Abstract
Mariner-like elements (MLE) are a super-family of DNA transposons widespread in animal and plant genomes. Based on their transposition characteristics, such as random insertions and high-frequency heterogeneous transpositions, several MLEs have been developed to be used as tools in gene tagging and gene therapy. Two active MLEs, Ppmar1 and Ppmar2, have previously been identified in moso bamboo (Phyllostachys edulis). Both of these have a preferential insertion affinity to AT-rich region and their insertion sites are close to random in the host genome. In Ppmar2 element, we studied the affinities of terminal inverted repeats (TIRs) to DNA binding domain (DBD) and their influence on the transposition activity. We could identify two putative boxes in the TIRs which play a significant role in defining the TIR’s affinities to the DBD. Seven mutated TIRs were constructed, differing in affinities based on similarities with those of other plant MLEs. Gel mobility shift assays showed that the TIR mutants with mutation sites G669A-C671A had significantly higher affinities than the mutants with mutation sites C657T-A660T. The high-affinity TIRs indicated that their transposition frequency was 1.5–2.0 times higher than that of the wild type TIRs in yeast transposition assays. The MLE mutants with low-affinity TIRs had relatively lower transposition frequency from that of wild types. We conclude that TIR affinity to DBD significantly affects the transposition activity of Ppmar2. The mutant MLEs highly active TIRs constructed in this study can be used as a tool for bamboo genetic studies.
Keywords: Transposon, Mariner-like elements, Terminal inverted repeat, DNA binding domain, Transposase, Transposition activity, Phyllostachys edulis, Moso bamboo
1. Introduction
Transposable elements (TEs) are genomic factors which can move around in the genome [1,2]. They have been employed as tools for genetic investigations in plants to improve their fitness and growth [3,4]. Based on the mechanism of transposition, there are two classes of TEs, Class I (RNA transposons) and Class II (DNA transposons) [5]. Among the Class II transposons, mariner-like elements (MLE) are a superfamily TEs widespread in diverse taxa, including higher animals, plants, fungi, insects, nematodes and fishes [6,7]. The structure of an MLE is simple, comprising of two target site duplicates (TSDs) of dinucleotide origin of thymine and adenine (TA rich), one open reading frame (ORF) encoding a transposase and two terminal inverted repeats (TIR) flanking the ORF region [8]. The TIRs are usually 10–40 bp long and composed of protein binding elements (PBE) which are specific for individual MLE. For example, in the synthetic transposon Sleeping Beauty (SB), PBE is located in the TIRs of 15 bp long direct repeats (DR) [9]. Also, the PBE of Frog Prince (FP) is located in the TIRs of 21 bp long DRs [10]. FP and SB are Tc1/MLEs superfamily members sharing ~50% sequence similarity and used for gene therapy in fish, amphibian and mammalian cell lines [10,11].
The transposition activities of MLE are regulated by the transposase [12], TIRs, and the flanking sequences [13,14,15]. Specific interaction of transposases with the TIRs and the affinity of TIRs to the transposase influence the activity of MLEs and their transposition efficiency [16,17,18,19]. Usually, the first step in transposition occurs with the binding of transposase to the PBE of TIRs. The binding occurs at the DNA binding domain (DBD) of the transposase. Since TIR sequences can vary at each end, the transposase binding has different affinities to these sequences [20]. For instance, the PBE of the rice MLE, Osmar5 is 17 bp long and composed of two boxes, Box I and Box II. Mutations of both the boxes have affected the affinity of TIRs to the transposase DBD [17]. Furthermore, the transposition frequency of the DNA-transposase complex is altered by the A and T content of TIRs. In the Drosophila MLE, mos1, the differential AT content of the right and left TIRs (64.3% and 50.0%, respectively), changes the affinity of the right TIR to be 5–10 times higher than that of the left. When the right TIR was replaced by the left, the affinity was found to decrease by more than 50 times. In contrast, when the left TIR was replaced by the right, the affinity was found to shift in the opposite direction by showing an increase by more than 26 times [21,22,23]. Likewise, when the guanines of 3′ TIRs of the ant (Messor bouvieri) transposon, Mboumar9 were replaced by adenines, the TIR affinity was found four times higher [20]. Additionally, the subterminal region, 30–35 bp flanking sequences of the TIR (Sub-TIR) can also affect the transposition efficiency. Longer the sub-TIRs, such as in the case of Mos1, transposition activity was found lowered [24,25]. Importance of sub-TIRs was further demonstrated by Yang et al. [18] through site-directed mutagenesis of the 3′ sub-TIRs of the rice MLE, Osm14NAS. They found that a mutation of some motifs could repress the transposition activity.
Based on the transposition characteristics, such as random insertions and high frequency of heterogeneous transpositions, several MLEs such as Hemar from the flatworm Himasthla elongata [26], Himar1 from the horn fly Haematobia irritans [27], Hsmar1 from humans [28], and Mos1 [29] have been used as tools for gene tagging and gene therapy. Lampe et al. [30] found that enhanced transposase activity of the mutated Himar1 was due to the increased affinity of DNA in general. Enhanced DNA-binding activity of hyperactive SB element was due to improved cleavage kinetics and increased element mobilisation from host cell chromosomes [31], which dramatically enhanced gene transfer capabilities in vivo in mice [32]. In order to improve Mos1 transposition frequency, hyperactive Mos1 transposase versions were generated by site-directed mutagenesis [13,14]. Although several MLEs have been found functional in yeast (Saccharomyces cerevisiae), their activities in general in plants are relatively less studied [18,33].
Among the commercially important plants, bamboos adorn a significant place for being a prime source of industrial raw material, contributing substantially to the South Asian economy [34,35,36]. Worldwide, there are more than 1642 cultivated bamboo species belonging to 75 genera adapted to diverse climatic regions, especially confined to China, India, Japan, Korea, Myanmar and Australia (https://www.inbar.int/, last access on: 24 July 2019). Among these, moso bamboo (Phyllostachys edulis) is an important commercial species in China that generates an equivalent of about 5 billion US dollars from industries including textile, timber and food [37]. Characterized by its rapid growth, unique biomass turnover and adaptation to temperate conditions, moso bamboo not only provides raw materials for paper and rayon industries but is also popular for its unique timber strength.
Reproductive behaviour in bamboos has invoked scientific curiosity for a long time because flowering intervals range from several years to more than a hundred years between the species, and plants often die after fruiting [38]. The moso bamboo has a long lifespan and shows sporadic flowering rather than the gregarious flowering as seen in other bamboo species. Understanding the genetic and molecular mechanism of flower development and rapid growth [39] remains the main challenge, with conclusive answers still elusive [37,40]. Similar to any cultivated species, moso bamboo suffers biotic and abiotic stresses under cultivation. Being predominantly clonally propagated with an unpredictable and extended breeding cycle, relatively little has taken place in terms of genetic improvements for yield, quality and stress tolerance. Presently, the whole genome information of moso bamboo is available, and a chromosome level genome database [41] and BambooNET database (http://bioinformatics.cau.edu.cn/bamboo, last access on: 24 July 2019) [42] have been developed. These novel tools can drive investigations in unravelling the reproductive and vegetative biology of moso bamboo. Although the genetic transformation system in moso bamboo remains under establishment, transposon-based mutagenesis can be employed as an additional tool to manipulate the moso bamboo genome [15] in the investigations of genetic routes of bamboo biology.
Among the reported transposons in bamboo, 82 MLEs from 44 bamboo species belonging to 38 genera have been characterized [43]. A few MLEs found in the moso bamboo genome [43,44] indicated highly conserved transposases [45]. Two active full-length MLEs, Ppmar1 and Ppmar2, isolated from moso bamboo that contained perfect TIRs and intact transposases [46], showed size differences, with Ppmar1 being longer and belonging to A2 subfamily, and the shorter Ppmar2 belonging to C subfamily [46]. Both these elements have shown transposition activity in Arabidopsis thaliana and established their preferential insertion to the TA rich regions with random insertions in the host genome [44]. Additionally, fourteen different hyperactive Ppmar1 transposase variants generated by single amino acid substitutions were shown to increase the transposition activity of Ppmar1 MLEs [15]. When a non-autonomous transposon Ppmar1NA system was generated by excising the transposase gene, Ppmar1 transposase variants were shown to promote element excision and reintegration of the non-autonomous Ppmar1NA in the yeast genome at TA dinucleotide sequences. The most hyperactive transposase variant S171A was found to induce 10-fold more active excisions in yeast [15].
Notwithstanding the effects of transposases, MLE transposition efficiency is also dependent on the binding affinities of TIRs to DBD [18,19,20], as well as to the length of sub-TIRs. However, information on the TIR variants and their influence on transposition in the moso bamboo MLEs are not yet available. This study aims to fill this gap by mutating the TIRs of Ppmar2 through site-directed mutagenesis to alter transposition activity and to quantify their affinities to DBD of Ppmar1 and Ppmar2 transposases. The affinity variants were investigated by electrophoretic mobility shift assay (EMSA), also known as “gel mobility shift assay”, while the transposition activity of shortened Ppmar2 mobilized by the mutant TIRs and catalysed by Ppmar1 and Ppmar2 transposases was investigated by yeast transposition assays.
2. Results
2.1. The Identification of Box I and II in Ppmar2-TIRs and Development of Mutant Sequences
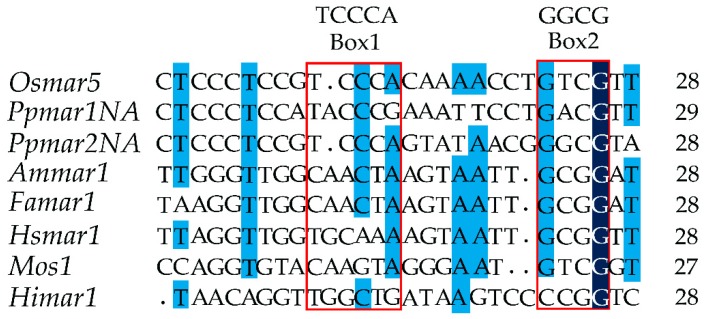
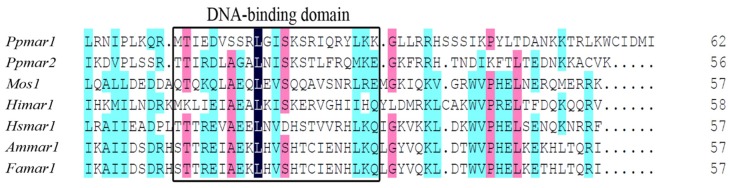
The non-autonomous Ppmar2NA-TIR1, lacking the transposase gene, was developed from the wild Ppmar2 sequences (Figure 1) and two conserved domains, Box I and Box II, were identified within the TIRs by comparative analysis with TIRs of six other MLE sequences (Osmar5, Ammar1, Famar1, Hsmar1, Mos1 and Himar1). The conserved domain sequence of Box I was TCCCA and of Box II was GGCG (Figure 2). It was found that in Box1, cytosine (C) was more conserved than T and A, whereas in Box II, guanine (G) was more conserved than C. Seven base mutations designed within the two boxes of Ppmar2NA-TIR1, four in Box I and three in Box II (Table 1), had 19 nucleotide substitutions in total. Ten substitutions were made to A, seven to T, one each to G and C. In Box I, one T was substituted by A, five C were substituted by T, two C by A and three A were substituted by one C, T and G. In Box II, four Gs were substituted by A and three C by three As.
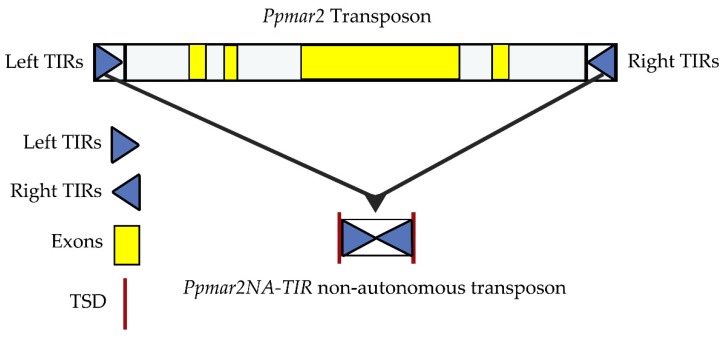
Figure 1.
Structure of the Ppmar2 transposon and the non-autonomous transposon Ppmar2NA-TIRs (TIRs—terminal inverted repeats). Yellow bars represent the exons of Ppmar2 transposase, blue triangles the TIRs of Ppmar2 transposon, and dark red vertical lines the target site duplications (TSD).
Figure 2.
The alignment of TIRs of Mariner-like elements (MLEs). Osmar5, AP008207 of Oryza sativa; Ammar1, U19902 of Apis mellifera; Famar1, AY226507 of Forficula auricularia; Hsmar1, U52077 of Homo sapiens; Mos1, X78906 of Drosophila mauritiana; Himar1, U11646 of Haematobia irritans.
Table 1.
The Ppmar2NA-TIRs and their sequences with mutations details. The mutated bases are marked by the red font. Bold and underlined sequences on the left and right within each TIR represent Box1 and Box2, respectively.
| S. No. | Name of TIR | Sequence of TIR Variants |
|---|---|---|
| 1 | Ppmar2NA-TIR1 | TACTCCCTCCGTCCCAGTATAACGGGCGTATAAAAAAATTT |
| 2 | Ppmar2NA-TIR2 | TACTCCCTCCGATACAGTATAACGGGCGTATAAAAAAATTT |
| 3 | Ppmar2NA-TIR3 | TACTCCCTCCGTTCCTGTATAACGGGCGTATAAAAAAATTT |
| 4 | Ppmar2NA-TIR4 | TACTCCCTCCGTTTCGGTATAACGGGCGTATAAAAAAATTT |
| 5 | Ppmar2NA-TIR5 | TACTCCCTCCGTACTCGTATAACGGGCGTATAAAAAAATTT |
| 6 | Ppmar2NA-TIR6 | TACTCCCTCCGTCCCAGTATAACGAGAGTATAAAAAAATTT |
| 7 | Ppmar2NA-TIR7 | TACTCCCTCCGTCCCAGTATAACGGGATTATAAAAAAATTT |
| 8 | Ppmar2NA-TIR8 | TACTCCCTCCGTCCCAGTATAACGAAAATATAAAAAAATTT |
2.2. Affinity Analysis of Ppmar2NA-TIRs to Ppmar1 and Ppmar2 Transposases
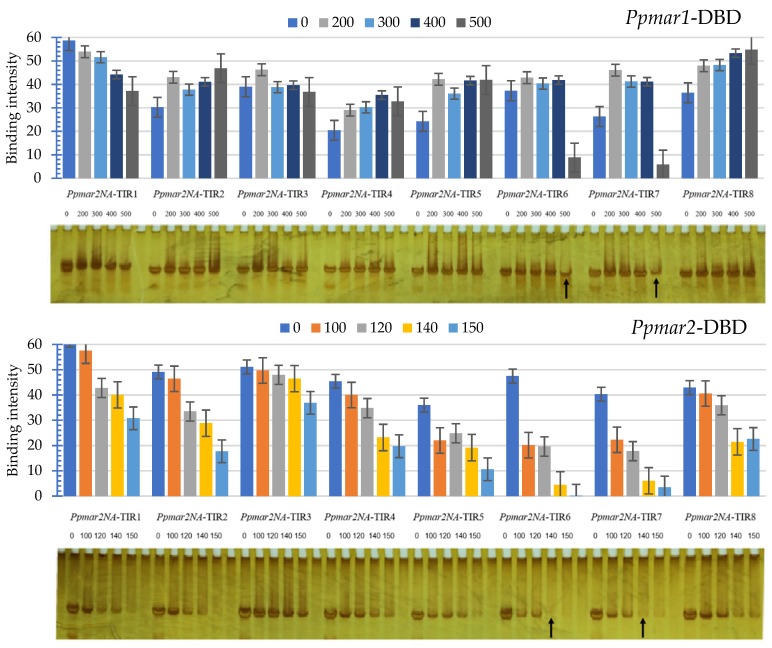
The EMSA analysis revealed that synthesized Ppmar2NA-TIRs and DBDs based on Ppmar1 and Ppmar2 sequences showed differential affinity, with the former showing less affinity and the latter having a high affinity (Figure 3). In general, the binding of Ppmar1NA-TIRs occurred at higher concentrations of Ppmar1-DBD than for Ppmar2-DBD. Therefore, the concentration of Ppmar1-DBD was varied as 0, 200, 300, 400 and 500 µM, while that of Ppmar2-DBD was set as 0, 100, 120, 140, and 150 µM. In both cases, Ppmar2NA-TIRs was kept at 1 µM.
Figure 3.
Electrophoretic mobility shift assays (EMSA) of mutated Ppmar2NA-TIRs 1–8, obtained with different concentrations of Ppmar1-DBD and Ppmar2-DBD. The concentration varied between 0 and 500 µM for the former and 0 and 150 µM for the latter. Black arrows indicate the highest interactions of mutated Ppmar2-TIRs and DBD forming the bound complex, as shown by the relative scantiness of Ppmar2NA-TIRs in the corresponding lanes. The error bars indicate standard errors.
Ppmar2NA-TIRs exhibited a very weak affinity to Ppmar1-DBD even at higher concentrations. In most of the TIRs, a non-specific pattern across concentrations was observed. A relatively strong affinity was observed in two mutants, Ppmar2NA-TIR6 and Ppmar2NA-TIR7, also at 500 µM concentration. Unlike with Ppmar1-DBD, we found that almost all Ppmar2NA-TIRs were bound to Ppmar2-DBD when the concentration of Ppmar2-DBD was above 150 µM. There was a regular pattern of affinity in this case, which increased with the concentration. This indicated that 0–200 µM concentrations of Ppmar2-DBD were suitable for detection of the affinity between Ppmar2NA-TIRs and Ppmar2-DBD.
The DBD affinities of mutant Ppmar2NA-TIRs varied significantly from the wild type (Ppmar2NA-TIR1) and indicated an increased binding in all the cases. The binding was highest in Ppmar2NA-TIR6 followed by Ppmar2NA-TIR7 at 150 µM concentrations. Among the mutants, Ppmar2NA-TIR3 was observed to have relatively less affinity shift towards elevated concentrations. In the remaining mutants, such as Ppmar2NA-TIR2, Ppmar2NA-TIR4 and Ppmar2NA-TIR8 the affinity gradient was almost similar to that of the wild type. In general, at the 150 µM concentration level of Ppmar2-DBD, the Ppmar2NA-TIRs showed the highest level of affinities in both wild types and mutants except for Ppmar2NA-TIR8. The highest affinity in Ppmar2NA-TIR8 was detected at 140 µM concentration.
Analysis of affinity shift in the mutants from the wild type, with respect to their individual mutations, indicated that Box I mutations were relatively more non-specific than Box II mutations (Table 2). The non-specificity was particularly apparent with Ppmar1-DBD compared to Ppmar2-DBD. The Box I mutations were predominantly C→T substitutions and C→A. However, Box II mutations were significantly different from those of Box I because at least two mutants, Ppmar2NA-TIR6 and Ppmar2NA-TIR7, exhibited affinity consistency across transposases. In Box II, there were four G→A substitutions, three C→A substitutions and one C→T substitution. Across Box I and Box II, it is evident that C→A substitutions have consistently resulted in higher affinity with Ppmar2 transposase. Furthermore, the G substitutions in Box II either to A or T seemed to improve the TIRs affinity for transposases from both the moso bamboo MLEs. However, when there were too many G substitutions in Box II, the affinity shift was found reversed for Ppmar1-DBD while it was reduced for Ppmar2-DBD, as found in the mutant Ppmar2NA-TIR8.
Table 2.
The details of the Ppmar2NA-TIRs interaction with different concentration of Ppmar2-DBD and Ppmar1-DBD transposase.
| Ppmar2-TRIs | Nucleotide Substitutions * | Affinity Shift (%) | ||
|---|---|---|---|---|
| Box I | Box II | Ppmar1-DBD @ 500 µM | Ppmar2-DBD @ 150 µM | |
| Ppmar2NA-TIR1 | ++ | ++ | 0.00 | 0.00 |
| Ppmar2NA-TIR2 | T→A, C→T, C→A | ++ | −26.10 | 42.38 |
| Ppmar2NA-TIR3 | C→T, A→T | ++ | 1.03 | –19.89 |
| Ppmar2NA-TIR4 | C→T (2), A→G | ++ | 11.81 | 35.93 |
| Ppmar2NA-TIR5 | C→A, C→T, A→C | ++ | −12.75 | 65.55 |
| Ppmar2NA-TIR6 | ++ | G→A, C→A | 76.20 | 99.55 |
| Ppmar2NA-TIR7 | ++ | C→A, G→T | 84.23 | 88.87 |
| Ppmar2NA-TIR8 | ++ | G→A (3), C→A | −47.47 | 26.64 |
++, wild type; * values in brackets show the number of times that particular nucleotide substitution occurred in the mutant; affinity shift is the change in affinity in mutants from the wild type and expressed as the percentage over wild type.
2.3. The Influence of Affinity Contrasting Mutant TIRs on the Transposition Frequency
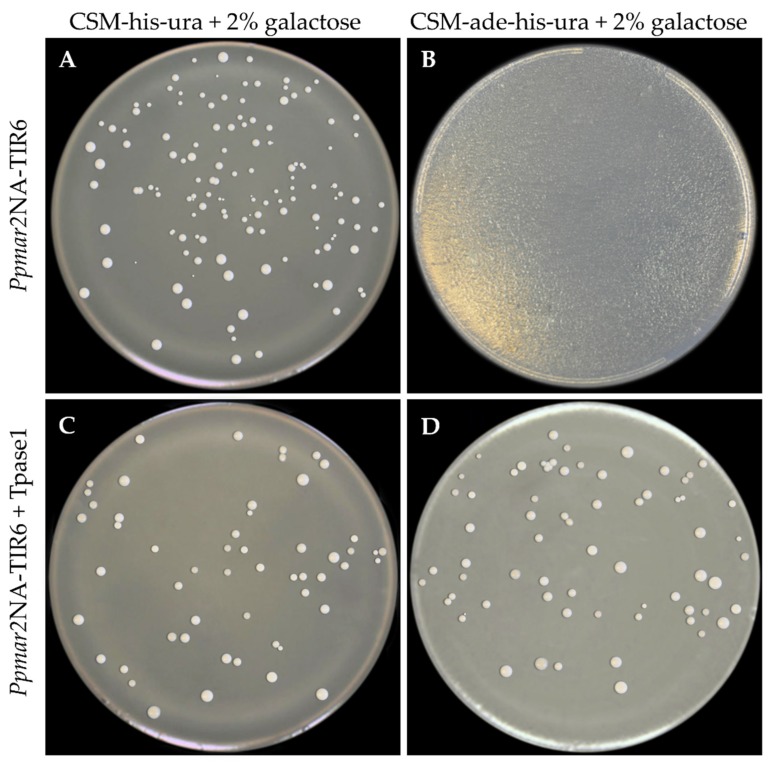
Considering that the sub-terminal sequences of TIRs are known to affect the transposition of MLEs in the yeast, sub-TIRs of Ppmar2NA were deleted to avoid ambiguity in the yeast transposition assay. Three TIRs were used for the transposition assay: Two mutants (Ppmar2NA-TIR3 (C657T-A660T) with a weak affinity; and Ppmar2NA-TIR6 (G669A-C671A) with a strong affinity); and the wild-type Ppmar2NA-TIR1. The vectors pWL89a-Ppmar2NA-TIR1, pWL89a-Ppmar2NA-TIR3 and pWL89a-Ppmar2NA-TIR6 were each co-transformed into yeast cells, together with pAG413gal-Tpase1 and pAG413gal-Tpase2. When the transformed colonies with TIR alone and without transposase were grown on a complete supplement mixture (CSM) medium without histidine and uracil, but adenine sufficient (CSM-his-ura) and adenine deficient (CSM-ade-his-ura), there was normal growth of colonies in the former and no colonies developed in the latter. Alternatively, when the double-transformed colonies possessing TIRs and transposase were grown on the same media, there was colony development in both the cases (Figure 4). Adenine is essential for colony growth, and it should be provided either from the medium or be synthesized internally by the yeast cells. In the transformed yeast cells, however, the adenine synthesis is arrested by silencing the key enzyme ADE2 coding for phosphoribosylaminoimidazole carboxylase, in the purine synthetic pathway. In this case, the silencing was achieved by placing the TIR sequences within the ADE2 gene of the plasmid vector pWL89a, which was used for transformation. The colony growth in the adenine deficient medium of the double transformants indicated a de novo ADE2 activity, resulting in systemic adenine synthesis. To become active, the TIR sequences need to be cleaved out of the ADE2 exons, which is possible only if transposase binds to TIRs initiating transposition activity. The proportion of ADE2 revertants in the yeast assay indicated the transposition efficiency of the transposases.
Figure 4.
Yeast transformation assay for the active mutant TIR Ppmar2NA-TIR6, showing active colony growth in the presence of adenine, irrespective of the presence of Ppmar1 transposase (Tpase1) ((A), transposase absent; (C), transposase present). When grown on adenine free medium, no colonies developed when transposase was absent (B), but colonies developed when transposase was present (D). The transposase could initiate the transposition of the TIR sequences out of the ADE2 gene, thereby reactivating the ADE2 resulting in de novo adenine synthesis.
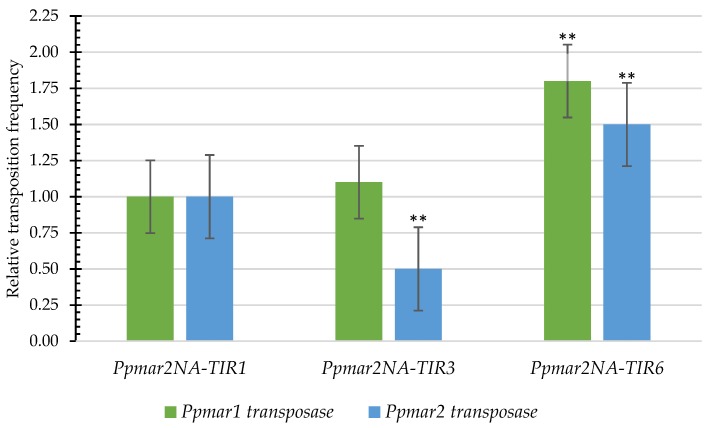
In the affinity contrasting mutant TIRs, Ppmar2NA-TIR3 (C657T-A660T) and Ppmar2NA-TIR6 (G669A-C671A), more frequent transposition was catalyzed by Ppmar1 transposase than Ppmar2 transposase, whereas for the wild type TIR Ppmar2NA-TIR1, the relative transposition frequency of both the transposases was similar (Figure 5). In the low-affinity TIR mutant Ppmar2NA-TIR3, the Ppmar1 transposase produced an almost similar frequency of transposition as that of the wild type, while the Ppmar2 transposase resulted in half the frequency of transposition than the wild type. Nevertheless, in the high-affinity mutant TIR, Ppmar2NA-TIR6, the transposases of Ppmar1 and Ppmar2 resulted in 1.5 and 1.8 times higher transposition activity than the wild type, respectively. In agreement with the in vitro affinity pattern (Figure 3), strong affinity Ppmar2NA-TIRs had a high transposition activity, while those with weak affinity indicated a low transposition activity.
Figure 5.
Relative transposition frequencies of mutated Ppmar2NA-TIRs triggered by Ppmar1 and Ppmar2 transposases. The Y-axis indicates the ratio of the mean excision frequency of six independent experiments with Ppmar2NA mutants, to that of the wild type Ppmar2NA-TIR1. The symbol ** indicates a statistically significant (p < 0.01) difference with the wild type Ppmar2NA-TIR1. Error bars indicate standard errors.
3. Discussion
Moso bamboo, the most valued bambusoid on earth, has a genome size of 2075 Mb and contains 59% of TEs [47], although very few TEs have been characterized so far. TEs are potential tools for genetic studies because of their ability to move around the genome creating random insertions. This behaviour of the TEs can be exploited to create mutations within genes to silence them, thereby enabling the studies of gene functions [1,2,3,4]. Transposition of the TEs from the silenced gene can reactivate them on a later stage. Since TEs are natural elements, they play a significant role in the evolution of crop species and drive the genomes to adapt themselves to unfavorable environments by creating beneficial mutations [48]. Among the TEs, MLEs are Class II DNA transposons abundantly available in the living organisms. Of the several MLEs available in the bamboo genomes, two MLEs are characterized in moso bamboo, namely, Ppmar1 and Ppmar2 [45]. To use them as a potential tool for genetic investigations in moso bamboo, there is a need to document the efficiency of these elements. There are several factors that determine the transposition efficiency of MLEs, such as internal and external factors [7,15]. Internal factors are primarily structural features such as the TIRs, sub-TIRs and the transposase sequences. There can be several external factors such as internal and external cellular environment, quantum and duration of accumulation transposases, age and metabolic state of the cell, and the abundance of active TEs in the cells [49].
TIRs of the MLEs vary significantly between different classes and families [9,10,11]. However, there are certain regions that remain highly conserved within TIRs, especially at the PBE site that plays an important role in transposition activity. Fundamentally, transposition requires a transposase to bind to the PBE of the TIRs triggering the activity [9,10,11]. Binding requires a mutual affinity between the PBE and the transposase DBD. Therefore, altering the nucleotide balance in the TIR conserved domain boxes could bring about variations in affinity. We were able to construct mutations in the conserved boxes I and II of the Ppmar2 element by random nucleotide substitutions. The effect of these mutations was tested by stripping the transposase off the TIRs and allowing them to bind to the DBD domain in vitro to assess the affinity. In the in vitro system, the binding affinities of transposons to the DBD of both Ppmar1 and Ppmar2 transposases varied with different mutation levels in the non-autonomous elements of Ppmar2NA. Ppmar1 transposase was less specific to mutations in Box I than that in Box II, while Ppmar2 transposase showed high affinity with both boxes. This is in agreement with the previous report [17,18], where multiple variants of the Osmar5 TIRs showed significant effects on the binding of Osmar5N with respect to the mutations they carried. The significant impact on the binding of Ppmar1 and Ppmar2 transposases to multiple variants of both Box I and Box II observed in our study indicates that both boxes are critical in prescribing the affinity. Since Ppmar2-DBD is the native transposase domain of the Ppmar2 element, it is reasonable to assume that this domain will have a preferably greater affinity than the Ppmar1-DBD.
Previously, we reported the development of Ppmar1 transposase variants by single amino acid substitutions based on homology analysis with the other functional MLEs [15]. We generated non-autonomous transposons, Ppmar1NA and Ppmar2NA, by deleting the transposase gene from Ppmar1 and Ppmar2 sequences, while retaining the 5′- and 3′-TIRs along with their corresponding sub-terminal sequences [15]. The Ppmar1 transposase variants catalysed Ppmar1NA to transpose in a yeast system, but Ppmar2NA showed false-positive insertions, even when transposase was absent. Transposition occurred in TA rich regions, and transposase variant S171A was more active than wild type transposase. Apart from the transposase, the TIRs and their flanking sequences, chromatin status, DNA methylation, and host proteins might affect the transposition activity [18,50]. In the current study, TIR variants were developed to increase the transposition frequency of transposon, based on the alignment of Ppmar2 TIRs with other TIRs such as Osmar5, Ammar1, Famar1, Himar1, Hsmar1 and Mos1.
Moreover, we deleted the sub-TIRs of Ppmar2NA to overcome the difficulty in the yeast transposition assay. With respect to the nucleotide substitutions, the substitution of G with A and T in Box II was found to improve the affinity tremendously, suppressing the transposase specificity between Ppmar1 and Ppmar2. Further, the C→A substitutions indicated a positive effect on binding affinity across the boxes and transposases. Three base-pair substitutions had reduced or no significant impact on the affinity, whereas two base-pair substitutions increased the interaction of DBD and Ppmar2NA-TIRs. This is in agreement with a previous report on rice [17], where two or more base-pair substitutions were found to decrease the level of interaction between TIRs and transposase. We have identified two high-affinity TIRs, Ppmar2NA-TIR6 and Ppmar2NA-TIR7, which showed almost equal affinity towards Ppmar1 and Ppmar2 transposases. Our data support a model of the cross-mobilisation of different MLEs [17,18]. In rice MLEs, one specific Osmar transposase can interact with the TIRs of other Osmar elements in the same clade. For example, Osmar5N was able to bind to the TIR sequences of rice MLEs from the same clade [17]. In our study, Ppmar2NA-TIRs could also be bound to the DBD of Ppmar1 transposase, which indicated that the Ppmar2NA might be mobilized by Ppmar1 transposase catalysis as well. It would be ideal to test their improved general affinity with other transposase systems. These elements can be further employed in genetic studies, not only in moso bamboo but also in other organisms.
In the yeast transposition system, the Ppmar2NA-TIR6 transposon showed 1.8 times higher transposition activity than the wild type Ppmar2NA-TIR1 with Ppmar1 transposase, which was 1.5 times higher with Ppmar2 transposase. Although there was a clear advantage of increased binding affinity for Ppmar2 transposase to the TIR variants indulged in this study, the efficiency of transposition was relatively higher with Ppmar1 transposase. The increased efficiency with Ppmar1 transposase could be attributed to its natural relative efficiency over the Ppmar2 enzyme. There are various factors that can affect the efficiency of the transposon, which may include affinity level, binding non-specificity and the size of the transposon itself [49]. Whether higher efficiency of Ppmar1 can be attributed to its size—Ppmar1 is a longer element than Ppmar2—is a question for further investigation. Among the multiple variants of the Ppmar2NA-TIRs, affinity towards Ppmar2-DBD was stronger for Ppmar2NA-TIR6 (G669A-C671A), while the same was weaker for Ppmar2NA-TIR3 (C657T-A660T). High-affinity TIRs could produce a high frequency of transposition, as observed with the Ppmar2NA-TIR6 element. Perhaps Ppmar2NA-TIR6 could have had a stronger interaction with DBD than the wild type, facilitating catalysis of more frequent transposition either by Ppmar1 transposase or Ppmar2 transposase. On the other hand, the weak affinity Ppmar2NA-TIR3 had a reduced activity than the wild type Ppmar2NA-TIR1 and resulted in poor catalysis by both the enzymes. The broader biological implications of Ppmar2NA-TIRs needs to be addressed in future investigations. Furthermore, to validate the hyperactivity of mutated Ppmar2NA-TIRs, they need to be examined in model plants such as foxtail millet, rice or Arabidopsis. This could help to develop actively modified transposons as tools for genetic manipulations and bamboo improvement.
4. Materials and Methods
4.1. The Synthesis of Ppmar2NA-TIRs
The Ppmar2 used was a full-length MLE, with perfect 27-bp TIRs, the 5’-TIR (CTC CCT CCG TCC CAG TAT AAC TTT TTT) and the 3’-TIR (AAA AAA GTT ATA CTG GGA CGG AGG GAG) [46]. The end-to-end TIRs were synthesized without the intervening transposase sequences by Sangon Biotech (Shanghai, China) and named as non-autonomous Ppmar2NA-TIR1. The synthesized sequence was 58 bp long with a structure of TA CTC CCT CCG TCC CAG TAT AAC TTT TTT AAA AAA GTT ATA CTG GGA CGG AGG GAG AT, including two target site duplications (TSD). The structure of Ppmar2 (autonomous) and Ppmar2NA-TIR1 (non-autonomous) is shown in Figure 1.
4.2. Identification of Conserved Domains and Mutations in Ppmar2NA-TIRs
The non-autonomous Ppmar2NA-TIR1 was aligned with TIRs sequences of Osmar5 (Accession No. GQ382183) from rice [33], Ammar1 (Accession No. AY155490) from European honey bee (Apis mellifera) [51], Famar1 (Accession No. AY155492) from the earwig (Forficula auricularia) [52], Himar1 (Accession No. U11646) from horn fly (Haematobia irritans) [50,53], Hsmar1 (Accession No. U52077) from human genes [54], and Mos1 (Accession No. X78906.1) from Drosophila mauritiana [55]. Two conserved domains were found in Ppmar2NA-TIR1 and named as Box I and Box II according to literature (Figure 2). Using the conserved domain sequences of Ppmar2NA-TIR1 and based on our previous reports of Ppmar1 and Ppmar2 transposition activity in Arabidopsis thaliana [44], seven base mutations were designed in the two Boxes of Ppmar2NA-TIR1 (Table 1). The mutated TIRs were synthesised by Sangon Biotech (Shanghai, China) and named Ppmar2NA-TIR2 to Ppmar2NA-TIR8, respectively.
4.3. Construction of Ppmar2NA Donor Vector
The Ppmar2NA was inserted into the XhoI site of pWL89a vector (kindly provided by Dr Susan R Wessler from University of Georgia, USA) resulting in pWL89a-Ppmar2NA. The inserted product was transformed into E. coli DH5α and positive clones were screened.
4.4. Mutagenesis of Ppmar2NA-TIRs
The mutagenesis of Ppmar2NA-TIRs was performed with the QuikChange lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) using four pairs of primers. The name of the primers and their sequences are given in Table 3. Mutagenesis reactions in 25 µL mix contained 100 ng of the template of Ppmar2NA donor vector, 2 µM of each primer, and 0.5 µL of QuikChange lightning buffer. The plasmid vector pWL89a-Ppmar2NA was used as the template for site-directed mutagenesis. To confirm the presence of the targeted mutation, all plasmids were sequenced with specific primers.
Table 3.
The primers and their sequences used for site-directed mutagenesis of Ppmar2NA-TIRs.
| S. No. | Primer | Sequence (5′-3′) |
|---|---|---|
| 1 | XT-C657T-A660T-F | CGAGTACTCCCTCCGTTCCTGTATAACGGGCGTATA |
| 2 | XT-C657T-A660T-R | TATACGCCCGTTATACAGGAACGGAGGGAGTACTCG |
| 3 | XT-G669A-C671A-F | CCCTCCGTCCCAGTATAACGAGAGTATAAAAAAATTTCAGAGAC |
| 4 | XT-G669A-C671A-R | GTCTCTGAAATTTTTTTATACTCTCGTTATACTGGGACGGAGGG |
| 5 | XT-T1459A-G1462A-F | TTTTTTATACGCCCGTTATACAGGAACGGAGGGAGTACTCG |
| 6 | XT-T1459A-G1462A-R | CGAGTACTCCCTCCGTTCCTGTATAACGGGCGTATAAAAAA |
| 7 | XT-G1448T-C1450T-F | GGACAGCAGAAATTTTTTTATACTCTCGTTATACTGGGACGGAGG |
| 8 | XT-G1448T-C1450T-R | CCTCCGTCCCAGTATAACGAGAGTATAAAAAAATTTCTGCTGTCC |
4.5. The Extraction and Synthesis of Transposases Ppmar1-DBD and Ppmar2-DBD
The DBDs of Ppmar1 and Ppmar2 transposases used in the current study contained 22 amino acids each, MTI EDV SSR LGI SKS RIQ RYL K for the former and TTI RDL AGA LNI SKS TLF RQM K for the latter. The homologous alignment of the DBD amino acid sequences of transposase (Figure 6) indicated a helix-turn-helix (HTH) motif [45]. The DBD sequences were synthesised by Sangon Biotech (Shanghai, China) and named as Ppmar1-DBD and Ppmar2-DBD, respectively.
Figure 6.
The homologous alignment of amino acid sequences of DNA binding domain (DBD) of Ppmar1 transposase and the DBD of Ppmar2 transposase in the following DBDs: Mos1 from Drosophila mauritiana, Himar1 from horn fly, Hsmar1 from human genes, Ammar1 from European honey bee, and Famar1 from earwig.
4.6. Construction of Transposase Expression Vectors
In order to construct the transposase expression vectors, full-length transposases of Ppmar1 and Ppmar2 were cloned between Not1 and EcoRV sites of the pMD-18-T vector (Takara, Japan). The Ppmar1 and Ppmar2 transposase ORFs were amplified respectively by primer pairs, Tpase1-F + Tpase1-R andTpase2-F + Tpase2-R, both containing Not1 and EcoRV restriction sites (Table 4). The PCR product was digested by Not1 and EcoRV. The pAG413gal-ccdB vector (kindly provided by Dr Susan R Wessler from University of Georgia, USA) was digested by Not1 and EcoRV and the digested PCR products were ligated to the large fragment of the pAG413gal-ccdB vector. The transposases of Ppmar1 and Ppmar2 sequences replaced the ccdB region in the pAG413gal-ccdB vector and were named as pAG413gal-Tpase1 and pAG413gal-Tpase2, respectively.
Table 4.
The detailed sequences of primers used in the study to amplify Ppmar1 and Ppmar2 transposases.
| S. No. | Primer Name | Sequence (5′-3′) |
|---|---|---|
| 1 | Tpase1-F | AGAATGCGGCCGCAAAAAAATGGCTGACCCAATAGATTCGCGGCCGC |
| 2 | Tpase1-R | GACTGATATCTGCTGCTGCAAAAGAGTAACGAT ATC |
| 3 | Tpase2-F | AGAATGCGGCCGCAAAAAAATGGCGAATTTGGACCTAAATCGCGGCCGC |
| 4 | Tpase2-R | GACTGATATCCTAATTGATGTACACAATTGGATATC |
4.7. Affinity Analysis of Ppmar2NA-TIRs to Ppmar1-DBD and Ppmar2-DBD
The single-strand Ppmar2NA-TIRs including their mutants (TIR1-TIR8) were dissolved in ultra-pure water to an initial concentration of 10 µM. The mix was then incubated at 100°C in a boiling water bath for 10 min for denaturation and renatured into double TIRs by natural cooling. The Ppmar1-DBD and Ppmar2-DBD were dissolved separately in protein buffer (100 mM NaCl, 10 mM Tris, 10% glycerol, pH 8.0) to a concentration of 1 mM. The concentration of different mutated Ppmar2NA-TIRs was set at 1 µM, while the concentrations of Ppmar1-DBD and Ppmar2-DBD were set at 0, 100, 120, 140, 150, 200, 300, 400, or 500 µM. The double TIRs (1 µM) and the diluted Ppmar1-DBD and Ppmar2-DBD with their corresponding concentrations were fully mixed (10 µL) and incubated at 65 °C for 20 min and then cooled down. The mixture was separated using 18% non-denaturing polyacrylamide gel electrophoresis (PAGE) (BioRad, Hercules, CA, USA), followed by silver staining [56]. The gel images were quantified for the band intensity using ImageJ v.1.52o [57].
4.8. Estimation of the Transposition Frequency
The transposition assay was performed using the yeast haploid strain DG2523 (MATalpha ura3-167 trp1-hisG leu2-hisG his3-del200 ade2-hisG) (kindly provided by David Garfinkel, University of Georgia, USA). Two plasmids, pWL89a-Ppmar2NA and pAG413gal-Tpase1 (or) pAG413gal-Tpase2 were co-transformed into DG2523 and the transformed colonies were selected by growing them for ten days at 30 °C in a complete supplement mix (CSM) devoid of histidine and uracil (CSM-his-ura) in presence of 2% galactose. Each galactose-induced yeast cell colony was suspended in 50 μl of water and plated on CSM-ade-his-ura medium which additionally lacks adenine. After a seven-day incubation at 30 °C, the ADE2 revertant colonies were counted for all mutant constructs [15]. The excision frequencies were computed as the number of revertant per live yeast cell, based on the density of live cells in the suspension used for plating. The transposition assay workflow is shown in Figure 7.
Figure 7.
The vector constructs and workflow of Ppmar2NA-TIRs transposition assay in yeast cells (Saccharomyces cerevisiae). The Ppmar2NA-TIRs were inserted to the pWL89a vector, resulting in the vector pWL89a-Ppmar2NA. The Ppmar1 and Ppmar2 transposases were separately inserted to the pAG413gal-ccdB vector, resulting in pAG413gal-Tpase1 and pAG413gal-Tpase2, respectively. The yeast cells were grown on medium CSM-his-ura with 2% galactose. The co-transformation of pWL89a-Ppmar2NA and pAG413gal-Tpase1 vectors and co-transformation of pWL89a-Ppmar2NA and pAG413gal-Tpase2 vectors were inserted into yeast strain DG2523. ADE2 revertant cells were grown on medium CSM-ade-his-ura with 2% galactose.
5. Conclusions
To conclude, we have found that the Ppmar2NA-TIRs developed in this study can be mobilized in the yeast cell. Two conserved domain boxes were identified in the TIRs with varying affinities to the DBDs of Ppmar1 and Ppmar2 transposases. The affinity of TIRs to DBD could significantly influence the transposition activity of MLEs in certain elements with non-specific affinity to transposases. Hyperactive Ppmar2NA-TIR mutants could be developed to use a genetic tool for the construction of bamboo mutant libraries for gene identification and subsequently for bamboo breeding. Further, they can also be used in mutagenesis studies in other organisms as well.
Acknowledgments
We are grateful to Susan R. Wessler (University of Georgia, USA) for providing pWL89a and pAG413gal-ccdB vectors.
Abbreviations
| TEs | Transposon elements |
| MLEs | Mariner-like elements |
| TIRs | Terminal Inverted Repeats |
| TSDs | Target site duplicates |
| ORF | Open Reading Frame |
| DBD | DNA binding domain |
| PBEs | Protein Binding Elements |
| SB | Sleeping Beauty |
| DRs | Direct repeats |
| FP | Frog Prince |
| EMSA | Electrophoretic mobility shift assay |
Author Contributions
Data curation, M.R., M.Z. and C.P.; Formal analysis, R.M., K.K.V. and M.Z.; Funding acquisition, M.Z.; Investigation, M.R., M.Z. and C.P.; Methodology, M.R., M.Z. and C.P.; Project administration, M.Z.; Resources, M.Z.; Validation, M.Z. and C.P.; Visualization, M.R. and C.P.; Writing—original draft, M.R. and C.P.; Writing—review and editing, M.R., K.K.V., M.Z., H.H., K.Y. and D.T.
Funding
This work was funded by a grant from Zhejiang Provincial Natural Science Foundation of China (No. LZ19C160001), the National Natural Science Foundation of China (No 31470615 and 31870656).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Cho J., Benoit M., Catoni M., Drost H.-G., Brestovitsky A., Oosterbeek M., Paszkowski J. Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants. Nat. Plants. 2019;5:26–33. doi: 10.1038/s41477-018-0320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupeyron M., Singh K.S., Bass C., Hayward A. Evolution of Mutator transposable elements across eukaryotic diversity. Mob. DNA. 2019;10:12. doi: 10.1186/s13100-019-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho J. Transposon-derived non-coding RNAs and their function in plants. Front. Plant. Sci. 2018;9:600. doi: 10.3389/fpls.2018.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths J., Catoni M., Iwasaki M., Paszkowski J. Sequence-independent identification of active LTR retrotransposons in Arabidopsis. Mol. Plant. 2018;11:508–511. doi: 10.1016/j.molp.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Tang D.Q., Zhou M.B. The Distribution, Evolution, Structural Characteristics, and Functional Analysis of the Mariner-Like Elements in Bamboo. In: Kumar A., Ogita S., Yau Y.Y., editors. Biofuels: Greenhouse Gas Mitigation and Global Warming: Next Generation Biofuels and Role of Biotechnology. Springer; New Delhi, India: 2018. pp. 387–406. [Google Scholar]
- 6.Xie L.Q., Wang P.L., Jiang S.H., Zhang Z., Zhang H.H. Genome-wide identification and evolution of TC1/Mariner in the silkworm (Bombyx mori) genome. Genes Genom. 2018;40:485–495. doi: 10.1007/s13258-018-0648-6. [DOI] [PubMed] [Google Scholar]
- 7.Sinzelle L., Jégot G., Brillet B., Rouleux-Bonnin F., Bigot Y., Augé-Gouillou C.J.B.M.B. Factors acting on Mos1 transposition efficiency. BMC Mol. Biol. 2008;9:106. doi: 10.1186/1471-2199-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson H.M., MacLeod E.G. Five major subfamilies of mariner transposable elements in insects, including the Mediterranean fruit fly, and related arthropods. Insect Mol. Biol. 1993;2:125–139. doi: 10.1111/j.1365-2583.1993.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 9.Ivics Z., Hackett P.B., Plasterk R.H., Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/S0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 10.Miskey C., Izsvak Z., Plasterk R.H., Ivics Z. The Frog Prince: A reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izsvak Z., Ivics Z. Sleeping beauty transposition: Biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Fan P.D., Narzisi G., Jayaprakash A.D., Venturini E., Robine N., Smibert P., Germer S., Yu H.A., Jordan E.J., Paik P.K., et al. YES1 amplification is a mechanism of acquired resistance to EGFR inhibitors identified by transposon mutagenesis and clinical genomics. Proc. Natl. Acad. Sci. USA. 2018;115:E6030–E6038. doi: 10.1073/pnas.1717782115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pledger D.W., Coates C.J. Mutant Mos1 mariner transposons are hyperactive in Aedes aegypti. Insect Biochem. Mol. Biol. 2005;35:1199–1207. doi: 10.1016/j.ibmb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Germon S., Bouchet N., Casteret S., Carpentier G., Adet J., Bigot Y., Auge-Gouillou C. Mariner Mos1 transposase optimization by rational mutagenesis. Genetica. 2009;137:265–276. doi: 10.1007/s10709-009-9375-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou M.B., Hu H., Miskey C., Lazarow K., Ivics Z., Kunze R., Yang G., Izsvak Z., Tang D.Q. Transposition of the bamboo Mariner-like element Ppmar1 in yeast. Mol. Phylogenet. Evol. 2017;109:367–374. doi: 10.1016/j.ympev.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Feschotte C., Swamy L., Wessler S.R. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feschotte C., Osterlund M.T., Peeler R., Wessler S.R. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 2005;33:2153–2165. doi: 10.1093/nar/gki509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang G., Nagel D.H., Feschotte C., Hancock C.N., Wessler S.R. Tuned for transposition: Molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–1394. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- 19.Lampe D.J., Walden K.K., Robertson H.M. Loss of transposase-DNA interaction may underlie the divergence of mariner family transposable elements and the ability of more than one mariner to occupy the same genome. Mol. Biol. Evol. 2001;18:954–961. doi: 10.1093/oxfordjournals.molbev.a003896. [DOI] [PubMed] [Google Scholar]
- 20.Trubitsyna M., Grey H., Houston D.R., Finnegan D.J., Richardson J.M. Structural Basis for the Inverted Repeat Preferences of mariner Transposases. J. Biol Chem. 2015;290:13531–13540. doi: 10.1074/jbc.M115.636704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auge-Gouillou C., Hamelin M.-H., Demattei M.-V., Periquet G., Bigot Y. The ITR binding domain of the Mariner Mos-1 transposase. Mol. Genet. Genom. 2001;265:58–65. doi: 10.1007/s004380000386. [DOI] [PubMed] [Google Scholar]
- 22.Auge-Gouillou C., Hamelin M.-H., Demattei M.-V., Periquet M., Bigot Y. The wild-type conformation of the Mos-1 inverted terminal repeats is suboptimal for transposition in bacteria. Mol. Genet. Genom. 2001;265:51–57. doi: 10.1007/s004380000385. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L., Dawson A., Finnegan D.J. DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 2001;29:3566–3575. doi: 10.1093/nar/29.17.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohe A.R., Hartl D.L. Reduced germline mobility of a mariner vector containing exogenous DNA: Effect of size or site? Genetics. 1996;143:1299–1306. doi: 10.1093/genetics/143.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohe A.R., Hartl D.L. Efficient mobilization of mariner in vivo requires multiple internal sequences. Genetics. 2002;160:519–526. doi: 10.1093/genetics/160.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galaktionov N.K., Solovyeva A.I., Fedorov A.V., Podgornaya O.I. Trematode Himasthla elongata mariner element (Hemar): Structure and applications. J. Exp. Zool. B Mol. Dev. Evol. 2014;322:142–155. doi: 10.1002/jez.b.22553. [DOI] [PubMed] [Google Scholar]
- 27.Bilyk B., Weber S., Myronovskyi M., Bilyk O., Petzke L., Luzhetskyy A. In vivo random mutagenesis of streptomycetes using mariner-based transposon Himar1. Appl. Microbiol. Biotechnol. 2013;97:351–359. doi: 10.1007/s00253-012-4550-x. [DOI] [PubMed] [Google Scholar]
- 28.Claeys Bouuaert C., Chalmers R. Hsmar1 transposition is sensitive to the topology of the transposon donor and the target. PLoS ONE. 2013;8:e53690. doi: 10.1371/journal.pone.0053690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert V.J. Engineering the Caenorhabditis elegans genome by Mos1-induced transgene-instructed gene conversion. Methods Mol. Biol. 2012;859:189–201. doi: 10.1007/978-1-61779-603-6_11. [DOI] [PubMed] [Google Scholar]
- 30.Lampe D.J., Akerley B.J., Rubin E.J., Mekalanos J.J., Robertson H.M. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riordan J.D., Drury L.J., Smith R.P., Brett B.T., Rogers L.M., Scheetz T.E., Dupuy A.J. Sequencing methods and datasets to improve functional interpretation of sleeping beauty mutagenesis screens. BMC Genom. 2014;15:1150. doi: 10.1186/1471-2164-15-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yant S.R., Park J., Huang Y., Mikkelsen J.G., Kay M.A. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: Critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang G., Weil C.F., Wessler S.R. A rice Tc1/mariner-like element transposes in yeast. Plant. Cell. 2006;18:2469–2478. doi: 10.1105/tpc.106.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramakrishnan M., Zhou M., Baskar K., Packiam S. Role of Bamboo in Ecosystem. Austin J. Env. Toxicol. 2018;4:1023. [Google Scholar]
- 35.Xu L., Shi Y., Zhou G., Xu X., Liu E., Zhou Y., Zhang F., Li C., Fang H., Chen L. Structural development and carbon dynamics of Moso bamboo forests in Zhejiang Province, China. For. Ecol. Manag. 2018;409:479–488. doi: 10.1016/j.foreco.2017.11.057. [DOI] [Google Scholar]
- 36.Pan F., Wu M., Hu W., Liu R., Yan H., Xiang Y. Genome-Wide Identification and Expression Analyses of the bZIP Transcription Factor Genes in moso bamboo (Phyllostachys edulis) Int. J. Mol. Sci. 2019;20:2203. doi: 10.3390/ijms20092203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L., Cheng Z., Ma Y., Bai Q., Li X., Cao Z., Wu Z., Gao J. The association of hormone signalling genes, transcription and changes in shoot anatomy during moso bamboo growth. Plant. Biotechnol. J. 2018;16:72–85. doi: 10.1111/pbi.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jing X., Cai C., Fan S., Wang L., Zeng X. Spatial and Temporal Calcium Signaling and Its Physiological Effects in Moso Bamboo under Drought Stress. Forests. 2019;10:224. doi: 10.3390/f10030224. [DOI] [Google Scholar]
- 39.Liu J., Cheng Z., Xie L., Li X., Gao J. Multifaceted Role of PheDof12-1 in the Regulation of Flowering Time and Abiotic Stress Responses in Moso Bamboo (Phyllostachys edulis) Int. J. Mol. Sci. 2019;20:424. doi: 10.3390/ijms20020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T., Liu L., Wang X., Liang L., Yue J., Li L. Comparative Analyses of Anatomical Structure, Phytohormone Levels, and Gene Expression Profiles Reveal Potential Dwarfing Mechanisms in Shengyin Bamboo (Phyllostachys edulis f. tubaeformis) Int. J. Mol. Sci. 2018;19:1697. doi: 10.3390/ijms19061697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H., Gao Z., Wang L., Wang J., Wang S., Fei B., Chen C., Shi C., Liu X., Zhang H., et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis) GigaScience. 2018;7:giy115. doi: 10.1093/gigascience/giy115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X., Zhao H., Xu W., You Q., Yan H., Gao Z., Su Z. Co-expression Gene Network Analysis and Functional Module Identification in Bamboo Growth and Development. Front. Genet. 2018;9:574. doi: 10.3389/fgene.2018.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou M.-B., Lu J.-J., Zhong H., Tang K.-X., Tang D.-Q. Distribution and polymorphism of mariner-like elements in the Bambusoideae subfamily. Plant. Syst. Evol. 2010;289:1–11. doi: 10.1007/s00606-010-0323-0. [DOI] [Google Scholar]
- 44.Zhou M., Hu H., Liu Z., Tang D. Two active bamboo mariner-like transposable elements (Ppmar1 and Ppmar2) identified as the transposon-based genetic tools for mutagenesis. Mol. Breed. 2016;36:163. doi: 10.1007/s11032-016-0588-2. [DOI] [Google Scholar]
- 45.Zhou M.-B., Zhong H., Tang D.-Q. Isolation and characterization of seventy-nine full-length mariner-like transposase genes in the Bambusoideae subfamily. J. Plant. Res. 2011;124:607–617. doi: 10.1007/s10265-010-0396-4. [DOI] [PubMed] [Google Scholar]
- 46.Zhou M.-B., Zhong H., Hu J.-L., Tang D.-Q. Ppmar1 and Ppmar2: The first two complete and intact full-length mariner-like elements isolated in Phyllostachys edulis. Acta Bot. Gall. 2015;162:127–137. doi: 10.1080/12538078.2014.999117. [DOI] [Google Scholar]
- 47.Peng Z., Lu Y., Li L., Zhao Q., Feng Q., Gao Z., Lu H., Hu T., Yao N., Liu K., et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) Nat. Genet. 2013;45:456–461. doi: 10.1038/ng.2569. [DOI] [PubMed] [Google Scholar]
- 48.Munoz-Lopez M., Garcia-Perez J.L. DNA transposons: Nature and applications in genomics. Curr. Genom. 2010;11:115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollister J.D., Gaut B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lampe D.J., Grant T.E., Robertson H.M. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lampe D.J., Churchill M.E., Robertson H.M. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. doi: 10.1002/j.1460-2075.1996.tb00930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barry E.G., Witherspoon D.J., Lampe D.J. A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics. 2004;166:823–833. doi: 10.1534/genetics.166.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson H.M., Lampe D.J. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol Evol. 1995;12:850–862. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- 54.Miskey C., Papp B., Mates L., Sinzelle L., Keller H., Izsvak Z., Ivics Z. The ancient mariner sails again: Transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bryan G., Garza D., Hartl D. Insertion and excision of the transposable element mariner in Drosophila. Genetics. 1990;125:103–114. doi: 10.1093/genetics/125.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo D., Zhang J., Shi C., Zhang G., Bai X., Wei G. A Simple, Fast and Efficient Method for DNA Silver Staining. J. Henan Agric. Sci. 2010;7:74–76. [Google Scholar]
- 57.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]