Abstract
Background
Peripheral blood mononuclear cells (PBMCs) are implicated in the pathogenesis of age-related macular degeneration (AMD). We here mapped the global gene transcriptome of PBMCs from patients with different clinical subtypes of late AMD.
Results
We sampled fresh venous blood from patients with geographic atrophy (GA) secondary to AMD without choroidal neovascularizations (n = 19), patients with neovascular AMD without GA (n = 38), patients with polypoidal choroidal vasculopathy (PCV) (n = 19), and aged control individuals with healthy retinae (n = 20). We isolated PBMCs, extracted RNA, and used microarray to investigate gene expression. Volcano plots identified statistically significant differentially expressed genes (P < 0.05) at a high magnitude (≥30% higher/lower) for GA (62 genes), neovascular AMD (41 genes), and PCV (41 genes). These clinical subtypes differed substantially across gene expression and the following pathways identified in enrichment analyses. In a subgroup analysis, we investigated presence vs. absence of subretinal fibrosis and found 826 differentially expressed genes (≥30% higher/lower, P < 0.05) with relation to mRNA splicing, endothelial migration, and interleukin-1 signaling.
Conclusions
We here map the global gene transcriptome of PBMCs related to clinical subtypes of late AMD and find evidence of subtype-specific immunological involvement. Our findings provide a transcriptomic insight into the systemic immunity associated with AMD.
Electronic supplementary material
The online version of this article (10.1186/s12979-019-0160-0) contains supplementary material, which is available to authorized users.
Keywords: Age-related macular degeneration, Choroidal neovascularization, Geographic atrophy, Polypoidal choroidal vasculopathy, Subretinal fibrosis, Peripheral blood mononuclear cells, Transcriptome
Background
Age-related macular degeneration (AMD) is the most frequent cause of visual impairment in the developed countries and the demographic developments towards an aging population are expected to continually and significantly increase the disease burden in the years to come [1–3]. Early stages of the disease are characterized by drusen formation. Drusen are deposits of extracellular material and lipoproteins under the retinal pigment epithelium (RPE) that to a certain extent are considered a normal aging phenomenon, and they are typically asymptomatic [4, 5]. While this stage characterizes the majority of individuals with AMD, an important group of patients experience progression of disease to the late form of AMD [4].
Late AMD manifests with different clinical features. In geographic atrophy (GA), fundus examination reveals the demarcated areas of atrophy of the neuroretina and the RPE, which gradually progress over time. This subtype of late AMD, GA without any choroidal neovascularizations (CNV), accounts for ~ 40% of all late AMD cases in Caucasian populations [3, 4, 6]. Currently, no treatment exists [4]. This is in contrast to neovascular AMD, where CNVs development is mediated by vascular endothelial growth factor (VEGF) expression, which is treatable through regular intravitreal injections with anti-VEGF antibodies [4].
When diagnosing neovascular AMD, retinal angiography is the gold standard and polypoidal choroidal vasculopathy (PCV) is a frequent differential diagnosis [7–9]. PCV is present in ~ 9% of White patients and ~ 50% of Asian patients suspected of neovascular AMD who undergo retinal angiography for diagnosis [7]. Interestingly, PCV is not strongly associated to drusen as otherwise observed in eyes with neovascular AMD [7], and studies increasingly deal with the question of whether PCV is just another clinical phenotype of neovascular AMD or a distinct clinical entity [10].
Intravitreal injection treatment with anti-VEGF has dramatically improved the prognosis of neovascular AMD [11, 12]. However, when left untreated, on average 1 line (defined as five letters constituting a line on the Early Treatment of Diabetic Retinopathy Study visual acuity chart) is lost at 3 months, 3 lines are lost at 12 months, and 4 lines are lost at 24 months; mainly due to progression of the neovascular lesion and through development of fibrotic scars [13]. In the era of modern anti-VEGF therapy, subretinal fibrosis is considered a harbinger of poor visual outcomes [14]. It may be present at time of diagnosis or develop gradually despite anti-VEGF treatment [14]. There is currently no treatment for subretinal fibrosis.
The exact pathogenesis of late AMD remains to be mapped, but consensus so far is that late AMD is a complex interplay of susceptibility through genetic background, chronic progressive degeneration of the macula, immunosenescence, and dysfunction of the immune system [4, 5, 15–19]. Emerging evidence suggest correlations between specific pathological mechanisms and different clinical phenotypes of late AMD [20–30]. Involvement of systemic immune cells is demonstrated in an increasing number of observational studies in human patients with clinical subtypes of late AMD [20–31]. In this study, we investigated the global gene transcriptome of peripheral blood mononuclear cells (PBMC) across different clinical subtypes of late AMD and compared to those in aged control individuals with healthy retinae. We also compared individuals with any subretinal fibrosis to those without subretinal fibrosis. This comparison provided important insight into a currently untreatable clinical subtype of neovascular AMD. Overall, we find that the PBMC in patients with late AMD differ from healthy controls, but that specific clinical subtypes associate with specific immunological changes.
Methods
Study design and ethics
This was a prospective clinic-based case-control study. We explained the nature of the study to potential participants prior to any participation and all participants gave oral and written informed consent. All aspects of this study follow the principles in the Declaration of Helsinki and ethical approval was obtained by the Regional Committee of Ethics in Research of the Region of Zealand (SJ-379).
Study participants and eligibility
Patients were recruited from our outpatient retinal clinic at Department of Ophthalmology, Zealand University Hospital, Roskilde, Denmark. We included three clinical subtypes of late AMD and one aged control group with healthy retina in both eyes. Participants with late AMD had: 1) GA secondary to AMD in one or both eyes with no signs of CNV, 2) neovascular AMD in one or both eyes with no signs of GA, 3) PCV in one or both eyes with no GA. Among patients with neovascular AMD, we included equally cases of patients who had no subretinal fibrosis in both eyes and patients who had subretinal fibrosis in one or both eyes. We did not include patients with recent onset of CNV because we previously found indication of possible acute immune activity in relation to recent onset of CNV [32]. In addition, we wanted to evaluate whether a patient would develop subretinal fibrosis, which would be difficult to evaluate in a treatment-naïve eye. Elderly healthy control individuals were recruited among biologically unrelated healthy relatives to the patients. This was an intentional strategy applied to better match the control group on environmental exposures.
We employed two different strategies in participant selection to avoid influence of non-AMD related immunology. Participants were only included if they did not have any active cancer, autoimmune disease, or any active immune, infectious or inflammatory diseases, or were in chemotherapy or immunomodulating therapy for any reason. We sampled blood from potential participants in lithium-heparin coated tubes for routine plasma C-reactive protein measurement and did not include any participant with plasma C-reactive protein > 15 mg/L, which is a sign of ongoing acute immune response [33].
To evaluate number of participants needed, we looked at the literature of the transcriptome of the PBMC. Grunin et al. were able to successfully evaluate the transcriptome of blood monocytes based on a sample of 14 patients with neovascular AMD and 15 healthy controls [34]. Thus, we aimed for 96 participants in total (analyses are made in four batches with 24 samples each) with at least 15 individuals in each group (patients with GA with no CNV; patients with neovascular AMD with no GA and no subretinal fibrosis; patients with neovascular AMD with no GA but with subretinal fibrosis; patients with PCV; healthy controls).
Retinal diagnosis and clinical data
Participants were examined using slit-lamp bio-microscopy and fundus examination, scanning laser imaging, spectral-domain optical coherence tomography (OCT), fundus autofluorescence, and retinal angiography (fluorescein and indocyanine green angiography (ICGA)) where CNV was suspected. We used the following definitions for participant grouping:
Healthy controls: participants with healthy retinae defined as less than 10 small drusen (diameter < 63 μm); no signs of choroidal abnormalities, atrophy, CNV, or pigment abnormalities; and no signs of other retinal diseases.
Patients with GA: participants who had GA secondary to AMD in one or both eyes without any signs of CNV. This was defined as a presence of drusen maculopathy with one or more well-defined atrophic area(s) with decreased retinal pigment seen on OCT corresponding to hypofluorescence on fundus autofluorescence.
Patients with neovascular AMD: participants who had fibrovascular detachment of the retinal pigment epithelium and choroidal neovascular membranes with subretinal or sub-RPE hemorrhages or fibrosis, and no signs of GA. Presence of subretinal fibrosis was evaluated on OCT and used for subgrouping into those with either no subretinal fibrosis in both eyes or those with any subretinal fibrosis in one or both eyes.
Patients with PCV: participants who had one or more polyps in early-phase ICGA with a hypofluorescent halo and with/without branching vascular networks, and without any GA. Other PCV stigmata used to support the diagnosis of PCV were orange-red focal subretinal polyp-like structures, pulsation of polyps on ICGA video, and a protrusion from the choroid elevating RPE from the Bruch’s membrane observed on OCT.
All participants were interviewed to obtain data on lifestyle and medical history. Medical data were crosschecked with the electronic patient record. Smoking habits were categorized in current smokers, previous smokers (smoked > 100 cigarettes during lifetime and ceased smoking > 12 months), or never smokers. Alcohol use was reported in units/week (1 unit = 12 mL ethanol), which is a measure widely used in Denmark by layman. Physical activity was assessed using a single question on regular activity validated in previous studies for patients with AMD in Denmark [33, 35]. Height and weight were measured to calculate body mass index (BMI).
Tissue sampling and preparation
Venous blood (10 mL) was sampled from antecubital veins in ethylenediaminetetraacetic acid (EDTA) coated tubes. The EDTA stabilized blood was prepared within 4 h. In a 15 mL centrifuge tube, we added 5 mL LymphoPrep™ (STEMCELL Technologies Inc., Vancouver, British Columbia, Canada) and then 10 mL blood carefully on top of the LymphoPrep layer. Lymphoprep™ is a density gradient medium that allows isolation of PBMC through centrifugation. We centrifuged the tube for 30 min at 4 °C at 1000 g with slow acceleration and deceleration. This process allowed following separation (in order from top to bottom): plasma, PBMCs, LymphoPrep, granulocytes, and erythrocytes. We carefully transferred the PBMC layer into a 2 mL tube, centrifuged (5 min at 4 °C at 1000 g), removed the supernatant, added 1.5 mL 4 °C sterile phosphate-buffered saline, centrifuged (5 min at 4 °C at 1000 g), removed the supernatant, and snap-froze the remaining pellet in liquid nitrogen where it was stored until ribonucleic acid (RNA) extraction and analysis. Transportation to RNA extraction was made using dry ice.
Microarray analysis, bioinformatics, and statistical analysis
RNA was extracted from PBMC using NuGEN Ovation® Pico WTA System V2 kit (NuGEN Technologies Inc., Redwood City, CA, USA) according to the manufacturer’s recommendations. The RNA was analyzed for gene expression with the Human Gene 2.0 ST array (Affymetrix, Santa Clara, CA, USA), which contains 1–2 probes/exon and ~ 26 probes/transcript in total containing 11,086 long intergenic non-coding RNA transcripts and a RefSeq gene count of 24,838. The RNA was labelled using the NuGen Ovation Kit (NuGEN Technologies, San Carlos, California, USA). The arrays were washed and strained with phycoerytrin-conjugated streptavidin using the Affymetrix Fluidics Station® 450 and the arrays were scanned in the Affymetrix GeneArray® 3000 scanner to generate fluorescent images, as described in the Affymetrix GeneChip® protocol. Cell intensity files (CEL files) were generated in the Affymetrix GeneChip® Commond Console® Software.
Participant characteristics were summarized using descriptive statistics and compared using parametric statistics or non-parametric statistics according to distribution characteristics of the variables. Categorical variables were summarized using numbers and percentages and compared using χ2-test or Fisher’s Exact test when categories were small. Statistics were made using SPSS 23 (IBM, Armonk, New York, USA). A p-value less than 0.05 was considered statistically significant. Microarray data are modelled using the Robust Multichip Average approach followed by mean one-step probe set summarization giving each gene a single expression value, all done using the software package Partek Gemonics Suite 6 (Partek, St. Louis, Missouri, USA). Gene annotation list is available as Additional file 1. Comparisons between groups were made using an analysis of covariance (ANCOVA) including run date, age, and sex as covariates and with clinical disease subtype versus healthy control individuals as contrast. For subgroup analyses comparing patients with neovascular AMD with or without subretinal fibrosis, we performed new ANCOVA analysis including run date, age, and sex as covariates and presence of subretinal fibrosis vs. absence of subretinal fibrosis as contrast. Differentially expressed genes were defined as those expressed either ≥30% or ≤ − 30% in a disease group compared to the healthy control individuals and where statistically significant. Volcano plots were used to illustrate differences between the groups. Heatmaps and hierarchical clustering were made using Partek Gemonics Suite 6. Differentially expressed genes were summarized in tables and analyzed using the Enrichr database (http://amp.pharm.mssm.edu/Enrichr/) to predict functional pathways and biological functions of the differentially expressed genes [36, 37].
Results
We included a total of 96 participants: 19 patients with GA with no CNV, 38 patients with neovascular AMD with no GA, 19 patients with PCV, and 20 healthy controls. Participant characteristics are summarized in Table 1. We obtained gene expression values from a total of 29,410 genes
Table 1.
Participant characteristics
| Patients with GA (n = 19) |
Patients with nAMD (n = 38) |
Patients with PCV (n = 19) |
Healthy controls (n = 20) |
P-value | |
|---|---|---|---|---|---|
| Age, years, mean (SD) | 80.4 (8.3) | 78.3 (7.8) | 71.9 (7.8) | 71.7 (8.9) | 0.001 |
| Females, n (%) | 8 (42) | 17 (45) | 13 (68) | 14 (70) | 0.11 |
| Smoking status, n (%) | 0.50 | ||||
| Active | 5 (26) | 10 (26) | 7 (37) | 3 (15) | |
| Ex-smoker | 10 (53) | 7 (18) | 3 (16) | 8 (40) | |
| Never smoker | 4 (21) | 21 (56) | 9 (47) | 9 (45) | |
| Alcohol consumption, units, median (IQR) | 3 (2 to 10) | 7 (1 to 10) | 4 (1 to 13) | 5 (1 to 8) | 0.96 |
| Body mass index, mean (SD) | 26.5 (6.7) | 25.3 (4.4) | 25.2 (3.1) | 24.7 (3.1) | 0.68 |
| Physically active, n (%) | 11 (58) | 21 (55) | 10 (53) | 12 (60) | 0.97 |
Abbreviations: GA Geographic atrophy, IQR Interquartile range, nAMD Neovascular age-related macular degeneration, PCV Polypoidal choroidal vasculopathy, SD Standard deviation
Differentially expressed genes in PBMCs across clinical subtypes of late AMD
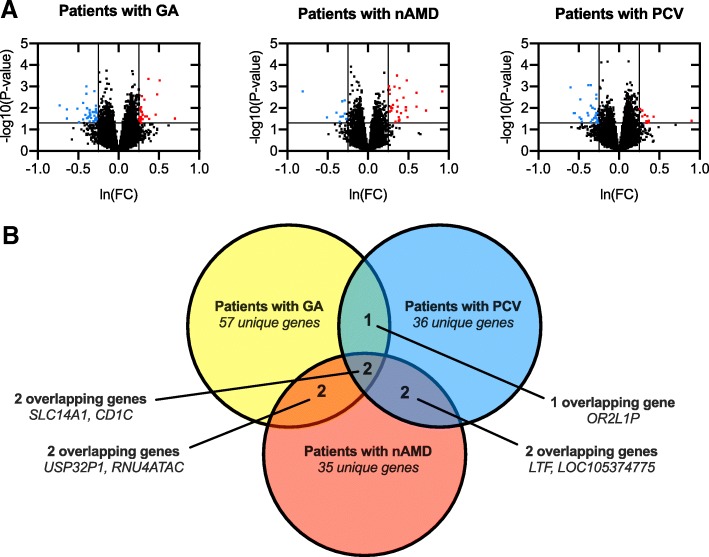
The total transcriptome across different clinical subtypes and heatmaps are available as Additional files 2 and 3. Volcano-plots illustrate differentially expressed genes across different clinical subtypes of late AMD (Fig. 1). In patients with GA, 817 (3%) genes were differentially expressed (P < 0.05); of these, 27 genes had ≥30% higher and 35 genes had ≥30% lower expression (Table 2). In patients with neovascular AMD, 644 (2%) genes were differentially expressed (P < 0.05); of these, 31 genes had ≥30% higher and 10 had ≥30% lower expression (Table 2). In patients with PCV, 806 (3%) genes were differentially expressed (P < 0.05); of these, 11 genes had ≥30% higher and 30 genes had ≥30% lower expression (Table 2). These significantly (both in terms of statistics and magnitude) differentially expressed genes overlapped to a minor degree and at a higher degree between GA and nAMD and between nAMD and PCV than between GA and PCV (Fig. 1).
Fig. 1.
Volcano plots of all quantified genes in the transcriptome analysis of peripheral blood mononuclear cells (PBMCs) and Venn diagram to illustrate similarities and differences. a Volcano plots the PBMC transcriptome for each of the different clinical subtype of late AMD when compared to the healthy controls. Significantly differentially expressed genes are defined as those with at least ±30% change (level of magnitude, vertical lines) and P < 0.05 (level of statistical significance, horizontal line) and illustrated in red (increased expression) or blue (decreased expression). P-values are obtained using analysis of covariance including run date, age, and sex as co-variates and with patients versus healthy controls as contrast. P-values are logarithmic transformed for plot construction. b Venn diagram illustrates the small overlap of differentially expressed genes across the different clinical subtypes
Table 2.
Significantly differentially expressed genes in peripheral blood mononuclear cells of patients with different clinical subtypes of late AMD
| Gene name | Description | FC % | P-value |
|---|---|---|---|
| Patients with GA | |||
| SNORA14A | small nucleolar RNA, H/ACA box 14A | 101 | 0.031 |
| USP32P1 | ubiquitin specific peptidase 32 pseudogene 1 | 67 | < 0.001 |
| SLC14A1 | solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | 62 | 0.002 |
| MIR4516 | microRNA 4516 | 59 | 0.020 |
| DDA1 | DET1 and DDB1 associated 1 | 46 | 0.033 |
| MIR3665 | microRNA 3665 | 44 | < 0.001 |
| SNORA49 | small nucleolar RNA, H/ACA box 49 | 43 | 0.025 |
| MIR4509–1 | microRNA 4509–1 | 39 | 0.024 |
| TMEM120A | transmembrane protein 120A | 38 | 0.004 |
| LY6E | lymphocyte antigen 6 complex, locus E | 35 | 0.021 |
| IFITM1 | interferon induced transmembrane protein 1 | 35 | 0.033 |
| CSNK1G2 | casein kinase 1, gamma 2 | 34 | 0.015 |
| NINJ1 | ninjurin 1 | 33 | 0.003 |
| HSH2D | hematopoietic SH2 domain containing | 33 | 0.009 |
| GIMAP1-GIMAP5 | GIMAP1-GIMAP5 readthrough | 32 | 0.030 |
| LINC01000 | long intergenic non-protein coding RNA 1000 | 32 | 0.026 |
| SNORD88A | small nucleolar RNA, C/D box 88A | 32 | 0.014 |
| CAMKK2 | calcium/calmodulin-dependent protein kinase kinase 2, beta | 32 | 0.049 |
| KRT73 | keratin 73, type II | 32 | 0.034 |
| SDF4 | stromal cell derived factor 4 | 31 | 0.012 |
| CD247 | CD247 molecule | 31 | 0.042 |
| TMSB4XP4 | thymosin beta 4, X-linked pseudogene 4 | 31 | 0.026 |
| CD200R1 | CD200 receptor 1 | 31 | 0.032 |
| CISH | cytokine inducible SH2-containing protein | 31 | 0.011 |
| MX2 | MX dynamin-like GTPase 2 | 30 | 0.035 |
| MICALCL | MICAL C-terminal like | 30 | 0.026 |
| CD5 | CD5 molecule | 30 | 0.037 |
| C15orf54 | chromosome 15 open reading frame 54 | − 108 | 0.008 |
| RNU4ATAC | RNA, U4atac small nuclear (U12-dependent splicing) | − 91 | 0.031 |
| LOC105370195 | uncharacterized LOC105370195 | − 89 | 0.012 |
| MYCT1 | myc target 1 | −67 | 0.011 |
| MS4A1 | membrane-spanning 4-domains, subfamily A, member 1 | −64 | 0.045 |
| LOC105377384 | uncharacterized LOC105377384 | −56 | 0.034 |
| OR2L1P | olfactory receptor, family 2, subfamily L, member 1 pseudogene | −55 | 0.006 |
| LOC105379818 | uncharacterized LOC105379818 | −51 | 0.021 |
| CD1C | CD1c molecule | −49 | 0.001 |
| LOC105369884 | uncharacterized LOC105369884 | −49 | 0.002 |
| LOC102724714 | uncharacterized LOC102724714 | −47 | 0.030 |
| RSU1 | Ras suppressor protein 1 | −47 | 0.010 |
| TNFSF4 | tumor necrosis factor (ligand) superfamily, member 4 | −46 | 0.028 |
| SNORD42A | small nucleolar RNA, C/D box 42A | −45 | 0.026 |
| WDR11-AS1 | WDR11 antisense RNA 1 | − 43 | 0.025 |
| LINC00989 | long intergenic non-protein coding RNA 989 | −43 | 0.043 |
| CRYZ | crystallin zeta | −43 | 0.016 |
| DNM3 | dynamin 3 | −43 | 0.012 |
| LINC00266–1 | long intergenic non-protein coding RNA 266–1 | −42 | 0.021 |
| SLFN14 | schlafen family member 14 | −40 | 0.029 |
| HIST1H2BC | histone cluster 1, H2bc | −38 | 0.016 |
| GFI1B | growth factor independent 1B transcription repressor | −37 | 0.016 |
| PF4 | platelet factor 4 | −37 | 0.015 |
| PPIAL4A | peptidylprolyl isomerase A (cyclophilin A)-like 4A | −36 | 0.041 |
| SENCR | smooth muscle and endothelial cell enriched migration/differentiation-associated lncRNA | −36 | 0.002 |
| LOC101928979 | uncharacterized LOC101928979 | −35 | 0.023 |
| MIR1321 | microRNA 1321 | −34 | 0.016 |
| CTTN | cortactin | −33 | 0.033 |
| GRB14 | growth factor receptor bound protein 14 | −32 | 0.008 |
| PCYT1B | phosphate cytidylyltransferase 1, choline, beta | −32 | 0.028 |
| P2RY1 | purinergic receptor P2Y, G-protein coupled, 1 | −32 | 0.011 |
| KCNQ5-IT1 | KCNQ5 intronic transcript 1 | −32 | 0.037 |
| FAM111B | family with sequence similarity 111, member B | −32 | 0.014 |
| GUSBP1 | glucuronidase, beta pseudogene 1 | −31 | 0.033 |
| LIMS1 | LIM and senescent cell antigen-like domains 1 | −31 | 0.016 |
| Patients with nAMD | |||
| OSBP2 | oxysterol binding protein 2 | 151 | 0.002 |
| IGHV1OR21–1 | immunoglobulin heavy variable 1/OR21–1 (non-functional) | 106 | 0.013 |
| ANK1 | ankyrin 1, erythrocytic | 83 | 0.002 |
| PITHD1 | PITH (C-Terminal Proteasome-Interacting Domain Of Thioredoxin-Like) Domain Containing 1 | 82 | 0.009 |
| MIR4644 | microRNA 4644 | 64 | 0.027 |
| MARCH8 | membrane associated ring finger 8 | 61 | 0.001 |
| PAGE2B | P antigen family, member 2B | 60 | 0.009 |
| TMOD1 | tropomodulin 1 | 60 | 0.013 |
| HBD | hemoglobin, delta | 56 | 0.003 |
| LOC401127 | WD repeat domain 5 pseudogene | 51 | 0.018 |
| RHD | Rh blood group, D antigen | 48 | 0.036 |
| TRBV7–4 | T cell receptor beta variable 7–4 (gene/pseudogene) | 48 | 0.001 |
| USP32P1 | ubiquitin specific peptidase 32 pseudogene 1 | 47 | 0.038 |
| FAM210B | family with sequence similarity 210, member B | 47 | 0.027 |
| BPGM | 2,3-bisphosphoglycerate mutase | 47 | 0.040 |
| FECH | ferrochelatase | 45 | 0.009 |
| LINC01291 | long intergenic non-protein coding RNA 1291 | 44 | < 0.001 |
| HBM | hemoglobin, mu | 43 | 0.015 |
| MMP8 | matrix metallopeptidase 8 | 43 | 0.004 |
| DCAF12 | DDB1 and CUL4 associated factor 12 | 41 | 0.044 |
| FAM46C | family with sequence similarity 46, member C | 40 | 0.043 |
| LTF | lactotransferrin | 39 | 0.001 |
| PGLYRP1 | peptidoglycan recognition protein 1 | 37 | 0.007 |
| CAMP | cathelicidin antimicrobial peptide | 35 | 0.010 |
| SLC25A39 | solute carrier family 25, member 39 | 34 | 0.010 |
| SLC14A1 | solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | 34 | 0.013 |
| SLC25A37 | solute carrier family 25 (mitochondrial iron transporter), member 37 | 32 | 0.009 |
| SLC4A1 | solute carrier family 4 (anion exchanger), member 1 (Diego blood group) | 31 | 0.001 |
| IFIT1B | interferon-induced protein with tetratricopeptide repeats 1B | 31 | 0.002 |
| AHSP | alpha hemoglobin stabilizing protein | 31 | 0.015 |
| ALAS2 | 5-aminolevulinate synthase 2 | 30 | 0.001 |
| RNU4ATAC | RNA, U4atac small nuclear (U12-dependent splicing) | − 125 | 0.002 |
| SNORD3C | small nucleolar RNA, C/D box 3C | −66 | 0.028 |
| ADAM28 | ADAM metallopeptidase domain 28 | − 48 | 0.017 |
| RNU5E-1 | RNA, U5E small nuclear 1 | −42 | 0.044 |
| SNORD12C | small nucleolar RNA, C/D box 12C | −41 | 0.026 |
| FRG1JP | FSHD region gene 1 family member J, pseudogene | −37 | 0.005 |
| MIR1537 | microRNA 1537 | −36 | 0.025 |
| CD1C | CD1c molecule | −34 | 0.004 |
| SNAR-G2 | small ILF3/NF90-associated RNA G2 | − 34 | 0.018 |
| LOC105374775 | uncharacterized LOC105374775 | −31 | 0.033 |
| Patients with PCV | |||
| TRIM48 | tripartite motif containing 48 | 146 | 0.040 |
| LOC105373103 | uncharacterized LOC105373103 | 54 | 0.026 |
| TRAJ13 | T cell receptor alpha joining 13 | 45 | 0.046 |
| SLC14A1 | solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | 44 | 0.040 |
| IGKV1–12 | immunoglobulin kappa variable 1–12 | 42 | 0.024 |
| LTF | lactotransferrin | 41 | 0.050 |
| TRAV8–4 | T cell receptor alpha variable 8–4 | 40 | 0.041 |
| IGHG4 | immunoglobulin heavy constant gamma 4 (G4 m marker) | 37 | 0.021 |
| IGKV2D-24 | immunoglobulin kappa variable 2D-24 (non-functional) | 34 | 0.024 |
| SNORA71D | small nucleolar RNA, H/ACA box 71D | 34 | 0.013 |
| FAR2P3 | fatty acyl-CoA reductase 2 pseudogene 3 | 30 | 0.011 |
| LOC101928215 | uncharacterized LOC101928215 | −81 | 0.001 |
| SNORD117 | small nucleolar RNA, C/D box 117 | − 76 | 0.017 |
| SNORD63 | small nucleolar RNA, C/D box 63 | −73 | 0.017 |
| SCARNA9L | small Cajal body-specific RNA 9-like | −63 | 0.028 |
| SNORD14E | small nucleolar RNA, C/D box 14E | −62 | 0.004 |
| SNORD59B | small nucleolar RNA, C/D box 59B | −61 | 0.032 |
| ZEB2 | zinc finger E-box binding homeobox 2 | −58 | 0.044 |
| LOC105370515 | uncharacterized LOC105370515 | −48 | 0.017 |
| H1F0 | H1 histone family, member 0 | −47 | 0.017 |
| USP12-AS1 | USP12 antisense RNA 1 | −47 | 0.023 |
| TRAJ56 | T cell receptor alpha joining 56 | −46 | 0.001 |
| CD1C | CD1c molecule | −42 | 0.001 |
| RNVU1–15 | RNA, variant U1 small nuclear 15 | −42 | 0.008 |
| LOC728093 | putative POM121-like protein 1-like | −41 | 0.045 |
| LOC105374775 | uncharacterized LOC105374775 | −40 | 0.010 |
| LOC105375566 | uncharacterized LOC105375566 | −39 | 0.035 |
| LOC101927770 | uncharacterized LOC101927770 | −36 | 0.030 |
| FLT3 | fms-related tyrosine kinase 3 | −35 | 0.011 |
| OR2L1P | olfactory receptor, family 2, subfamily L, member 1 pseudogene | −34 | 0.030 |
| LOC100272216 | uncharacterized LOC100272216 | −34 | 0.028 |
| MERTK | MER proto-oncogene, tyrosine kinase | −34 | 0.023 |
| MIR486–2 | microRNA 486–2 | −33 | 0.029 |
| NFKBIZ | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | −33 | 0.046 |
| LOC105378488 | uncharacterized LOC105378488 | −33 | 0.017 |
| MIR3160–1 | microRNA 3160–1 | −32 | 0.004 |
| YPEL5 | yippee like 5 | −32 | 0.002 |
| NLRP3 | NLR family, pyrin domain containing 3 | −31 | 0.035 |
| SNORA7B | small nucleolar RNA, H/ACA box 7B | −31 | 0.045 |
| HIST1H2BN | histone cluster 1, H2bn | −31 | 0.018 |
| LOC101929516 | uncharacterized LOC101929516 | −30 | 0.049 |
Abbreviations: FC Fold change, GA Geographic atrophy, nAMD Neovascular age-related macular degeneration, PCV Polypoidal choroidal vasculopathy
Pathways analysis across clinical subtypes of late AMD
Identifying a set of genes with higher or lower expression do provide important data into mechanisms of disease, but their collective pattern also provides important insight of a general trend. One popular and powerful method is to analyze expression patterns to prior knowledge gene-set libraries—the gene set enrichment analysis approach. Using the Gene Ontology (GO) resource, it is possible to predict these functional pathways in the three domains: biological process, molecular function, and cellular component. Results are presented in Table 3 for each clinical subtype of late AMD. Interestingly, although GO Biological Process revealed involvement of the immune system in all three clinical subtypes, different pathways were predicted across the different clinical subtypes: GA was associated with pathways in type I interferon signaling and memory T cell differentiation; neovascular AMD was associated with pathways in oxygen gas homeostasis and T cell activation; and PCV was associated with pathways in immune regulation and T-helper 2 cell related functions.
Table 3.
Enrichr-based Gene Ontology (GO) enrichment analysis of genes significantly increased or decreased in expression in peripheral blood mononuclear cells of patients with different clinical subtypes of late AMD. Listed are the three strongest in each category (ranked using the Enrichr computed combined score (CS) which multiplies log p-value to z-score; only CS > 5 were considered to only consider strong signals) within the three GO-terms: Biological Process (BP), Molecular Function (MF), and Cellular Component (CC)
| Increased | Decreased | ||||
|---|---|---|---|---|---|
| GO | Description | CS | GO | Description | CS |
| Patients with GA vs. Healthy controls | |||||
| BP | response to interferon-alpha | 15.10 | BP | positive regulation of memory T cell differentiation | 14.05 |
| BP | positive regulation of autophagy of mitochondrion | 13.71 | BP | regulation of memory T cell differentiation | 13.18 |
| BP | type I interferon signaling pathway | 13.08 | BP | positive regulation of T-helper 2 cell differentiation | 13.16 |
| MF | urea transmembrane transporter activity | 15.87 | MF | NADPH binding | 12.91 |
| MF | acetylcholine receptor regulator activity | 15.48 | MF | G-protein coupled nucleotide receptor activity | 12.90 |
| MF | amide transmembrane transporter activity | 12.40 | MF | G-protein coupled purinergic nucleotide receptor activity | 12.70 |
| CC | T cell receptor complex | 10.27 | CC | – | – |
| CC | Cul4-RING E3 ubiquitin ligase complex | 6.62 | CC | – | – |
| CC | – | – | CC | – | – |
| Patients with nAMD vs. Healthy controls | |||||
| BP | oxygen gas homeostasis | 21.30 | BP | lymphocyte activation involved in immune response | 10.65 |
| BP | gas homeostasis | 18.33 | BP | T cell activation involved in immune response | 8.71 |
| BP | regulation of cellular carbohydrate catabolic process | 17.35 | BP | – | – |
| MF | urea transmembrane transporter activity | 15.36 | MF | peptidase activity, acting on L-amino acid peptides | 5.27 |
| MF | iron ion transmembrane transporter activity | 15.27 | MF | – | – |
| MF | sodium:bicarbonate symporter activity | 13.78 | MF | – | – |
| CC | specific granule lumen | 26.32 | CC | – | – |
| CC | tertiary granule lumen | 25.98 | CC | – | – |
| CC | endocytic vesicle lumen | 20.12 | CC | – | – |
| Patients with PCV vs. Healthy controls | |||||
| BP | regulator of immune effector process | 18.39 | BP | negative regulation of acute inflammatory response | 13.62 |
| BP | positive regulation of toll-like receptor 4 signaling pathway | 18.14 | BP | positive regulation of T-helper 2 cell differentiation | 13.62 |
| BP | negative regulation by host of viral process | 15.92 | BP | regulation of T-helper 2 cell cytokine production | 12.56 |
| MF | urea transmembrane transporter activity | 18.92 | MF | vascular endothelial growth factor-activated receptor activity | 13.97 |
| MF | amide transmembrane transporter activity | 14.94 | MF | MAP kinase kinase kinase activity | 11.30 |
| MF | signal recognition particle binding | 13.89 | MF | transmembrane receptor protein tyrosine kinase activity | 9.35 |
| CC | endocytic vesicle lumen | 11.94 | CC | NLRP3 inflammasome complex | 9.85 |
| CC | specific granule lumen | 6.87 | CC | nuclear euchromatin | 9.33 |
| CC | tertiary granule lumen | 6.77 | CC | euchromatin | 8.98 |
Abbreviations: BP Biological process, CC Cellular component, CS Combined score, GA Geographic atrophy, GO Gene ontology, MF Molecular function, nAMD Neovascular age-related macular degeneration, PCV Polypoidal choroidal vasculopathy
Differences in the PBMC transcriptome among patients with neovascular AMD between those with and without subretinal fibrosis
Characteristics of patients with neovascular AMD with (n = 21) or without subretinal fibrosis (n = 17) are summarized in Table 4. We observed a trend towards higher age in those with subretinal fibrosis, which did not reach statistical significance (mean difference: 4.7 years; 95% confidence interval: 9.9 to − 0.4 years; P = 0.071). However, patients with subretinal fibrosis had been followed in the retinal clinic for a significantly longer time than those without subretinal fibrosis (mean difference: 18 months; 95% confidence interval: 2 to 35 months; P = 0.028). These circumstances follow the general clinical experience that 1) subretinal fibrosis occur if time to diagnosis occur with delay—a problem among the very old Danes who seem more settled with poor vision because of age and do not always react upon subtle symptoms—and 2) subretinal fibrosis can develop with time despite treatment.
Table 4.
Characteristics of patients with neovascular AMD stratified according to whether subretinal fibrosis was absent or present
| Subretinal fibrosis absent (n = 17) |
Subretinal fibrosis present (n = 21) |
P-value | |
|---|---|---|---|
| Age, years, mean (SD) | 75.7 (8.6) | 80.4 (6.5) | 0.071 |
| Females, n (%) | 7 (41) | 10 (48) | 0.69 |
| Smoking status, n (%) | 0.23 | ||
| Active | 5 (29) | 5 (24) | |
| Ex-smoker | 7 (29) | 14 (10) | |
| Never smoker | 5 (41) | 2 (67) | |
| Alcohol consumption, units, median (IQR) | 7 (1 to 14) | 7 (1 to 10) | 0.28 |
| Body mass index, mean (SD) | 25.1 (3.4) | 25.5 (5.1) | 0.75 |
| Physically active, n (%) | 8 (47) | 13 (62) | 0.36 |
| Time from diagnosis to samplinga, mean (SD) | 19 (14) | 38 (33) | 0.028 |
Abbreviations: IQR Interquartile range, SD Standard deviation
aIn cases with bilateral disease, time from diagnosis to sampling was defined as the time from diagnosis of the first eye
The gene list of differentially expressed genes between patients with neovascular AMD with or without subretinal fibrosis and heat-maps are available as Additional files 4 and 5. Volcano-plots illustrate these differentially expressed genes (Fig. 2). A total of 3344 (13%) genes were differentially expressed (P < 0.05); of these, 663 genes had ≥30% higher and 163 genes had ≥30% lower expression (see Additional file 6). Genes differentially expressed were included for enrichment analyses (Table 5). Enrichment analyses suggested the involvement of mRNA splicing mechanisms, regulation of interleukin-1 secretion, and endothelial cell migration.
Fig. 2.
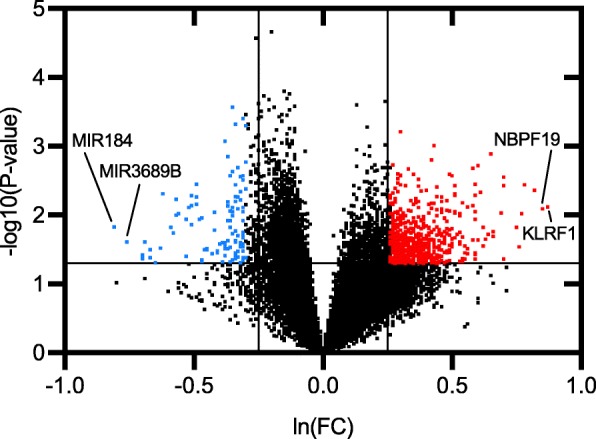
Volcano plots of all quantified genes in the transcriptome analysis of peripheral blood mononuclear cells of patients with neovascular AMD with subretinal fibrosis compared to those without. Significantly differentially expressed genes are defined as those with at least ±30% change (level of magnitude, vertical lines) and P < 0.05 (level of statistical significance, horizontal line) and illustrated in red (increased expression) or blue (decreased expression). The two genes with highest increase or decrease are stated with names. P-values are obtained using analysis of covariance including run date, age, and sex as co-variates and with patients with subretinal fibrosis versus patients without subretinal fibrosis as contrast. P-values are logarithmic transformed for plot construction
Table 5.
Enrichr-based Gene Ontology (GO) enrichment analysis of genes significantly increased or decreased in expression in peripheral blood mononuclear cells of patients with neovascular AMD with subretinal fibrosis compared to those without subretinal fibrosis. Listed are the three strongest in each category (ranked using the Enrichr computed combined score (CS) which multiplies log p-value to z-score; only CS > 5 were considered to only consider strong signals) within the three GO-terms: Biological Process (BP), Molecular Function (MF), and Cellular Component (CC)
| Subretinal fibrosis present vs. Subretinal fibrosis absent | |||||
|---|---|---|---|---|---|
| Increased | Decreased | ||||
| GO | Description | CS | GO | Description | CS |
| BP | mRNA splicing, via spliceosome | 12.17 | BP | negative regulation of interleukin-1 beta secretion | 6.71 |
| BP | mRNA splice site selection | 12.17 | BP | endothelial cell migration | 6.65 |
| BP | regulation of mRNA splicing, via spliceosome | 11.93 | BP | negative regulation of interleukin-1 secretion | 6.36 |
| MF | ubiquitin-like protein ligase activity | 29.03 | MF | dipeptidyl-peptidase activity | 7.76 |
| MF | RNA binding | 28.09 | MF | caspase binding | 7.07 |
| MF | ubiquitin-specific protease binding | 27.92 | MF | – | – |
| CC | nuclear speck | 21.62 | CC | invadopodium | 5.53 |
| CC | nuclear body | 21.52 | CC | – | – |
| CC | ribonucleoprotein granule | 14.72 | CC | – | – |
Abbreviations: BP Biological process, CC Cellular component, CS Combined score, GO Gene ontology, MF Molecular function
Discussion
Defining clinical features of late AMD—choroidal neovascularization, geographically demarcated areas of atrophy, and choroidal polyps—illustrate the wide pathophysiological heterogeneity that is included in the term late AMD. Acknowledging this heterogeneity, studies are increasingly focusing on explaining disease mechanisms of these specific clinical features [20–31]. Peripheral immunological changes in human patients with AMD are gaining increasingly attention, and in this study, we shed important light into how different clinical features of late AMD correlate to the PBMC transcriptome. We find that only a small number of genes (< 0.1–0.2%) are differentially expressed at a high magnitude (≥ 30%) in patients with GA, neovascular AMD, and PCV, and that only a few genes overlap between these clinical subtypes. Our findings underscore the increasing notion that different mechanisms may be involved in these different clinical subtypes.
Pathophysiological mechanisms of GA are incompletely understood, but a range of findings in the immune system have been reported. In Cx3cr1-deficient mice, where atrophic lesions develop upon aging and light-challenge, Sennlaub et al. demonstrated that the degeneration of the retina leads to CCL2 expression recruiting CCR2+ monocytes from the systemic circulation [38]. These CCR2+ monocytes infiltrate, accumulate, and participate in development of retinal atrophy; and the authors showed that pharmaceutical inhibition of the CCL2-CCR2 axis halts atrophy [38]. Studies on human patients with GA confirm the involvement of the immune system [24, 29, 39, 40]. Monocytes in patients with GA express CD200 more than healthy aged individuals, and patients who experience a fast progression of their atrophy have a higher CD200 expression level [40]. The glycoprotein CD200 is a ligand to the CD200 receptor (CD200R), and the CD200-CD200R interaction between peripheral monocytes and retinal microglia is thought to play a role in modulating neuroinflammatory activity [41–44]. Interestingly, in this study we find that the gene CD200R1, which encodes the CD200 receptor CD200R, is increasingly expressed in patients with GA, which further suggests the involvement of neuroinflammation in GA and possible contribution to this from the systemic circulation. In addition to this, another interesting finding was reported by Faber et al. who found a higher proportion of aged, differentiated memory CD8+ T-cells in a group of patients with AMD which consisted of early AMD, GA, and neovascular AMD [45]. In line with these findings were the enrichment analysis in this study, which suggested most strong signals related to type I interferon pathway and memory T cell differentiation. In an in vitro setting, Juel et al. demonstrated that when activated T cells are cocultured with RPE cells, RPE cells increase their expression of chemokines, which provides one explanation for the complex interplay involving both the innate and the adaptive immune system [46]. Taken together, these findings collectively suggest that both the innate and the adaptive immune system may play a role in GA; however, the picture remains unclear and further studies are warranted to investigate immunity in human patients stratified according to clinical subtype.
Unlike GA, both experimental and clinical studies have extensively documented the important contribution of the systemic immune system in neovascular AMD. Experimental laser-based induction of CNV in mice show strong recruitment of peripheral immune cells, and the inhibition of this process lead to a smaller CNV-size indicating that these immune cells play an important role in this process [47–49]. Observational studies in human patients with neovascular AMD confirm the involvement of both the innate and adaptive immune system [20–31]. Findings include higher proportion of pro-angiogenic CD11b+ and CCR2+ monocytes [25], dysregulation of CXCR3 expression in T-cells [24], and a higher proportion of aged cytotoxic CD8+CD56+ T-cells [26]. The latter is an interesting phenomenon thought to be an immunological ageing phenomenon that is accelerated in patients with neovascular AMD [26]. Results so far suggest that fibroblast growth factor receptor-1 (FGFR-1) may be a ligand for CD56 [50, 51]. FGFR-1 is expressed in human specimens of neovascular AMD and its role in retinal disease is also validated in an animal model of retinal injury [52, 53]. Our enrichment analysis suggest the involvement of immune response and lymphocyte activation, which is in line with otherwise reported observations in human patients with neovascular AMD. Interestingly, when Grunin et al. investigated the transcriptome in monocytes of human patients with neovascular AMD, the highest DAVID enrichment score was attributed to leukocyte/lymphocyte activation [34]. In this aspect, we confirm the findings of the Grunin et al. study. However, our enrichment analyses also suggest a strong involvement of gas homeostasis, including oxygen gas homeostasis. Considering that there is, to some extent, an overlap in the immunological findings in patients with neovascular AMD and GA, these findings collectively give rise to an interesting hypothesis: is the clinical manifestation of late AMD a question of how an aged immune system is phenotypically tailored to handle the abundant reactive oxygen species in the aged and stressed retina? To answer this question, further experimental studies and functional immunological studies are warranted.
Although anti-VEGF therapy of neovascular AMD has dramatically changed the prognosis for the patients, some still develop subretinal fibrosis, which is considered the untreatable end stage of the disease. Contributing mechanisms for subretinal fibrosis remain incompletely mapped but shares common mechanisms with fibrosis elsewhere in the human body, such as the lung and the skin [54]. Histopathological studies ofsurgical specimens have reported presence of fibroblasts, endothelium, and activated inflammatory cells in relation to the lesions [54]. Singh et al. reported that 25-hydroxyvitamin D, which is a circulating vitamin acting as a steroid hormone that suppresses inflammation, oxidative stress, angiogenesis, and fibrosis, is significantly lower in the plasma of patients with neovascular AMD that has subretinal fibrosis compared to those without [55]. Lechner et al. sampled plasma from patients with neovascular AMD with and without subretinal fibrosis, and found increased levels of C3a, C4a, and C5a, which are complement fragments measurable after complement activation through the classical pathway, mannose-binding lectine pathway, and the alternative pathway [21]. Our results in this study show that a large number of genes are differentially expressed between those with and without subretinal fibrosis, indicating that a complex range of immunological activities may contribute to this process. Enrichment analyses gives another perspective on this broad immune activation when suggesting activities related to mRNA splicing as well as ubiquitin- and caspase-activity. Interestingly, we also found signals related to endothelial migration and interleukin-1 signaling; these findings are in line with mechanisms and findings reported by other groups suggesting that elastin-mediated choroidal endothelial cell migration may play a role [56] as well as interleukin-1 signaling that contribute to the inflammatory response upon tissue stress that eventually leads to fibrosis [28, 57]. However, studies specific to subretinal fibrosis are lacking, and our findings suggest that these aspects should be studied further in a specific context of subretinal fibrosis.
Unlike neovascular AMD, PCV is far from being extensively studied, and much less in known regarding possible pathological mechanisms. Kumar et al. recently published a mouse-model of PCV and demonstrated that expression of the protease HTRA1 in RPE lead to degeneration of elastin in choroid mimicking features of PCV, which then progressed through infiltration of immune cells [58]. Details and characteristics of these immune cells remains unmapped; however, Sasaki et al. found increased IL-4 levels in the aqueous humor of eyes with PCV and Yu et al. found that cultured PBMCs from patients with PCV increasingly secreted IL-4 upon PHA-stimulation [59, 60]. We recently found that patients with PCV have a lower percentage of regulatory T cells that are increasingly Th2-like polarized [61]. Additionally, in patients with PCV, we recently found significantly higher plasma levels of IL-33 [61], which is a contributor to Th2-like polarization. Halim et al. mapped Th-like regulatory T cells and found that Th2-like regulatory T cells have increased migratory ability and higher viability and blasting capacity, which the authors linked to increased STAT5 phosphorylation that is known to promote angiogenesis [62, 63]. In this study, PBMCs of patients with PCV showed many differentially expressed genes related to T cell receptors and our enrichment analysis suggested involvement of immune effectors cells and Th2, as well as VEGF-activated receptor activity; which further suggests the involvement of recruited immune cells in PCV and that Th2 related immunity may be an important contributing factor.
Genetic susceptibility is a strong risk factor for developing AMD [4, 64, 65]. Heritability of late AMD was previously investigated in a twin study and estimated to 71% [64]. A range of single nucleotide polymorphisms (SNPs) has been identified that indicate involvement of the immune system with strongest associations in SNPs related to the complement pathways [4, 64, 65]. These findings are interesting in light with recent findings by Schemeidel et al. who demonstrated that SNPs associate with specific expression patterns in protein-coding as well as non-coding RNA regions in different immune cell types [66]. Hence, there may be an important link between the strong genetic susceptibility associated with AMD development and the transcriptomic profile of specific immune cell types which we find in this study that may contribute to disease development [66]. An interesting topic for a future investigation is to investigate the correlation between disease activity, genetic background, and the transcriptomic profile of specific immune cell populations. Comparison of the transcriptome of healthy retinae vs. retinae with AMD provide another important insight into the mechanisms of disease [67, 68]. In a transcriptomic study of retinae from eight pairs of postmortem retinae, Kim et al. found that the transcriptomic profile of retinae with AMD had alterations in pathways related to regulation of protein translational, mTOR signaling, phototransduction, and mitochondrial dysfunction [67]. The authors found differentially expressed anti-sense RNA, complement and apolipoprotein genes [67]. When looking at pathways, the anti-sense expression pattern in AMD was involved in pathways related to apoptosis, mitochondrial function, and oxidative stress response [67]. In a larger study of 453 postmortem retinae, Ratnapriya et al. investigated different stages of AMD and found that pathways related to immune regulation and cholesterol metabolism were upregulated in late AMD [68]. The authors also identified altered expression of genes related to extracellular matrix stability and protein degeneration [68]. Taken together, these findings underscore that AMD is a disease with a complex pathogenesis that involve local and systemic factors.
Study limitations
When interpreting the results of this study, important limitations must be considered. This is a cross-sectional observational study; hence, we can only speculate on whether correlations identified in this study are indicative of a causal relationship. Experimental studies are needed to confirm any causality. In line with this limitation, it is important to understand that this study is exploratory. Our sample tissue was PBMCs, which consists of a range of immune cells with different functions. Another approach could be to focus specifically on individual cell types, which would give more detailed insight into mechanisms in specific cells. The gradient centrifugation-based separation of human blood to isolate PBMCs was performed at 4 °C, which is our standard setting for handling human blood. The manufacturer recommends room temperature, which yields a good compromise between time of separation (which is longer with lower temperatures) avoiding aggregation of erythrocytes which decreases the yield of lymphocytes (which is increased at higher temperatures). When we developed our protocols, we found satisfactory results upon centrifuging at 30 min (longer than the recommended 20 min) at 1000 g (faster than the recommended 800 g). However, it is important to note that this approach deviates from the recommended protocol by the manufacturer. Confirming and validating our findings warrant additional studies, e.g. confirmatory qPCR expression studies, testing of protein levels, and functional immunological studies. These studies could further establish specific roles in pathology and their implications. Finally, although we sampled an elderly healthy control group to better match the patient group (mean age 72 years), our groups of patients with GA (mean age 80 years) and patients with neovascular AMD (mean age 78 years) were significantly older. However, we accommodated to this issue with statistical adjustments.
Conclusions
Increasing evidence find involvement of PBMC in late AMD. In this study, we present the global gene transcriptome of such PBMC across the different clinical subtypes of late AMD. We find that clinical subtypes of late AMD differ in PBMC gene expression profile. Our data support the findings of previous studies in patients with AMD and give rise to additional topics that warrants further investigation.
Additional files
Gene annotation list. (XLSX 1932 kb)
Complete data of gene expression comparison across the three subtypes of late AMD. (XLSX 2968 kb)
Heatmaps of differentially expressed genes in the three subtypes of late AMD. (DOCX 7945 kb)
Complete data of gene expression comparison between neovascular AMD with and without subretinal fibrosis. (XLSX 1443 kb)
Heatmaps of differentially expressed genes between neovascular AMD with and without subretinal fibrosis. (PNG 189 kb)
List of significantly differentially expressed genes between neovascular AMD with and without subretinal fibrosis. (XLSX 47 kb)
Acknowledgements
Not applicable.
Abbreviations
- AMD
Age-related macular degeneration
- ANCOVA
Analysis of covariance
- BMI
Body mass index
- BP
Biological process
- CC
Cellular component
- CD200R
CD200 receptor
- CNV
Choroidal neovascularization
- CS
Combined score
- EDTA
Ethylenediaminetetraacetic acid
- FC
Fold change
- GA
Geographic atrophy
- GO
Gene ontology
- ICGA
Indocyanine green angiography
- IQR
Interquartile range
- MF
Molecular function
- OCT
Optical coherence tomography
- PBMC
Peripheral blood mononuclear cells
- PCV
Polypoidal choroidal vasculopathy
- RNA
Ribonucleic acid
- RPE
Retinal pigment epithelium
- SD
Standard deviation
- VEGF
Vascular endothelial growth factor
Authors’ contributions
YS, MKN, HBS, FS, and TLS contributed to the conception and design of the work; YS, MKN, CRM, and CL acquired the data; YS and HBS analyzed the data; all authors (YS, MKN, CRM, CL, HBS, FS, and TLS) interpreted the data; YS drafted the work. All authors (YS, MKN, CRM, CL, HBS, FS, and TLS) revised the manuscript critically for important intellectual content and approved publication of the manuscript.
Funding
This study was funded by the Danish Eye Research Foundation, Fight for Sight Denmark, the Velux Foundation, the University of Copenhagen, and Bayer AG. The sponsors had no role in the design, execution, interpretation, or writing of the study.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
We explained the nature of the study to potential participants prior to any participation and all participants gave oral and written informed consent. Ethical approval was obtained by the Regional Committee of Ethics in Research of the Region of Zealand (SJ-379).
Consent for publication
Not applicable.
Competing interests
Authors YS and TLS are co-applicants of a pending patent application regarding uses of regulatory T-cells in polypoidal choroidal vasculopathy. Authors MKN, CRM, CL, HBS, and FS declare that no potential conflicts of interests exist in relation to this work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yousif Subhi, Phone: +45 4732 3900, Email: ysubhi@gmail.com.
Marie Krogh Nielsen, Email: mrrm@regionsjaelland.dk.
Christopher Rue Molbech, Email: cmop@regionsjaelland.dk.
Charlotte Liisborg, Email: clih@regionsjaelland.dk.
Helle Bach Søndergaard, Email: helle.bach.soendergaard@regionh.dk.
Finn Sellebjerg, Email: finn.thorup.sellebjerg@regionh.dk.
Torben Lykke Sørensen, Email: tlso@regionsjaelland.dk.
References
- 1.Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY, Wong TY. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Colijn JM, Buitendijk GHS, Prokofyeva E, Alves D, Cachulo ML, Khawaja AP, Cougnard-Gregoire A, Merle BMJ, Korb C, Erke MG, Bron A, Anastasopoulos E, Meester-Smoor MA, Segato T, Piermarocchi S, de Jong PTVM, Vingerling JR, Topouzis F, Creuzot-Garcher C, Bertelsen G, Pfeiffer N, Fletcher AE, Foster PJ, Silva R, Korobelnik JF, Delcourt C, Klaver CCW, EYE-RISK consortium. European Eye Epidemiology (E3) consortium Prevalence of age-related macular degeneration in Europe: the past and the future. Ophthalmology. 2017;124:1753–1763. doi: 10.1016/j.ophtha.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sedeh FB, Scott DAR, Subhi Y, Sørensen TL. Prevalence of neovascular age-related macular degeneration and geographic atrophy in Denmark. Dan Med J. 2017;64:A5422. [PubMed] [Google Scholar]
- 4.Mitchell P, Liew G, Gopinath B, Wong TY. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 5.Subhi Y, Forshaw T, Sørensen TL. Macular thickness and volume in the elderly: a systematic review. Ageing Res Rev. 2016;29:42–49. doi: 10.1016/j.arr.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Jonasson F, Fisher DE, Eiriksdottir G, Sigurdsson S, Klein R, Launer LJ, Harris T, Gudnason V, Cotch MF. Five-year incidence, progression, and risk factors for age-related macular degeneration: the age, gene/environment susceptibility study. Ophthalmology. 2014;121:1766–1772. doi: 10.1016/j.ophtha.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorentzen TD, Subhi Y, Sørensen TL. Prevalence of polypoidal choroidal vasculopathy in white patients with exudative age-related macular degeneration: systematic review and meta-analysis. Retina. 2018;38:2363–2371. doi: 10.1097/IAE.0000000000001872. [DOI] [PubMed] [Google Scholar]
- 8.Alasil T, Munoz N, Keane PA, Tufail A, Coady PA, Novais E, de Carlo TE, Baumal CR, Waheed NK, Duker JS, Adelman RA. Characteristics and racial variations of polypoidal choroidal vasculopathy in tertiary centers in the United States and United Kingdom. Int J Retina Vitreous. 2017;3:9. doi: 10.1186/s40942-017-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorentzen TD, Subhi Y, Sørensen TL. Presenting characteristics and prevalence of polypoidal choroidal vasculopathy in Scandinavian patients with treatment-naïve exudative age-related macular degeneration. Acta Ophthalmol. 2018;96:475–480. doi: 10.1111/aos.13646. [DOI] [PubMed] [Google Scholar]
- 10.Laude A, Cackett PD, Vithana EN, Yeo IY, Wong D, Koh AH, Wong TY, Aung T. Polypoidal choroidal vasculopathy and neovascular age-related macular degeneration: same or different disease? Prog Retin Eye Res. 2010;29:19–29. doi: 10.1016/j.preteyeres.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Bloch SB, Larsen M, Munch IC. Incidence of legal blindness from age-related macular degeneration in Denmark: year 2000 to 2010. Am J Ophthalmol. 2012;153:209–213. doi: 10.1016/j.ajo.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Skaat A, Chetrit A, Belkin M, Kinori M, Kalter-Leibovici O. Time trends in the incidence and causes of blindness in Israel. Am J Ophthalmol. 2012;153:214–221. doi: 10.1016/j.ajo.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 13.Wong TY, Chakravarthy U, Klein R, Mitchell P, Zlateva G, Buggage R, Fahrbach K, Probst C, Sledge I. The natural history and prognosis of neovascular age-related macular degeneration: a systematic review of the literature and meta-analysis. Ophthalmology. 2008;115:116–126. doi: 10.1016/j.ophtha.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Casalino G, Stevenson MR, Bandello F, Chakravarthy U. Tomographic biomarkers predicting progression to fibrosis in treated neovascular age-related macular degeneration: a multimodal imaging study. Ophthalmol Retina. 2018;2:451–461. doi: 10.1016/j.oret.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarthy U, Wong TY, Fletcher A, Piault E, Evans C, Zlateva G, Buggage R, Pleil A, Mitchell P. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 2010;10:31. doi: 10.1186/1471-2415-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Mei, Luo Chang, Zhao Jiawu, Devarajan Gayathri, Xu Heping. Immune regulation in the aging retina. Progress in Retinal and Eye Research. 2019;69:159–172. doi: 10.1016/j.preteyeres.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillonneau X, Eandi CM, Paques M, Sahel JA, Sapieha P, Sennlaub F. On phagocytes and macular degeneration. Prog Retin Eye Res. 2017;61:98–128. doi: 10.1016/j.preteyeres.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Chen M, Xu H. Parainflammation, chronic inflammation, and age-related macular degeneration. J Leukoc Biol. 2015;98:713–725. doi: 10.1189/jlb.3RI0615-239R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton T. Genetic research provides insights into age-related macular degeneration. JAMA. 2010;304:1541–1543. doi: 10.1001/jama.2010.1411. [DOI] [PubMed] [Google Scholar]
- 20.Lechner J, Chen M, Hogg RE, Toth L, Silvestri G, Chakravarthy U, Xu H. Alterations in circulating immune cells in neovascular age-related macular degeneration. Sci Rep. 2015;5:16754. doi: 10.1038/srep16754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lechner J, Chen M, Hogg RE, Toth L, Silvestri G, Chakravarthy U, Xu H. Higher plasma levels of complement C3a, C4a and C5a increase the risk of subretinal fibrosis in neovascular age-related macular degeneration: complement activation in AMD. Immun Ageing. 2016;13:4. doi: 10.1186/s12979-016-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Lechner J, Zhao J, Toth L, Hogg R, Silvestri G, Kissenpfennig A, Chakravarthy U, Xu H. STAT3 activation in circulating monocytes contributes to Neovascular age-related macular degeneration. Curr Mol Med. 2016;16:412–423. doi: 10.2174/1566524016666160324130031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner J, Chen M, Hogg RE, Toth L, Silvestri G, Chakravarthy U, Xu H. Peripheral blood mononuclear cells from neovascular age-related macular degeneration patients produce higher levels of chemokines CCL2 (MCP-1) and CXCL8 (IL-8) J Neuroinflammation. 2017;14(1):42. doi: 10.1186/s12974-017-0820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A, Subhi Y, Krogh Nielsen M, Falk MK, Matzen SMH, Sellebjerg F, Sørensen TL. Systemic frequencies of T helper 1 and T helper 17 cells in patients with age-related macular degeneration: a case-control study. Sci Rep. 2017;7(1):605. doi: 10.1038/s41598-017-00741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subhi Y, Krogh Nielsen M, Molbech CR, Oishi A, Singh A, Nissen MH, Sørensen TL. CD11b and CD200 on circulating monocytes differentiate two angiographic subtypes of Polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci. 2017;58(12):5242–5250. doi: 10.1167/iovs.17-22479. [DOI] [PubMed] [Google Scholar]
- 26.Subhi Y, Nielsen MK, Molbech CR, Oishi A, Singh A, Nissen MH, Sørensen TL. T-cell differentiation and CD56+ levels in polypoidal choroidal vasculopathy and neovascular age-related macular degeneration. Aging (Albany NY) 2017;9:2436–2452. doi: 10.18632/aging.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subhi Y, Krogh Nielsen M, Molbech CR, Sørensen TL. Altered proportion of CCR2+ and CX3CR1+ circulating monocytes in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Clin Exp Ophthalmol. 2018;46:661–669. doi: 10.1111/ceo.13152. [DOI] [PubMed] [Google Scholar]
- 28.Subhi Y, Krogh Nielsen M, Molbech CR, Oishi A, Singh A, Nissen MH, Sørensen TL. Plasma markers of chronic low-grade inflammation in polypoidal choroidal vasculopathy and neovascular age-related macular degeneration. Acta Ophthalmol. 2019;97:99–106. doi: 10.1111/aos.13886. [DOI] [PubMed] [Google Scholar]
- 29.Krogh Nielsen M, Subhi Y, Rue Molbech C, Nilsson LL, Nissen MH, Sørensen TL. Imbalances in tissue inhibitors of metalloproteinases differentiate choroidal neovascularization from geographic atrophy. Acta Ophthalmol. 2019;97:84–90. doi: 10.1111/aos.13894. [DOI] [PubMed] [Google Scholar]
- 30.Niazi Siar, Krogh Nielsen Marie, Sørensen Torben Lykke, Subhi Yousif. Neutrophil-to-lymphocyte ratio in age-related macular degeneration: a systematic review and meta-analysis. Acta Ophthalmologica. 2019;97(6):558–566. doi: 10.1111/aos.14072. [DOI] [PubMed] [Google Scholar]
- 31.Grunin M, Burstyn-Cohen T, Hagbi-Levi S, Peled A, Chowers I. Chemokine receptor expression in peripheral blood monocytes from patients with neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012;53:5292–5300. doi: 10.1167/iovs.11-9165. [DOI] [PubMed] [Google Scholar]
- 32.Subhi Y, Lykke ST. New neovascular age-related macular degeneration is associated with systemic leucocyte activity. Acta Ophthalmol. 2017;95:472–480. doi: 10.1111/aos.13330. [DOI] [PubMed] [Google Scholar]
- 33.Subhi Y, Singh A, Falk MK, Sørensen TL. In patients with neovascular age-related macular degeneration, physical activity may influence C-reactive protein levels. Clin Ophthalmol. 2014;8:15–21. doi: 10.2147/OPTH.S55080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grunin M, Hagbi-Levi S, Rinsky B, Smith Y, Chowers I. Transcriptome analysis on monocytes from patients with neovascular age-related macular degeneration. Sci Rep. 2016;6:29046. doi: 10.1038/srep29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subhi Y, Sørensen TL. Physical activity patterns in patients with early and late age-related macular degeneration. Dan Med J. 2016;63:A5303. [PubMed] [Google Scholar]
- 36.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sennlaub F, Auvynet C, Calippe B, Lavalette S, Poupel L, Hu SJ, Dominguez E, Camelo S, Levy O, Guyon E, Saederup N, Charo IF, Rooijen NV, Nandrot E, Bourges JL, Behar-Cohen F, Sahel JA, Guillonneau X, Raoul W, Combadiere C. CCR2(+) monocytes infiltrate atrophic lesions in age-related macular disease and mediate photoreceptor degeneration in experimental subretinal inflammation in Cx3cr1 deficient mice. EMBO Mol Med. 2013;5:1775–1793. doi: 10.1002/emmm.201302692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogh Nielsen M, Subhi Y, Molbech CR, Falk MK, Nissen MH, Sørensen TL. Systemic levels of Interleukin-6 correlate with progression rate of geographic atrophy secondary to age-related macular degeneration. Invest Ophthalmol Vis Sci. 2019;60:202–208. doi: 10.1167/iovs.18-25878. [DOI] [PubMed] [Google Scholar]
- 40.Krogh Nielsen M, Subhi Y, Molbech CR, Falk MK, Singh A, Nissen MH, Sørensen TL. Patients with a fast progression profile in geographic atrophy have increased CD200 expression on circulating monocytes. Clin Exp Ophthalmol. 2019;47:69–78. doi: 10.1111/ceo.13362. [DOI] [PubMed] [Google Scholar]
- 41.Wright GJ, Puklavec MJ, Willis AC, Hoek RM, Sedgwick JD, Brown MH, Barclay AN. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity. 2000;13:233–242. doi: 10.1016/S1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- 42.Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH, Barclay AN, Sedgwick JD. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 43.Hernangómez M, Mestre L, Correa FG, Loría F, Mecha M, Iñigo PM, Docagne F, Williams RO, Borrell J, Guaza C. CD200-CD200R1 interaction contributes to neuroprotective effects of anandamide on experimentally induced inflammation. Glia. 2012;60:1437–1450. doi: 10.1002/glia.22366. [DOI] [PubMed] [Google Scholar]
- 44.Broderick C, Hoek RM, Forrester JV, Liversidge J, Sedgwick JD, Dick AD. Constitutive retinal CD200 expression regulates resident microglia and activation state of inflammatory cells during experimental autoimmune uveoretinitis. Am J Pathol. 2002;161:1669–1677. doi: 10.1016/S0002-9440(10)64444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faber C, Singh A, Krüger Falk M, Juel HB, Sørensen TL, Nissen MH. Age-related macular degeneration is associated with increased proportion of CD56(+) T cells in peripheral blood. Ophthalmology. 2013;120:2310–2316. doi: 10.1016/j.ophtha.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Juel HB, Faber C, Udsen MS, Folkersen L, Nissen MH. Chemokine expression in retinal pigment epithelial ARPE-19 cells in response to coculture with activated T cells. Invest Ophthalmol Vis Sci. 2012;53:8472–8480. doi: 10.1167/iovs.12-9963. [DOI] [PubMed] [Google Scholar]
- 47.Espinosa-Heidmann DG, Suner IJ, Hernandez EP, Monroy D, Csaky KG, Cousins SW. Macrophage depletion diminishes lesion size and severity in experimental choroidal neovascularization. Invest Ophthalmol Vis Sci. 2003;44:3586–3592. doi: 10.1167/iovs.03-0038. [DOI] [PubMed] [Google Scholar]
- 48.Sakurai E, Anand A, Ambati BK, van Rooijen N, Ambati J. Macrophage depletion inhibits experimental choroidal neovascularization. Invest Ophthalmol Vis Sci. 2003;44:3578–3585. doi: 10.1167/iovs.03-0097. [DOI] [PubMed] [Google Scholar]
- 49.Ma W, Zhang Y, Gao C, Fariss RN, Tam J, Wong WT. Monocyte infiltration and proliferation reestablish myeloid cell homeostasis in the mouse retina following retinal pigment epithelial cell injury. Sci Rep. 2017;7:8433. doi: 10.1038/s41598-017-08702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michel JJ, Griffin P, Vallejo AN. Functionally diverse NK-like T cells are effectors and predictors of successful aging. Front Immunol. 2016;7:530. doi: 10.3389/fimmu.2016.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kos FJ, Chin CS. Costimulation of T cell receptor-triggered IL-2 production by Jurkat T cells via fibroblast growth factor receptor 1 upon its engagement by CD56. Immunol Cell Biol. 2002;80:364–369. doi: 10.1046/j.1440-1711.2002.01098.x. [DOI] [PubMed] [Google Scholar]
- 52.Rosenthal R, Malek G, Salomon N, Peill-Meininghaus M, Coeppicus L, Wohlleben H, Wimmers S, Bowes Rickman C, Strauss O. The fibroblast growth factor receptors, FGFR-1 and FGFR-2, mediate two independent signalling pathways in human retinal pigment epithelial cells. Biochem Biophys Res Commun. 2005;337:241–247. doi: 10.1016/j.bbrc.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 53.Ozaki S, Radeke MJ, Anderson DH. Rapid upregulation of fibroblast growth factor receptor 1 (flg) by rat photoreceptor cells after injury. Invest Ophthalmol Vis Sci. 2000;41:568–579. [PubMed] [Google Scholar]
- 54.Ishikawa K, Kannan R, Hinton DR. Molecular mechanisms of subretinal fibrosis in age-related macular degeneration. Exp Eye Res. 2016;142:19–25. doi: 10.1016/j.exer.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh A, Falk MK, Subhi Y, Sørensen TL. The association between plasma 25-hydroxyvitamin D and subgroups in age-related macular degeneration: a cross-sectional study. PLoS One. 2013;8:e70948. doi: 10.1371/journal.pone.0070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skeie JM, Mullins RF. Elastin-mediated choroidal endothelial cell migration: possible role in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:5574–5580. doi: 10.1167/iovs.08-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolb M, Margetts PJ, Anthony DC, Pitossi F, Gauldie J. Transient expression of IL-1beta induces acute lung injury and chronic repair leading to pulmonary fibrosis. J Clin Invest. 2001;107:1529–1536. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S, Nakashizuka H, Jones A, Lambert A, Zhao X, Shen M, Parker M, Wang S, Berriochoa Z, Fnu A, VanBeuge S, Chévez-Barrios P, Tso M, Rainier J, Fu Y. Proteolytic degradation and inflammation play critical roles in Polypoidal choroidal vasculopathy. Am J Pathol. 2017;187:2841–2857. doi: 10.1016/j.ajpath.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasaki S, Miyazaki D, Miyake K, Terasaka Y, Kaneda S, Ikeda Y, Funakoshi T, Baba T, Yamasaki A, Inoue Y. Associations of IL-23 with polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci. 2012;53:3424–3430. doi: 10.1167/iovs.11-7913. [DOI] [PubMed] [Google Scholar]
- 60.Yu Y, Ren XR, Wen F, Chen H, Su SB. T-helper-associated cytokines expression by peripheral blood mononuclear cells in patients with polypoidal choroidal vasculopathy and age-related macular degeneration. BMC Ophthalmol. 2016;16:80. doi: 10.1186/s12886-016-0251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subhi Y, Krogh Nielsen M, Molbech CR, Oishi A, Singh A, Nissen MH, Sørensen TL. Polypoidal choroidal vasculopathy associate with diminished regulatory T cells that are polarized into a T helper 2-like phenotype. Invest Ophthalmol Vis Sci. 2019;60:2583–2590. doi: 10.1167/iovs.19-26882. [DOI] [PubMed] [Google Scholar]
- 62.Halim L, Romano M, McGregor R, Correa I, Pavlidis P, Grageda N, Hoong SJ, Yuksel M, Jassem W, Hannen RF, Ong M, Mckinney O, Hayee B, Karagiannis SN, Powell N, Lechler RI, Nova-Lamperti E, Lombardi G. An atlas of human regulatory T helper-like cells reveals features of Th2-like Tregs that support a tumorigenic environment. Cell Rep. 2017;20:757–770. doi: 10.1016/j.celrep.2017.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dudley AC, Thomas D, Best J, Jenkins A. A VEGF/JAK2/STAT5 axis may partially mediate endothelial cell tolerance to hypoxia. Biochem J. 2005;390:427–436. doi: 10.1042/BJ20050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 65.Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HP, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YT, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GH, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, von Strachwitz CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Saïd S, Sahel JA, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanché H, Deleuze JF, Igo RP Jr, Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA Jr, Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NT, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CC, Hayward C, Gorin MB, Klein ML, Baird PN, den Hollander AI, Fauser S, Yates JR, Allikmets R, Wang JJ, Schaumberg DA, Klein BE, Hagstrom SA, Chowers I, Lotery AJ, Léveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BH, Abecasis GR, Heid IM. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–43. [DOI] [PMC free article] [PubMed]
- 66.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, Seumois G, Rao A, Kronenberg M, Peters B, Vijayanand P. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1705. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim EJ, Grant GR, Bowman AS, Haider N, Gudiseva HV, Chavali VRM. Complete transcriptome profiling of normal and age-related macular degeneration eye tissues reveals dysregulation of anti-sense transcription. Sci Rep. 2018;8:3040. doi: 10.1038/s41598-018-21104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ratnapriya R, Sosina OA, Starostik MR, Kwicklis M, Kapphahn RJ, Fritsche LG, Walton A, Arvanitis M, Gieser L, Pietraszkiewicz A, Montezuma SR, Chew EY, Battle A, Abecasis GR, Ferrington DA, Chatterjee N, Swaroop A. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 2019;51:606–610. doi: 10.1038/s41588-019-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene annotation list. (XLSX 1932 kb)
Complete data of gene expression comparison across the three subtypes of late AMD. (XLSX 2968 kb)
Heatmaps of differentially expressed genes in the three subtypes of late AMD. (DOCX 7945 kb)
Complete data of gene expression comparison between neovascular AMD with and without subretinal fibrosis. (XLSX 1443 kb)
Heatmaps of differentially expressed genes between neovascular AMD with and without subretinal fibrosis. (PNG 189 kb)
List of significantly differentially expressed genes between neovascular AMD with and without subretinal fibrosis. (XLSX 47 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.