Abstract
Genetic resistance against biotic stress is a major goal in many wheat breeding programs. However, modern wheat cultivars have a limited genetic variation for disease and pest resistance and there is always a possibility of the evolution of new diseases and pests to overcome previously identified resistance genes. A total of 125 synthetic hexaploid wheats (SHWs; 2n = 6x = 42, AABBDD, Triticum aestivum L.) were characterized for resistance to fungal pathogens that cause wheat rusts (leaf; Puccinia triticina, stem; P. graminis f.sp. tritici, and stripe; P. striiformis f.sp. tritici) and crown rot (Fusarium spp.); cereal cyst nematode (Heterodera spp.); and Hessian fly (Mayetiola destructor). A wide range of genetic variation was observed among SHWs for multiple (two to five) biotic stresses and 17 SHWs that were resistant to more than two stresses. The genomic regions and potential candidate genes conferring resistance to these biotic stresses were identified from a genome-wide association study (GWAS). This GWAS study identified 124 significant marker-trait associations (MTAs) for multiple biotic stresses and 33 of these were found within genes. Furthermore, 16 of the 33 MTAs present within genes had annotations suggesting their potential role in disease resistance. These results will be valuable for pyramiding novel genes/genomic regions conferring resistance to multiple biotic stresses from SHWs into elite bread wheat cultivars and providing further insights on a wide range of stress resistance in wheat.
Keywords: disease and pest resistance, rusts, pyramiding novel genes, marker-trait associations, candidate gene
1. Introduction
Wheat (Triticum aestivum L.) is one of the most widely grown cereal crops with a production of more than 756 million tonnes in 2017–2018 [1] and it feeds more than one-third of the world’s population [2]. Global population is increasing at an alarming rate and is estimated to reach 9.8 billion by 2050 [3]. To meet global food demands, cereal production needs to be increased by 50% by 2030 [4]. However, biotic stresses caused by diseases and pests will impose significant constraints on agricultural production and productivity [5,6].
Wheat is grown in a wide range of environments and exposed to multiple diseases and pests throughout the growing season [7]. Biotic stresses observed in crops are divided into four groups: foliar and stem diseases, seed transmitted diseases, soilborne diseases, and pests. Foliar and stem diseases such as a leaf or brown rust (Lr; incited by Puccinia triticina), stem or black rust (Sr; incited by P. graminis f.sp. tritici), and stripe or yellow rust (Yr; incited by P. striiformis f.sp. tritici) are major threats to wheat production throughout the world [5,6]. Global wheat yield losses due to these rusts range from 20% to 100% [5,8,9,10] and were estimated at $5 billion per year since the 1990s [11]. Similarly, soilborne pathogens such as the cereal cyst nematode (CCNs; Heterodera spp.) cause significant cereal crop losses [12,13,14]. For instance, global crop losses due to nematodes have been estimated to be up to $80 billion [12]. Crown rot (Cr; caused by Fusarium spp.) is an important soilborne disease of wheat and severe Cr can cause significant yield losses in many wheat growing regions [15,16]. For example, the grain yield losses at different locations due to Cr have been reported to be up to 43% in Turkey [17], 46% in Iran [18], 61% in the United States [19], and 89% in Australia [20]. Hessian fly (HF; Mayetiola destructor), is one of the major destructive pests of wheat that causes grain yield loss of up to 80% [21] and may cause an average annual loss of 5% in the United States wheat production [22]. In Morocco, wheat grain yield loss due to Hessian fly was estimated to be up to 36% [23] and 42% [24]. The damage caused by diseases and pests can be reduced through crop rotation, cultural practices, and application of chemicals. Genetic resistance is the most economical, environment-friendly, and sustainable method of controlling crop losses. However, breeding for resistance to multiple diseases and pests requires identification of genetic sources of resistance and novel genes [7].
Synthetic hexaploid wheat (SHW; a cross between T. turgidum L. × Aegilops tauschii Coss.) is reported to have a novel source of genetic diversity for wheat genetic improvement especially for the D genome diversity [25,26] and is a potential source for resistance to abiotic [27] and biotic stresses [7,28,29]. For example, SHWs were found to have resistance to leaf rust [7,26,30], stem rust [7,31], stripe rust [7,31,32], Fusarium head blight (incited by Fusarium graminearum) [30], yellow spot (incited by Pyrenophora tritici-repentis) [7,31], Septoria glume blotch (incited by Parastagonospora nodorum) [7,31], Septoria leaf blotch (incited by Zymoseptoria tritici) [30,31], cereal cyst nematode [31], crown rot [7], root-lesion nematode (incited by Pratylenchus thornei and P. neglectus) [31], and Karnal bunt (incited by Tilletia indica) [32]. Additionally, SHWs have been reported to be resistant to multiple pests [7,28,30].
Currently, more than 110 leaf rust, 86 stem rust, and 83 stripe rust resistance genes have been reported in wheat or wild relatives, most conferring race-specific resistance [33]. Some of the important stem rust resistance genes are Sr2, Sr23, Sr24, Sr25, Sr31, Sr33, Sr35, Sr36, Sr38, Sr45, Sr50, SrTmp, and Sr1RSAmigo [10]; the stripe rust resistance genes are Yr2, Yr6, Yr7, Yr8, Yr9, Yr17, and Yr27; and leaf rust resistance genes are Lr9, Lr14a, Lr16, Lr17a, Lr21, Lr22, Lr24, Lr26, Lr32, Lr39, and Lr41 [8,10,34], where Lr21, Lr22, Lr32, Lr39, and Lr41 were originated from Ae. tauschii [35]. To date, more than 35 Hessian fly resistance genes have been reported where H13, H22, H23, H24, H26, and H32 were originated from Ae. tauschii [33,36]. However, most of the resistance genes are short-lived due to the result of genetic recombination by sexual reproduction or somatic hybridization and mutation in the absence of alternative hosts [37], resulting in a modern variety with an inadequate level of insect-pest resistance. Therefore, exploiting genetic variation in SHWs for multiple biotic stresses and identifying genomic regions controlling their resistance is necessary for their utilization in an elite wheat breeding program. The objectives of this study were to (i) characterize unique sets of SHWs for resistance to CCN, Cr, HF, Lr, Sr, and Yr, (ii) identify lines resistant to multiple biotic stresses; (iii) identify genomic regions controlling their resistance using genome-wide association study (GWAS); and (iv) identify the potential genes underlying significant single nucleotide polymorphisms (SNPs). These data will provide useful information for pyramiding favorable alleles and multiple disease resistance genes from SHWs into elite bread wheat germplasm.
2. Results and Discussion
2.1. Phenotypic Distribution of Disease and Pest
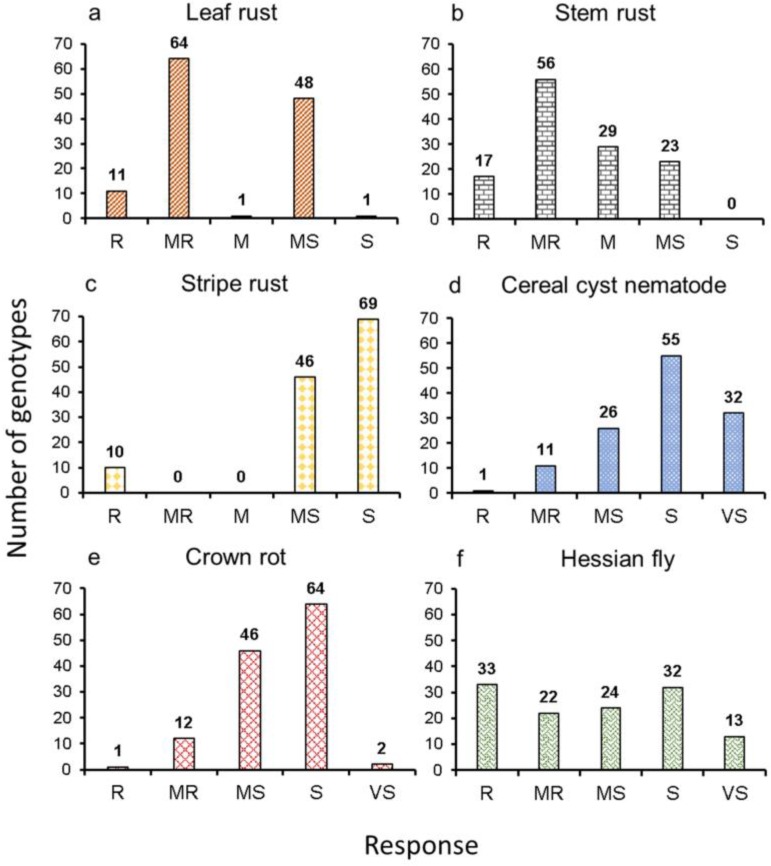
A wide range of genetic variation for multiple biotic stresses was observed in 125 SHWs (Figure 1 and Supplementary Table S1). As expected, seedling and adult plant resistance screening for susceptible checks showed a moderately susceptible to very susceptible response and resistant checks showed a resistant to very resistant response, indicating that the screening methods were effective (Supplementary Table S1). More than 50% of the lines had a resistant (11) to moderately resistant (64) reaction to Lr at the adult plant stage (Figure 1a). For Lr seedling resistance screening, we identified 109 lines as immune (no symptoms) and 16 lines as very resistant. Of the 11 resistant lines at the adult plant stage, ten lines showed an immune response and one line showed a very resistant response at the seedling stage. These results indicated that 11 lines may possess major Lr resistance genes.
Figure 1.
Frequency distribution of response to (a) leaf rust, (b) stem rust, and (c) stripe rust at the adult plant stage, (d) cereal cyst nematode, (e) crown rot, and (f) Hessian fly in 125 synthetic hexaploid wheat. R, Resistant; MR, Moderately resistant; M, Intermediate; MS, Moderately susceptible; S, Susceptible; and VS, Very susceptible.
Approximately 60% of the lines (73 lines) were resistant (17 lines) to moderately resistant (56 lines) to stem rust at the adult plant stage (Figure 1b). Seedling resistance screening against Sr identified 51, 52, 13, seven, and two lines as immune, very resistant, resistant, moderately resistant, and moderately susceptible, respectively against race TPMK (Supplementary Table S1). These results indicated that most of the lines may have Sr genes Sr6, Sr9a, Sr9b, or Sr30 based on the TMPK race used. Of the 17 adult plant resistant lines, nine were immune, seven were resistant to very resistant, and one line was moderately resistant at the seedling stage, indicating that these lines may possess major Sr resistance genes.
The adult plant stage Yr screening identified 10 resistant lines (Figure 1c). Similarly, seedling resistance screening identified one very resistant, 106 resistant, and 18 moderately resistant lines against the mixture of three Pst races (Pstv-14, Pstv-37, and Pstv-40) (Supplementary Table S1). This indicates that most of these synthetic genotypes may have Yr genes Yr1, Yr5, Yr10, Yr15, Yr24, Yr17, Yr32, YrSP, or YrTye or other unknown genes as they were resistant against three races (Pstv-14, Pstv-37, and Pstv-40). Of the 10 adult plant resistant lines, seven lines were resistant, and three lines were moderately resistant at the seedling stage, indicating that these lines may possess major Yr resistance genes.
We have also screened SHW lines for CCN, Cr, and HF resistance. This study identified one resistant and 11 moderately resistant lines for CCN (Figure 1d and Supplementary Table S1), one resistant and 12 moderately resistant lines for Cr (Figure 1e), and 33 resistant and 22 moderately resistant lines for HF (Figure 1f).
2.2. Genome-Wide Association Study
A GWAS identified a total of 124 marker-trait associations (MTAs) for Hessian fly infestation (%), field-based infection type (IT) for Yr, and field disease severity (SEV), IT, and coefficient of infection (COI) for Lr and Sr (Figure 2 and Supplementary Table S2). The phenotypic variance explained (PVE) by these MTAs ranged from 0.3% to 27.4% (Supplementary Table S2). The MTAs identified in this study were year specific because of the significant influence of genotype × year interaction as can be seen by weak phenotypic correlation between years (r = 0.18; p = 0.06 to 0.42, p = 0.02). Also, we ran GWAS analysis for CCN and Cr, but we were unable to identify significant MTAs in our population; hence, the GWAS results for these traits will not be discussed further. Additionally, we did not run GWAS analysis on seedling rust resistance data because most of the genotypes were resistant and data were highly skewed (Supplementary Table S1).
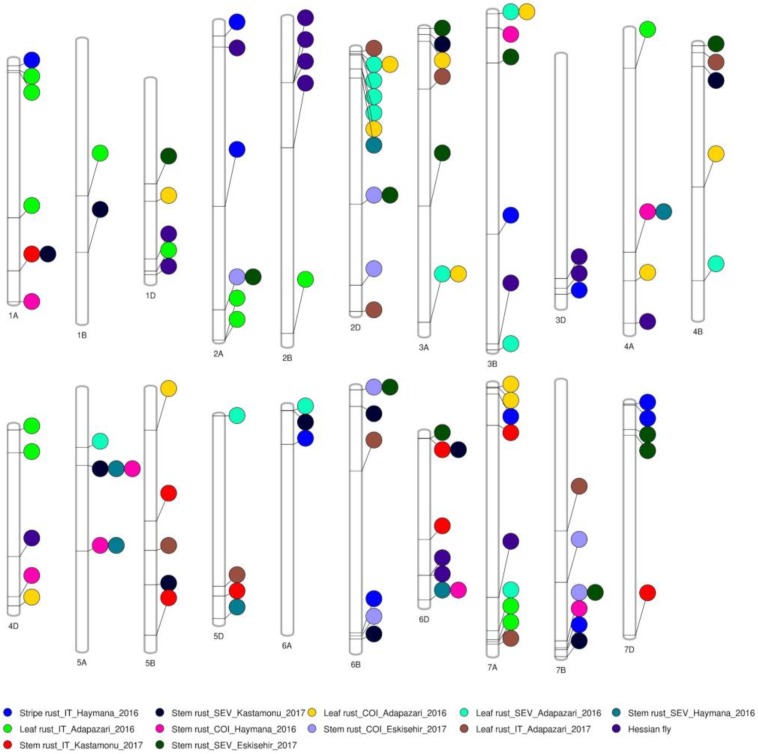
Figure 2.
Significant marker-trait associations (p = 9.2 × 10−5 (−log10p = 4.04)) identified on each chromosome for leaf rust, stem rust, stripe rust, and Hessian fly from a genome-wide association study using 35,798 single nucleotide polymorphisms in 125 synthetic hexaploid wheats grown in 2016 and 2017 in Turkey (Adapazari, Eskisehir, Haymana, and Kastamonu) and in 2017 in Morocco (Settat; Hessian fly experiment). COI, Coefficient of infection; IT, Infection type; and SEV, Field disease severity.
At the adult plant stage for Lr, a total of 46 MTAs for SEV (12), IT (22), and COI (12) were identified on chromosomes 1A (3), 1B (1), 1D (2), 2A (2), 2B (1), 2D (8), 3A (4), 3B (3), 4A (2), 4B (3), 4D (3), 5A (1), 5B (2), 5D (2), 6A (1), 6B (1), 7A (6), and 7B (1) (Figure 2) and PVE by these MTAs ranged from 0.3% to 27.4% (Supplementary Table S2). To date, over 110 Lr resistance genes have been identified and they were distributed across all chromosomes in wheat except 5A and 6D [33]. Leaf rust resistance genes such as Lr21, Lr22, Lr32, Lr39, and Lr41 were derived from Ae. tauschii [35]. The identification of 15 MTAs on the D genome clearly represent variation from the Ae. tauschii and show the potential of SHW for its utilization in a marker-assisted breeding program after being validated in an independent genetic background. This study identified three co-located MTAs (S2D_13778898, S3A_721353764, S3B_4553494) and a haplotype block (perfect linkage disequilibrium with R2 = 1) for Lr consisting of two SNPs (S2D_50105731 and S2D_50105753) of size 22 base pairs for COI and SEV on chromosomes 2D, 3A, and 3B (Supplementary Table S2). This was expected as these traits (SEV and COI) had a high correlation (ranging from 0.65 to 0.87, p < 0.001).
At the adult plant stage for Sr, a total of 52 MTAs for field SEV (29), IT (8), and COI (15) were identified on chromosomes 1A (3), 1B (1), 1D (1), 2A (2), 2D (4), 3A (3), 3B (2), 4A (2), 4B (2), 4D (1), 5A (5), 5B (3), 5D (2), 6A (1), 6B (5), 6D (6), 7A (1), 7B (5), and 7D (3) (Figure 2) with PVE ranging from 1.0 to 27.0% (Supplementary Table S3). To date, over 86 Sr genes have been identified, which were distributed across all chromosomes in wheat [33]. Several MTAs (17) identified on the D genome showed the potential of SHWs for improving Sr resistance in modern wheat cultivars. Additionally, this study identified seven co-located MTAs (S2D_381953220, S4A_545611013, S5A_208152827, S5A_440734987, S6B_7943502, S6D_456325441, and S7B_7067875454) for COI and SEV on chromosomes 2D, 4A, 5A, 6B, 6D, and 7B) and one co-located MTA (S6D_16745888) for IT and SEV on chromosome 6D. Identification of co-located MTAs was supported by the high correlation among these traits (r > 0.64, p < 0.001).
Similarly, at the adult plant stage for Yr, a total of 11 MTAs for IT were identified on chromosomes 1A (1), 2A (2), 3B (1), 3D (1), 6A (1), 6B (1), 7A (1), 7B (1), and 7D (2) (Figure 2) with PVE ranging from 0.4% to 15.5% (Supplementary Table S3). To date, over 83 Yr resistance genes have been reported, which were distributed across all chromosomes in wheat [33]. Most of the genes are race-specific and are often short-lived due to the frequent change in the pathogen population [32]. The MTAs identified in the current study showed the quantitative nature of the resistance to Yr in SHWs. Previous studies have identified MTAs for Yr on chromosomes 2A, 3B, 6A, and 7B in an association mapping panel of 181 SHWs [32]. The MTAs identified on the D genome (chromosomes 3D and 7D) showed the potential of SHWs for improving Yr resistance in modern wheat cultivars.
For HF, 15 MTAs were identified on chromosomes 1D (2), 2A (1), 2B (4), 3B (1), 3D (2), 4A (1), 4D (1), 6D (2), and 7A (1) (Figure 2) and PVE by these MTAs ranged from 0.4% to 15.5% (Supplementary Table S3). A previous study comprised of 134 wheat genotypes (landrace, SHW, and elite germplasm) had identified seven MTAs responsible for HF resistance on chromosomes 1B, 2D, 3A, 3D, 5D, and 7D [36]. The 13 MTAs identified for HF resistance in this study have not been previously reported and they are potentially novel MTAs responsible for HF resistance.
2.3. Genes Underlying Marker-Trait Associations and Their Functional Annotations
The functional annotation of genes underlying MTAs was available through IWGSC RefSeq v1.0 annotations. A total of 33 of 124 MTAs were found within genes and of these, 16 were present in seven genes whose functional annotations suggested their involvement in disease resistance in wheat based on previous literature (Table 1 and Supplementary Table S3). The identification of underlying genes with annotations related to the trait provides further reliability for the MTAs identified. For instance, four genes (TraesCS4D01G096900: Lr, TraesCS7A01G517300: Lr, TraesCS4B01G007000: Sr, and TraesCS3A01G495400: Lr) were annotated as the leucine-rich repeat protein family, which was previously reported to have a potential role in Yr [38,39] and Sr resistance [40]. Similarly, five genes (TraesCS2A01G076800: Yr, TraesCS7B01G473300: Yr, TraesCS1A01G325300: Sr, TraesCS6B01G394600: Yr, and TraesCS5B01G199700: Sr) were annotated as the F-box family protein, which were reported to have potential role in Lr [41], Sr [42], and Yr [43] resistance in wheat. These two classes of genes are interesting genes for future study in wheat as they were also reported to be involved in abiotic stress tolerance such as drought [27,44,45]. However, further validation studies are needed for the MTAs identified and underlying gene functions prior to their utilization in a marker-assisted selection program. Additional examples where the underlying genes had annotations matching the trait of interest are provided in Table 1.
Table 1.
List of potential candidate genes found in regions identified by marker-trait associations for disease resistance, favorable allele (bold), and SNP effects in 125 SHWs.
| Trait a | SNP_ID b | PVE c | Allele d | Effect | −Log10(p) | Gene-ID | Potential Candidate Gene | Trait e | References f |
|---|---|---|---|---|---|---|---|---|---|
| Yr | S3B_544810753 | 6.4 | G/A | −0.09 | 8.21 | TraesCS3B01G338200 | Calcium-binding protein | Yr | [46] |
| Lr | S4D_73425349 | 18.5 | C/T | 0.27 | 10.14 | TraesCS4D01G096900 | Leucine-rich repeat transmembrane neuronal protein 3 | Yr, Sr | [38,39,40] |
| Sr | S7B_706787545 | 15.1 | A/G | 15.54 | 7.95 | TraesCS7B01G441400 | ARM repeat superfamily protein | Tanspot | [47] |
| Yr | S2A_34483654 | 12.4 | T/G | −0.13 | 9.63 | TraesCS2A01G076700 | Elongation factor 1 alpha a | Yr | [43] |
| Yr | S2A_34483654 | 12.4 | T/G | −0.13 | 9.63 | TraesCS2A01G076800 | F-box protein | Lr, Sr, Yr | [41,42,43] |
| Yr | S7B_730117633 | 3.8 | C/A | −0.09 | 7.54 | TraesCS7B01G473300 | F-box protein | Lr, Sr, Yr | [41,42,43] |
| Sr | S1A_516079891 | 9.9 | G/C | −0.09 | 6.29 | TraesCS1A01G325300 | F-box protein | Lr, Sr, Yr | [41,42,43] |
| Yr | S6B_669910684 | 0.9 | C/G | 0.15 | 8.03 | TraesCS6B01G394600 | F-box protein | Lr, Sr, Yr | [41,42,43] |
| Sr | S5B_360428982 | 10.8 | T/C | −0.07 | 6.95 | TraesCS5B01G199700 | F-box protein | Lr, Sr, Yr | [41,42,43] |
| Lr | S5A_158912458 | 8.7 | G/A | −5.51 | 4.96 | TraesCS5A01G103000 | F-box protein | Lr, Sr, Yr | [41,42,43] |
| Lr | S7A_701569874 | 8.5 | A/G | 0.15 | 8.60 | TraesCS7A01G517300 | Leucine-rich repeat protein kinase family protein | Lr, Sr, Yr | [41,42,43] |
| Sr | S4B_4791631 | 3.0 | G/C | −6.10 | 7.43 | TraesCS4B01G007000 | Leucine-rich repeat receptor-like protein kinase family protein | Sr, Yr | [38,39,40] |
| Sr | S2D_381953220 | 8.4 | G/T | 8.33 | 7.48 | TraesCS2D01G299700 | Peroxidase | Sr, Tanspot | [47,48] |
| Lr | S3A_721353764 | 14.6 | G/T | 3.51 | 7.78 | TraesCS3A01G495400 | Leucine rich repeat N-terminal domain | Sr, Yr | [38,39,40] |
| Sr | S1A_592532938 | 6.4 | G/A | −7.50 | 5.54 | TraesCS1A01G444500 | Leucine rich repeat N-terminal domain | Sr, Yr | [38,39,40] |
| Lr | S2D_644835165 | 1.4 | G/T | 0.07 | 5.89 | TraesCS2D01G586500 | WAT1-related protein/EamA-like transporter family | Fungal pathogen | [49] |
| Lr | S7A_708435571 | 19.4 | T/C | −0.18 | 8.20 | TraesCS7A01G526900 | WAT1-related protein/EamA-like transporter family | Fungal pathogen | [49] |
a Lr, leaf rust; Sr, stem rust; Yr, stripe rust. b S+chromosome_position in bp, c PVE, phenotypic variance explained (%), d Allele that is in bold text is the favorable allele that increases the disease resistance, e Trait identified in the past, f References for the association of annotation with traits.
2.4. Multiple Stress Resistant Lines
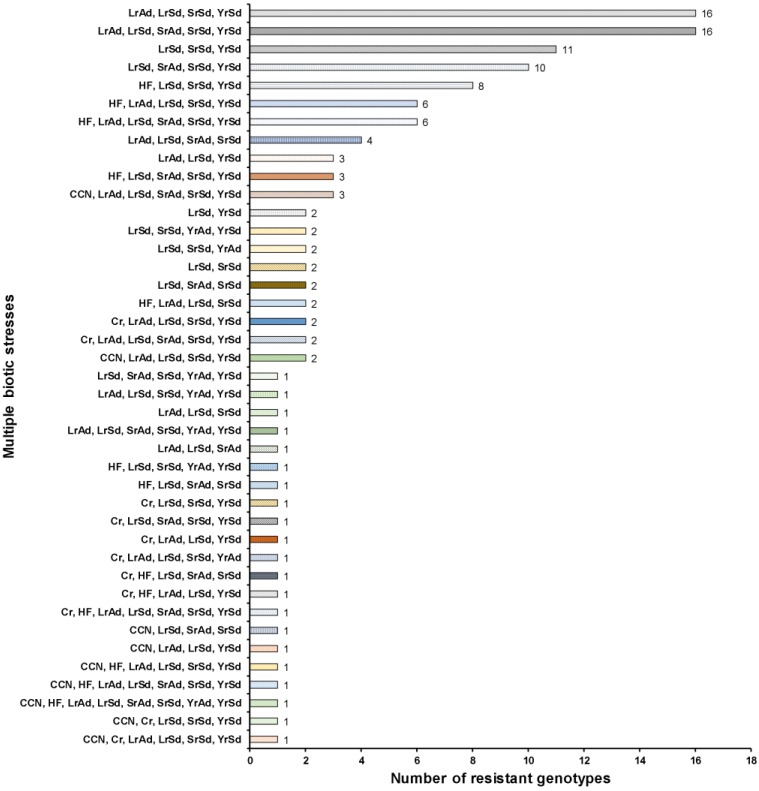
There were six biotic stresses (three rusts assessed at both seedling and adult plant stages, CCN, Cr, and HF) assessed to identify multiple stress resistant lines. All the SHW lines showed resistance to multiple biotic stresses ranging from two to five biotic stresses (Figure 3 and Supplementary Table S1). Specifically, 14, 67, 38, and 6 lines showed resistance against two, three, four, and five biotic stresses, respectively (Figure 3). From this study, we can recommend 17 lines (15.3% of the total resistant lines) that were resistant to more than two stresses and had a large number of favorable alleles (alleles that increased its resistance against multiple stresses based on significant markers identified in GWAS) conferring resistance (Supplementary Table S4). These lines can be used as parents in breeding programs with the goal of reducing disease and pest damage. The results indicated the usefulness of these lines for pyramiding multiple genes for durable resistance to multiple diseases and pests.
Figure 3.
Number of synthetic hexaploid wheat lines showing resistance to multiple biotic stresses. CCN, cereal cyst nematode; Cr, crown rot; HF, Hessian fly; Lr, leaf rust; Sr, stem rust; Yr, stripe rust; Sd, seedling stage; and Ad, adult stage.
3. Materials and Methods
3.1. Genetic Resources
A total of 125 synthetic hexaploid wheat (110 winter synthetics and 15 spring synthetics) lines derived from crosses between seven durum (T. turgidum) parents and 25 different Ae. tauschii accessions were used in this study (Supplementary Table S1). These lines were distributed by the International Winter Wheat Improvement Program (http://www.iwwip.org) and details were provided previously [25,27,50].
3.2. Evaluation of Disease and Insect Resistance
3.2.1. Seedling Resistance Screening for Leaf, Stem, and Stripe Rusts
Seedlings of 125 SHWs were evaluated in the greenhouse for Lr disease reaction using mixed spores collected from Nebraska wheat fields, against Sr race “TPMK” (Isolate 74MN1409, avirulence/virulence formula Sr6, Sr9a, Sr9b, Sr24, Sr30, Sr31, Sr38, Sr1A.1R./Sr5, Sr7b, Sr8a, Sr9a, Sr9d, Sr9e, Sr10, Sr11, Sr17, Sr21, Sr36, SrMcN, SrTmp) [51], and against Yr using a mixture of three races (Pstv-14: avirulence/virulence formula on Yr genes Yr5, Yr10, Yr15, Yr24, Yr32, YrSP/Yr1, Yr6, Yr7, Yr8 Yr 9, Yr17, Yr27, Yr43, Yr44, YrTr1, YrExp2, YrTye; Pstv-37: avirulence/virulence formula on Yr genes Yr1, Yr5, Yr10, Yr15, Yr24, Yr32, YrSP, YrTye/Yr6, Yr7, Yr8 Yr 9, Yr17, Yr27, Yr43, Yr44, YrTr1, YrExp2; and Pstv-40: avirulence/virulence formula on Yr genes Yr1, Yr5, Yr15, Yr17, YrSP, YrTye/Yr6, Yr7, Yr8, Yr9, Yr10, Yr24, Yr27, Yr32, Yr43, Yr44, YrTr1, YrExp2). The experimental design used in these studies was a randomized complete block design with six replications. Seedling inoculation procedures were performed using previously described protocols for Lr (http://plantsciences.montana.edu/facultyorstaff/faculty/huang/Rust%20Inoculation%20Protocols.pdf), Yr (https://s3.wp.wsu.edu/uploads/sites/2151/2015/12/Greenhouse-Protocols-for-Testing-and-Handling-Stripe-Rust.pdf), and Sr [51]. At 14–18 days after inoculation, leaf and stem rust infection types were scored based on a scale of 0–4 [52,53]. The infection type score of 0 was considered as immune (I) or as very resistant, 1 as resistant (R), 2 as moderately resistant (MR), 3 as moderately susceptible (MS), and 4 as susceptible (S). Stripe rust infection types were scored based on a scale of 0 (very resistant) to 9 (very susceptible) as described previously [54]. For Sr resistance screening, the cultivar “Goodstreak” was used as a resistant check whereas “Cheyenne” and “McNair 701” were used as susceptible checks. For Lr resistance screening, the cultivar “Karahan” was used as a resistant check whereas “Goodstreak” and “McNair 701” were used as susceptible checks. For Yr resistance screening, the cultivar ‘Summit 515’ was used as a resistant check whereas “Morocco” was used a susceptible check.
3.2.2. Adult Plant Resistance Screening for Leaf, Stem, and Stripe Rusts
For the adult plant rust screening, all the genotypes were tested under field conditions for Lr, Sr, and Yr resistance in an augmented design with replicated checks (“Gerek” and “Karahan”) in two rows (distance between rows: 22.5 cm) of a meter-long plot. Leaf rust was tested under natural conditions in Adapazarı, Turkey in the 2016 and 2017 growing seasons. Stem rust was tested against artificial inoculation of race “TTTTF” (avirulent on: Sr24 and Sr31; virulent on: Sr5, Sr6, Sr7b, Sr8a, Sr9a, Sr9b, Sr9d, Sr9e, Sr9g, Sr10, Sr11, Sr17, Sr21, Sr30, Sr36, Sr38, and SrMcN) at Haymana in Turkey in 2016 under natural conditions using natural races in Kastamonu in Turkey in 2016 and 2017, Eskisehir in Turkey, and at Njoro, Kenya in 2017. Similarly, Yr was tested against the artificial inoculation of “Warrior race” (avirulent on: Yr8 and Yr27; virulent on: Yr1, Yr2, Yr3, Yr4, Yr6, Yr7, Yr9, Yr17, Yr25, Yr32, YrSp, YrAvs, and YrAmb) under field conditions at Haymana research station in Turkey in 2016, whereas in 2017, the genotypes were tested under natural conditions using natural races of Yr at Kenya Agricultural and Livestock Research Organization (KALRO) in Njoro, Kenya. Yellow rust disease severity on the flag leaf was recorded three times during the growing season and the final score was taken as a representative score for the analysis. “Gerek” was a susceptible check and “Karahan” was a resistant check for the Lr, Sr, and Yr (Warrior race) used in this study. Rust disease intensities at the adult plant stage were recorded using a modified cobb scale [55] based upon severity (percentage of the plant infected) and response (a type of disease reaction). Field disease severity (SEV) was recorded as a percentage using the following scores: Trace, 5, 10, 20, 40, 60, and 100% infection, whereas a disease response or an infection type (IT) was recorded by dividing the infection type into six groups: 0, resistant (R), moderately resistant (MR), moderately resistant to susceptible (M), moderately susceptible (MS), and susceptible (S) [55]. An average coefficient of infection (COI) was calculated as the multiplication of severity and assigned a constant value based on IT. Constant values were 0.2, 0.4, 0.6, 0.8, and 1.0 for field responses of R, MR, MR–MS (M), MS, and S genotypes, respectively.
3.2.3. Cereal Cyst Nematode Resistance Screening
The cereal cyst nematode resistance screening was carried out at the Soil Borne Diseases Program at International Maize and Wheat Improvement Center (CIMMYT)-Turkey facilities located at the Transitional Zone Agriculture Research Institute (TZARI) Eskisehir, Turkey. The screening was performed in 2016 and 2017 in a randomized complete block design with three replications in the growth chamber as per [14]. Briefly, the population of H. filipjevi was collected from a field in Yozgat, Turkey and freshly hatched second stage juveniles (J2s) were used as inoculum in the screening tests. Three replicates per genotype were inoculated after emergence with 400 freshly hatched J2 by making three holes around the stem base under controlled conditions. Nine weeks after inoculation, cysts from both root and soil extractions were counted under a stereomicroscope. The mean number of cysts was recorded and classed into five groups: Resistant (R), equal or fewer cysts than in a known resistant check; moderately resistant (MR), slightly more cysts than in a resistant check; moderately susceptible (MS), slightly more cysts than in an MR check whereas less than in a susceptible (S) check; S, cyst number similar to S check; and highly susceptible (VS), more cysts than in the S check [14]. Wheat cultivars “Katea”, “Sonmez”, and “Yelken” were used as resistant checks whereas “Bezostaja” and “Kutluk” were used as susceptible checks for CCN.
3.2.4. Crown Rot Resistance Screening
Crown rot resistance screening was performed in a randomized complete block design with three replicates under greenhouse conditions at the Transitional Zone Agricultural Research Institute, Eskisehir, Turkey in 2016 and 2017. The crown rot resistance screening protocol used in this study was provided in detail in Erginbas-Orakci et al. [56]. In brief, the pathogenic F. culmorum Isolate 18 and Isolate 41 were isolated from a mature wheat crowns near Usak city in the Aegean region and Konya, Central Anatolia Plateau of Turkey, respectively. Three replicates of each genotype were grown in a plastic tube (3 cm diameter × 12.5 cm in length) in the greenhouse for about two months with a day/night photoperiod of 16/8 h at a temperature of 25/15 °C and a relative humidity of 60/80%. Around 0.5 g fungus colonized wheat bran was added to the tubes at the time of seed sowing. Around six weeks after inoculation (Zadoks growth stage 14), plants were scored using a 1–5 scale: 1, Resistant; 2, Moderately Resistant; 3, Moderately Susceptible; 4, Susceptible; and 5, Very Susceptible [56]. Wheat cultivars “Altay”,”Carisma”, “Katea”, “Mace”, “Spitfire”, “Suntop”, “Wallup”, “249”, were used as R–MR checks; “Scout”, “Sonmez”, and “Sunco” were used as MR–MS checks; “Bezostaya”,”Ega wylie”, “Emu Rock”, and “Janz” were used as MS checks; whereas “Dumlupinar”, “Kiziltan”, “Kutluk”, “Puseas”, “Seri”, and “Suzen97” were used as susceptible checks for Cr.
3.2.5. Hessian Fly Resistance Screening
The Hessian fly resistance screening using a population from Morocco was performed in 2017 in the greenhouse (20 °C and 70% relative humidity) at the International Center for Agricultural Research the Dry Areas (ICARDA) in Settat, Morocco [28]. A genotype that was resistant to this population may have Hessian fly resistance genes H5, H11, H13, H14, H15, H21, H22, H23, H25, or H26 [28]. The detailed protocol for Hessian fly resistance screening used in this study was provided previously [57]. Briefly, 10 plants per genotype were grown in a greenhouse flat (54 × 36 × 8 cm). Then the seedlings at the one leaf stage were placed under a cheesecloth tent along with infested plants containing mature Hessian fly pupae from which adult Hessian flies emerged and infected the seedling plants. Plant reactions to larval feeding were determined after 20 days. Susceptible genotypes were stunted, dark green, and contained live larvae whereas resistant genotypes were not stunted, light green, and contained dead or live larvae. Percentage of resistant plants was calculated as the number of resistant plants divided by a total number of plants evaluated. The percentage score was divided into five groups: 0%, very susceptible (VS); 10–40%, susceptible; 50–70%, moderately susceptible; 80–90% moderately resistant (MR); and 100% as resistant (R) genotype [57]. Wheat cultivar ‘Nesma’ was used as a susceptible check [57] and resistant plants were checked (under a microscope; 40x) for the presence of dead larvae to confirm the antibiosis effect [36].
3.3. Genotyping and SNP Discovery
The details on genotyping and SNP discovery were described previously [25,27]. Briefly, the genotyping was performed using genotyping-by-sequencing (GBS) approach and SNP discovery was made using TASSEL v. 5.2.40 GBS v2 Pipeline [58] with physical alignment to the Chinese Spring genome sequence, RefSeq v1.0 [59]. The SNPs with minor allele frequency less than 5% and missing information greater than 20% were excluded [50]. All genotypes in this study met the filtering criteria of missing sites less than 20% and were retained in this study [25,27].
3.4. Population Structure and Genome-Wide Association Study
The population structure analysis of 125 SHWs used in this study was performed using a Bayesian clustering algorithm in STRUCURE v.2.3.4 [60] and principal component analysis in TASSEL [61], and has been described previously [29,50]. A GWAS was conducted using 35,798 GBS-derived SNPs and best linear unbiased estimates (BLUEs) of HF infestation score, CCN score, Cr score, and field-based rust SEV, IT, and COI for each location and year. BLUEs were estimated using PROC MIXED in SAS 9.4 (SAS Institute Inc., Cary, NC) by assuming genotype as a fixed effect and replication/block as a random effect. A multi-locus mixed linear model using FarmCPU (Fixed and random model Circulating Probability Unification) algorithm with the coefficient of ancestries of the first three population structure subgroups as covariates and FarmCPU calculated kinship [62] as a random effect were implemented in the Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool (MVP R software package—https://github.com/XiaoleiLiuBio/MVP) for GWAS. The marker-trait associations (MTAs) were selected with a genome-wide significance threshold level of p = 9.18 × 10−5 (−log10p = 4.04) considering the deviation of the observed test statistics value from the test statistics values in the quantile–quantile plots [27]. Haplotype block analysis was performed using default parameters settings of Haploview software (https://www.broadinstitute.org/haploview/haploview). The gene underlying MTAs and their annotation was retrieved from the IWGSC RefSeq v1.0 annotation. The potential candidate genes associated with the trait of interest were further examined using published literature.
4. Conclusions
The present study identified SHWs resistant to diseases and pests that can be used for breeding for resistance to multiple biotic stresses. Additionally, this study identified several lines having resistance to multiple rusts (Lr, Sr, and Yr) at both the seedling and adult plant stage. Resistance genes from these lines can be transferred to bread wheat cultivars using back-crossing.
From GWAS, this study identified 124 genomic regions associated with various diseases that can be used in a marker-assisted breeding program upon validation in an independent population. Furthermore, several genes in those significant genomic regions had gene annotations suggesting their involvement in disease resistance, which provided confidence on the reliability of the identified genomic regions. Furthermore, this study identified several MTAs (42) on the D genome which demonstrates the potential of SHWs to expand the genetic resources of the D genome and for their utilization in wheat breeding programs. Finally, this study also provided information towards further understanding of resistance to multiple biotic stresses in wheat.
Acknowledgments
This work was funded by the Monsanto Beachell-Borlaug International Scholarship Program. This project is a collaborative effort between the University of Nebraska-Lincoln, Lincoln, USA, the International Maize and Wheat Improvement Center (CIMMYT) in Turkey (supported by CRP WHEAT; Ministry of Food, Agriculture and Livestock of Turkey; Bill and Melinda Gates Foundation, and UK Department for International Development, grant OPP1133199), and Omsk State Agrarian University (supported by the Russian Science Foundation project No. 16-16-10005). We would like to thank CIMMYT-Turkey for providing seeds of synthetic hexaploid wheat. We would like to acknowledge CIMMYT-Turkey staffs including Adem Urglu and Ibrahim Ozturk. Partial funding for P.S. Baenziger is from Hatch project NEB-22-328, AFRI/2011-68002-30029, the USDA National Institute of Food and Agriculture as part of the International Wheat Yield Partnership award number 2017-67007-25939, the CERES Trust Organic Research Initiative, and USDA under Agreement No. 59-0790-4-092 which is a cooperative project with the U.S. Wheat and Barley Scab Initiative. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the USDA. Cooperative investigations of the Nebraska Agric. Res. Div., Univ. of Nebraska, and USDA-ARS.
Abbreviations
| SHW | Synthetic hexaploid wheat |
| CCN | Cereal cyst nematode |
| Cr | Crown rot |
| HF | Hessian fly |
| Lr | Leaf rust |
| Sr | Stem rust |
| Yr | Stripe rust |
| GBS | Genotyping-by-sequencing |
| SNP | Single nucleotide polymorphism |
| GWAS | Genome-wide association study |
| MTA | Marker-trait association |
| BLUE | Best linear unbiased estimate |
| PVE | Phenotypic variance explained |
| SEV | Field disease severity |
| COI | Coefficient of infection |
| IT | Infection type |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/20/15/3667/s1.
Author Contributions
Conceptualization, M.B., A.M., and P.S.B.; methodology, M.B.; validation, M.B.; formal analysis, M.B.; investigation, M.B.; resources, M.B., A.M., P.S.B., A.A.D., G.E.-O., S.N.W., M.E.B., P.G., J.P., N.A., L.D., and R.W.; data curation, M.B., A.M., and V.B.; writing of the original draft preparation, M.B.; writing of review and editing, M.B., A.M., V.B., P.S.B., A.A.D., G.E.-O., S.N.W., M.E.B., P.G., J.P., N.A., L.D., and R.W.; supervision, A.M. and P.S.B.; project administration, A.M. and P.S.B.
Conflicts of Interest
The authors declare that they have no conflict of interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.World Food Situation. [(accessed on 8 August 2018)]; Available online: http://www.fao.org/worldfoodsituation/csdb/en/
- 2.Poudel R., Bhatta M. Review of Nutraceuticals and Functional Properties of Whole Wheat. J. Nutr. Food Sci. 2017;7:1. doi: 10.4172/2155-9600.1000571. [DOI] [Google Scholar]
- 3.World Population Prospects. [(accessed on 8 August 2018)];2017 Available online: https://esa.un.org/unpd/wpp/Download/Standard/Population/
- 4.Alexandratos N., Bruinsma J. World Agriculture Towards 2030/2050: The 2012 Revision. FAO; Rome, Italy: 2012. p. 160. [Google Scholar]
- 5.Bhatta M., Regassa T., Wegulo S.N., Baenziger P.S. Foliar Fungicide Effects on Disease Severity, Yield, and Agronomic Characteristics of Modern Winter Wheat Genotypes. Agron. J. 2018;110:602–610. doi: 10.2134/agronj2017.07.0383. [DOI] [Google Scholar]
- 6.Bhatta M., Regassa T. Seed Treatment Affected Yield and Economic Returns of Nebraska Winter Wheat Genotypes. J. Agric. Sci. 2018;9:5–10. doi: 10.31901/24566535.2018/09.1-2.116. [DOI] [Google Scholar]
- 7.Jighly A., Alagu M., Makdis F., Singh M., Singh S., Emebiri L.C., Ogbonnaya F.C. Genomic regions conferring resistance to multiple fungal pathogens in synthetic hexaploid wheat. Mol. Breed. 2016;36:127. doi: 10.1007/s11032-016-0541-4. [DOI] [Google Scholar]
- 8.Huerta-Espino J., Singh R.P., Germán S., McCallum B.D., Park R.F., Chen W.Q., Bhardwaj S.C., Goyeau H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica. 2011;179:143–160. doi: 10.1007/s10681-011-0361-x. [DOI] [Google Scholar]
- 9.Wellings C.R. Global status of stripe rust: A review of historical and current threats. Euphytica. 2011;179:129–141. doi: 10.1007/s10681-011-0360-y. [DOI] [Google Scholar]
- 10.Singh R.P., Hodson D.P., Jin Y., Lagudah E.S., Ayliffe M.A., Bhavani S., Rouse M.N., Pretorius Z.A., Szabo L.J., Huerta-Espino J., et al. Emergence and Spread of New Races of Wheat Stem Rust Fungus: Continued Threat to Food Security and Prospects of Genetic Control. Phytopathology. 2015;105:872–884. doi: 10.1094/PHYTO-01-15-0030-FI. [DOI] [PubMed] [Google Scholar]
- 11.Schumann G.L., D’Arcy C.J. Essential Plant Pathology. 2nd ed. American Phytopathological Society; Saint Paul, MN, USA: 2009. [Google Scholar]
- 12.Nicol J.M., Turner S.J., Coyne D.L., den Nijs L., Hockland S., Maafi Z.T. Current Nematode Threats to World Agriculture. In: Jones J., Gheysen G., Fenoll C., editors. Genomics and Molecular Genetics of Plant-Nematode Interactions. Springer Netherlands; Dordrecht, The Netherlands: 2011. pp. 21–43. [Google Scholar]
- 13.Dababat A.A., Erginbaş-Orakci G., Toktay H., Imren M., Akin B., Braun H.-J., Dreisigacker S., ElekcioğLu İ.H., Morgounov A. Resistance of winter wheat to Heterodera filipjevi in Turkey. Turk. J. Agric. For. 2014;38:180–186. doi: 10.3906/tar-1305-47. [DOI] [Google Scholar]
- 14.Dababat A.A., Ferney G.B.H., Erginbas-Orakci G., Dreisigacker S., Imren M., Toktay H., Elekcioglu H.I., Mekete T., Nicol J.M., Ansari O., et al. Association analysis of resistance to cereal cyst nematodes (Heterodera avenae) and root lesion nematodes (Pratylenchus neglectus and P. thornei) in CIMMYT advanced spring wheat lines for semi-arid conditions. Breed. Sci. 2016;66:692–702. doi: 10.1270/jsbbs.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erginbas-Orakci G., Sehgal D., Sohail Q., Ogbonnaya F., Dreisigacker S., Pariyar S., Dababat A., Erginbas-Orakci G., Sehgal D., Sohail Q., et al. Identification of Novel Quantitative Trait Loci Linked to Crown Rot Resistance in Spring Wheat. Int. J. Mol. Sci. 2018;19:2666. doi: 10.3390/ijms19092666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X., Pan Y., Singh P.K., He X., Ren Y., Zhao L., Zhang N., Cheng S., Chen F. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat. BMC Plant Biol. 2019;19:153. doi: 10.1186/s12870-019-1758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tunali B., Nicol J.M., Hodson D., Uçkun Z., Büyük O., Erdurmuş D., Hekimhan H., Aktaş H., Akbudak M.A., Bağci S.A. Root and Crown Rot Fungi Associated with Spring, Facultative, and Winter Wheat in Turkey. Plant Dis. 2008;92:1299–1306. doi: 10.1094/PDIS-92-9-1299. [DOI] [PubMed] [Google Scholar]
- 18.Saremi H., Ammarellou A., Jafary H. Incidence of crown rot disease of wheat cuased by Fusarium pseudograminearum as a new soil born fungal species in North West Iran. Pak. J. Biol. Sci. 2007;10:3606–3612. doi: 10.3923/pjbs.2007.3606.3612. [DOI] [PubMed] [Google Scholar]
- 19.Smiley R.W., Gourlie J.A., Easley S.A., Patterson L.M. Pathogenicity of Fungi Associated with the Wheat Crown Rot Complex in Oregon and Washington. Plant Dis. 2005;89:949–957. doi: 10.1094/PD-89-0949. [DOI] [PubMed] [Google Scholar]
- 20.Klein T.A., Burgess L.W., Ellison F.W. The incidence and spatial patterns of wheat plants infected by Fusarium graminearum group 1 and the effect of crown rot on yield. Aust. J. Agric. Res. 1991;42:399–407. doi: 10.1071/AR9910399. [DOI] [Google Scholar]
- 21.Wise I.L., Lamb R.J., McKenzie R.I.H., Whistlecraft J.W. Resistance to Hessian fly (Diptera: Cecidomyiidae) in a Canadian spring wheat cultivar1. Can. Entomol. 2006;138:638–646. doi: 10.4039/n05-051. [DOI] [Google Scholar]
- 22.Buntin G.D. Hessian Fly (Diptera: Cecidomyiidae) Injury and Loss of Winter Wheat Grain Yield and Quality. J. Econ. Entomol. 1999;92:1190–1197. doi: 10.1093/jee/92.5.1190. [DOI] [Google Scholar]
- 23.Amri A., Bouhssini M.E., Lhaloui S., Cox T.S., Hatchett J.H. Estimates of yield loss due to the Hessian fly (Diptera: Cecidomyiidae) on bread wheat using near-isogenic lines. Al Awamia. 1992;77:75–88. [Google Scholar]
- 24.Lhaloui S., Buschman L., Bouhssini’ M.E., Starks K., Kfith D., Houssaini K.E. Control of Mayetiola species (Diptera: Cecidomyiidae) with carbofuran in bread wheat, durum wheat and barley with yield loss assessment and its economic analysis. Al Awamia. 1992;77:55–73. [Google Scholar]
- 25.Bhatta M., Morgounov A., Belamkar V., Poland J., Baenziger P.S. Unlocking the novel genetic diversity and population structure of synthetic hexaploid wheat. BMC Genom. 2018;19:591. doi: 10.1186/s12864-018-4969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogbonnaya F.C., Abdalla O., Mujeeb-Kazi A., Kazi A.G., Xu S.S., Gosman N., Lagudah E.S., Bonnett D., Sorrells M.E., Tsujimoto H. Synthetic Hexaploids: Harnessing Species of the Primary Gene Pool for Wheat Improvement. In: Janick J., editor. Plant Breeding Reviews. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2013. pp. 35–122. [Google Scholar]
- 27.Bhatta M., Morgounov A., Belamkar V., Baenziger P. Genome-Wide Association Study Reveals Novel Genomic Regions for Grain Yield and Yield-Related Traits in Drought-Stressed Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018;19:3011. doi: 10.3390/ijms19103011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgounov A., Abugalieva A., Akan K., Akın B., Baenziger S., Bhatta M., Dababat A.A., Demir L., Dutbayev Y., Bouhssini M.E., et al. High-yielding winter synthetic hexaploid wheats resistant to multiple diseases and pests. Plant Genet. Resour. 2018;16:273–278. doi: 10.1017/S147926211700017X. [DOI] [Google Scholar]
- 29.Bhatta M., Morgounov A., Belamkar V., Yorgancılar A., Baenziger P.S. Genome-wide association study reveals favorable alleles associated with common bunt resistance in synthetic hexaploid wheat. Euphytica. 2018;214:200. doi: 10.1007/s10681-018-2282-4. [DOI] [Google Scholar]
- 30.Das M.K., Bai G., Mujeeb-Kazi A., Rajaram S. Genetic diversity among synthetic hexaploid wheat accessions (Triticum aestivum) with resistance to several fungal diseases. Genet. Resour. Crop. Evol. 2016;63:1285–1296. doi: 10.1007/s10722-015-0312-9. [DOI] [Google Scholar]
- 31.Ogbonnaya F.C., Imtiaz M., Ye G., Hearnden P.R., Hernandez E., Eastwood R.F., van Ginkel M., Shorter S.C., Winchester J.M. Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor. Appl. Genet. 2008;116:891–902. doi: 10.1007/s00122-008-0712-8. [DOI] [PubMed] [Google Scholar]
- 32.Zegeye H., Rasheed A., Makdis F., Badebo A., Ogbonnaya F.C. Genome-Wide Association Mapping for Seedling and Adult Plant Resistance to Stripe Rust in Synthetic Hexaploid Wheat. PLoS ONE. 2014;9:e105593. doi: 10.1371/journal.pone.0105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIntosh R.A., Dubcovsky J., Rogers J.W., Morris C.F., Appels R., Xia X. Catalogue of gene symbols for wheat: 2013-14 Supplement. Annu. Wheat Newsl. 2014;58:52. [Google Scholar]
- 34.Ali S., Rodriguez-Algaba J., Thach T., Sørensen C.K., Hansen J.G., Lassen P., Nazari K., Hodson D.P., Justesen A.F., Hovmøller M.S. Yellow Rust Epidemics Worldwide Were Caused by Pathogen Races from Divergent Genetic Lineages. Front. Plant Sci. 2017;8:1057. doi: 10.3389/fpls.2017.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox T.S., Raupp W.J., Gill B.S. Leaf Rust-Resistance Genes Lr41, Lr42, and Lr43 Transferred from Triticum tauschii to Common Wheat. Crop Sci. 1994;34:339–343. doi: 10.2135/cropsci1994.0011183X003400020005x. [DOI] [Google Scholar]
- 36.Joukhadar R., El-Bouhssini M., Jighly A., Ogbonnaya F.C. Genome-wide association mapping for five major pest resistances in wheat. Mol. Breed. 2013;32:943–960. doi: 10.1007/s11032-013-9924-y. [DOI] [Google Scholar]
- 37.Lei Y., Wang M., Wan A., Xia C., See D.R., Zhang M., Chen X. Virulence and Molecular Characterization of Experimental Isolates of the Stripe Rust Pathogen (Puccinia striiformis) Indicate Somatic Recombination. Phytopathology. 2016;107:329–344. doi: 10.1094/PHYTO-07-16-0261-R. [DOI] [PubMed] [Google Scholar]
- 38.Yuan C., Wu J., Yan B., Hao Q., Zhang C., Lyu B., Ni F., Caplan A., Wu J., Fu D. Remapping of the stripe rust resistance gene Yr10 in common wheat. Theor. Appl. Genet. 2018;131:1253–1262. doi: 10.1007/s00122-018-3075-9. [DOI] [PubMed] [Google Scholar]
- 39.Wu J., Zeng Q., Wang Q., Liu S., Yu S., Mu J., Huang S., Sela H., Distelfeld A., Huang L., et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018;131:1481–1496. doi: 10.1007/s00122-018-3092-8. [DOI] [PubMed] [Google Scholar]
- 40.Chen S., Zhang W., Bolus S., Rouse M.N., Dubcovsky J. Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet. 2018;14:e1007287. doi: 10.1371/journal.pgen.1007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manickavelu A., Joukhadar R., Jighly A., Lan C., Huerta-Espino J., Stanikzai A.S., Kilian A., Singh R.P., Ban T. Genome wide association mapping of stripe rust resistance in Afghan wheat landraces. Plant Sci. 2016;252:222–229. doi: 10.1016/j.plantsci.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Mourad A.M.I., Sallam A., Belamkar V., Wegulo S., Bowden R., Jin Y., Mahdy E., Bakheit B., El-Wafaa A.A., Poland J., et al. Genome-Wide Association Study for Identification and Validation of Novel SNP Markers for Sr6 Stem Rust Resistance Gene in Bread Wheat. Front. Plant Sci. 2018;9:380. doi: 10.3389/fpls.2018.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bozkurt O., Unver T., Akkaya M.S. Genes associated with resistance to wheat yellow rust disease identified by differential display analysis. Physiol. Mol. Plant Pathol. 2007;71:251–259. doi: 10.1016/j.pmpp.2008.03.002. [DOI] [Google Scholar]
- 44.Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Shumayla, Sharma S., Kumar R., Mendu V., Singh K., Upadhyay S.K. Genomic Dissection and Expression Profiling Revealed Functional Divergence in Triticum aestivum Leucine Rich Repeat Receptor Like Kinases (TaLRRKs) Front. Plant Sci. 2016;7:1374. doi: 10.3389/fpls.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng H., Wang X., Sun Y., Wang X., Chen X., Guo J., Duan Y., Huang L., Kang Z. Cloning and characterization of a calcium binding EF-hand protein gene TaCab1 from wheat and its expression in response to Puccinia striiformis f. sp. tritici and abiotic stresses. Mol. Biol. Rep. 2011;38:3857–3866. doi: 10.1007/s11033-010-0501-8. [DOI] [PubMed] [Google Scholar]
- 47.Juliana P., Singh R.P., Singh P.K., Poland J.A., Bergstrom G.C., Huerta-Espino J., Bhavani S., Crossa J., Sorrells M.E. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor. Appl. Genet. 2018;131:1405–1422. doi: 10.1007/s00122-018-3086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seevers P.M., Daly J.M., Catedral F.F. The Role of Peroxidase Isozymes in Resistance to Wheat Stem Rust Disease 1. Plant Physiol. 1971;48:353–360. doi: 10.1104/pp.48.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayana G.T., Ali S., Sidhu J.S., Gonzalez Hernandez J.L., Turnipseed B., Sehgal S.K. Genome-Wide Association Study for Spot Blotch Resistance in Hard Winter Wheat. Front. Plant Sci. 2018;9:926. doi: 10.3389/fpls.2018.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhatta M., Baenziger P.S., Waters B.M., Poudel R., Belamkar V., Poland J., Morgounov A. Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018;19:3237. doi: 10.3390/ijms19103237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward R.A. Master’s Thesis. University of Nebraska; Lincoln, NE, USA: 2012. Identification of Stem Rust Resistance in Three Synthetic Wheat Populations. [Google Scholar]
- 52.Stakman E.C., Stewart D.M., Loegering W.Q. Identification of Physiologic Races of Puccinia Graminis var. Tritici. Vol. 617. USDA; Washington, DC, USA: 1962. pp. 8–51. [Google Scholar]
- 53.Roelfs A.P. Resistance to leaf and stem rusts in wheat. Chapter 2; Proceedings of the Breeding Strategies for Resistance to the Rusts of Wheat; El Batan, Mexico. 29 June–1 July 1987. [Google Scholar]
- 54.McNeal F.H., Konzak C.F., Smith E.P., Tate W.S., Russell T.S. A Uniform System for Recording and Processing Cereal Research Data. USDA Bulletin Agric. Res. Serv.; Beltsville, MD, USA: 1971. pp. 34–121. [Google Scholar]
- 55.Peterson R.F., Campbell A.B., Hannah A.E. A Diagrammatic Scale for Estimating Rust Intensity on Leaves and Stems of Cereals. Can. J. Res. 1948;26:496–500. doi: 10.1139/cjr48c-033. [DOI] [Google Scholar]
- 56.Erginbas-Orakci G., Poole G., Nicol J.M., Paulitz T., Dababat A.A., Campbell K. Assessment of inoculation methods to identify resistance to Fusarium crown rot in wheat. J. Plant Dis. Prot. 2016;123:19–27. doi: 10.1007/s41348-016-0001-8. [DOI] [Google Scholar]
- 57.El Bouhssini M., Chen M., Lhaloui S., Zharmukhamedova G., Rihawi F. Virulence of Hessian fly (Diptera: Cecidomyiidae) in the Fertile Crescent. J. Appl. Entomol. 2009;133:381–385. doi: 10.1111/j.1439-0418.2008.01339.x. [DOI] [Google Scholar]
- 58.Glaubitz J.C., Casstevens T.M., Lu F., Harriman J., Elshire R.J., Sun Q., Buckler E.S. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS ONE. 2014;9:e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.International Wheat Genome Sequencing Consortium Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 60.Pritchard J.K., Stephens M., Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 62.Liu X., Huang M., Fan B., Buckler E.S., Zhang Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016;12:e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.