Abstract
Objective
We aimed to address deficiencies in structured electronic health record (EHR) data for race and ethnicity by identifying black and Hispanic patients from unstructured clinical notes and assessing differences between patients with or without structured race/ethnicity data.
Materials and Methods
Using EHR notes for 16 665 patients with encounters at a primary care practice, we developed rule-based natural language processing (NLP) algorithms to classify patients as black/Hispanic. We evaluated performance of the method against an annotated gold standard, compared race and ethnicity between NLP-derived and structured EHR data, and compared characteristics of patients identified as black or Hispanic using only NLP vs patients identified as such only in structured EHR data.
Results
For the sample of 16 665 patients, NLP identified 948 additional patients as black, a 26%increase, and 665 additional patients as Hispanic, a 20% increase. Compared with the patients identified as black or Hispanic in structured EHR data, patients identified as black or Hispanic via NLP only were older, more likely to be male, less likely to have commercial insurance, and more likely to have higher comorbidity.
Discussion
Structured EHR data for race and ethnicity are subject to data quality issues. Supplementing structured EHR race data with NLP-derived race and ethnicity may allow researchers to better assess the demographic makeup of populations and draw more accurate conclusions about intergroup differences in health outcomes.
Conclusions
Black or Hispanic patients who are not documented as such in structured EHR race/ethnicity fields differ significantly from those who are. Relatively simple NLP can help address this limitation.
Keywords: race, ethnicity, natural language processing, electronic health record
BACKGROUND AND SIGNIFICANCE
Racial and ethnic disparities in the quality of health care and outcomes have been extensively documented across diseases and care settings in the United States.1,2 To identify, understand, and reduce these pervasive disparities, high-quality data collection on race, ethnicity, and other social determinants of health is imperative.3–5 Most medical centers in the United States use electronic health records (EHRs) to collect demographic information from their patients.3 Since the rollout of Meaningful Use by the Center for Medicare and Medicaid Services in 2011, collection of race/ethnicity data in the EHR in a structured format has been required.6 However, despite this regulation, there remains a large degree of missing or misclassified data for both race and ethnicity data fields.4 This inaccuracy hinders an institution’s ability to examine potential gaps in quality of care for their racial and ethnic minorities.4 Furthermore, inaccurate reporting on race and ethnicity in the EHR also limits the impact of disparities research, which is critical to the reduction of health disparities, in that patients with no structured EHR race data, or uninformative structured EHR race data may differ from patients with structured, informative data in clinically or statistically significant ways, rendering analyses dependent solely on structured data subject to bias that that may adversely impact the accuracy and value of results.
At our institution (Weill Cornell Medicine [WCM]), where clinicians have used an EHR system for nearly 2 decades, structured data for race and ethnicity according to federal Office of Management and Budget standards7 is available for roughly one-third and two-thirds of patients with at least 1 encounter, respectively. Critically, structured values of “declined” or “not specified” for race and ethnicity in our local EHR complicate efforts to identify black/African American (“black”) and Hispanic/Latino/a (“Hispanic”) patients, who are historically underrepresented in clinical trials,8 and clinical research at large. Furthermore, race and ethnicity values recorded in a structured fashion by registration clerks may differ from those recorded by clinicians in unstructured notes as part of patient care.9 To address deficiencies in structured EHR race and ethnicity data, researchers and practitioners have applied natural language processing (NLP), a host of computational techniques that automate identification and extraction of a wide range of structured clinical concepts from unstructured clinical notes, for identification of these patient demographic characteristics.10–13
OBJECTIVE
The goals of this study were to develop an NLP method for extracting race and ethnicity from unstructured clinical notes, determine the extent to which the NLP-extracted race and ethnicity values improved identification of black and Hispanic patients compared with structured EHR data, and compare characteristics of patients identified as black or Hispanic in structured EHR race and ethnicity data as compared with NLP-derived data for patient demographics.
MATERIALS AND METHODS
Setting
WCM is an academic medical center in New York City with more than 1600 attending physicians conducting 1.7 million annual patient visits across more than 20 outpatient sites. WCM physicians hold admitting privileges at NewYork-Presbyterian Hospital. To document outpatient care, WCM physicians have used the EpicCare Ambulatory EHR system since 2000. The WCM Institutional Review Board approved this study.
Study design and data collection
We performed a cross-sectional observational study of 16 665 adult patients with 2 or more office visits with a physician, physician’s assistant, or nurse practitioner between January 1, 2017, and August 1, 2018, at a specific WCM internal medicine practice with a diverse patient population. For these patients, we obtained from the EHR all clinical notes (n = 4.7 million) authored by clinicians from all outpatient practices across the institution regardless of clinician specialty and date of authorship.14
System development
To automatically obtain black race and Hispanic ethnicity from clinical notes, we first manually reviewed a convenience sample of clinical notes (n = 1000) to identify textual patterns and then iteratively developed rules to extract terms based on the patterns.15 As shown in Supplementary Appendix 1, the notes demonstrated considerable lexical variation in expression of black race and Hispanic ethnicity. Formatting and content of notes varied, although most contained race and ethnicity documentation in designated subsections denoted as “history of present illness (HPI),” “general,” “appearance,” “subjective,” “objective,” “race/ethnicity,” “mental state examination (MSE),” and “assessment and plan.” A substantial number of clinical notes had no mention of race or ethnicity. For notes containing race and ethnicity descriptions, mostly there was only a single mention of race or ethnicity. However, we observed notes in which race and ethnicity information could be extracted from patient descriptions available at different sections of the note, and the values inferred from these descriptions may or may not match each other.
Using the Apache Unstructured Information Management Architecture–based Leo NLP system maintained by the U.S. Veterans Administration,16,17 we developed a rule-based algorithm called CIREX (Clinical Information Race Ethnicity eXtractor) to extract African American race and Hispanic ethnicity from clinical notes. Previously we demonstrated use of Leo to extract ejection fraction13 and Patient Health Questionnaire-9 values.18 We employed a similar approach to identify all instances of race and ethnicity in notes and classify each note as positive or negative for containing black race as well as Hispanic ethnicity.
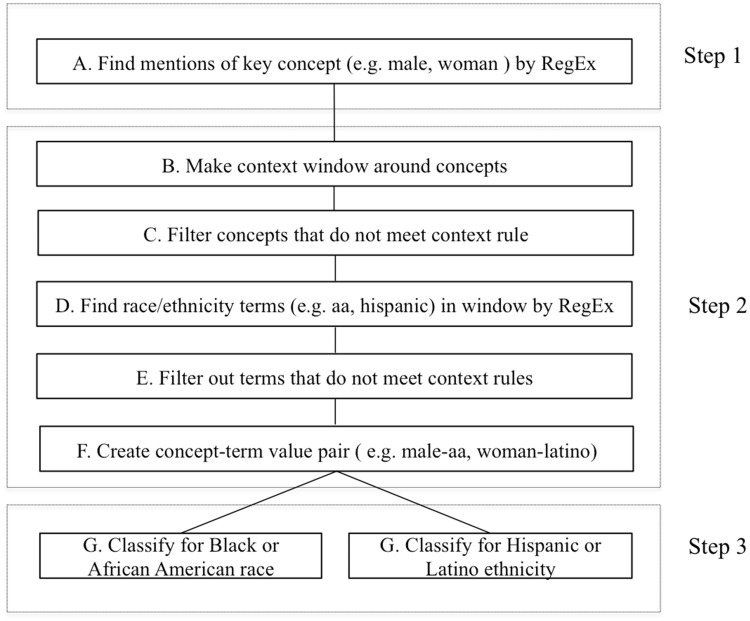
The creation of extraction logic was an iterative 3-step process that included concept and term definitions, context analysis, rule definition, system application, error analysis, and classification (Figure 1).
Figure 1.
Natural language processing pipeline logic implemented in classifying notes for race/ethnicity. RegEx: regular expressions.
The first step involved identifying a set of core concepts that co-existed with a term for race and ethnicity in narrative notes. For example, “The patient is a 71 year old African-American male alert, cooperative, no distress” related the main concept “patient” with age, race, and general appearance. Similarly, “Generally healthy-appearing AA male, +moderate facial wasting” described the appearance, race, and gender of the “male.” Table 1 lists all the concepts that we used to find possible mentions of race or ethnicity. Terms such as male, female, man, or lady were frequent sentinels indicating the presence of a referent term for race or ethnicity (eg, “Spanish-speaking lady” or “AA male”). Regular expressions, string matching, and filters were used to extract the concepts. In step 2, we used iterative context analysis to determine if concepts were mentioned in the context of race and ethnicity. A window of appropriate surrounding words was then defined for finding specific terms that referred to race or ethnicity as defined by terms in Table 1.
Table 1.
List of terms used to identify the concepts of race and ethnicity in clinical notes and reports
| Terms | |
|---|---|
| A. Identifying concepts of race or ethnicity | Male, female, man, woman, boy, girl, lady, gentleman, patient, pt, infant, young, old, elderly, aged, ethnicity, race, background, language, interpreter, descent, origin, nationality, identity, racial |
| B. Describing any race or ethnicity | aa, afghan, African American, african, alaskan native, alaskan nation, algeria, american indian,anglo saxon, asian, austronesian, arab, arabian, black, bangladeshi, bengali, burmese, bi-race, bi-racial, cameroon, caucasian, canadian, caucasoid, cambodian, central american, chinese, congo, cuban, cuban american, dominican, danish, dutch, european, egyptian, eskimo, ethiopia, french, german, ghana, gujarati, haitian, hawaiian, hispanic, irish, indian, israeli, jamaican, japanese, jewish, kenya, korean, latina, latino, libyan, laotian, malayalam, malaysian, mexican, mixed race, mixed racial, multi race, multi racial, moroccan, morocco, native american, native, alaskan, nigeria, north american, oriental, pacificislander, pakistani, philipino, philippine, polish, polynesian, puerto rican, russian, scandinavian, spanish, south american, sri lankan, sudan, swedish, swiss, tamil, telungu, thai, uganda, vietnamese, white, zambia |
| C. Describing black/African American race specifically | African American, black, aa, haitian, african, sudan, nigeria, algeria, ethiopia, kenya, ghana, uganda, cameroon, congo, zambia, carribean, jamaican |
| D. Describing Hispanic or Latino ethnicity specifically | hispanic, latina, latino, south american, central american, cuban, mexican, puerto rican, dominican, spanish |
While the terms may not represent a complete lexicon used by physicians to refer to the concepts in question, they covered the majority of notes that we examined during the initial exploratory phase of this study. We defined regular expressions that allowed for variations in these terms, then applied various context analysis, validation, and filtering rules to these terms to identify a given race or ethnicity, thus improving the overall detection of true positive (TP) cases of race and ethnicity instances. Each concept–term relation identified through this process was then mapped into 1 of 6 categories of race and 2 categories of ethnicity.7
In step 3, we classified a note as positive or negative for African American race or Hispanic ethnicity based on identified concept–term pairs. If multiple race concept–term pairs existed in a note, then we classified that note as positive if at least 1 of the concept–term pairs corresponded to black race. We applied a similar heuristic for classifying a note with multiple ethnicity concept–term pairs as positive for Hispanic ethnicity. As illustrated in Figure 1, we iteratively developed and adjusted rules using Leo until the NLP method reliably extracted race and ethnicity values from the convenience sample of notes.
Evaluation
Creation of gold standard and assessment of interrater reliability
To construct a gold standard for evaluating the NLP method, we manually prescreened notes to generate a sample of 400 documents where 80% contained reference of black race or Hispanic ethnicity and 20% did not. Two reviewers (SAL and CI) manually categorized 400 documents according to their race and ethnicity values, and a third reviewer (LCP) resolved any disagreements. To assess interrater reliability, we determined Krippendorff’s alpha, a metric ranging from –1 to 1, where 1 represents perfect reliability, –1 represents total systematic disagreement, and 0 represents the absence of reliability. We also calculated Cohen’s kappa to determine pairwise interrater reliability for each permutation of the 3 reviewers.
We used federal categories for race and ethnicity19 to define concepts of interest in this study. If a note contained a value for race, reviewers determined whether the note described black race. If a note contained a value for ethnicity, reviewers determined whether the note described Hispanic ethnicity. For a note containing multiple values of race, reviewers classified the note as positive for black race if at least 1 value indicated black race. Similarly, for a note containing multiple values of ethnicity, reviewers classified the note as positive for Hispanic ethnicity if at least 1 value indicated Hispanic ethnicity.
Measurement of NLP method performance
We compared the reference standard’s classification of black race and Hispanic ethnicity against output of the NLP method using the same 400 notes. We classified each note into 1 of 4 categories for both race and ethnicity. First, a TP was defined as an instance in which the note contained a mention of black race or Hispanic ethnicity (as determined by manual review) and the NLP method successfully identified the note as containing a mention of black race or Hispanic ethnicity. Second, a false positive (FP) was defined as an instance in which the note either did not contain a mention of race or ethnicity or contained a mention of nonblack race or non-Hispanic ethnicity, but the NLP method classified the note as containing a mention of black race or Hispanic ethnicity. Third, a true negative (TN) was defined as an instance in which the note did not contain mention of black race or Hispanic ethnicity and the NLP method did not classify a note as containing a mention of black race or Hispanic ethnicity. Finally, a false negative (FN) was defined as an instance in which the note contained a mention of black race or Hispanic ethnicity and the NLP method did not classify the note as containing a mention of race or ethnicity. We used counts of the 4 possible cases to construct 2 confusion matrices to calculate precision (TP/(TP+FP)), accuracy (TP+TN/(TP+TN+FP+FN)), recall (TP/(TP+FN)), and F score (harmonic mean of recall and precision) with the R statistical software package.20
Comparison of NLP method with structured EHR race data
For all 16 665 patients in the study sample, we compared black race and Hispanic ethnicity extracted via the NLP method vs structured values stored in the EHR. We then determined the proportion of the patients identified as black or Hispanic in structured EHR race data. We also calculated the total number of patients that the NLP method identified as black or Hispanic that were not identified as such in the EHR.
Comparison of characteristics of black and Hispanic patients identified by structured EHR data only and NLP only
We compared black patients identified by structured EHR race data vs black patients identified only by NLP with respect to age, sex, insurance status, and number of medical conditions (as defined by the number of active entries on each patient’s problem list in the EHR). Additionally, we conducted the same comparison on patients identified as Hispanic in structured EHR ethnicity data to patients identified as Hispanic only by NLP. To perform the comparisons, we used independent samples t tests and chi-square tests, considering a P value of <.05 as statistically significant.
RESULTS
For the 16 665 patients meeting study inclusion criteria, we obtained 4.7 million clinical notes. Table 2 describes characteristics of the study sample. Software for the NLP method is available publicly (https://github.com/wcmc-research-informatics/CIREX).
Table 2.
Characteristics of patients included in sample (N = 16 665)
| Age | |
|---|---|
| 18-44 years of age | 3245 (19.5) |
| 45-64 years of age | 5733 (34.4) |
| 65+ years of age | 7687 (46.1) |
| Female | 10 900 (65.4) |
| Structured race | |
| White | 5756 (34.5) |
| Asian | 1277 (7.7) |
| American Indian/Alaska Native | 68 (0.4) |
| Native Hawaiian/Pacific Islander | 46 (0.3) |
| Declined | 1295 (7.8) |
| Other combinations not described | 4501 (27) |
| Null | 62 (0.4) |
| Black/African American | 3660 (22) |
| Structured ethnicity | |
| Hispanic/Latino | 3298 (19.8) |
| Null | 159 (1) |
| Unknown | 1019 (6.1) |
| Declined | 3741 (22.4) |
| Multiracial | 29 (0.2) |
| Not Hispanic/Latino | 8419 (50.5) |
| Active problem list entries | 8 (5-14) |
| Progress notes | 848 (204-3162) |
Values are n (%) or median (interquartile range).
Assessment of interrater reliability
We observed a Krippendorff’s alpha of .50 for whether a note described race and .55 for whether a note describing race indicated black race. However, values differed for ethnicity, with an alpha of .89 for whether a note described ethnicity and .93 for whether a note described a patient as being of Hispanic ethnicity. For whether a note describing race indicated black race, we observed a Cohen’s kappa of 0.88 between reviewers 1 and 3, a Cohen’s kappa of 0.42 between reviewers 1 and 2, and a Cohen’s kappa of 0.456 between reviewers 2 and 3.
NLP method performance
As illustrated in Table 3, for classifying patients as black or not black based on 400 notes, the NLP method achieved precision of 0.885, recall of 0.939, and F score of 0.911 (Table 4). An example of a FP was a note that contained a checklist value for hypertensive risk factors with the value “Sex: Male Is an African American: No” and an example of a FN was a note that contained a reference to a patient as a “female of American Indian and AA background.”
Table 3.
Confusion matrix for performance of CIREX on race (n = 400)
| NLP predicted: black | NLP predicted: not black | |
|---|---|---|
| Gold standard: black | True positive: 92 | False negative: 6 |
| Gold standard: Not black | False positive: 12 | True negative: 290 |
NLP: natural language processing.
Table 4.
Confusion matrix for performance of CIREX on ethnicity (n = 400)
| NLP predicted: Hispanic | NLP predicted: Not Hispanic | |
|---|---|---|
| Gold standard: Hispanic | True positive: 126 | False negative: 2 |
| Gold standard: Not Hispanic | False positive: 2 | True negative: 270 |
NLP: natural language processing.
As illustrated in Table 4, for classifying patients as Hispanic or non-Hispanic based on 400 notes, the NLP method achieved precision of 0.984, recall of 0.984, and F score of 0.984 (Table 5). An example of a FN was a patient described as a “Spanish speaking AA woman in NAD.” An example of a FP was a note that contained a checklist value for hypertensive risk factors with the value “Is Non-Hispanic African American: Yes.”
Table 5.
Characteristics of patients identified as black via NLP alone compared with patients identified as black in structured EHR race data
| Characteristic | Recorded as black in structured EHR data (n = 3660) | Identified as black via NLP alone (n = 948) |
|---|---|---|
| Age as of today (years) | 59.0 (46.9-69.3), 19.0-103.9 | 64.9 (55.1-73.6), 19.9-95.7 |
| Sex | ||
| Male | 1027 (28.1) | 359 (37.9) |
| Female | 2633 (71.9) | 589 (62.1) |
| Active problem list entries | 11.2, 0-84 | 14.5, 0-73 |
| Insurance | ||
| Commercial | 1277 (34.9) | 283 (29.9) |
| Medicaid | 132 (3.6) | 43 (4.5) |
| Medicare | 765 (20.9) | 319 (33.6) |
| Self-pay | 62 (1.7) | 10 (1.0) |
| Managed Medicaid/Medicare | 1424 (38.9) | 293 (30.9) |
Values are median (interquartile range), range; n (%); or mean, range.
EHR: electronic health record; NLP: natural language processing.
Comparison with structured EHR race data
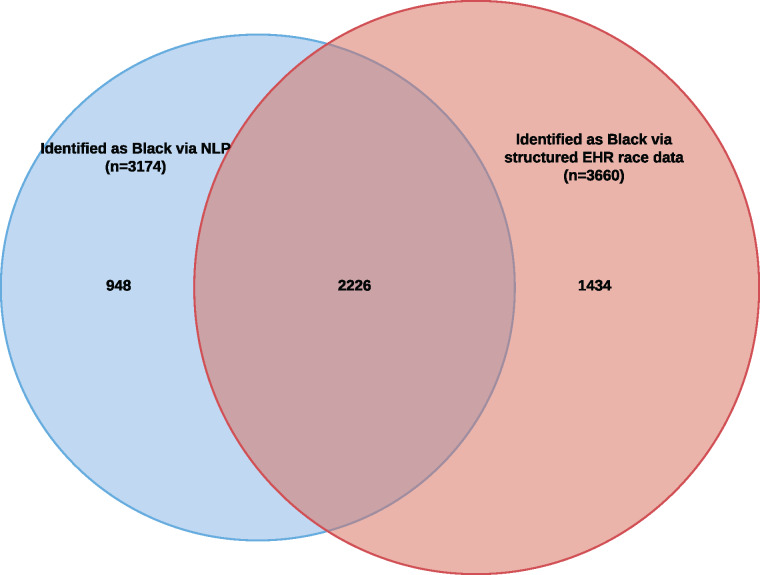
As described in Figure 2, when applied to 4.7 million notes for 16 665 patients, the NLP method identified 948 patients as black beyond the 3660 identified as such in structured EHR race data, a relative percentage increase of 26%. A total of 48% of all patients identified by either NLP or structured EHR race data as black were identified by both methods.
Figure 2.
Venn diagram illustrating overlap of patients identified as black using natural language processing (NLP) with patients identified as black using structured electronic health record (EHR) race data (n = 4608).
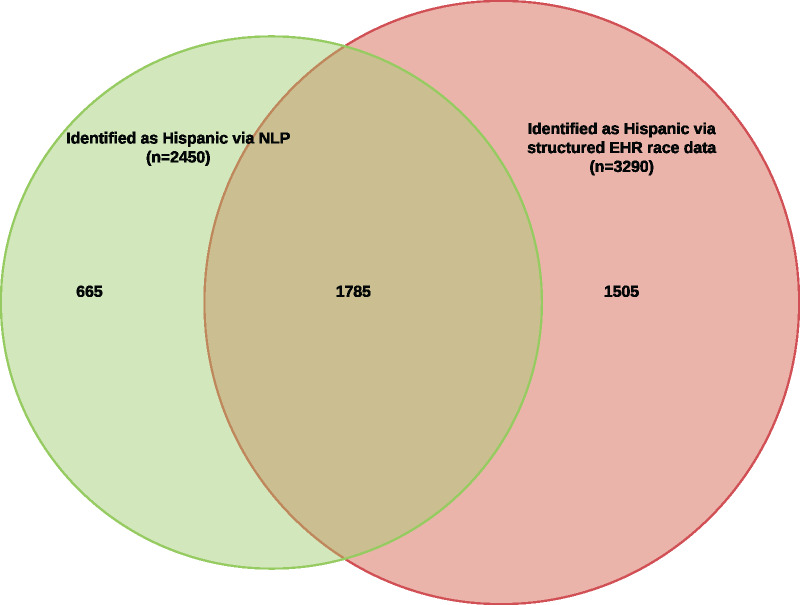
Similarly, as shown in Figure 3, the NLP method identified 665 patients as Hispanic beyond the 3290 identified as such in structured EHR ethnicity data, a relative percentage increase of 20%. 45% of all patients identified by either NLP or structured EHR ethnicity data as Hispanic were identified by both.
Figure 3.
Venn diagram illustrating overlap of patients identified as Hispanic using natural language processing (NLP) with patients identified as Hispanic using structured electronic health (EHR) record ethnicity data (n = 3955).
While only 128 patients were identified as both black and Hispanic in structured EHR race and ethnicity data, incorporating the results of the NLP method allowed us to identify 591 patients as black and Hispanic, a relative percentage increase of 462%.
Characteristics of black and Hispanic patients identified by structured EHR race data only and NLP only
As described in Table 5, the sample of patients identified in structured EHR race data as black differed from the sample of patients identified as black via NLP alone. Likewise, as described in Table 6, the sample of patients identified in structured EHR data as Hispanic differed from the sample of patients identified as Hispanic via NLP alone. Notably, patients identified as black via NLP alone were more likely to be male, older, and had more active problem list entries in the EHR; additionally, they were less likely to have commercial insurance. The same associations were present in patients identified as Hispanic via NLP alone. Independent sample t tests demonstrated that the differences in mean age and number of active problem list entries between the 2 patient samples were statistically significant (P < .001). Likewise, Chi square tests indicated that differences in the distribution of insurance status and sex were also statistically significant (P < .001).
Table 6.
Characteristics of patients identified as Hispanic via NLP alone compared with patients identified as Hispanic in structured EHR race data
| Characteristic | Recorded as Hispanic in structured EHR data (n = 3290) | Identified as Hispanic via NLP alone (n = 665) |
|---|---|---|
| Age as of today (years) | 59.0 (46.9-69.3), 19.0-103.9 | 63.1 (51.8-73.0), 18.9-97.0 |
| Sex | ||
| Male | 1027 (28.1) | 207 (31.1) |
| Female | 2633 (71.9) | 458 (68.9) |
| Active problem list entries | 11.2, 0-84 | 13.1, 0-63 |
| Insurance | ||
| Commercial | 1277 (34.9) | 155 (23.3) |
| Medicaid | 132 (3.6) | 34 (5.1) |
| Medicare | 765 (20.9) | 193 (29.0) |
| Self-pay | 62 (1.7) | 7 (1.1) |
| Managed Medicaid/Medicare | 1424 (38.9) | 276 (41.5) |
Values are median (interquartile range), range; n (%); or mean, range.
EHR: electronic health record; NLP: natural language processing.
DISCUSSION
This study demonstrated the successful development and validation of an NLP method to identify black and Hispanic patients based on clinical notes, as well as its impact: namely, that without the incorporation of data elements abstracted from free text notes, researchers may both underestimate the number of black or Hispanic patients in a given cohort and underestimate their age, disease burden, poverty level, and proportion of male sex. Overall, combining NLP and structured EHR race data increased the number of patients identified in the sample as black by 948 (5.7%) and the number of patients identified in the sample as Hispanic by 665 (4.0%). Patients identified as black only through NLP differed significantly from patients identified as black in structured EHR data, with higher proportions of male sex, lower rates of commercial insurance, higher average number of active problem list entries, and higher average age. Patients identified as Hispanic only through NLP differed significantly from patients identified as Hispanic in structured EHR ethnicity data in a substantially similar fashion.
Our results highlight the potential of NLP techniques to improve the identification of race and ethnicity in EHR data to enable researchers to conduct large-scale, population-based analyses. Without knowing patient demographics in a given cohort, it is impossible to accurately assess healthcare disparities and intergroup differences in health service utilization and outcomes. Underestimating the portion of a given patient population that is black or Hispanic by relying solely on structured EHR race data may also lead to biased study conclusions, especially given the differences observed in our study between patients identified as black/Hispanic in structured EHR race/ethnicity data vs NLP-derived data. By implementing our study’s approach, researchers can more accurately ascertain the true demographic makeup of their intended patient population, which may differ substantially from estimates using structured EHR race or ethnicity data fields alone.
To date, few studies have assessed the possibility of supplementing existing structured data on race and ethnicity in the EHR with data derived from NLP techniques. As such, our study, while methodologically reliant on a rule-based approach, holds the potential to advance the field by offering one of the first examples of a clinically validated NLP approach for extracting race and ethnicity that compares the output of a given pipeline to the results of expert manual adjudication. Additionally, while some previous research21 has focused on specific populations with lexically distinct sentinel phrases (eg, Somali immigrants), our study differs in that it relies on specific phrases not only commonly used in other contexts within medicine (eg “white lesions”) but also on phrases that may occur not in the context of the patient’s race, but rather within the context of a diagnostic threshold (eg “African-American/Non-Hispanic: no” in a list of risk factors). The NLP method described herein, while tailored specifically toward identifying black race and Hispanic ethnicity, also extracts other mentions of race and ethnicity and can be used to determine patient populations belonging to other races, such as Asian Americans.
The primary limitation of this study is the relatively small sample size of manually annotated notes, especially given the relative rarity of FNs in the dataset. The generalizability of this sample is also worth considering, given that the study sample comprised patients treated at a single institution with relatively high data density and recent contacts with the healthcare system. An additional limitation we observed was relatively low interrater reliability for race, which was mostly driven by one reviewer’s discordance with the other 2. While we were in part able to mitigate this by relying on an adjudication system, whereby reviewer 3 resolved differences between reviewers 1 and 2, this suggests that careful training of reviewers is crucial to ensure the validity of ground truth data in this area.
Future work may serve to further address these limitations by pursuing validation of the methodology at other sites, which holds the potential to mitigate issues related to interrater reliability, increase the size of the annotated dataset, and determine whether similar differences exist among black/Hispanic patients with and without structured documentation of race and ethnicity in other geographic contexts. In addition, investigators can potentially make use of NLP-derived data on race and ethnicity to further accrual of minority populations to clinical trials requiring specific demographic makeups in patient cohorts, as well as to further large-scale retrospective observational research on EHR data.
CONCLUSION
To address incomplete structured race and ethnicity data in an EHR system, we developed and validated an NLP approach for identifying black race and Hispanic ethnicity from unstructured clinical notes. We found that the NLP method exhibited high precision, recall, and F score, and allowed us to successfully increase the number of patients identified as black or Hispanic in a sample of patients.
FUNDING
This work was supported by Weill Cornell Medicine Clinical and Translational Science Center Grant No. UL1 TR000457 and National Institutes of Health Grant Nos. R01 GM105688 and R01 MH105384.
AUTHOR CONTRIBUTORS
PA developed the NLP pipeline and drafted the Methods section of the manuscript. ES extracted structured data to compare against the output of the NLP pipeline, conducted all statistical analyses, drafted the Results and Discussion section of the manuscript, and participated in revisions of the paper. LP, with ES, drafted the Background section of the manuscript. LP, CL, and SL conducted manual annotation to derive the gold standard against which the NLP pipeline was measured. SS supervised the annotation process and contributed to the final version of the manuscript. SJ, LP, and JP contributed to the revision of the manuscript. MD identified the need to use NLP to extract race and ethnicity data, and pioneered initial trials of regular expressions. MS and TC supervised the project and contributed extensively to the revision and final development of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Nelson A. Unequal treatment: confronting racial and ethnic disparities in health care. J Natl Med Assoc 2002; 94 (8): 666–8. [PMC free article] [PubMed] [Google Scholar]
- 2. U.S. Department of Health and Human Services. National Healthcare Disparities Report 2011. Publication 12-0006. Rockville, MD: Agency for Healthcare Research and Quality; 2012. [Google Scholar]
- 3. Hasnain-Wynia R, Baker DW.. Obtaining data on patient race, ethnicity, and primary language in health care organizations: current challenges and proposed solutions. Health Serv Res 2006; 41: 1501–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klinger EV, Carlini SV, Gonzalez I, et al. Accuracy of race, ethnicity, and language preference in an electronic health record. J Gen Intern Med 2015; 30 (6): 719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hasnain-Wynia R, Van Dyke K, Youdelman M, et al. Barriers to collecting patient race, ethnicity, and primary language data in physician practices: an exploratory study. J Natl Med Assoc 2010; 102 (9): 769–75. [DOI] [PubMed] [Google Scholar]
- 6. Blumenthal D, Tavenner M.. The “meaningful use” regulation for electronic health records. N Engl J Med 2010; 363 (6): 501–4. [DOI] [PubMed] [Google Scholar]
- 7. Office of Management and Budget. Revisions to the Standards for the Classification of Federal Data on Race and Ethnicity. Federal Register. 1997. https://www.whitehouse.gov/wp-content/uploads/2017/11/Revisions-to-the-Standards-for-the-Classification-of-Federal-Data-on-Race-and-Ethnicity-October30-1997.pdf. Accessed December 13, 2018. [Google Scholar]
- 8. McGarry ME, McColley SA.. Minorities are underrepresented in clinical trials of pharmaceutical agents for cystic fibrosis. Ann Am Thorac Soc 2016; 13 (10): 1721–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson SB, Friedman C.. Integrating data from natural language processing into a clinical information system. Proc AMIA Annu Fall Symp 1996; 1996: 537–41. [PMC free article] [PubMed] [Google Scholar]
- 10. Nadkarni PM, Ohno-Machado L, Chapman WW.. Natural language processing: an introduction. J Am Med Inform Assoc 2011; 18 (5): 544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bellows BK, LaFleur J, Kamauu AWC, et al. Automated identification of patients with a diagnosis of binge eating disorder from narrative electronic health records. J Am Med Inform Assoc 2014; 21 (e1): e163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heintzelman NH, Taylor RJ, Simonsen L, et al. Longitudinal analysis of pain in patients with metastatic prostate cancer using natural language processing of medical record text. J Am Med Inform Assoc 2013; 20 (5): 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson SB, Adekkanattu P, Campion TR, et al. From sour grapes to low-hanging fruit: a case study demonstrating a practical strategy for natural language processing portability. AMIA Jt Summits Transl Sci Proc 2018; 2017: 104–12. [PMC free article] [PubMed] [Google Scholar]
- 14. Sholle ET, Kabariti J, Johnson SB, et al. Secondary use of patients’ electronic records (SUPER): an approach for meeting specific data needs of clinical and translational researchers. AMIA Annu Symp Proc 2017; 2017: 1581–8. [PMC free article] [PubMed] [Google Scholar]
- 15. Carrell DS, Cronkite D, Palmer RE, et al. Using natural language processing to identify problem usage of prescription opioids. Int J Med Inform 2015; 84 (12): 1057–64. [DOI] [PubMed] [Google Scholar]
- 16. Patterson OV, Freiberg MS, Skanderson M, et al. Unlocking echocardiogram measurements for heart disease research through natural language processing. BMC Cardiovasc Disord 2017; 17: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc 2012; 19 (5): 859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adekkanattu P, Sholle ET, DeFerio J, et al. Ascertaining Depression Severity by ExtractingPatient Health Questionnaire-9 (PHQ-9) scores from clinical notes. AMIA Annu Symp Proc 2018; 2018: 147–56. [PMC free article] [PubMed] [Google Scholar]
- 19. Office of Management and Budget. Standards for maintaining, collecting, and presenting federal data on race and ethnicity. Federal Register 1997. https://www.doi.gov/pmb/eeo/Data-Standards. Accessed December 13, 2018. [Google Scholar]
- 20.R version 3.5.0. Vienna Austria, R Project for Statistical Computing; 2018.
- 21. Wieland ML, Wu ST, Kaggal VC, et al. Tracking health disparities through natural-language processing. Am J Public Health 2013; 103 (3): 448–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.