Abstract
In the era of precision medicine, radiotherapy strategies should be determined based on genetic profiles that predict tumor radiosensitivity. Accordingly, pre-clinical research aimed at discovering clinically applicable genetic profiles is needed. However, how a given genetic profile affects cancer cell radiosensitivity is unclear. To address this issue, we performed a pilot in vitro study by utilizing EGFR mutational status as a model for genetic profile. Clonogenic assays of EGFR mutant (n = 6) and wild-type (n = 9) non-small cell lung carcinoma (NSCLC) cell lines were performed independently by two oncologists. Clonogenic survival parameters SF2, SF4, SF6, SF8, mean inactivation dose (MID), D10, D50, α, and β were obtained using the linear quadratic model. The differences in the clonogenic survival parameters between the EGFR mutant and wild-type cell lines were assessed using the Mann–Whitney U test. As a result, for both datasets, the p values for SF2, SF4, D50, α, and α/β were below 0.05, and those for SF2 were lowest. These data indicate that a genetic profile of NSCLC cell lines might be predictive for their radiation response; i.e., EGFR mutant cell lines might be more sensitive to low dose- and low fraction sized-irradiation.
Keywords: precision medicine, radiation therapy, radiosensitivity, clonogenic assays, gene mutations
1. Introduction
Radiation therapy is one of the most important treatments for cancer. In the clinical practice of definitive radiation therapy, the doses required for clinical tumor control vary widely even for tumors arising in the same anatomical sites. For example, an integrated analysis of 50 single-institution studies performed by Okunieff et al. revealed that the dose required for 50% clinical tumor control ranged from 21.4 to 90.3 Gy for breast cancer, 50.4 to 83.4 Gy for supraglottic cancer, and 24.3 to 64.4 Gy for cervical cancer [1]. These data highlight the need to establish predictive biomarkers of tumor radioresponsiveness that could advance personalization of radiation therapy.
In recent years, the concept of precision medicine, i.e., stratification of treatment strategy based on individual patients’ genetic profiles, has become widespread in cancer treatment, in part due to technological advances in next-generation sequencing [2]. Drugs that target tumors carrying specific genetic profiles have yielded favorable outcomes in the clinic [3,4,5,6]. This indicates that a given genetic profile may affect cancer cell radiosensitivity as well.
Clonogenic assays are the gold standard for assessing cancer cell radiosensitivity in pre-clinical settings [7]. Evidence compiled over the past few decades suggests that the radiosensitivity of cancer cells, as determined by clonogenic assays, is relevant to tumor response to radiation therapy [8,9,10,11]. Multiple clonogenic survival parameters have been used as radiosensitivity endpoints for clonogenic assays [12]. However, how a given genetic profile affects cancer cell radiosensitivity as assessed by various clonogenic survival parameters is unclear. To address this issue, we performed a pilot study by utilizing EGFR mutational status as a model for genetic profile.
2. Results
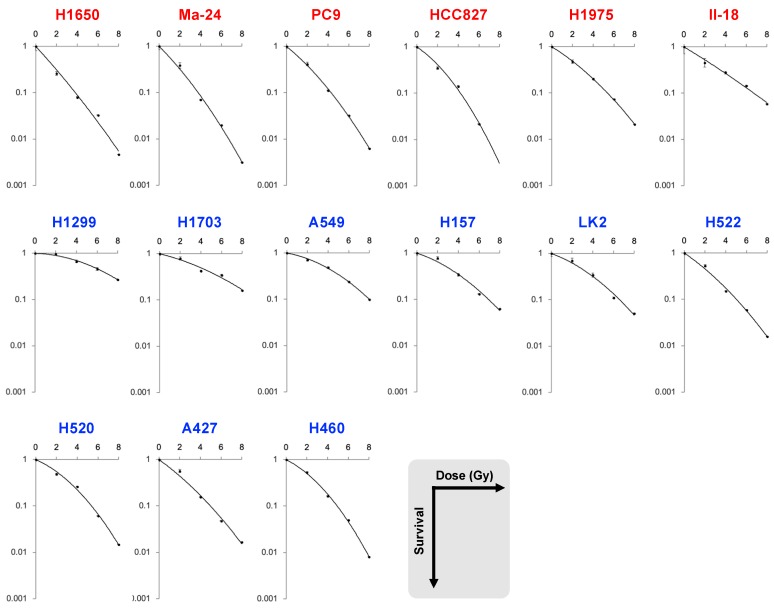
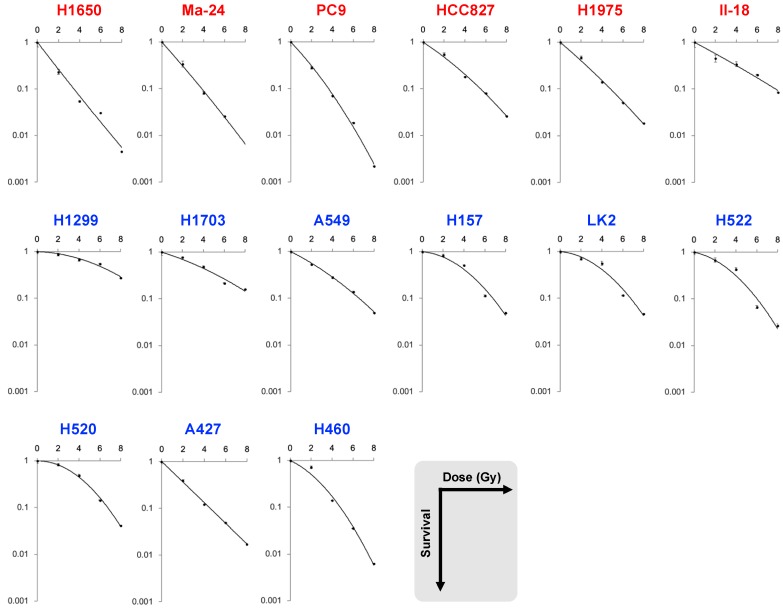
To analyze the association of a given genetic profile with clonogenic survival parameters, we chose to analyze EGFR mutation status in non-small cell lung carcinoma (NSCLC); EGFR status is associated with the response of NSCLCs to radiation therapy [13]. Two independent oncologists performed clonogenic assays after X-ray irradiation using EGFR mutant (n = 6) and wild-type (n = 9) cell lines; hereafter, the two datasets obtained are referred to as datasets A and B (Figure 1 and Figure 2). The following clonogenic survival parameters were obtained: SF2, SF4, SF6, SF8, α, and β of the linear quadratic (LQ) model, D10, D50, and the mean inactivation dose (MID) (Table 1). Radiosensitivities for the individual cell lines showed significantly high correlation between dataset A and B in terms of all the clonogenic survival parameters (Table 2), indicating technical robustness of the experiments.
Figure 1.
Dataset A: Survival curves for EGFR mutant (red) and wild-type (blue) non-small cell lung carcinoma cell lines treated with X-rays.
Figure 2.
Dataset B: Survival curves for EGFR mutant (red) and wild-type (blue) non-small cell lung carcinoma cell lines treated with X-rays.
Table 1.
Clonogenic survival parameters for datasets A and B.
| Dataset | Cell Line | EGFR | SF2 | SF4 | SF6 | SF8 | MID | D10 | D50 | α | β | α/β |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | H1650 | mutant | 0.26 | 0.079 | 0.033 | 0.0047 | 1.7 | 3.8 | 1.2 | 0.56 | 0.012 | 46.4 |
| Ma-24 | 0.39 | 0.070 | 0.020 | 0.0031 | 1.7 | 3.8 | 1.3 | 0.51 | 0.026 | 19.7 | ||
| PC9 | 0.42 | 0.11 | 0.032 | 0.0063 | 2.0 | 4.3 | 1.5 | 0.42 | 0.027 | 15.4 | ||
| HCC827 | 0.35 | 0.14 | 0.022 | NA | 2.0 | 4.2 | 1.6 | 0.34 | 0.048 | 7.2 | ||
| H1975 | 0.47 | 0.20 | 0.073 | 0.021 | 2.5 | 5.4 | 1.9 | 0.32 | 0.021 | 15.5 | ||
| II-18 | 0.45 | 0.28 | 0.15 | 0.058 | 3.0 | 6.7 | 2.2 | 0.31 | 0.0047 | 65.9 | ||
| H1299 | wild-type | 0.97 | 0.67 | 0.46 | 0.27 | 6.1 | 10.7 | 5.7 | 0.015 | 0.019 | 0.82 | |
| H1703 | 0.79 | 0.43 | 0.35 | 0.16 | 4.7 | 9.5 | 4.0 | 0.13 | 0.012 | 10.3 | ||
| A549 | 0.71 | 0.49 | 0.24 | 0.098 | 4.2 | 8.0 | 3.8 | 0.092 | 0.025 | 3.7 | ||
| H157 | 0.78 | 0.34 | 0.13 | 0.062 | 3.4 | 6.9 | 2.9 | 0.17 | 0.024 | 7.2 | ||
| LK2 | 0.70 | 0.34 | 0.11 | 0.051 | 3.2 | 6.6 | 2.7 | 0.19 | 0.025 | 7.5 | ||
| H522 | 0.53 | 0.15 | 0.059 | 0.016 | 2.3 | 5.0 | 1.8 | 0.35 | 0.021 | 17.0 | ||
| H520 | 0.49 | 0.26 | 0.061 | 0.015 | 2.6 | 5.4 | 2.3 | 0.22 | 0.038 | 5.8 | ||
| A427 | 0.57 | 0.15 | 0.047 | 0.017 | 2.2 | 5.0 | 1.7 | 0.37 | 0.019 | 19.3 | ||
| H460 | 0.53 | 0.16 | 0.050 | 0.0081 | 2.4 | 4.9 | 2.0 | 0.26 | 0.043 | 6.0 | ||
| p value | <0.001 | 0.018 | 0.049 | 0.082 | 0.054 | 0.025 | 0.0076 | 0.012 | 0.95 | 0.036 | ||
| B | H1650 | mutant | 0.23 | 0.054 | 0.031 | 0.0072 | 1.3 | 3.4 | 0.95 | 0.75 | −0.018 | NA |
| Ma-24 | 0.34 | 0.081 | 0.026 | NA | 1.7 | 3.8 | 1.2 | 0.58 | 0.0068 | 84.4 | ||
| PC9 | 0.28 | 0.070 | 0.019 | 0.0022 | 1.6 | 3.6 | 1.2 | 0.53 | 0.028 | 19.2 | ||
| HCC827 | 0.55 | 0.18 | 0.081 | 0.026 | 2.5 | 5.5 | 1.9 | 0.34 | 0.015 | 22.8 | ||
| H1975 | 0.47 | 0.14 | 0.051 | 0.018 | 2.1 | 4.8 | 1.5 | 0.44 | 0.0082 | 53.8 | ||
| II-18 | 0.45 | 0.34 | 0.20 | 0.085 | 3.4 | 7.8 | 2.5 | 0.27 | 0.0029 | 93.3 | ||
| H1299 | wild-type | 0.87 | 0.67 | 0.55 | 0.28 | 6.3 | 11.1 | 5.9 | 0.015 | 0.017 | 0.85 | |
| H1703 | 0.77 | 0.49 | 0.22 | 0.16 | 4.3 | 9.1 | 3.6 | 0.16 | 0.010 | 15.5 | ||
| A549 | 0.53 | 0.28 | 0.14 | 0.049 | 3.0 | 6.6 | 2.4 | 0.26 | 0.014 | 17.9 | ||
| H157 | 0.83 | 0.51 | 0.11 | 0.049 | 3.8 | 6.8 | 3.5 | 0.042 | 0.044 | 1.0 | ||
| LK2 | 0.70 | 0.56 | 0.12 | 0.046 | 3.8 | 6.7 | 3.5 | 0.044 | 0.044 | 1.0 | ||
| H522 | 0.68 | 0.43 | 0.066 | 0.027 | 3.2 | 6.0 | 2.9 | 0.11 | 0.046 | 2.3 | ||
| H520 | 0.83 | 0.49 | 0.14 | 0.041 | 3.9 | 6.8 | 3.7 | −0.005 | 0.051 | NA | ||
| A427 | 0.39 | 0.12 | 0.049 | 0.021 | 1.9 | 4.5 | 1.3 | 0.54 | −0.0068 | NA | ||
| H460 | 0.72 | 0.14 | 0.036 | 0.0062 | 2.4 | 4.8 | 2.0 | 0.24 | 0.051 | 4.7 | ||
| p value | 0.005 | 0.012 | 0.087 | 0.14 | 0.017 | 0.0660 | 0.0076 | 0.0048 | 0.049 | 0.0016 |
MID, mean inactivation dose.
Table 2.
Correlation of clonogenic survival parameters between datasets A and B.
| Parameters | R Values | p Values |
|---|---|---|
| SF2 | 0.73 | 0.003 |
| SF4 | 0.81 | <0.001 |
| SF6 | 0.86 | <0.001 |
| SF8 | 0.91 | <0.001 |
| MID | 0.87 | <0.001 |
| D10 | 0.90 | <0.001 |
| D50 | 0.88 | <0.001 |
| α | 0.79 | <0.001 |
| β | 0.63 | 0.014 |
| α/β | 0.62 | 0.037 |
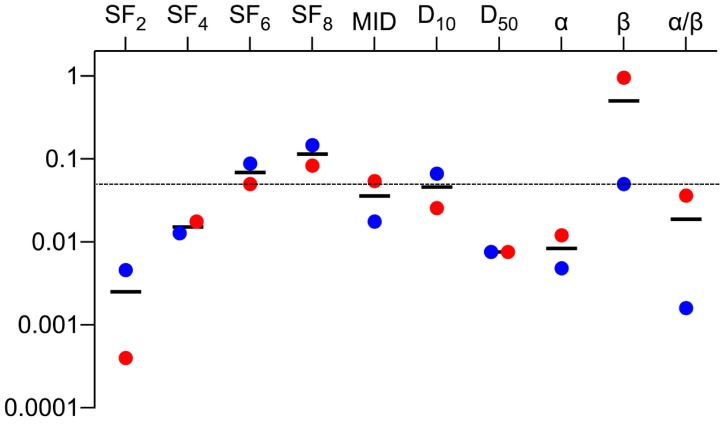
We then examined differences in the clonogenic survival parameters between the EGFR mutant and wild-type cell lines. In both datasets, the p values for SF2, SF4, D50, α, and α/β were below 0.05, and those for SF2 were the lowest (Table 1 and Figure 3). These data indicate that EGFR mutant cell lines might be more sensitive to low dose- and low fraction sized-irradiation.
Figure 3.
Summary of the p values for datasets A and B. MID, mean inactivation dose. Black lines indicate the mean values.
There were no significant differences in SF2 between the cell lines carrying Δ746_750 (n = 3) and those carrying L858R (n = 3) for both datasets A and B (p = 0.20 and 0.70, respectively). This is reasonable in light of the clinical situation where these two mutations are used without functional distinction as indicators of EGFR-upregulated tumors targeted with its tyrosine kinase inhibitors.
3. Discussion
A large body of evidence supports the ability of EGFR status to predict tumor radioresponsiveness. In the pre-clinical setting, Amornwichet et al. [14] found that radiosensitivity determined by D10 from clonogenic assays was significantly higher for EGFR mutant cells than for EGFR wild-type cells in a panel of NSCLC cell lines, as well as in genetically engineered isogenic NSCLC cells. On the other hand, in a clinical setting, Yagishita et al. [13] retrospectively analyzed the outcomes of non-squamous NSCLC patients, and showed that the frequency of local relapse after chemo-radiation therapy was significantly lower in patients with EGFR mutations than in those with wild-type EGFR (4% versus 21%). Importantly, those two studies, as well as this study, analyzed the same activating mutations in EGFR (i.e., exon 19 in-frame deletion and exon 21 L858R missense mutation); therefore, the data in these studies are comparable. From the standpoint of mechanism, multiple groups have shown that mutations in EGFR cause defects in EGFR translocation to the nucleus and binding of EGFR to the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), a protein that plays pivotal roles in non-homologous end joining (NHEJ) of DNA double-strand breaks (DSBs) induced by ionizing irradiation [15,16,17]. Amornwichet et al. [14] showed that the increase in X-ray-induced γH2AX foci, a marker of DSBs, upon addition of NU7441, an inhibitor of NHEJ, is significantly smaller in EGFR mutant cells than in wild-type cells. These findings suggest that the high radiosensitivity of EGFR mutant cancer cells is at least in part based on reduced NHEJ activity associated with dysfunction of DNA-PKcs in response to ionizing irradiation. Together, these data support the scientific validity of our study design, which analyzed EGFR status as a model for a candidate genetic profile associated with cancer cell radiosensitivity as assessed by clonogenic assays.
In this study, the p values for the comparison between EGFR mutant and wild-type cell lines had the same relationship (SF2 < SF4 < SF6 < SF8) in datasets A and B. In addition, the differences in α values between the two groups reached statistical significance in both datasets, whereas the differences in β values did not. Thus, for assessment of cancer cell radiosensitivity using clonogenic assays, surviving fractions in the low dose range, where the linear component of the LQ model is dominant, better predict genetic profiles associated with clinical tumor radioresponsiveness. Over the past few decades, multiple groups have investigated the correlation between radiosensitivity, as determined by various clonogenic survival parameters, and the clinical responses of tumors to radiation therapy [12]. Fertil and Malaise [8] analyzed 59 survival curves derived from human cell lines, and found that SF2 was associated with the clinical dose required for tumor control. The same group analyzed an additional 101 survival curves derived from human cell lines, and found that radiosensitivity expressed in terms of SF2, α, and MID reflected the clinical responsiveness of the tumor from which a cell line was derived [11]. Daecon et al. analyzed the data on 51 human tumor cell lines, and found that the steepness of the initial slope of the survival curve is associated with the clinical response of a tumor to radiation therapy [9]. Although these previous studies stratified cells by histological type or primary tumor site, but not genetic profile, they share key findings with the present study. Based on these findings, we can conclude that the importance of parameters related to the initial slope of the survival curve has been re-confirmed in the context of precision medicine. From another aspect, NSCLC patients are nowadays often treated with hypofractionation schemes. Our data showing no radiosensitivity differences between EGFR mutant and wild-type groups at high doses (i.e., SF8 and SF6) suggest that hypofractionated irradiation employing high fraction size is likely to achieve comparable tumor control regardless of EGFR status.
Previous studies show that the mean SF2 differs among different cancer types (e.g., the mean SF2 is higher for glioblastoma cell line group compared to that for lymphoma cell line group). Nevertheless, SF2 show large variation among the cell lines within the same cancer type. From this perspective, SF2 should not be used as a clinical test to predict the response of an individual tumor to radiotherapy. On the other hand, evidence suggests clinical applicability of genetic profile-based algorithms generated from correlation analyses of in vitro SF2 data and the genetic profile counterparts [18,19,20,21]. Together, our data support the notion that SF2 is useful for pre-clinical study aiming at establishment of genetic profiles that predict tumor radioresponsiveness.
We previously analyzed the inter-study precision of clonogenic survival parameters in a given cell line [22]; in that study we found that SF2 and D10 have acceptable inter-study precision as a measure for radiosensitivity assessment. The data support the usefulness of SF2 from another aspect, i.e., technical robustness of the assay.
A limitation of this study is that we only analyzed the association with clonogenic survival parameters using a single gene (i.e., EGFR) because this study was a pilot attempt. Therefore, the results of this study cannot be applied directly to the clinic. Realistically, radiosensitivity of the tested cell lines must be influenced not only by EGFR but also by other genetic aberrations; therefore, clinical tumor radioresponsiveness will be more accurately predicted by genetic profiles comprising multiple genes, which warrants further investigation.
In summary, we showed that radiosensitivity differences between EGFR mutant and wild-type lung cancer cells are larger at lower doses. These findings may be useful for optimization of radiotherapy schemes to treat NSCLCs.
4. Materials and Methods
4.1. Cell Lines
A427, A549, H1299, H1650, H1703, H1975, H460, H520, H522, and HCC827 were obtained from ATCC (Manassas, VA, USA). II-18 and LK2 were obtained from the Japanese Cancer Resources Bank. H157 was provided by Dr. Harris (National Cancer Institute, Bethesda, Rockville, MD, USA). Ma-24 was provided by Dr. Shimizu (Tokushima University, Tokushima, Japan). PC9 was provided by Dr. Kato (Tokyo Medical College, Tokyo, Japan). The EGFR status of these cell lines is summarized in Table 3. All cell lines were cultured in RPMI-1640 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (Life Technologies, Carlsbad, CA, USA).
Table 3.
EGFR mutation status in non-small cell lung carcinoma cell lines.
| Cell Line | Histopathology | EGFR Status | Reference |
|---|---|---|---|
| H1650 | Adenocarcinoma | ΔE746_A750 | [23,24] |
| Ma-24 | Adenocarcinoma | L858R, E709G | [23,25] |
| PC9 | Adenocarcinoma | ΔE746_A750 | [23,26] |
| HCC827 | Adenocarcinoma | ΔE746_A750 | [24,27] |
| H1975 | Adenocarcinoma | L858R, T790M | [24,26,27] |
| II-18 | Adenocarcinoma | L858R | [23,26,28] |
| H1299 | Large cell carcinoma | Wild-type | [23] |
| H1703 | Adenocarcinoma | Wild-type | [23,24] |
| A549 | Adenocarcinoma | Wild-type | [23,24,27,28] |
| H157 | Squamous cell carcinoma | Wild-type | [23,24,28] |
| LK2 | Squamous cell carcinoma | Wild-type | [23,26] |
| H522 | Adenocarcinoma | Wild-type | [23] |
| H520 | Squamous cell carcinoma | Wild-type | [23,24,27] |
| A427 | Adenocarcinoma | Wild-type | [23,28] |
| H460 | Large cell carcinoma | Wild-type | [23,24,27,28] |
4.2. Clonogenic Assays
For each of the 15 NSCLC cell lines, clonogenic survival parameters were obtained independently by two radiation oncologists (datasets A and B, respectively).
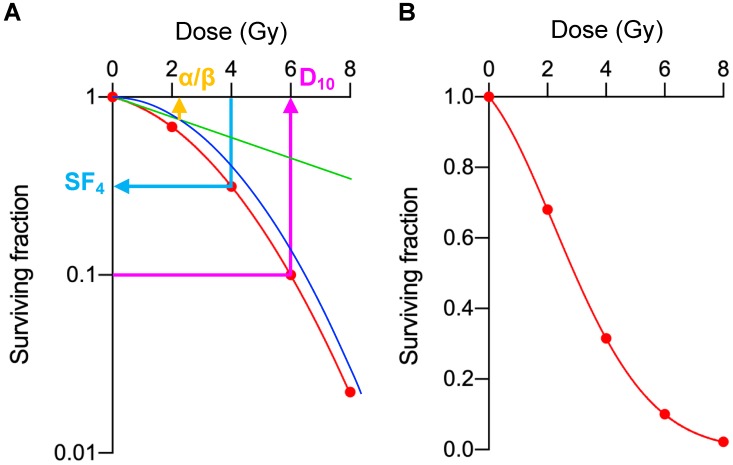
The experimental procedure was standardized among practitioners using the following protocol. Cells were passaged at least two times after thawing from freeze stocks, and the cells in log phase growth were used for the experiments. Cells were detached from culture dishes using trypsin (Sigma-Aldrich, St. Louis, MO, USA) and prepared as single-cell suspension in culture media. The cells were counted using a hemocytometer under an inverted microscope. The cell suspension was subjected to two serial dilutions at 1:10 (i.e., 1:100 dilution in total), from which the cell suspension containing the intended number of cells was prepared. The cells were seeded in 6-well plates; the number of seeded cells was determined based on the plating efficiency and the intrinsic radiosensitivities estimated by preparatory experiments (Table 4). After incubation for the minimum possible period of time for the seeded cells to attach on the plates (6–12 h, depending on cell lines), the cells were exposed to 2, 4, 6, or 8 Gy X-rays using a Faxitron RX-650 irradiator (100 kVp, 1.14 Gy/min; Faxitron Bioptics, Tucson, AZ, USA). After incubation for an additional 6–10 days, the cells were fixed with methanol and stained with crystal violet. Colonies comprising at least 50 cells were counted under an inverted microscope. The experiments were performed at least in triplicate, and the mean values for the surviving fractions were calculated after normalization to the corresponding un-irradiated controls. These survival data were fitted to the LQ model [29], and then α, β, D10, and D50 were calculated (Figure 4A). MID was calculated as previously described (Figure 4B) [10].
Table 4.
Plating efficiency (PE) and the number of seeded cells.
| Cell Line | PE (%) | 0 Gy | 2 Gy | 4 Gy | 6 Gy | 8 Gy |
|---|---|---|---|---|---|---|
| H1650 | 12 | 500 | 500 | 1000 | 2000 | 3000 |
| Ma-24 | 24 | 500 | 500 | 1000 | 2000 | 3000 |
| PC9 | 25 | 500 | 500 | 1000 | 2000 | 3000 |
| HCC827 | 39 | 300 | 300 | 500 | 1000 | 2000 |
| H1975 | 35 | 300 | 300 | 500 | 1000 | 2000 |
| II-18 | 21 | 500 | 500 | 1000 | 2000 | 3000 |
| H1299 | 71 | 200 | 200 | 300 | 500 | 500 |
| H1703 | 34 | 300 | 300 | 500 | 1000 | 2000 |
| A549 | 72 | 200 | 200 | 300 | 500 | 500 |
| H157 | 65 | 200 | 200 | 300 | 500 | 500 |
| LK2 | 40 | 300 | 300 | 500 | 1000 | 2000 |
| H522 | 43 | 300 | 300 | 500 | 1000 | 2000 |
| H520 | 28 | 500 | 500 | 1000 | 2000 | 3000 |
| A427 | 32 | 300 | 300 | 500 | 1000 | 2000 |
| H460 | 58 | 300 | 300 | 500 | 1000 | 2000 |
Figure 4.
Exemplary presentation of the analyzed parameters for clonogenic survival. (A) Surviving fractions for the cells irradiated with 2, 4, 6, or 8 Gy (SF2, SF4, SF6, and SF8, respectively) are plotted on a semi-logarithmic-scaled graph (indicated as red dots). The survival data were fitted to the LQ model: S = exp(−(αD + βD2)), where S is the surviving fraction and D is the dose (red line indicates the LQ curve; green and blue line indicates its linear and quadratic component, respectively). D10 and D50 are calculated from the LQ model formula, where DX indicates the dose that decreases the surviving fraction to X%. As an example, α/β (2.1), SF4 (0.32), and D10 (6.0) are shown as orange, light blue, and violet arrow, respectively. (B) The same exemplary survival data were plotted on a linear-scaled graph. MID equals the area under the curve (indicated as light red) [11].
4.3. Statistical Analysis
Clonogenic survival parameters were compared between EGFR mutant and wild-type cell lines by non-parametric two-sided Mann–Whitney U test. Correlation of the survival data between datasets was examined by Spearman’s rank order test. Differences were considered statistically significant at p < 0.05. Analyses were performed using Prism8 (GraphPad, San Diego, CA, USA).
Abbreviations
| NSCLC | non-small cell lung carcinoma |
| LQ model | linear quadratic model |
| MID | mean inactivation dose |
| DNA-PKcs | DNA-dependent protein kinase, catalytic subunit |
| NHEJ | non-homologous end joining |
Author Contributions
Conceptualization, T.O.; data acquisition, N.A., D.K. and Y.H.; formal analysis, M.A. and A.N.; writing—original draft preparation, M.A. and A.N.; writing—review and editing, T.O. and A.S.; supervision, T.N.; funding acquisition, M.A.
Funding
This research was funded by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan for programs for Leading Graduate Schools, “Cultivating Global Leaders in Heavy Ion Therapeutics and Engineering.”
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Okunieff P., Morgan D., Niemierko A., Suit H.D. Radiation dose-response of human tumors. Int. J. Radiat. Oncol. Biol. Phys. 1995;32:1227–1237. doi: 10.1016/0360-3016(94)00475-Z. [DOI] [PubMed] [Google Scholar]
- 2.Hinkson I.V., Davidsen T.M., Klemm J.D., Kerlavage A.R., Kibbe W.A. A Comprehensive Infrastructure for Big Data in Cancer Research: Accelerating Cancer Research and Precision Medicine. Front. Cell Dev. Biol. 2017;2:83. doi: 10.3389/fcell.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan E.J., Kim H.R., Arcila M.E., Barron D., Chakravarty D., Gao J., Chang M.T., Ni A., Kundra R., Jonsson P., et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robson M., Im S.A., Senkus E., Xu B., Domchek S.M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 5.Lamba J.K., Chauhan L., Shin M., Loken M.R., Pollard J.A., Wang Y.C., Ries R.E., Aplenc R., Hirsch B.A., Raimondi S.C., et al. CD33 Splicing Polymorphism Determines Gemtuzumab Ozogamicin Response in De Novo Acute Myeloid Leukemia: Report from Randomized Phase III Children’s Oncology Group Trial AAML0531. J. Clin. Oncol. 2017;35:2674–2682. doi: 10.1200/JCO.2016.71.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris M.H., DuBois S.G., Glade Bender J.L., Kim A., Crompton B.D., Parker E., Dumont I.P., Hong A.L., Guo D., Church A., et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol. 2016;2:608–615. doi: 10.1001/jamaoncol.2015.5689. [DOI] [PubMed] [Google Scholar]
- 7.Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;2:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 8.Fertil B., Malaise E.P. Inherent cellular radiosensitivity as a basic concept for human tumor radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 1981;7:621–629. doi: 10.1016/0360-3016(81)90377-1. [DOI] [PubMed] [Google Scholar]
- 9.Deacon J., Peckham M.J., Steel G.G. The radioresponsiveness of human tumours and the initial slope of the cell survival curve. Radiother. Oncol. 1984;2:317–323. doi: 10.1016/S0167-8140(84)80074-2. [DOI] [PubMed] [Google Scholar]
- 10.Fertil B., Dertinger H., Courdi A., Malaise E.P. Mean inactivation dose: A useful concept for intercomparison of human cell survival curves. Radiat. Res. 1984;99:73–84. doi: 10.2307/3576448. [DOI] [PubMed] [Google Scholar]
- 11.Fertil B., Malaise E.P. Intrinsic radiosensitivity of human cell lines is correlated with radioresponsiveness of human tumors: Analysis of 101 published survival curves. Int. J. Radiat. Oncol. Biol. Phys. 1985;11:1699–1707. doi: 10.1016/0360-3016(85)90223-8. [DOI] [PubMed] [Google Scholar]
- 12.Steel G.G. The radiobiology of tumours. In: Steel G.G., editor. Basic Clinical Radiobiology. 3rd ed. CRC Press; Boca Raton, FL, USA: 2002. pp. 182–191. [Google Scholar]
- 13.Yagishita S., Horinouchi H., Taniyama K.T., Nakamichi S., Kitazono S., Mizugaki H., Kanda S., Fujiwara Y., Nokihara H., Yamamoto N., et al. Epidermal growth factor receptor mutation is associated with longer local control after definitive chemoradiotherapy in patients with stage III nonsquamous non-small-cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2015;91:140–148. doi: 10.1016/j.ijrobp.2014.08.344. [DOI] [PubMed] [Google Scholar]
- 14.Amornwichet N., Oike T., Shibata A., Nirodi C.S., Ogiwara H., Makino H., Kimura Y., Hirota Y., Isono M., Yoshida Y., et al. The EGFR mutation status affects the relative biological effectiveness of carbon-ion beams in non-small cell lung carcinoma cells. Sci. Rep. 2015;5:11305. doi: 10.1038/srep11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A.K., Chen B.P., Story M.D., Sato M., Minna J.D., Chen D.J., Nirodi C.S. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–5274. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 16.Dittmann K., Mayer C., Fehrenbacher B., Schaller M., Raju U., Milas L., Chen D.J., Kehlbach R., Rodemann H.P. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J. Biol. Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 17.Huang S.M., Harari P.M. Modulation of radiation response after epidermal growth factor receptor blockade in squamous cell carcinomas: Inhibition of damage repair, cell cycle kinetics, and tumor angiogenesis. Clin. Cancer Res. 2000;6:2166–2174. [PubMed] [Google Scholar]
- 18.Strom T., Harrison L.B., Giuliano A.R., Schell M.J., Eschrich S.A., Berglund A., Fulp W., Thapa R., Coppola D., Kim S., et al. Tumour radiosensitivity is associated with immune activation in solid tumours. Eur. J. Cancer. 2017;84:304–314. doi: 10.1016/j.ejca.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Roca J.F., Fulp W.J., Caudell J.J., Servant N., Bollet M.A., van de Vijver M., Naghavi A.O., Harris E.E., Eschrich S.A. Integration of a radiosensitivity molecular signature into the assessment of local recurrence risk in breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2015;93:631–638. doi: 10.1016/j.ijrobp.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed K.A., Chinnaiyan P., Fulp W.J., Eschrich S., Torres-Roca J.F., Caudell J.J. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget. 2015;6:34414–34422. doi: 10.18632/oncotarget.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strom T., Hoffe S.E., Fulp W., Frakes J., Coppola D., Springett G.M., Malafa M.P., Harris C.L., Eschrich S.A., Torres-Roca J.F., et al. Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother. Oncol. 2015;117:159–164. doi: 10.1016/j.radonc.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nuryadi E., Permata T.B.M., Komatsu S., Oike T., Nakano T. Inter-assay presicion of clonogenic assay for radiosensitivity in cancer cell line A549. Oncotarget. 2018;9:13706–13712. doi: 10.18632/oncotarget.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco R., Iwakawa R., Tang M., Kohno T., Angulo B., Pio R., Montuenga L.M., Minna J.D., Yokota J., Sanchez-Cespedes M. A gene-alteration profile of human lung cancer cell lines. Hum. Mutat. 2009;30:1199–1206. doi: 10.1002/humu.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helfrich B.A., Raben D., Varella-Garcia M., Gustafson D., Chan D.C., Bemis L., Coldren C., Barón A., Zeng C., Franklin W.A., et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin. Cancer Res. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- 25.Sakai K., Yokote H., Murakami-Murofushi K., Tamura T., Saijo N., Nishio K. Pertuzumab, a novel HER dimerization inhibitor.; inhibits the growth of human lung cancer cells mediated by the HER3 signaling pathway. Cancer Sci. 2007;98:1498–1503. doi: 10.1111/j.1349-7006.2007.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawahara A., Yamamoto C., Nakashima K., Azuma K., Hattori S., Kashihara M., Aizawa H., Basaki Y., Kuwano M., Kage M., et al. Molecular diagnosis of activating EGFR mutations in non-small cell lung cancer using mutation-specific antibodies for immunohistochemical analysis. Clin. Cancer Res. 2010;16:3163–3170. doi: 10.1158/1078-0432.CCR-09-3239. [DOI] [PubMed] [Google Scholar]
- 27.Spoerke J.M., O’Brien C., Huw L., Koeppen H., Fridlyand J., Brachmann R.K., Haverty P.M., Pandita A., Mohan S., Sampath D., et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin. Cancer Res. 2012;18:6771–6783. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- 28.Oike T., Ogiwara H., Tominaga Y., Ito K., Ando O., Tsuta K., Mizukami T., Shimada Y., Isomura H., Komachi M., et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 29.Oike T., Ogiwara H., Torikai K., Nakano T., Yokota J., Kohno T. Garcinol, a histone acetyltransferase inhibitor, radiosensitizes cancer cells by inhibiting non-homologous end joining. Int. J. Radiat. Oncol. Biol. Phys. 2012;84:815–821. doi: 10.1016/j.ijrobp.2012.01.017. [DOI] [PubMed] [Google Scholar]