017). Recently, Yap et al. (2019), after verifying the higher expression of laminin-221 (LN-221) in adult cardiac tissue, developed a LN-221-based cardiac differentiation protocol, which reached more than 80% TNNT2+ cells and presented high reproducibility with 2 different hESC lines. Additionally, the cardiac progenitors generated during this process were able to improve cardiac function in mice after myocardial infarction (Yap et al., 2019).
In addition to affecting PSC cardiac differentiation, niche components and structure could also be used to stimulate the maturation of PSC-CM. Then, studies have attempted to use both 2D or 3D cultures with synthetic or natural matrices to reach this goal. To generate a microenvironment favorable to CM maturation, hPSC-CM and non-CM cells (as FB, SMC, and EC) were seeded in a Col I gel localized around a template suture in a poly(dimethylsiloxane) (PDMS) channel – a platform called “biowire.” Using this 3D structure associated with electrical stimulation, CMs increased their size, acquired a characteristic rod-like shape and an organized sarcomeric banding, presented a lower proliferative rate and improved Ca+2 handling properties. This finding indicates that the “biowire” platform with electrical stimulus (6 Hz) was able to induce hPSC-CM to a more mature phenotype (Nunes et al., 2013).
Comparisons of 2D and 3D cultures were performed by other groups. Using a 3D fibrin-based cardiac patch, differentiated EBs were dissociated and seeded with different percentages of SIRPA+ cells (CM marker) on the patch. Compared to monolayer cultures, 3D scaffolds augmented the conduction velocity, the size of sarcomeres and the expression of cardiac genes in hESC-CM (Zhang et al., 2013). Also, 3D cardiac strips produced through encapsulation of hESC-CM – with and without other niche cells (FB or MSC) – in Matrigel and Col I, associated with mechanical cyclic stretch, demonstrated potential in mature CM. Although all the conditions could generate a level of maturation in CM, those cultured with MSC or FB presented a more mature phenotype. Additionally, the use of cyclic stretching improved the sarcomere length and gene expression of maturation markers (Zhang et al., 2017).
Another platform of 3D construct is the “cardiopatch,” a hydrogel composed of fibrinogen, Matrigel and cardiac media mixed with CM. During 5 weeks of differentiation, hPSC-CM presented sarcomeric banding (including M-band and T-tubules), enhanced expression of markers related to the cardiac maturation process (e.g., TNNI3, MYL2, CASQ2, and CKM), progressive decrease in cell proliferation, higher conduction velocity, among others (Shadrin et al., 2017). Comparing a 3D EHT generated with hiPSC-CM in a fibrin matrix with 2D monolayer cultures, Lemoine et al. (2017) demonstrated that the EHT strategy increased sodium current density and upstroke velocity, both values more similar to those from adult human CM (Lemoine et al., 2017). Furthermore, early-stage hiPSC-CM incorporated into fibrin hydrogels subjected to stretch and electrical stimulation (increase intensity training) accelerated CM maturation, confirmed by expression of mature cardiac makers, increase cell size and sarcomere length and mature-like electrophysiological responses (Ronaldson-Bouchard et al., 2018).
The Matrigel®, a commercially available protein mixture used in maintenance of PSC and cardiac differentiation protocol (Zhang J. et al., 2012), was also applied to different strategies, alone or in combination with specific ECM proteins. For example, Herron et al. (2016) tested different ECM combinations to improve the maturation of hPSC-CM: coating of FN or Matrigel in glass coverslips or PDMS membranes. Matrigel + PDMS promoted greater CM maturation. Parameters such as conduction and upstroke velocities, electrophysiological, number of binucleated and proliferated cells, expression of mature sarcolemal and myofilament markers resemble that of mature CM under optimal conditions (Herron et al., 2016). Another strategy developed was the “Matrigel Mattress.” In this method, 0.4- to 0.8-mm-thick undiluted Matrigel® was prepared, and the hiPSC-CM cultured on this substrate developed a more rod-shaped morphology, increased sarcomere length, upstroke velocity and expression of cardiac markers (Feaster et al., 2015).
This type of methodology could be combined with in vivo maturation. For example, hiPSC-CM was diluted on Matrigel with pro-survival factors or on a hyaluronan-based hydrogel associated or not with pro-survival factors and transplanted into an infarcted rat. After 4 weeks, the CM cultured on Matrigel + factors presented a more mature phenotype than the other groups (Ogasawara et al., 2017). Kadota et al. (2017) compared cells transplanted in neonatal rat hearts uninjured and infarcted adult rat hearts. In all situations, the hiPSC-CM engrafted and survived in rat hearts, but it was the adult tissue that promoted faster CM maturation.
Among the most recent strategies to produce an environment for CM maturation is the use of 3D printing technologies. For instance, 3D printing allows the building of specific patterns of 3D constructs (Ma et al., 2018) and the use of solubilized decellularized ECM as bioink to culture with CPC (Jang et al., 2017) or iPSC-CM (Noor et al., 2019; Yu et al., 2019) to improve maturation of the cells. Considering the results discussed in this study, we showed that mimic ECM microenvironmental signals using 3D cultures, combinations of ECM proteins or a natural scaffold are important modulators that influence cardiac differentiation or maturation.
Cardiac Soluble Factors: Effects on Cardiomyocyte Fate and Behavior
To maintain their functions properly, chemical signaling in the heart should be orchestrated by a variety of signals released by myocyte and non-myocyte cells. As mentioned previously, the set of those signals constituted by secreted soluble factors comprises the CS. CS include several bioactive molecules, such as growth factors, endocrine hormones, cytokines and peptides (Doroudgar and Glembotski, 2012). In addition, CS may also contain extracellular vesicles (EVs) that load different types of cargo, which may vary with biogenesis, cell type, and physiological conditions (Abels and Breakefield, 2016). Because of the complex interplay between cardiac factors, cell communication in the heart has not been fully elucidated to date. However, extensive evidence indicates that the secretome from cardiac cells may influence CM development and behavior.
One approach to studying and decipher the actors of cellular communication in the heart is the analysis of the conditioned medium from in vitro cultures of cardiac cells (Figure 3), such as cardiospheres (Sharma et al., 2015), cardiac resident cells (Zhang et al., 2015; Reus et al., 2016) and heart explant tissues (Schittini et al., 2010). In the last several years, mass spectrometry approaches have been used for signaling investigation. According to Lindoso et al. (2016), this approach provides an overview of proteins present in media and enables a measurement of protein level changes during normal or pathophysiological conditions. Recently, a study analyzed the secretome collected at seven different time points during in vitro CM differentiation. The authors found 1802 proteins significantly regulated during differentiation of which 431 are annotated as secreted. Numerous proteins that remarkably vary during the differentiation process affect the Wnt, TGFβ, Activin A, Nodal, BMP and FGF signaling pathways (Wolling et al., 2018). The identification of paracrine factors that modulate CM differentiation, proliferation and maturation may help the development or improvement of protocols for the in vitro differentiation of CM from PSC. In this section, we provide a broad overview of recent investigations of paracrine factors that affect CM with an emphasis on those released by the main types of cardiac cells. We also explore studies involving EVs and its influence in cardiac cell behavior.
Signaling From Cardiac Resident Cells
Endothelial Cells
EC constitute an important component of the heart. Recently, ECs were identified as one of the most abundant cell types in this organ (Pinto et al., 2016). Anatomically, these cells form a monolayer that covers the heart cavities and compose the vascular network that perfuses the myocardium. Therefore, factors secreted by ECs, such as NO, endothelin-1, angiotensin II (Ang II), prostaglandins, natriuretic peptides, adenyl purines, neuregulin-1, FGF, and VEGF, are able to affect heart function (Shah, 1996; Tirziu et al., 2010). Recently, the mediators of the EC-CM interaction involved in cardiac remodeling and regeneration were summarized in a review (Talman and Kivelä, 2018). In fact, the communication between ECs and CM is crucial for the maintenance of cardiac homeostasis, and the disruption in this signalization can result in pathophysiological conditions (Gogiraju et al., 2019). Furthermore, CM generation was demonstrated to be affected by the niche provided by ECs (Chen et al., 2010), and the ability of ECs to enhance the maturity of hPSC-CM was already verified (Caspi et al., 2007; Tulloch et al., 2011; Ravenscroft et al., 2016). Kivelä et al. (2019) also demonstrated that ECs regulate physiological CM growth via VEGFR2. Recently, a study generated a human cardiac microtissue through the co-differentiation of CM and ECs from PSC (Giacomelli et al., 2017). Interestingly, the inclusion of ECs (generated with a cardiac identity) and prolonged time in culture induced changes in CM gene expression associated with CM maturation. Among the EC-CM mediators, neuregulin-1 (NRG-1) is one of the key players in CM development (Rupert and Coulombe, 2015). Previous studies reported that NRG-1β/ErbB signaling was able to increase cardiomyogenesis in mESC and participate in cardiac subtype (“working-type” atrial or nodal) selection (Chen et al., 2013). Beyond the individual effects of NRG-1, this molecule also acts synergistically with IGF-1 to enhance proliferation and metabolic maturity in CM (Rupert and Coulombe, 2017). Other soluble factors released by ECs are able to drive cardiac differentiation. Endothelin-1 added to Nkx2.5 + CPC culture induced CPC differentiation into cardiac pacemaking cells (Zhang X. et al., 2012). This molecule has also been suggested to stimulate terminal differentiation of CM (Paradis et al., 2014). In addition, brain natriuretic peptide (BNP), a cardiac hormone secreted by EC, CM and cFB, stimulated CPC proliferation and CM differentiation, and these effects were demonstrated to occur via BNP binding to NPR-A and NPR-B, respectively (Bielmann et al., 2015).
Cardiac Fibroblasts
The cFBs are cells that produce connective tissues and are recognized as modulators of cardiac function, development and homeostasis (Zhang P. et al., 2012; Ivey and Tallquist, 2017). As mentioned previously, cFB are responsible for the synthesis of the major part of ECM proteins in the heart. Therefore, the main interplay between cFB-CM communication appears to occur through ECM molecules secreted by cFBs. These cells also secrete proteins related to cardiac development, such as FGFs, TGF, Ang II, interleukin-6 (IL-6), and IL-33 (Kakkar and Lee, 2010; Takeda and Manabe, 2011). In fact, conditioned medium from ventricular cFBs was demonstrated to induce Nkx2.5+ CPC differentiation in CM through Wnt pathway activation (Zhang et al., 2015). Similarly, our group investigated the effects of conditioned medium from CRSCs (a population that includes DDR-2+ cells) on progenitor cell (H9c2) behavior. The CRSC secretome was able to drive the proliferation and cardiac differentiation of H9c2 cells (Reus et al., 2016). Furthermore, the cFB conditioned medium was also reported to influence CM phenotype, including hypertrophy, expression of vimentin and electrophysiological changes (LaFramboise et al., 2007; Pedrotty et al., 2009). Recently, the secretome of cFB was investigated in normal and stressed (hypoxic) conditions (Cosme et al., 2017). The conditioned media was separated to obtain exosome and exosome-depleted fractions, and the results revealed almost 494 proteins differentially expressed between fractions and oxygen conditions. Indeed, culture conditions interfere with cFB secretion and consequently in the CM response. Furthermore, cFB can be activated in response to tissue injury and present a phenotype characterized by the expression of alpha-smooth muscle actin (α-SMA) (Hu and Phan, 2013). Cartledge et al. (2015) demonstrated hypertrophic effects on CM cocultivated with cFB, myofibroblast and myofibroblast-conditioned medium, which were related to TGF-β released from cFB. In fact, CM hypertrophy induced by cFB-paracrine factors has been extensively described (Booz et al., 1999; LaFramboise et al., 2007). In addition, in damaged myocardium, activated cFB also releases proinflammatory cytokines, such as cardiotrophin-1 (CT-1), a member of the IL-6 family. This molecule could enhance mouse IPS cardiomyogenic differentiation partly via the JAK2/STAT3/Pim-1 pathway and stimulate CM maturation (Liu et al., 2015).
Cardiac Progenitor Cells
Cardiac progenitor cells are a heterogeneous group of cells distributed throughout the heart. These cells are extensively studied mainly because of their potential effects on injured cardiac tissue (reviewed by Le and Chong, 2017). Originally, it was believed that after transplantation, CPC would be able to restore cardiac function through differentiation in CM, EC, and SMC (Torella et al., 2006; Bearzi et al., 2007). However, recent reports suggest that the regenerative effects induced by CPC occur through secreted paracrine factors (Khanabdali et al., 2016; Le and Chong, 2017). Many subtypes of cardiac stem cells have been reported (Chong et al., 2014), and apparently, the paracrine factors released are also diversified between them; however, the therapeutic effects occur with the different CPC populations. Recently, Torán et al. (2019) aimed to define a set of proteins specifically secreted by CPC. Authors isolated human CPC from myocardial samples and conducted a proteomic assay. The analysis identified a group of factors expressed at high to medium levels by CPC that included IL-1, GROa (CXCL1), CXCL6 (GCP2), and IL-8 (Torán et al., 2019). Similarly, considering the effects of CPC in myocardial recovery, Sharma et al. (2017) isolated cardiac progenitor cells from neonatal (nCPC) and adult patients (aCPC) and compared the functionality of their conditioned medium. After extensive characterization of CPC paracrine factors, differences were found in the secretion profile during development that affected the ability of conditioned medium to recover myocardial function. Both secretomes significantly induced proliferation and reduced apoptosis in CM; however, nCPC conditioned medium was more effective than aCPC (Sharma et al., 2017). A recent study also profiled the proteins that were secreted by CPC (Sca−1+) derived from healthy and transgenic heart failure mice aiming to define the factors that are modulated in a failing heart microenvironment. The results showed that proteins usually associated with tissue regeneration, such as CSF1, COCA, IBP6, and TCPG, were found to be more abundant in transgenic samples than in healthy samples (Samal et al., 2018).
Extracellular Vesicles Signaling: Effects on Cardiomyocytes and Cardiac Tissue
In the heart, as well as in other tissues, protein secretion occurs in different ways: (1) direct release in the extracellular space through membrane-derived secretory vesicles; (2) translocation of cytosolic proteins across the plasma membrane; and (3) packaging in EVs (Reviewed by Doroudgar and Glembotski, 2012). The last mechanism is becoming more studied over the last several years. Since the discovery that EVs may harbor a varied content that includes not only proteins but also other active molecules (miRNAs, mRNA, DNA, and lipids) (Abels and Breakefield, 2016), the understanding of cell signaling was brought to a higher level of complexity. EVs have been isolated from different sources, including the previously mentioned cells: EC (Piryani et al., 2019), cFB (Borosch et al., 2017) and CPC (Barile et al., 2014), as well as other important components of the cardiac microenvironment, such as ECM (An et al., 2017), MSCs (Angulski et al., 2017), and CM (Liu et al., 2018). Recently, we isolated and characterized EVs from different regions of human heart tissue. Our results demonstrated that cardiac EVs contain a set of proteins advantageous for tissue regeneration approaches, and we verified their potential to modulate proliferation, wound healing, adhesion and angiogenesis differently depending on the target cell type (Leitolis et al., 2019). A systematic review compiled studies that investigated the cardioprotective characteristics of EVs (Wendt et al., 2018). In fact, EVs are able to regulate many biological activities, including those related to CM. Cardiac progenitor cell-derived EVs have been shown to stimulate migration and proliferation in AC16 CM (Hocine et al., 2019) and inhibit CM apoptosis (Barile et al., 2014) through a mechanism that involves PAPP-A (pregnancy-associated plasma protein-A) (Barile et al., 2018). Similarly, Liu et al. (2018) showed that CM-derived EVs not only decreased CM apoptosis 24 h after rat infarction but also reduced arrhythmia and hypertrophy postinfarction (Liu et al., 2018). The hypertrophic effect on CM was also verified through cFB-derived exosomes in a paracrine mechanism by which Ang II intensifies its own signaling in CM (Lyu et al., 2013). In addition, apoptotic bodies (AB) also appear to modulate myocyte activity. CM-derived AB were able to stimulate proliferation and differentiation of CM precursors, as well as their frequency of contraction (Tyukavin et al., 2015). Beyond the studies conducted with EVs derived from the sources mentioned above, many studies have shown the biological effects of MSC-derived EVs (MSC-EVs). To date, many reports have demonstrated that MSC-EVs are able to exert protective effects in cardiovascular diseases, especially by the delivery of miRNA content to recipient cells. Recently, Moghaddam et al. (2019) summarized the cardioprotective exosomal miRNAs that include those secreted by MSCs. For instance, Wang et al. (2018) demonstrated that cardiac stem cells treated with MSC-derived exosomes containing miR-214 showed decreased apoptosis and reactive oxygen species (ROS) production after oxidative stress injury (Wang et al., 2018). Similarly, a recent study showed that exosomal miR-21-5p increased cardiac calcium handling and thereby contractility via the PI3K signaling cascade in human engineered cardiac tissue (Mayourian et al., 2018). Furthermore, a novel study showed that the coculture of iPSC-CM with MSCs modulates the functionality and maturation of CMs. These effects can be partially explained by the miRNA content in MSC-derived EVs (Yoshida et al., 2018). In fact, CM differentiation and maturation are closely related to miRNA regulation (White et al., 2016; Lock et al., 2018). For instance, let-7 family miRNAs were found to be highly upregulated during CM maturation, and let-7 members control CM metabolism, cell size and force contractility (Kuppusamy et al., 2015).
Perspectives
The elements from microenvironmental signaling, such as ECM proteins and paracrine factors, both secreted by cardiac resident cells, are biologically active molecules capable of affecting CM commitment, subtype specification, proliferation and maturation. These molecules are being tested in coculture experiments (CM plus non-myocyte cells), as well as in assays that use total secretome (conditioned medium), EVs, decellularized heart ECM or combinations of isolated ECM proteins. In most cases, the molecular mechanisms by which these molecules act have not been fully decoded.
Despite the progress in understanding the cardiac niche in different development stages, the modulation of extracellular signaling and how it governs cardiac commitment has not been thoroughly elucidated to date. The factors mentioned in this review, as well as others not explored to date, may be important tools for modulation of in vitro cardiomyogenesis, mainly regarding maturation of the developed CM. Hence, improving the functional characteristics of CM could potentiate their use in regenerative medicine and facilitate further advances in cell therapy and tissue engineering.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the staff of the Carlos Chagas Institute (FIOCRUZ-PR) for the administrative support and Wagner Nagib de Souza Birbeire for visual design of the illustrations.
References
- Abels E. R., Breakefield X. O. (2016). Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 36 301–312. 10.1007/s10571-016-0366-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Sanchez C., Michael M., Pennings S. (2018). Cardiac stem cells in the postnatal heart: lessons from development. Stem Cells Int. 2018 1–13. 10.1155/2018/1247857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M., Kwon K., Park J., Ryu D. R., Shin J. A., Lee Kang J., et al. (2017). Extracellular matrix-derived extracellular vesicles promote cardiomyocyte growth and electrical activity in engineered cardiac atria. Biomaterials 146 49–59. 10.1016/j.biomaterials.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Andrés-Delgado L., Mercader N. (2016). Interplay between cardiac function and heart development. Biochim. Biophys. Acta Mol. Cell Res. 1863 1707–1716. 10.1016/j.bbamcr.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulski A. B. B., Capriglione L. G., Batista M., Marcon B. H., Senegaglia A. C., Stimamiglio M. A., et al. (2017). The protein content of extracellular vesicles derived from expanded human umbilical cord blood-derived CD133+ and human bone marrow-derived mesenchymal stem cells partially explains why both sources are advantageous for regenerative medicine. Stem Cell Rev. Reports 13 244–257. 10.1007/s12015-016-9715-z [DOI] [PubMed] [Google Scholar]
- Arslan Y. E., Galata Y. F., Sezgin Arslan T., Derkus B. (2018). Trans-differentiation of human adipose-derived mesenchymal stem cells into cardiomyocyte-like cells on decellularized bovine myocardial extracellular matrix-based films. J. Mater. Sci. Mater. Med. 29:127. 10.1007/s10856-018-6135-4 [DOI] [PubMed] [Google Scholar]
- Banerjee I., Fuseler J. W., Price R. L., Borg T. K., Baudino T. A. (2007). Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. Physiol. 293 1883–1891. 10.1152/ajpheart.00514.2007 [DOI] [PubMed] [Google Scholar]
- Barallobre-Barreiro J., Didangelos A., Schoendube F. A., Drozdov I., Yin X., Fernández-Caggiano M., et al. (2012). Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation 125 789–802. 10.1161/CIRCULATIONAHA.111.056952 [DOI] [PubMed] [Google Scholar]
- Barile L., Cervio E., Lionetti V., Milano G., Ciullo A., Biemmi V., et al. (2018). Cardioprotection by cardiac progenitor cell-secreted exosomes: role of pregnancy-associated plasma protein-A. Cardiovasc. Res. 114 992–1005. 10.1093/cvr/cvy055 [DOI] [PubMed] [Google Scholar]
- Barile L., Lionetti V., Cervio E., Matteucci M., Gherghiceanu M., Popescu L. M., et al. (2014). Extracellular vesicles fromhuman cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function aftermyocardial infarction. Cardiovasc. Res. 103 530–541. 10.1093/cvr/cvu167 [DOI] [PubMed] [Google Scholar]
- Bearzi C., Rota M., Hosoda T., Tillmanns J., Nascimbene A., De Angelis A., et al. (2007). Human cardiac stem cells. PNAS 104 14068–14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beqqali A., Kloots J., Ward-van Oostwaard D., Mummery C., Passier R. (2006). Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells 24 1956–1967. 10.1634/stemcells.2006-0054 [DOI] [PubMed] [Google Scholar]
- Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., et al. (2015). Dynamics of cell generation and turnover in the human heart. Cell 161 1566–1575. 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- Bertero A., Fields P. A., Ramani V., Bonora G., Yardimci G. G., Reinecke H., et al. (2019). Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat. Commun. 10:1538. 10.1038/s41467-019-09483-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser R. R., Ishahak M., Mayo V., Carbonero D., Claure I., Agarwal A. (2018). Engineered microenvironments for maturation of stem cell derived cardiac myocytes. Theranostics 8 124–140. 10.7150/thno.19441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielmann C., Rignault-Clerc S., Liaudet L., Li F., Kunieda T., Sogawa C., et al. (2015). Brain natriuretic peptide is able to stimulate cardiac progenitor cell proliferation and differentiation in murine hearts after birth. Basic Res. Cardiol. 110:455. 10.1007/s00395-014-0455-454 [DOI] [PubMed] [Google Scholar]
- Bonnans C., Chou J., Werb Z. (2014). Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15 786–801. 10.1038/nrm3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booz G. W., Dostal D. E., Baker K. M. (1999). Paracrine actions of cardiac fibroblasts on cardiomyocytes: implications for the cardiac renin-angiotensin system. Am. J. Cardiol. 83 44–47. 10.1016/S0002-9149(99)00257-X [DOI] [PubMed] [Google Scholar]
- Borosch S., Dahmen E., Beckers C., Stoppe C., Buhl E. M., Denecke B., et al. (2017). Characterization of extracellular vesicles derived from cardiac cells in an in vitro model of preconditioning. J. Extracell. Vesicles 6:1390391. 10.1080/20013078.2017.1390391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade T., Pane L. S., Moretti A., Chien K. R., Laugwitz K. (2013). Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 3 1–18. 10.1101/cshperspect.a013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. (2003). Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 258 1–19. 10.1016/S0012-1606(03)00112-X [DOI] [PubMed] [Google Scholar]
- Brizzi M. F., Tarone G., Defilippi P. (2012). Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr. Opin. Cell Biol. 24 645–651. 10.1016/j.ceb.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Keller G., Gold J. D., Wu J. C. (2012). Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10 16–28. 10.1016/j.stem.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P. W., Matsa E., Shukla P., Lin Z. C., Churko J. M., Ebert A. D., et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11 855–860. 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Wagner R. A., Wilson K. D., Xie X., Fu J., Drukker M., et al. (2008). Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One 3:e3474. 10.1371/journal.pone.0003474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartledge J. E., Kane C., Dias P., Tesfom M., Clarke L., Mckee B., et al. (2015). Functional crosstalk between cardiac fibroblasts and adult cardiomyocytes by solublemediators. Cardiovasc. Res. 105 260–270. 10.1093/cvr/cvu264 [DOI] [PubMed] [Google Scholar]
- Carvalho J. L., de Carvalho P. H., Gomes D. A., Goes A. M. (2012). Characterization of decellularized heart matrices as biomaterials for regular and whole organ tissue engineering and initial in-vitro recellularization with ips cells. J. Tissue Sci. Eng. 11:2. 10.4172/2157-7552.S11-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi O., Lesman A., Basevitch Y., Gepstein A., Arbel G., Huber I., et al. (2007). Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ. Res. 100 263–272. 10.1161/01.RES.0000257776.05673.ff [DOI] [PubMed] [Google Scholar]
- Chan C. K., Rolle M. W., Potter-Perigo S., Braun K. R., Van Biber B. P., Laflamme M. A., et al. (2010). Differentiation of cardiomyocytes from human embryonic stem cells is accompanied by changes in the extracellular matrix production of versican and hyaluronan. J. Cell. Biochem. 111 585–596. 10.1002/jcb.22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. W., Dalgliesh A. J., López J. E., Griffiths L. G. (2016). Cardiac extracellular matrix proteomics: challenges, techniques, and clinical implications. Prot. Clin. Appl. 10 39–50. 10.1002/prca.201500030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Bai H., Arzigian M., Gao Y. X., Bao J., Wu W. S., et al. (2010). Endothelial cells regulate cardiomyocyte development from embryonic stem cells. J. Cell. Biochem. 111 29–39. 10.1002/jcb.22680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Bi L. L., Wang Z. Q., Zhao F., Gan X. D., Wang Y. G. (2013). Time-dependent regulation of neuregulin-1β/ErbB/ERK pathways in cardiac differentiation of mouse embryonic stem cells. Mol. Cell. Biochem. 380 67–72. 10.1007/s11010-013-1658-y [DOI] [PubMed] [Google Scholar]
- Chong J. J. H., Forte E., Harvey R. P. (2014). Developmental origins and lineage descendants of endogenous adult cardiac progenitor cells. Stem Cell Res. 13 592–614. 10.1016/j.scr.2014.09.008 [DOI] [PubMed] [Google Scholar]
- Christalla P., Hudson J. E., Zimmermann W.-H. (2012). The cardiogenic niche as a fundamental building block of engineered myocardium. Cells Tissues Organs 195 82–93. 10.1159/000331407 [DOI] [PubMed] [Google Scholar]
- Churko J. M., Garg P., Treutlein B., Venkatasubramanian M., Wu H., Lee J., et al. (2018). Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat. Commun. 9:4906. 10.1038/s41467-018-07333-7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civitarese R. A., Kapus A., McCulloch C. A., Connelly K. A. (2017). Role of integrins in mediating cardiac fibroblast–cardiomyocyte cross talk: a dynamic relationship in cardiac biology and pathophysiology. Basic Res. Cardiol. 112:6. 10.1007/s00395-016-0598-6 [DOI] [PubMed] [Google Scholar]
- Cosme J., Guo H., Hadipour-Lakmehsari S., Emili A., Gramolini A. O. (2017). Hypoxia-induced changes in the fibroblast secretome. exosome, and whole-cell proteome using cultured, cardiac-derived cells isolated from neonatal mice. J. Proteome Res. 16 2836–2847. 10.1021/acs.jproteome.7b00144 [DOI] [PubMed] [Google Scholar]
- Crapo P. M., Gilbert T. W., Badylak S. F. (2011). An overview of tissue and whole organ decellularization processes. Biomaterials 32 3233–3243. 10.1016/j.biomaterials.2011.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro Brás L. E., Ramirez T. A., DeLeon-Pennell K. Y., Chiao Y. A., Ma Y., Dai Q., et al. (2013). Texas 3-Step decellularization protocol: looking at the cardiac extracellular matrix. J. Proteomics 86 43–52. 10.1016/j.jprot.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hartogh S. C., Schreurs C., Monshouwer-Kloots J. J., Davis R. P., Elliott D. A., Mummery C. L., et al. (2015). Dual reporter MESP1 mCherry/w -NKX2-5 eGFP/w hESCs enable studying early human cardiac differentiation. Stem Cells 33 56–67. 10.1002/stem.1842 [DOI] [PubMed] [Google Scholar]
- den Hartogh S. C., Wolstencroft K., Mummery C. L., Passier R. (2016). A comprehensive gene expression analysis at sequential stages of in vitro cardiac differentiation from isolated MESP1-expressing-mesoderm progenitors. Sci. Rep. 6:19386. 10.1038/srep19386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning C., Borgdorff V., Crutchley J., Firth K. S. A., George V., Kalra S., et al. (2016). Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. Biochim. Biophys. Acta Mol. Cell Res. 1863 1728–1748. 10.1016/j.bbamcr.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla H. D., Schwach V., Ford J. W., Milnes J. T., El-Haou S., Jackson C., et al. (2015). Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 7 394–410. 10.15252/emmm.201404757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroudgar S., Glembotski C. C. (2012). The cardiomyokine story unfolds: investigating stress-induced protein secretion in the heart. Trends Mol. Med. 17 207–214. 10.1016/j.molmed.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Liu Z., O’Neill J., Wan L. Q., Freytes D. O., Vunjak-Novakovic G. (2011). Hybrid gel composed of native heart matrix and collagen induces cardiac differentiation of human embryonic stem cells without supplemental growth factors. J. Cardiovasc. Transl. Res. 4 605–615. 10.1007/s12265-011-9304-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K. K., Palecek S. P. (2018). Engineering scalable manufacturing of high-quality stem cell-derived cardiomyocytes for cardiac tissue repair. Front. Med. 5:110. 10.3389/fmed.2018.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand C., Charbord P., Jaffredo T. (2018). The crosstalk between hematopoietic stem cells and their niches. Curr. Opin. Hematol. 25 285–289. 10.1097/MOH.0000000000000438 [DOI] [PubMed] [Google Scholar]
- Elliott D. A., Braam S. R., Koutsis K., Ng E. S., Jenny R., Lagerqvist E. L., et al. (2011). NKX2-5eGFP/w hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods 8 1037–1040. 10.1038/nmeth.1740 [DOI] [PubMed] [Google Scholar]
- Feaster T. K., Cadar A. G., Wang L., Williams C. H., Chun Y. W., Hempel J. E., et al. (2015). Matrigel mattress. Circ. Res. 117 995–1000. 10.1161/CIRCRESAHA.115.307580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferng A. S., Connell A. M., Marsh K. M., Qu N., Medina A. O., Bajaj N., et al. (2017). Acellular porcine heart matrices: whole organ decellularization with 3D-bioscaffold & vascular preservation. J. Clin. Transl. Res. 3 260–270. 10.18053/jctres.03.201702.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro F., Celso C. L., Scadden D. (2010). Adult Stem Cels and Their Niches. Boston, MA: Springer, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyka A. A., Passiera R., Rochata A., Mummery C. L. (2007). Regulation of cardiomyocyte differentiation of embryonic stem cells by extracellular signalling. Cell. Mol. Life Sci. 64 704–718. 10.1002/dvdy.20684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong A. H., Romero-López M., Heylman C. M., Keating M., Tran D., Sobrino A., et al. (2016). Three-dimensional adult cardiac extracellular matrix promotes maturation of human induced pluripotent stem cell-derived cardiomyocytes. Tissue Eng. Part A 22 1016–1025. 10.1089/ten.tea.2016.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountoulaki K., Dagres N., Iliodromitis E. K. (2015). Cellular communications in the heart. Card. Fail. Rev. 1 64–68. 10.15420/cfr.2015.1.2.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M. P., Breathwaite E., Bulysheva A. A., Varghese F., Rodriguez R. U., Dutta S., et al. (2017). Human placenta hydrogel reduces scarring in a rat model of cardiac ischemia and enhances cardiomyocyte and stem cell cultures. Acta Biomater. 52 92–104. 10.1016/j.actbio.2016.12.027 [DOI] [PubMed] [Google Scholar]
- Frangogiannis N. G. (2012). Matricellular proteins in cardiac adaptation and disease. Physiol. Rev. 92 635–688. 10.1152/physrev.00008.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis N. G. (2017). The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Invest. 127 1600–1612. 10.1172/JCI87491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman C. E., Nguyen Q., Lukowski S. W., Helfer A., Chiu H. S., Miklas J., et al. (2018). Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell 23 586.e–598.e. 10.1016/j.stem.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu K., Nakano H., Morselli M., Chen T., Pappoe H., Nakano A., et al. (2018). A temporal transcriptome and methylome in human embryonic stem cell-derived cardiomyocytes identifies novel regulators of early cardiac development. Epigenetics 13 1013–1026. 10.1080/15592294.2018.1526029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado M. B., Nim H. T., Boyd S. E., Rosenthal N. A. (2016). View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development 143 387–397. 10.1242/dev.120576 [DOI] [PubMed] [Google Scholar]
- Garreta E., de Oñate L., Fernández-Santos M. E., Oria R., Tarantino C., Climent A. M., et al. (2016). Myocardial commitment from human pluripotent stem cells: rapid production of human heart grafts. Biomaterials 98 64–78. 10.1016/j.biomaterials.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Giacomelli E., Bellin M., Sala L., van Meer B. J., Tertoolen L. G. J., Orlova V. V., et al. (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144 1008–1017. 10.1242/dev.143438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogiraju R., Bochenek M. L., Schäfer K. (2019). Angiogenic endothelial cell signaling in cardiac hypertrophy and heart failure. Front. Cardiovasc. Med. 6:20. 10.3389/fcvm.2019.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyette J. P., Charest J. M., Mills R. W., Jank B. J., Moser P. T., Gilpin S. E., et al. (2016). Bioengineering human myocardium on native extracellular matrix. Circ. Res. 118 56–72. 10.1161/CIRCRESAHA.115.306874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron T. J., Rocha A. M., Da Campbell K. F., Ponce-Balbuena D., Willis B. C., Guerrero-Serna G., et al. (2016). Extracellular matrix–mediated maturation of human pluripotent stem cell–derived cardiac monolayer structure and electrophysiological function. Circ. Arrhythmia Electrophysiol. 9 1–23. 10.1161/CIRCEP.113.003638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi S., Lin Q., Wang J., Lim T. K., Joshi S. B., Anand G. S., et al. (2013). Heart extracellular matrix supports cardiomyocyte differentiation of mouse embryonic stem cells. J. Biosci. Bioeng. 115 320–325. 10.1016/j.jbiosc.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt M. N., Hansen A., Eschenhagen T. (2014). Cardiac tissue engineering: state of the art. Circ. Res. 114 354–367. 10.1161/CIRCRESAHA.114.300522 [DOI] [PubMed] [Google Scholar]
- Hocine H. R., Brunel S., Chen Q., Giustiniani J., San Roman M. J., Ferrat Y. J., et al. (2019). Extracellular vesicles released by allogeneic human cardiac stem/progenitor cells as part of their therapeutic benefit. Stem Cells Transl. Med. 10.1002/sctm.18-0256 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsteen P., Robitaille A. M., Chapman D. P., Moon R. T., Murry C. E. (2016). Quantitative proteomics identify DAB2 as a cardiac developmental regulator that inhibits WNT/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 113 1002–1007. 10.1073/pnas.1523930113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X., Yuan Y., Sun X., Zhou M., Guo G., Zhang Q., et al. (2018). Skeletal extracellular matrix supports cardiac differentiation of embryonic stem cells: a potential scaffold for engineered cardiac tissue. Cell. Physiol. Biochem. 45 319–331. 10.1159/000486813 [DOI] [PubMed] [Google Scholar]
- Hu B., Phan S. H. (2013). Myofibroblasts. Curr. Opin. Rheumatol. 25 71–77. 10.1097/BOR.0b013e32835b1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O., Naba A. (2012). Overview of the matrisome–an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 4:a004903. 10.1101/cshperspect.a004903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., et al. (2000). Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 6 88–95. 10.1007/bf03401776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey M. J., Tallquist M. D. (2017). Defining the Cardiac fibroblast: a new hope. Circ. J. 80 2269–2276. 10.1253/circj.CJ-16-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J., Park H. J., Kim S. W., Kim H., Park J. Y., Na S. J., et al. (2017). 3D printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials 112 264–274. 10.1016/j.biomaterials.2016.10.026 [DOI] [PubMed] [Google Scholar]
- Johnson T. D., Hill R. C., Dzieciatkowska M., Nigam V., Behfar A., Christman K. L., et al. (2016). Quantification of decellularized human myocardial matrix: a comparison of six patients. Proteomics Clin. Appl. 10 75–83. 10.1002/prca.201500048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. L., Wagers A. J. (2008). No place like home: anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 9 11–21. 10.1038/nrm2319 [DOI] [PubMed] [Google Scholar]
- Jung J. P., Hu D., Domian I. J., Ogle B. M. (2015). An integrated statistical model for enhanced murine cardiomyocyte differentiation via optimized engagement of 3D extracellular matrices. Sci. Rep. 5 1–13. 10.1038/srep18705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota S., Pabon L., Reinecke H., Murry C. E. (2017). In Vivo maturation of human induced pluripotent stem cell-derived cardiomyocytes in neonatal and adult rat hearts. Stem Cell Reports 8 278–289. 10.1016/j.stemcr.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkar R., Lee R. T. (2010). Intramyocardial fibroblast myocyte communication. Circ. Res. 106 47–57. 10.1161/CIRCRESAHA.109.207456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I., Senyei G. D., Hansen J., Kong C.-W., Azeloglu E. U., Stillitano F., et al. (2014). Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Karakikes 3 18–31. 10.5966/sctm.2013-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S. J., Witty A. D., Gagliardi M., Dubois N. C., Niapour M., Hotta A., et al. (2011). Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8 228–240. 10.1016/j.stem.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Kehat I., Kenyagin-Karsenti D., Snir M., Segev H., Amit M., Gepstein A., et al. (2001). Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J. Clin. Invest. 108 407–414. 10.1172/JCI200112131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanabdali R., Rosdah A. A., Dusting G. J., Lim S. Y. (2016). Harnessing the secretome of cardiac stem cells as therapy for ischemic heart disease. Biochem. Pharmacol. 113 1–11. 10.1016/j.bcp.2016.02.012 [DOI] [PubMed] [Google Scholar]
- Kim S., Jeon J. M., Kwon O. K., Choe M. S., Yeo H. C., Peng X., et al. (2019). Comparative proteomic analysis reveals the upregulation of ketogenesis in cardiomyocytes differentiated from induced pluripotent stem cells. Proteomics 1800284 1–12. 10.1002/pmic.201800284 [DOI] [PubMed] [Google Scholar]
- Kita-Matsuo H., Barcova M., Prigozhina N., Salomonis N., Wei K., Jacot J. G., et al. (2009). Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One 4:e5046. 10.1371/journal.pone.0005046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivelä R., Hemanthakumar K. A., Vaparanta K., Robciuc M., Izumiya Y., Kidoya H., et al. (2019). Endothelial cells regulate physiological cardiomyocyte growth via VEGFR2 -mediated paracrine signaling. Circulation 139 2570–2584. 10.1161/CIRCULATIONAHA.118.036099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas F., Mahmoud A. I., Sosic D., Porrello E. R., Chen R., Garcia J. A., et al. (2012). The hypoxic epicardial and subepicardial microenvironment. J. Cardiovasc. Transl. Res. 5 654–665. 10.1007/s12265-012-9366-9367 [DOI] [PubMed] [Google Scholar]
- Kolanowski T. J., Antos C. L., Guan K. (2017). Making human cardiomyocytes up to date: derivation, maturation state and perspectives. Int. J. Cardiol. 241 379–386. 10.1016/j.ijcard.2017.03.099 [DOI] [PubMed] [Google Scholar]
- Konze S. A., Cajic S., Oberbeck A., Hennig R., Pich A., Rapp E., et al. (2017a). Quantitative assessment of sialo-glycoproteins and n-glycans during cardiomyogenic differentiation of human induced pluripotent stem cells. ChemBioChem 18 1317–1331. 10.1002/cbic.201700100 [DOI] [PubMed] [Google Scholar]
- Konze S. A., Werneburg S., Oberbeck A., Olmer R., Kempf H., Jara-Avaca M., et al. (2017b). Proteomic analysis of human pluripotent stem cell cardiomyogenesis revealed altered expression of metabolic enzymes and PDLIM5 isoforms. J. Proteome Res. 16 1133–1149. 10.1021/acs.jproteome.6b00534 [DOI] [PubMed] [Google Scholar]
- Kular J. K., Basu S., Sharma R. I. (2014). The extracellular matrix: Structure, composition, age-related differences, tools for analysis and applications for tissue engineering. J. Tissue Eng. 5:204173141455711. 10.1177/2041731414557112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K. T., Jones D. C., Sperber H., Madan A., Fischer K. A., Rodriguez M. L., et al. (2015). Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 112 E2785–E2794. 10.1073/pnas.1424042112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M. A., Chen K. Y., Naumova A. V., Muskheli V., Fugate J. A., Dupras S. K., et al. (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 25 1015–1024. 10.1038/nbt1327 [DOI] [PubMed] [Google Scholar]
- LaFramboise W. A., Scalise D., Stoodley P., Graner S. R., Guthrie R. D., Magovern J. A., et al. (2007). Cardiac fibroblasts influence cardiomyocyte phenotype in vitro. Am. J. Physiol. Physiol. 292 C1799–C1808. 10.1152/ajpcell.00166.2006 [DOI] [PubMed] [Google Scholar]
- Le T., Chong J. (2017). Cardiac progenitor cells for heart repair. Cell Death Discov. 2 1–4. 10.1038/cddiscovery.2016.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Protze S. I., Laksman Z., Backx P. H., Keller G. M. (2017). Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21 179.e–194.e. 10.1016/j.stem.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Lee P.-F., Chau E., Cabello R., Yeh A. T., Sampaio L. C., Gobin A. S., et al. (2017). Inverted orientation improves decellularization of whole porcine hearts. Acta Biomater. 49 181–191. 10.1016/j.actbio.2016.11.047 [DOI] [PubMed] [Google Scholar]
- Leitolis A., Suss P. H., Roderjan J. G., Angulski A. B. B., da Costa F. D. A., Stimamiglio M. A., et al. (2019). Human heart explant-derived extracellular vesicles: characterization and effects on the in vitro recellularization of decellularized heart valves. Int. J. Mol. Sci. 20:1279. 10.3390/ijms20061279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine M. D., Mannhardt I., Breckwoldt K., Prondzynski M., Flenner F., Ulmer B., et al. (2017). Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci. Rep. 7:5464. 10.1038/s41598-017-05600-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Yang H., Wang X., Zhan Y., Sheng W., Cai H., et al. (2017). Engineering human ventricular heart muscles based on a highly efficient system for purification of human pluripotent stem cell-derived ventricular cardiomyocytes. Stem Cell Res. Ther. 8 1–18. 10.1186/s13287-017-0651-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lin B., Yang L. (2015). Comparative transcriptomic analysis of multiple cardiovascular fates from embryonic stem cells predicts novel regulators in human cardiogenesis. Sci. Rep. 5:9758. 10.1038/srep09758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S. M., Zhu K., Hazeltine L. B., Bao X., et al. (2012). Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8 162–175. 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Kim J., Li Y., Pan H., Carvajal-Vergara X., Salama G., et al. (2012). High-purity enrichment of functional cardiovascular cells from human iPS cells. Cardiovasc. Res. 95 327–335. 10.1093/cvr/cvs185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindoso R. S., Sandim V., Collino F., Carvalho A. B., Dias J., da Costa M. R., et al. (2016). Proteomics of cell-cell interactions in health and disease. Proteomics 16 328–344. 10.1002/pmic.201500341 [DOI] [PubMed] [Google Scholar]
- Lindsey M. L., Jung M., Hall M. E., DeLeon-Pennell K. Y. (2018). Proteomic analysis of the cardiac extracellular matrix: clinical research applications. Expert Rev. Proteomics 15 105–112. 10.1080/14789450.2018.1421947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey S. E., Butcher J. T., Yalcin H. C. (2014). Mechanical regulation of cardiac development. Front. Physiol. 5:318. 10.3389/fphys.2014.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Lee B. W., Nakanishi K., Villasante A., Williamson R., Metz J., et al. (2018). Cardiac recovery via extended cell-free delivery of extracellular vesicles secreted by cardiomyocytes derived from induced pluripotent stem cells. Nat. Biomed. Eng. 2 293–303. 10.1038/s41551-018-0229-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Jiang C., Xu J., Zhao M. T., Van Bortle K., Cheng X., et al. (2017). Genome-wide temporal profiling of transcriptome and open chromatin of early cardiomyocyte differentiation derived from hiPSCs and hESCs. Circ. Res. 121 376–391. 10.1161/CIRCRESAHA.116.310456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zhang R., Guo T., Ma S., Han D., Li X. J., et al. (2015). Cardiotrophin-1 promotes cardiomyocyte differentiation from mouse induced pluripotent stem cells via JAK2/STAT3/Pim-1 signaling pathway. J. Geriatr. Cardiol. 12 591–599. 10.11909/j.issn.1671-5411.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Foley A. C. (2011). Signaling pathways in early cardiac development. Wiley Interdiscip. Rev. Syst. Biol. Med. 3 191–205. 10.1002/wsbm.112 [DOI] [PubMed] [Google Scholar]
- Lock M. C., Tellam R. L., Botting K. J., Wang K. C. W., Selvanayagam J. B., Brooks D. A., et al. (2018). The role of miRNA regulation in fetal cardiomyocytes, cardiac maturation and the risk of heart disease in adults. J. Physiol. 596 5625–5640. 10.1113/JP276072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart M., Wirrig E., Phelps A., Wessels A. (2011). Extracellular matrix and heart development. Birth Defects Res. A Clin. Mol. Teratol. 91 535–550. 10.1002/bdra.20810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Kaestle K., Huang J., Liu Q., Zhang P., Gao L., et al. (2017). Interactions of human embryonic stem cell-derived cardiovascular progenitor cells with immobilized extracellular matrix proteins. J. Biomed. Mater. Res. Part A 105 1094–1104. 10.1002/jbm.a.36005 [DOI] [PubMed] [Google Scholar]
- Lu T.-Y., Lin B., Kim J., Sullivan M., Tobita K., Salama G., et al. (2013). Repopulation of decellularized mouse heart with human induced pluripotent stem cell-derived cardiovascular progenitor cells. Nat. Commun. 4:2307. 10.1038/ncomms3307 [DOI] [PubMed] [Google Scholar]
- Lundy S. D., Zhu W.-Z., Regnier M., Laflamme M. A. (2013). Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 22 1991–2002. 10.1089/scd.2012.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu L., Wang H., Li B., Qin Q., Qi L., Nagarkatti M., et al. (2013). A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J. Mol. Cell Cardiol. 82 34–44. 10.1016/j.yjmcc.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Yu C., Wang P., Xu W., Wan X., Lai C. S. E., et al. (2018). Rapid 3D bioprinting of decellularized extracellular matrix with regionally varied mechanical properties and biomimetic microarchitecture. Biomaterials 185 310–321. 10.1016/J.BIOMATERIALS.2018.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiraju P., Greenway S. C. (2019). Current methods for the maturation of induced pluripotent stem cell-derived cardiomyocytes. World J. Stem Cells 11 33–43. 10.4252/wjsc.v11.i1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkut S., Dingal P. C. D. P., Discher D. E. (2014). Stress Sensitivity and mechanotransduction during heart development. Curr. Biol. 24 R495–R501. 10.1016/j.cub.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. L., Blaxall B. C. (2012). Cardiac intercellular communication: are myocytes and fibroblasts fair-weather friends? J. Cardiovasc. Transl. Res. 5 768–782. 10.1007/s12265-012-9404-9405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin M. J., Di Rocco G., Gardiner A., Bush S. M., Lassar A. B. (2001). Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 15 316–327. 10.1101/gad.855501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayourian J., Ceholski D. K., Gorski P. A., Mathiyalagan P., Murphy J. F., Salazar S. I., et al. (2018). Exosomal microRNA-21-5p mediates mesenchymal stem cell paracrine effects on human cardiac tissue contractility. Circ. Res. 122 933–944. 10.1161/CIRCRESAHA.118.312420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam A. S., Afshari J. T., Esmaeili S.-A., Saburi E., Joneidi Z., Momtazi-Borojeni A. A. (2019). Cardioprotective microRNAs: lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis 285 1–9. 10.1016/j.atherosclerosis.2019.03.016 [DOI] [PubMed] [Google Scholar]
- Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R., et al. (2003). Differentiation of human embryonic stem cells to cardiomyocytes. Circulation 107 2733–2740. 10.1161/01.CIR.0000068356.38592.68 [DOI] [PubMed] [Google Scholar]
- Mummery C. L., Ward D., Passier R. (2007). Differentiation of human embryonic stem cells to cardiomyocytes by coculture with endoderm in serum-free medium. Curr. Protoc. Stem Cell Biol. 2:1F.2.1-1F.2.14. 10.1002/9780470151808.sc01f02s2 [DOI] [PubMed] [Google Scholar]
- Mummery C. L., Zhang J., Ng E. S., Elliott D. A., Elefanty A. G., Kamp T. J. (2012). Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 111 344–358. 10.1161/CIRCRESAHA.110.227512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C. E., Keller G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132 661–680. 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Naba A., Clauser K. R., Ding H., Whittaker C. A., Carr S. A., Hynes R. O. (2016). The extracellular matrix: tools and insights for the “omics” era. Matrix Biol. 49 10–24. 10.1016/j.matbio.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A., Clauser K. R., Hoersch S., Liu H., Carr S. A., Hynes R. O. (2012). The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics 11 M111.014647. 10.1074/mcp.M111.014647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. L. J., Narayanan K., Gao S., Wan A. C. A. (2011). Lineage restricted progenitors for the repopulation of decellularized heart. Biomaterials 32 7571–7580. 10.1016/j.biomaterials.2011.06.065 [DOI] [PubMed] [Google Scholar]
- Nguyen D. T., O’Hara M., Graneli C., Hicks R., Miliotis T., Nyström A. C., et al. (2018). Humanizing miniature hearts through 4-flow cannulation perfusion decellularization and recellularization. Sci. Rep. 8 1–10. 10.1038/s41598-018-25883-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor N., Shapira A., Edri R., Gal I., Wertheim L., Dvir T. (2019). 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 6:1900344. 10.1002/advs.201900344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes S. S., Miklas J. W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., et al. (2013). Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes. Nat. Methods 10 781–787. 10.1038/nmeth.2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberwallner B., Brodarac A., Aniæ P., Šariæ T., Wassilew K., Neef K., et al. (2015). Human cardiac extracellular matrix supports myocardial lineage commitment of pluripotent stem cells. Eur. J. Cardio-thoracic Surg. 47 416–425. 10.1093/ejcts/ezu163 [DOI] [PubMed] [Google Scholar]
- Ogasawara T., Okano S., Ichimura H., Kadota S., Tanaka Y., Minami I., et al. (2017). Impact of extracellular matrix on engraftment and maturation of pluripotent stem cell-derived cardiomyocytes in a rat myocardial infarct model. Sci. Rep. 7:8630. 10.1038/s41598-017-09217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle B. M., Bursac N., Domian I., Huang N. F., Menasché P., Murry C. E., et al. (2016). Distilling complexity to advance cardiac tissue engineering. Sci. Transl. Med. 8 s13–s342. 10.1126/scitranslmed.aad2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott H. C., Matthiesen T. S., Goh S.-K., Black L. D., Kren S. M., Netoff T. I., et al. (2008). Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat. Med. 14 213–221. 10.1038/nm1684 [DOI] [PubMed] [Google Scholar]
- Paige S. L., Plonowska K., Xu A., Wu S. M. (2015). Molecular regulation of cardiomyocyte differentiation. Circ. Res. 116 341–353. 10.1161/CIRCRESAHA.116.302752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige S. L., Thomas S., Stoick-Cooper C. L., Wang H., Maves L., Sandstrom R., et al. (2012). A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151 221–232. 10.1016/j.cell.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis A., Xiao D., Zhou J., Zhang L. (2014). Endothelin-1 promotes cardiomyocyte terminal differentiation in the developing heart via heightened DNA methylation. Int. J. Med. Sci. 11 373–380. 10.7150/ijms.7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotty D. M., Klinger R. Y., Kirkton R. D., Bursac N. (2009). Cardiac fibroblast paracrine factors alter impulse conduction and ion channel expression of neonatal rat cardiomyocytes. Cardiovasc. Res. 83 688–697. 10.1093/cvr/cvp164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gil I., Gálvez-Montón C., Prat-Vidal C., Jorba I., Segú-Vergés C., Roura S., et al. (2018). Head-to-head comparison of two engineered cardiac grafts for myocardial repair: from scaffold characterization to pre-clinical testing. Sci. Rep. 8 1–13. 10.1038/s41598-018-25115-25112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira I. T., Spangenberg L., Robert A. W., Amorín R., Stimamiglio M. A., Naya H., et al. (2018). Polysome profiling followed by RNA-seq of cardiac differentiation stages in hESCs. Sci. Data 5 1–11. 10.1038/sdata.2018.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira I. T., Spangenberg L., Robert A. W., Amorín R., Stimamiglio M. A., Naya H., et al. (2019). Cardiomyogenic differentiation is fine-tuned by differential mRNA association with polysomes. BMC Genomics 20:219. 10.1186/s12864-019-5550-5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. R., Ilinykh A., Ivey M. J., Kuwabara J. T., D’antoni M. L., Debuque R., et al. (2016). Revisiting cardiac cellular composition. Circ. Res. 118 400–409. 10.1161/CIRCRESAHA.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piryani S. O., Jiao Y., Kam A. Y. F., Liu Y., Vo-Dinh T., Chen B. J., et al. (2019). Endothelial cell-derived extracellular vesicles mitigate radiation-induced hematopoietic injury. Int. J. Radiat. Oncol. Biol. Phys. 104 291–301. 10.1016/j.ijrobp.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E., Keung W., Liang Y., Ramalingam R., Yan B., Zhang S., et al. (2015). Proteomic analysis of human pluripotent stem cell-derived, fetal, and adult ventricular cardiomyocytes reveals pathways crucial for cardiac metabolism and maturation. Circ. Cardiovasc. Genet. 8 427–436. 10.1161/CIRCGENETICS.114.000918 [DOI] [PubMed] [Google Scholar]
- Protze S. I., Liu J., Nussinovitch U., Ohana L., Backx P. H., Gepstein L., et al. (2016). Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 35 56–68. 10.1038/nbt.3745 [DOI] [PubMed] [Google Scholar]
- Pursani V., Pethe P., Bashir M., Sampath P., Tanavde V., Bhartiya D. (2017). Genetic and epigenetic profiling reveals EZH2-mediated down regulation of OCT-4 involves NR2F2 during cardiac differentiation of human embryonic stem cells. Sci. Rep. 7 1–14. 10.1038/s41598-017-13442-13449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft S. M., Pointon A., Williams A. W., Cross M. J., Sidaway J. E. (2016). Cardiac non-myocyte cells show enhanced pharmacological function suggestive of contractile maturity in stem cell derived cardiomyocyte microtissues. Toxicol. Sci. 152 99–112. 10.1093/toxsci/kfw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Lee M. Y., Schliffke S., Paavola J., Amos P. J., Ge X., et al. (2011). Small molecule Wnt inhibitors enhance the efficiency of BMP-4-directed cardiac differentiation of human pluripotent stem cells. J. Mol. Cell. Cardiol. 51 280–287. 10.1016/j.yjmcc.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus T. L., Robert A. W., Da Costa M. B. A., de Aguiar A. M., Stimamiglio M. A. (2016). Secretome from resident cardiac stromal cells stimulates proliferation, cardiomyogenesis and angiogenesis of progenitor cells. Int. J. Cardiol. 221 396–403. 10.1016/j.ijcard.2016.06.199 [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S., Salza R. (2014). Matricryptins and matrikines: biologically active fragments of the extracellular matrix. Exp. Dermatol. 23 457–463. 10.1111/exd.12435 [DOI] [PubMed] [Google Scholar]
- Rienks M., Papageorgiou A.-P. (2016). Novel regulators of cardiac inflammation: matricellular proteins expand their repertoire. J. Mol. Cell. Cardiol. 91 172–178. 10.1016/j.yjmcc.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Rienks M., Papageorgiou A.-P., Frangogiannis N. G., Heymans S. (2014). Myocardial extracellular matrix: an ever-changing and diverse entity. Circ. Res. 114 872–888. 10.1161/CIRCRESAHA.114.302533 [DOI] [PubMed] [Google Scholar]
- Robert A. W., Schittini A. V., Marchini F. K., Batista M., Affonso Da Costa M. B., Senegaglia A. C., et al. (2017). Tissue-derived signals for mesenchymal stem cell stimulation: role of cardiac and umbilical cord microenvironments. Cells Tissues Organs 203 173–182. 10.1159/000450600 [DOI] [PubMed] [Google Scholar]
- Robertson C., Tran D. D., George S. C. (2013). Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells 31 829–837. 10.1002/stem.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S. P., Yeager K., Chen T., Song L., Sirabella D., et al. (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556 239–243. 10.1038/s41586-018-0016-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozario T., DeSimone D. W. (2010). The extracellular matrix in development and morphogenesis: a dynamic view. Dev. Biol. 341 126–140. 10.1016/j.ydbio.2009.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C. E., Coulombe K. L. K. (2015). The roles of neuregulin-1 in cardiac development, homeostasis, and disease. Biomark. Insights 10 1–9. 10.4137/BMIMI.S20061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C. E., Coulombe K. L. K. (2017). IGF1 and NRG1 enhance proliferation, metabolic maturity, and the force-frequency response in hESC-derived engineered cardiac tissues. Stem Cells Int. 2017 1–13. 10.1155/2017/7648409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa S., Wong L., McCloskey K. E. (2014). Combinatorial fibronectin and laminin signaling promote highly efficient cardiac differentiation of human embryonic stem cells. Biores. Open Access 3 150–161. 10.1089/biores.2014.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal R., Sappa P. K., Gesell Salazar M., Wenzel K., Reinke Y., Völker U., et al. (2018). Global secretome analysis of resident cardiac progenitor cells from wild-type and transgenic heart failure mice: why ambience matters. J. Cell. Physiol. 234 10111–10122. 10.1002/jcp.27677 [DOI] [PubMed] [Google Scholar]
- Sánchez P. L., Fernández-Santos M. E., Costanza S., Climent A. M., Moscoso I., Gonzalez-Nicolas M. A., et al. (2015). Acellular human heart matrix: a critical step toward whole heart grafts. Biomaterials 61 279–289. 10.1016/j.biomaterials.2015.04.056 [DOI] [PubMed] [Google Scholar]
- Sartiani L., Bettiol E., Stillitano F., Mugelli A., Cerbai E., Jaconi M. E. (2007). Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells 25 1136–1144. 10.1634/stemcells.2006-2466 [DOI] [PubMed] [Google Scholar]
- Scarrit M. E., Pashos N. C., Bunnell B. A. (2015). A review of cellularization strategies for tissue engineering of whole organs. Front. Bioeng. Biotechnol. 3:43. 10.3389/fbioe.2015.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenke-Layland K., Nsair A., Van Handel B., Angelis E., Gluck J. M., Votteler M., et al. (2011). Recapitulation of the embryonic cardiovascular progenitor cell niche. Biomaterials 32 2748–2756. 10.1016/j.biomaterials.2010.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittini A. V., Celedon P. F., Stimamiglio M. A., Krieger M., Hansen P., Da Costa F. D. A., et al. (2010). Human cardiac explant-conditioned medium: soluble factors and cardiomyogenic effect on mesenchymal stem cells. Exp. Biol. Med. 235 1015–1024. 10.1258/ebm.2010.010003 [DOI] [PubMed] [Google Scholar]
- Schofield R. (1978). The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4 7–25. [PubMed] [Google Scholar]
- Schwan J., Kwaczala A. T., Ryan T. J., Bartulos O., Ren Y., Sewanan L. R., et al. (2016). Anisotropic engineered heart tissue made from laser-cut decellularized myocardium. Sci. Rep. 6 1–12. 10.1038/srep32068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrin I. Y., Allen B. W., Qian Y., Jackman C. P., Carlson A. L., Juhas M. E., et al. (2017). Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat. Commun. 8:1825. 10.1038/s41467-017-01946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. M. (1996). Paracrine modulation of heart cell function by endothelial cells. Cardiovasc. Res. 31 847–867. 10.1016/0008-6363(96)00025-29 [DOI] [PubMed] [Google Scholar]
- Sharma S., Mishra R., Bigham G. E., Wehman B., Khan M. M., Xu H., et al. (2017). A deep proteome analysis identifies the complete secretome as the functional unit of human cardiac progenitor cells. Circ. Res. 120 816–834. 10.1161/CIRCRESAHA.116.309782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Mishra R., Simpson D., Wehman B., Colletti E. J., Deshmukh S., et al. (2015). Cardiosphere-derived cells from pediatric end-stage heart failure patients have enhanced functional activity due to the heat shock response regulating the secretome. Stem Cells 33 1213–1229. 10.1002/stem.1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. S. T., Macadangdang J., Leung W., Laflamme M. A., Kim D.-H. (2017). Human iPSC-derived cardiomyocytes and tissue engineering strategies for disease modeling and drug screening. Biotechnol. Adv. 35 77–94. 10.1016/J.BIOTECHADV.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stik G., Crequit S., Petit L., Durant J., Charbord P., Jaffredo T., et al. (2017). Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells. J Cell Biol. 216 2217–2230. 10.1083/JCB.201601109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel W. L., Kaplan D. L., Black L. D. (2016). Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv. Drug Deliv. Rev. 96 135–155. 10.1016/j.addr.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Kontaridis M. I. (2018). Physiology of cardiac development: from genetics to signaling to therapeutic strategies. Curr. Opin. Physiol. 1 123–139. 10.1016/j.cophys.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylva M., van den Hoff M. J. B., Moorman A. F. M. (2014). Development of the human heart. Am. J. Med. Genet. Part A 164 1347–1371. 10.1002/ajmg.a.35896 [DOI] [PubMed] [Google Scholar]
- Takeda N., Manabe I. (2011). Cellular interplay between cardiomyocytes and nonmyocytes in cardiac remodeling. Int. J. Inflam. 2011 1–13. 10.4061/2011/535241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist M. D., Molkentin J. D. (2017). Redefining the identity of cardiac fibroblasts. Nat. Rev. Cardiol. 14 484–491. 10.1038/nrcardio.2017.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talman V., Kivelä R. (2018). Cardiomyocyte—endothelial cell interactions in cardiac remodeling and regeneration. Front. Cardiovasc. Med. 5:101. 10.3389/fcvm.2018.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang-Quan K. R., Mehta N. A., Sampaio L. C., Taylor D. A. (2018). “Whole cardiac tissue bioscaffolds,” in Advances in Experimental Medicine and Biology, eds Schmuck E. G., Hematti P., Raval A. N. (Cham: Springer International Publishing; ), 85–114. 10.1007/978-3-319-97421-7_5 [DOI] [PubMed] [Google Scholar]
- Thimm T. N., Squirrell J. M., Liu Y., Eliceiri K. W., Ogle B. M. (2015). Endogenous optical signals reveal changes of elastin and collagen organization during differentiation of mouse embryonic stem cells. Tissue Eng. Part C Methods 21 995–1004. 10.1089/ten.tec.2014.0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirziu D., Giordano F. J., Simons M. (2010). Cell communications in the heart. Circulation 122 928–937. 10.1161/CIRCULATIONAHA.108.847731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., et al. (2013). Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12 127–137. 10.1016/j.stem.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Tompkins J. D., Jung M., Chen C. Y., Lin Z., Ye J., Godatha S., et al. (2016). Mapping human pluripotent-to-cardiomyocyte differentiation: methylomes, transcriptomes, and exon DNA methylation “memories. EBioMedicine 4 74–85. 10.1016/j.ebiom.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torán J. L., López J. A., Gomes-Alves P., Aguilar S., Torroja C., Trevisan-Herraz M., et al. (2019). Definition of a cell surface signature for human cardiac progenitor cells after comprehensive comparative transcriptomic and proteomic characterization. Sci. Rep. 9 1–16. 10.1038/s41598-019-39571-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torella D., Ellison G. M., Méndez-Ferrer S., Ibanez B., Nadal-Ginard B. (2006). Resident human cardiac stem cells: role in cardiac cellular homeostasis and potential for myocardial regeneration. Nat. Clin. Pract. Cardiovasc. Med. 3 8–13. 10.1038/ncpcardio0409 [DOI] [PubMed] [Google Scholar]
- Tulloch N. L., Muskheli V., Razumova M. V., Korte F. S., Regnier M., Hauch K. D., et al. (2011). Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ. Res. 109 47–59. 10.1161/CIRCRESAHA.110.237206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyukavin A. I., Belostotskaya G. B., Golovanova T. A., Galagudza M. M., Zakharov E. A., Burkova N. V., et al. (2015). Stimulation of proliferation and differentiation of rat resident myocardial cells with apoptotic bodies of cardiomyocytes. Bull. Exp. Biol. Med. 159 138–141. 10.1007/s10517-015-2909-6 [DOI] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A. D., Golob J. L., Pabon L., et al. (2007). Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104 9685–9690. 10.1073/pnas.0702859104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D., Dormeyer W., Braam S. R., Passier R., Monshouwer-kloots J., Oostwaard D. W., et al. (2010). Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res. 9 1610–1618. 10.1021/pr901138a [DOI] [PubMed] [Google Scholar]
- van Laake L. W., van Donselaar E. G., Monshouwer-Kloots J., Schreurs C., Passier R., Humbel B. M., et al. (2010). Extracellular matrix formation after transplantation of human embryonic stem cell-derived cardiomyocytes. Cell. Mol. Life Sci. 67 277–290. 10.1007/s00018-009-0179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidarsson H., Hyllner J., Sartipy P. (2010). Differentiation of human embryonic stem cells to cardiomyocytes for in vitro and in vivo applications. Stem Cell Rev. Reports 6 108–120. 10.1007/s12015-010-9113-x [DOI] [PubMed] [Google Scholar]
- Wagner M., Siddiqui M. A. (2007). Signal transduction in early heart development (I): cardiogenic induction and heart tube formation. Exp. Biol. Med. 232 852–865. [PubMed] [Google Scholar]
- Wang Y., Zhao R., Liu D., Deng W., Xu G., Liu W., et al. (2018). Exosomes derived from miR-214-enriched bone marrow-derived mesenchymal stem cells regulate oxidative damage in cardiac stem cells by targeting CaMKII. Oxid. Med. Cell. Longev. 2018 1–21. 10.1155/2018/4971261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Long D. W., Huang Y., Chen W. C. W., Kim K., Wang Y. (2019). Decellularized neonatal cardiac extracellular matrix prevents widespread ventricular remodeling in adult mammals after myocardial infarction. Acta Biomater. 87 140–151. 10.1016/j.actbio.2019.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt S., Goetzenich A., Goettsch C., Stoppe C., Bleilevens C., Kraemer S., et al. (2018). Evaluation of the cardioprotective potential of extracellular vesicles – a systematic review and meta-analysis. Sci. Rep. 8 1–14. 10.1038/s41598-018-33862-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann A., Patil N. P., Sabashnikov A., Jungebluth P., Korkmaz S., Li S., et al. (2014). Bioartificial heart: a human-sized porcine model – the way ahead. PLoS One 9:e111591. 10.1371/journal.pone.0111591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. C., Pang L., Yang X. (2016). MicroRNA-mediated maturation of human pluripotent stem cell-derived cardiomyocytes: towards a better model for cardiotoxicity? Food Chem. Toxicol. 98 17–24. 10.1016/j.fct.2016.05.025 [DOI] [PubMed] [Google Scholar]
- Wolling H., Konze S. A., Höfer A., Erdmann J., Pich A., Zweigerdt R., et al. (2018). Quantitative secretomics reveals extrinsic signals involved in human pluripotent stem cell cardiomyogenesis. Proteomics 18 1–12. 10.1002/pmic.201800102 [DOI] [PubMed] [Google Scholar]
- Xin T., Greco V., Myung P. (2016). Hardwiring stem cell communication through tissue structure. Cell 164 1212–1225. 10.1016/j.cell.2016.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Police S., Rao N., Carpenter M. K. (2002). Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ. Res. 91 501–508. 10.1161/01.RES.0000035254.80718.91 [DOI] [PubMed] [Google Scholar]
- Xu Q. X., Zweigerdt R., Soo S. Y., Ngoh Z. X., Tham S. C., Wang S. T., et al. (2008). Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy 10 376–389. 10.1080/14653240802105307 [DOI] [PubMed] [Google Scholar]
- Xu X. Q., Soo S. Y., Sun W., Zweigerdt R. (2009). Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cells 27 2163–2174. 10.1002/stem.166 [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M. H., Adler E. D., Roepke T. K., Kattman S. J., Kennedy M., et al. (2008). Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453 524–528. 10.1038/nature06894 [DOI] [PubMed] [Google Scholar]
- Yang X., Pabon L., Murry C. E. (2014). Engineering adolescence: maturation of human pluripotent stem cell–derived cardiomyocytes. Circ. Res. 114 511–523. 10.1161/CIRCRESAHA.114.300558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap L., Wang J.-W., Moreno-Moral A., Chong L. Y., Sun Y., Harmston N., et al. (2019). In Vivo generation of post-infarct human cardiac muscle by laminin-promoted cardiovascular progenitors. Cell Rep. 26 3231.e–3245.e. 10.1016/j.celrep.2019.02.083 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Miyagawa S., Fukushima S., Kawamura T., Kashiyama N., Ohashi F., et al. (2018). Maturation of human induced pluripotent stem cell-derived cardiomyocytes by soluble factors from human mesenchymal stem cells. Mol. Ther. 26 2681–2695. 10.1016/j.ymthe.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Ma X., Zhu W., Wang P., Miller K. L., Stupin J., et al. (2019). Scanningless and continuous 3D bioprinting of human tissues with decellularized extracellular matrix. Biomaterials 194 1–13. 10.1016/J.BIOMATERIALS.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Shadrin I. Y., Lam J., Xian H.-Q., Snodgrass H. R., Bursac N. (2013). Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 34 5813–5820. 10.1016/j.biomaterials.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Klos M., Wilson G. F., Herman A. M., Lian X., Raval K. K., et al. (2012). Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res. 111 1125–1136. 10.1161/CIRCRESAHA.112.273144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Su J., Mende U. (2012). Cross talk between cardiac myocytes and fibroblasts: from multiscale investigative approaches to mechanisms and functional consequences. Am. J. Physiol. Circ. Physiol. 303 H1385–H1396. 10.1152/ajpheart.01167.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Guo J. P., Chi Y. L., Liu Y. C., Zhang C. S., Yang X. Q., et al. (2012). Endothelin-induced differentiation of Nkx2.5 + cardiac progenitor cells into pacemaking cells. Mol. Cell. Biochem. 366 309–318. 10.1007/s11010-012-1309-8 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wilson G. F., Soerens A. G., Koonce C. H., Yu J., Palecek S. P., et al. (2009). Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 104 30–41. 10.1161/CIRCRESAHA.108.192237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., et al. (2011). Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 21 579–587. 10.1038/cr.2010.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kong C. W., Tong M. H., Chooi W. H., Huang N., Li R. A., et al. (2017). Maturation of human embryonic stem cell-derived cardiomyocytes (hESC-CMs) in 3D collagen matrix: effects of niche cell supplementation and mechanical stimulation. Acta Biomater. 49 204–217. 10.1016/j.actbio.2016.11.058 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sivakumaran P., Newcomb A. E., Hernandez D., Harris N., Khanabdali R., et al. (2015). Cardiac repair with a novel population of mesenchymal stem cells resident in the human heart. Stem Cells 33 3100–3113. 10.1002/stem.2101 [DOI] [PubMed] [Google Scholar]
- Zhu R., Blazeski A., Poon E., Costa K. D., Tung L., Boheler K. R. (2014). Physical developmental cues for the maturation of human pluripotent stem cell-derived cardiomyocytes. Stem Cell Res. Ther. 5:117. 10.1186/scrt507 [DOI] [PMC free article] [PubMed] [Google Scholar]