Abstract
Type 1 diabetes (T1D) is characterized by pancreatic islet infiltration by autoreactive immune cells and a near-total loss of β-cells1. Restoration of insulin-producing β-cells coupled with immunomodulation to suppress the autoimmune attack has emerged as a potential approach to counter T1D2–4. Here we report that enhancing β-cell mass early in life, in two models of female NOD mice, results in immunomodulation of T-cells, reduced islet infiltration and lower β-cell apoptosis, that together protect them from developing T1D. The animals displayed altered β-cell antigens, and islet transplantation studies showed prolonged graft survival in the NOD-LIRKO model. Adoptive transfer of splenocytes from the NOD-LIRKOs prevented development of diabetes in pre-diabetic NOD mice. A significant increase in the splenic CD4+CD25+FoxP3+ regulatory T-cell (Treg) population was observed to underlie the protected phenotype since Treg depletion rendered NOD-LIRKO mice diabetic. The increase in Tregs coupled with activation of TGF-β/SMAD3 signaling pathway in pathogenic T-cells favored reduced ability to kill β-cells. These data support a previously unidentified observation that initiating β-cell proliferation, alone, prior to islet infiltration by immune cells alters the identity of β-cells, decreases pathologic self-reactivity of effector cells and increases Tregs to prevent progression of T1D.
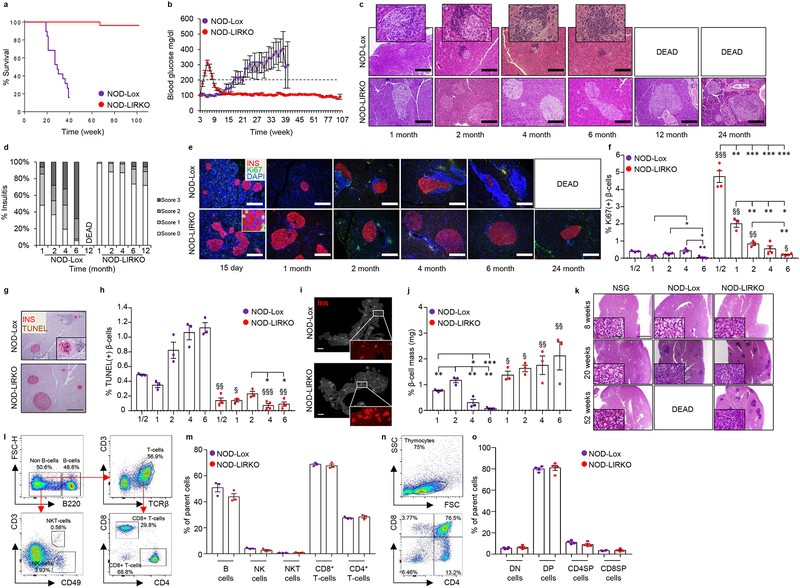
To determine whether enhanced β-cell proliferation, starting before an immune attack would provide protection against type 1 diabetes (T1D) development, we backcrossed the liver-specific insulin receptor knockout (LIRKO) mouse5, a model characterized by robust β-cell proliferation, onto the non-obese diabetic (NOD)6 background. Achieving >99.5% isogenicity while maintaining key NOD modifiers intact, we followed only the females (NOD-Lox and NOD-LIRKO hereafter) for up to 24 months (Supplementary Fig. 1a,b) since traditionally the NOD female exhibits a higher incidence of diabetes7. While most of the NOD-Lox (IRlox control) mice developed severe diabetes between 20–35 weeks of age, surprisingly, virtually all NOD-LIRKO mice survived through the follow-up period (Fig. 1a). Moreover, the NOD-Lox animals exhibited progressive hyperglycemia starting at age 16–18 weeks and began to succumb similarly to wild-type NOD mice (Fig. 1b and Supplementary Fig. 1c); however, the NOD-LIRKO mice exhibited transient hyperglycemia at the age of ~4–5 weeks that reverted to normoglycemia from ~10 weeks and during the entire follow-up period (Fig. 1b). The transient increase in blood glucose was also observed in LIRKO animals on the original background (Supplementary Fig 1c).
Figure 1|. NOD-LIRKO mice are protected from progression to develop diabetes.
a, Kaplan-Meier survival curve showing NOD-Lox and NOD-LIRKO mice monitored for mortality rates (NOD-Lox: n=19; NOD-LIRKO: n=26; P<0.0001, log-rank (Mantel-cox) test. b, Weekly blood glucose values of mice in a. The dashed line shows the upper limit of normoglycemia. c, d, Immunohistochemistry (from three or four mice per genotype from two independent experimental cohorts) showing mononuclear cell infiltration in pancreatic islets from 1, 2, 4, 6, 12 and 24 month-old mice (insets show enlarged islets) (c), and quantification of insulitis score of pancreatic sections from c (scale bar, 200 μm) (d).. e, Representative immunofluorescence images (from three or four mice per genotype from a single experimental cohort) showing proliferation in 15-day-old or 1, 2, 4, 6 or 24 month-old NOD-Lox and NOD-LIRKO mice (scale bar, 200 μm). f, Quantification of Ki67+ β-cells in e (NOD-Lox: 1/2, 1, 2, 4, and 6 months; n=3 per group; NOD-LIRKO: 1/2, 1, 2, 4, and 6 months; n=4, 3, 3, 4, and 4 per group respectively). g, h, Representative TUNEL staining (from three or four mice per genotype from a single experimental cohort) showing β-cell apoptosis (scale bar, 200 μm) (g) and quantification of % β-cell apoptosis (NOD-Lox: 1/2, 1, 2, 4, and 6 months; n=3 per group; NOD-LIRKO: 1/2, 1, 2, 4, and 6 months; n=4, 3, 3, 4, and 4 per group respectively) (h) in NOD-Lox and NOD-LIRKO mice. i, Representative pancreas sections (from three mice per genotype from a single experimental cohort) with insets showing insulin (red) positive islets (Scale bar, 2 mm). j, Morphometric analysis of β-cell mass as described in Methods. n=3 mice each group. k, Representative H&E staining (from four or five mice per genotype from a single experimental cohort) showing immune cell infiltration in salivary glands in NSG, NOD-Lox and NOD-LIRKO mice (scale bar, 1 mm). l, Representative images (from three mice per genotype from a single experimental cohort) of FACS analyses for splenic B, NK, NKT, CD4+ and CD8+ T-cells from NOD-Lox and NOD-LIRKO mice. m, Percent of immune cell subsets shown in l. (NOD-Lox: n=3; NOD-LIRKO: n=3). n, Representative images of FACS analyses for thymic DN, DP, CD4SP, and CD8SP cells from NOD-Lox and NOD-LIRKO mice. o, Percent of immune cell subsets shown in n. (NOD-Lox: n=4; NOD-LIRKO: n=4). All samples in each panel are biologically independent. §, NOD-Lox vs NOD-LIRKO. Data were expressed as means ± s.e.m. *, §P<0.01, **, §§P<0.01, ***, §§§P<0.01. Statistical analysis were performed by two-tailed, unpaired Student’s t-test.
To exclude the potential interactions between insertion of ‘loxP’ sites and T1D susceptibility loci, we phenotyped age-matched, Cre+ and Cre- female mice heterozygous for the floxed insulin receptor (NOD-IRLoxHET). The presence of hyperglycemia starting at ~16 weeks of age, in both the NOD-IRLoxHET Cre+ and Cre- mice similar to NOD-Lox controls indicated that the phenotype in the NOD-LIRKO mice is independent of potential epistatic interactions due to the backcrossing (Supplementary Fig. 1d).
Starting at age 1 month, female NOD-LIRKO mice exhibited elevated insulin and C-peptide levels that were consistent with increased insulin secretion (Supplementary Fig. 1e,f). Glucose challenge at age 2 months showed an impaired ability to dispose of the glucose load and resistance to glucose-lowering effects of insulin in NOD-LIRKO mice compared to IRlox controls (Supplementary Fig. 2a–g), a phenotype that was similar to previous reports in the LIRKOs5. One contribution to the elevated insulin and C-peptide levels could be impaired clearance in the LIRKOs. Pancreas morphology revealed hyperplastic islets and distribution of non-β-cells within the islet core in NOD-LIRKO mice, which was prominent at 2 months of age (Supplementary Fig. 3a). A notable feature was the presence of significantly increased number of small islet clusters (< 10 endocrine cells, Supplementary Fig. 3b,c) and single β-cells (Supplementary Fig. 3d,e) scattered throughout the exocrine pancreas that likely contributed significantly to the maintenance of β-cell mass in NOD-LIRKO mice.
The inflammatory profile of islets revealed invasive insulitis in control mice starting as early as 1 month compared to minimal infiltration, if any, in well-preserved islets in NOD-LIRKO animals. While the infiltration continued to be minimal even at 4 months in the NOD-LIRKOs, it increased to 80% in islets of female NOD-Lox mice (Fig. 1c,d). Evaluation of infiltration in non-pancreatic tissues showed mononuclear-cell accumulation in fat depots of IRLox controls at 6 months of age compared to normal tissue morphology in NOD-LIRKO mice (Supplementary Fig. 4a–c).
To examine the consequences of immune infiltration on changes in β-cells8, we measured proliferation and observed significantly enhanced β-cell mitosis in NOD-LIRKO mice at all ages. A significant increase in proliferation was evident at age 15 days (~12-fold) and 1 month (~17-fold) (Fig. 1e,f) without evidence of a significant increase in cell death (Fig. 1g,h). Instead, the numbers of TUNEL+ β-cells was significantly lower in 15-day-old (~76%) NOD-LIRKO mice. The concomitant altered balance between β-cell gain and loss led to a relative decrease in β-cell mass at 4 months of age in the NOD-Lox while the NOD-LIRKO mice either maintained their mass or showed an increase throughout the experimental period (Fig. 1i,j). The expected infiltration in salivary glands observed in 20 week-old NOD-Lox mice was also present in the NOD-LIRKO group suggesting that protection from immune infiltration is limited to pancreatic islets (Fig. 1k). Furthermore, evaluation of key immune cell subsets harvested from the spleen and draining lymph nodes such as B, CD4+, CD8+, Natural Killer (NK), NKT, and regulatory T (Treg) cells, especially at the time point when NOD-LIRKO mice show normoglycemia (~16–20 weeks of age), revealed no differences except for Tregs (discussed below) between NOD-Lox and NOD-LIRKO groups (Fig. 1 l,m). In addition, the cell populations observed during T-cell maturation in the thymus, including double negative (DN), double positive (DP) and single positive (CD4SP and CD8SP) were also similar between NOD-Lox and NOD-LIRKO mice (Fig. 1 n,o).
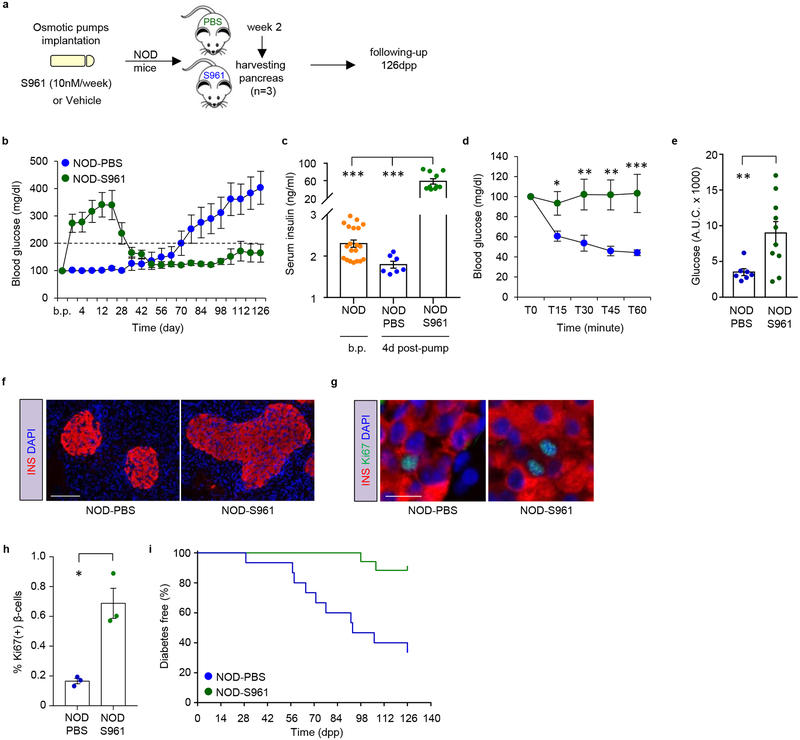
To explore an independent model to replicate the observations in the NOD-LIRKO mouse, we took advantage of S961, an insulin receptor antagonist9, that is known to cause hyperinsulinemia and promote compensatory β-cell proliferation (Fig 2a). While 4-week-old NOD mice implanted with two-week osmotic pumps carrying PBS (NOD-PBS) showed hyperglycemia around ~16 weeks of age, age-matched NOD mice implanted with pumps releasing S961 (NOD-S961) exhibited a transient increase in blood glucose that peaked on day 10, followed by normoglycemia 4 weeks after pump implantation (Fig. 2b). As expected, serum insulin levels were significantly increased (> 30-fold) in NOD-S961 mice compared to NOD-PBS animals as early as day 4 post-pump (p.p.) implantation (Fig. 2c) consistent with resistance to glucose-lowering effects of exogenous insulin in the former group (Fig. 2d,e). Examination of pancreas morphology revealed hyperplastic islets in NOD-S961 animals characterized by a significant increase in β-cell proliferation (Fig. 2f–h). Finally, an 18-week follow-up demonstrated that only ~10% of NOD-S961 mice developed diabetes compared to >60% in the NOD-PBS group (Fig. 2i). These data in the NOD-S961 mice together with observations in the NOD-LIRKOs indicate that boosting β-cell replication prior to immune inflitration protects from development of diabetes. All subsequent experiments were focused on the NOD-LIRKO model.
Figure 2|. S961 treatment enhanced β-cell replication and protects NOD mice from progression to develop diabetes.
a, Experimental design depicting implantation of osmotic pumps releasing either PBS or S961(10 nM/week) into 4-week-old NOD mice. Mice were followed-up for 126 days post-pump (p.p.) implantation. b, Blood glucose values of mice in a. (NOD-PBS: n=15; NOD-S961: n=17). The dashed line shows the upper limit of normoglycemia. c, Serum insulin levels measured in NOD mice before pump (b.p.) implantation in 4 week-old NOD mice (n=20) and 4 days p.p. implantation in NOD-PBS (n=7) and NOD-S961 (n=10) mice. d, Blood glucose levels plotted as % of basal values, following intraperitoneal injection of insulin (1 U/kg./b.wt) (NOD-PBS: n=7; NOD-S961: n=9). e, Quantification of area under the curve (A.U.C) from d. P=0.0075 f, Representative immunofluorescence images (from three mice per genotype from a single experimental cohort) of pancreas sections showing islet morphology of NOD-PBS and NOD-S961 mice. Scale bar: 100 μm. g, Representative pancreas sections (from three mice per genotype from a single experimental cohort) obtained from NOD-PBS and NOD-S961 two weeks p.p. implantation and stained for insulin (red), proliferation marker Ki67 (green) and nuclear dye DAPI (blue). Scale bar: 100 μm. h, Quantification of Ki67+ β-cells in g (NOD-PBS: n=3; NOD-S961: n=3). P=0.026i, % diabetes-free NOD-PBS and NOD-S961 at the end of the 126 day follow-up period (NOD-PBS: n=15; NOD-S961: n=17; P<0.0006). Log-rank (Mantel-cox) test was used for survival plots. All samples in each panel are biologically independent. Data were expressed as means ± s.e.m. *P<0.01, **P<0.01, ***P<0.01. Statistical analysis were performed by two-tailed, unpaired Student’s t-test.
To address whether the absence of diabetes in NOD-LIRKOs is secondary to the high levels of circulating insulin, we transplanted bone marrow obtained from age-matched NOD-Lox or BDC2.5 TCR transgenic (tg) NOD mice10 (the tg T-cells in these mice respond to hybrid insulin peptides (HIPs) from chromogranin-A and insulin11) donors into irradiated12 NOD-LIRKO recipients. Bone marrow transfer from 6–8 week old donors (pre-diabetic NOD-Lox or BDC2.5 mice) led to development of diabetes in hyperinsulinemic NOD-LIRKO recipients suggesting that insulin, either as a β-cell antigen or as a signaling molecule acting on the T-cell insulin receptor, is unlikely to contribute to the protection from infiltration observed in NOD-LIRKO mice (Supplementary Fig. 5a–c).
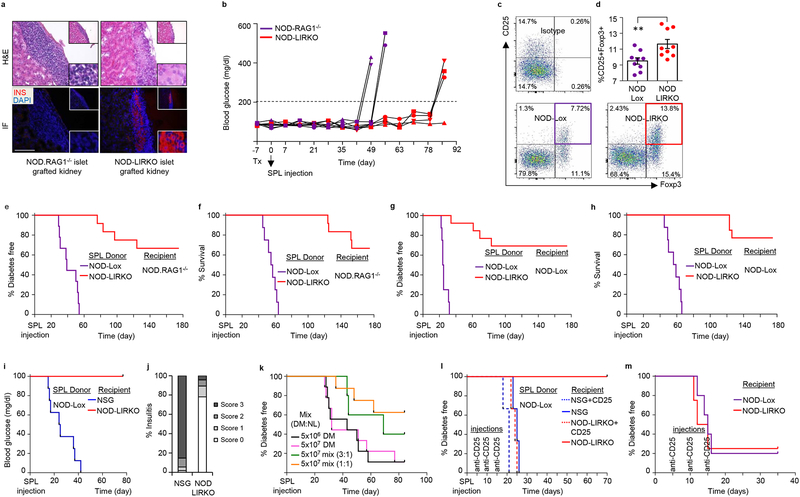
These observations point to several factors that could potentially contribute to the poor infiltration of islets in the NOD-LIRKO mice. First, immunofluorescent staining of the pancreas and mass spectrometry-based proteomics analyses of FACS-sorted islet β-cells revealed a significant decrease in β-cell antigens such as IA-2β/phogrin, ZnT8 and Chromogranin-A in the NOD-LIRKO mice (Supplementary Fig. 6a–f). The low expression of β-cell antigens in this group was supported by a significantly greater number of insulin and glucagon double+ cells (Supplementary Fig. 6g) which are potentially less susceptible to immune-cell attack. In this context it would be informative to explore the effects of metabolic stressors on β-cell function and survival in the NOD-LIRKOs. Next, to test whether NOD-LIRKO islets are less vulnerable to autoimmune attack, we transplanted islets from normoglycemic 16–20 week-old NOD-LIRKO mice under the kidney capsule of immunodeficient NOD-RAG1−/− mice followed by an intravenous injection of diabetogenic splenocytes (SPLs) harvested from multiple low-dose streptozotocin (MLDS) induced hyperglycemic NOD-Lox mice (Supplementary Fig. 7a). Since it is not possible to predict the onset of spontaneous diabetes in the NOD/NOD-Lox and collect diabetogenic splenocytes one week after islet transplantation, we chose to use splenocytes from MLDS treated mice. A ~2.5-fold prolongation in graft survival in the recipients bearing NOD-LIRKO islets compared to those receiving healthy NOD-RAG1−/− islets (Fig. 3a,b and Supplementary Fig. 7b–e) suggested protection potentially due to low levels of β-cell antigens and/or islet derived factors (such as FGF21, TGFβ, etc.) inducing β-cell survival13,14. Moreover, proteomic analysis of FACS-sorted β-cells revealed that beta-2-microglobulin (β2M), a component of the MHC class I molecule which plays a critical role in antigen presentation15, was the most downregulated protein in both 8 and 12-week old NOD-LIRKO mice compared to NOD-Lox animals (Supplementary Fig. 8a,b). Immunohistochemical analyses confirmed an increase in β2M in NOD-Lox mice in contrast to barely detectable expression in NOD-LIRKO mouse pancreas sections (Supplementary Fig. 8c). Similarly, β2M showed reduced expression in islets in the NOD-S961 group compared to non-infiltrated islets in the NOD-PBS group (Supplementary Fig. 8d). In contrast, we did not detect differences in β2M expression in salivary glands between the groups (Supplementary Fig. 8e). The low expression of β2M, which is essential for initiation of insulitis16, coupled with reduced expression of β-cell antigens might contribute, in part, to the protected phenotype in NOD-LIRKO mice.
Figure 3|. NOD-LIRKO islets and splenocytes prevent/delay progression to diabetes.
a, Representative H&E and immunofluorescence staining (from four mice per genotype from a single experimental cohort) of kidney sections showing islet infiltration and presence of insulin in control 6–8 week old NOD.RAG1−/− mice that received control (NOD.RAG1−/−) or NOD-LIRKO islet grafts (16–20 week-old). b, Weekly blood glucose values of NOD.RAG1−/− mice that received control (NOD.RAG1−/−) or NOD-LIRKO islet grafts before and after splenocyte injection. (NOD.RAG1−/−: n=4; NOD-LIRKO: n=4. c, Representative images (from nine mice per genotype from a single experimental cohort) of FACS analyses for CD4+CD25+FoxP3+ Regulatory T cells (Treg) obtained from the spleen and draining lymph nodes (inguinal, axillary, and brachial) of 16–20 weeks old NOD-Lox and NOD-LIRKO mice. d, Percent CD4+CD25+Foxp3+ Tregs shown in c. (NOD-Lox: n=9; NOD-LIRKO: n=9; P=0.0081). e,f, % of diabetes-free (P<0.0001) (e) and surviving (P<0.0001) (f) immunodeficient NOD.RAG1−/− mice post-SPL transfer from new onset diabetic NOD-Lox mice and normoglycemic NOD-LIRKO mice (NOD-Lox: n=16; NOD-LIRKO: n=12). g,h, % of diabetes-free (P<0.0001) (g) and surviving (P<0.0001) (h) pre-diabetic NOD-Lox mice injected with SPLs from new onset diabetic NOD-Lox mice and normoglycemic NOD-LIRKO mice (NOD-Lox: n=8; NOD-LIRKO: n=13). i, % of diabetes-free NSG and NOD-LIRKO mice after diabetogenic SPL transfer (NOD-Lox: n=9; NOD-LIRKO: n=9; P<0.0001). j, Quantification of insulitis score of pancreas sections obtained from recipient mice in i. k, % of diabetes-free NSG recipients receiving either total splenocytes from 16–20 week-old NOD-Lox (DM) (n=9 for both 5 ×106, black line; or 107, pink line) alone or together with total splenocytes from NOD-LIRKO mice (NL) mixed at 1:1 (n=8, orange line) or 1:3 (n=5, green line) DM:NL ratio. l, % of diabetes-free NSG or NOD-LIRKO recipients transferred with total splenocytes from 16–20 week-old new onset diabetic NOD-Lox followed by treatment with or without anti-CD25 mAb. Splenocytes from the same donor were used to inject each recipient per group. A total of 3 donors were used to inject 3 mice each group. m, Percentage of diabetes-free NOD-Lox and NOD-LIRKO mice (10–12 week-old) treated with anti-CD25 mAb on three occasions within fifteen days. Log-rank (Mantel-cox) test was used for adoptive transfer experiments. All samples in each panel are biologically independent. Data were expressed as means ± s.e.m. *, NOD-Lox vs NOD-LIRKO. **, P<0.01. Statistical analysis were performed by two-tailed, unpaired Student’s t-test.
Second, although the percentage of key immune cell subsets were similar between NOD-Lox and NOD-LIRKO mice (Fig. 1l,m), FACS analyses showed a significant increase in the splenic CD4+CD25+Foxp3+ Treg population, a master regulator to maintain tolerance to self-antigens17,18, in the NOD-LIRKO mice (Fig. 3c,d) and a trend to be increased in the thymus (Supplementary Fig. 9a). This was consistent with elevated serum levels of IL-17E/IL-25 (Supplementary Fig. 9b) which have been reported to promote the function of Tregs19. Therefore, to test the hypothesis that total splenocytes, containing an increased Treg population, act to suppress/prevent the autoimmune-cell attack, we performed adoptive transfer of diabetes experiments and transferred total splenocytes from either new onset diabetic NOD-Lox mice (16–20 weeks of age) or age-matched normoglycemic NOD-LIRKO mice into 6–8 week-old female immunodeficient NOD-RAG1−/− animals and followed them for diabetes development (Supplementary Fig. 9c). While all recipients receiving SPLs from NOD-Lox mice developed diabetes within 50 days and succumbed ~70 days after SPL transfer, ~65% of the NOD.RAG1−/− mice that received SPLs from NOD-LIRKO mice remained normoglycemic and survived the 180 day follow-up period (Fig. 3e,f and Supplementary Fig. 9d,e) suggesting that the protective effect is transferrable. These results prompted us to test a preventive strategy wherein we used 16–20 week-old new onset diabetic NOD-Lox mice and normoglycemic NOD-LIRKO mice as SPL donors and 10–12 week old pre-diabetic NOD-Lox mice as recipients (Supplementary Fig. 9f). While mice receiving SPLs from NOD-Lox controls induced progression of T1D in pre-diabetic animals, more than 65% of mice receiving SPLs from NOD-LIRKO mice remained diabetes-free and survived the 180 day follow-up period (Fig. 3g,h). Together, these results suggest one possible explanation for the protective effect seen in the NOD-LIRKO mice is attributable to immune-cell-intrinsic effects, or secondary changes of the T-cell repertoire due to changes in immunogenicity in NOD-LIRKO mouse β-cells. This prompted us to examine whether antigens for CD4 T cell clones, BDC-2.5 (2.5HIP-reactive) and BDC-4.38 (insulin-reactive) are present in NOD-LIRKO islet cells. ELISA for interferon-gamma (IFN-γ) showed immunogenicity in both NOD-Lox and NOD-LIRKO islet cells (Supplementary Fig. 9g). Although we observed that β-cell antigens were lower in NOD-LIRKO mice, the functional effects as determined by the production of IFN-γ did not show differences between groups suggesting that Tregs are potentially important drivers in the protective phenotype in NOD-LIRKO mice.
Next, we undertook studies to explore diabetes development in intact NOD-LIRKO mice. To this end we injected diabetogenic SPLs from new onset diabetic NOD-Lox mice (Supplementary Fig. 9h). All control immunodeficient non-obese diabetic (NOD)-scid IL2Rgnull (NSG)20, developed diabetes (Fig. 3i) consistent with our previous experiments (Fig. 3e and Supplementary Fig. 9d,e). Surprisingly, however, even when the NOD-LIRKO mice received the same number of SPLs from the same diabetic donors, H&E staining revealed barely infiltrated islets, if any, and a diabetes-free phenotype throughout the 11 week follow-up period (Fig. 3i,j and Supplementary Fig. 9i). Interestingly, examination of the very rare infiltrated islets for β2M expression showed a remarkably low expression even when immune cells were infiltrating these islets in the NOD-LIRKO group (Supplementary Fig. 9j) consistent with our previous observations (Supplementary Figure 8c,d). It is possible that NOD-LIRKOs exhibit a benign infiltrate that do not activate inflammatory mediators leading to low β2M expression in the islets.
Based on these results we surmised there is a minimum dose of NOD-LIRKO SPLs required for disease protection. To test this possibility, we transferred a mixture of cells, made up of SPLs from 16–20 week-old new onset diabetic NOD-Lox mice with SPLs from NOD-LIRKOs that showed protection from diabetes, into NSG mice (Supplementary Fig. 9k). While all the NSG mice in the two control groups, injected with either 107 or 5×106 NOD-Lox SPLs (to test whether this lower dose is adequate to transfer diabetes) developed diabetes, ~40% of NSG mice that received a 1:3 ratio (NOD-LIRKO:NOD-Lox SPLs) and more than 60% of NSG mice that received a 1:1 ratio of the SPLs remained diabetes free (Fig. 3k).
Moving forward, we proceeded to confirm whether the protective phenotype is mainly driven by the increased Treg population in NOD-LIRKO mice, by using 2 different Treg depletion approaches in vivo. First, we injected new onset diabetic NOD-Lox SPLs into 10–12 weeks-old female NSG and NOD-LIRKO mice and treated these animals with anti-CD25 mAb or vehicle control (Supplementary Fig. 9l). While all NSG mice regardless of anti-CD25 mAb treatment, as well as NOD-LIRKOs treated with anti-CD25, developed diabetes, the NOD-LIRKO mice that were untreated remained normoglycemic (Fig. 3l). Similar results were obtained when anti-CD25 mAb was injected into NOD-LIRKO mice (Fig. 3m) and suggest that CD25+ Tregs play an important role in protecting NOD-LIRKO mice from T1D. Second, treating mice with cyclophosphamide, which preferentially depletes Treg cells, broke the tolerance in the NOD-LIRKOs and promoted the development of diabetes (Supplementary Fig 9m). Although we did not directly examine Treg function, their surface phenotype and cytokine production, the two independent approaches described above indicate the NOD-LIRKOs possess potential effector T-cells and that their protected phenotype can be reversed by Treg depletion.
The recognition that both CD4+ and CD8+ T-cells play a role in diabetes progression21,22 motivated us to explore the possibility of activation of these T-cell subsets in the NOD-LIRKOs. To investigate insulin-reactive CD4+ T-cells, we immunized both NOD-Lox and NOD-LIRKO mice with peptide emulsified in complete Freunds’ adjuvant, and harvested the spleen and draining lymph nodes following immunization. While total tetramer positive (Tet+) cell count was not changed between untreated NOD-Lox and NOD-LIRKO mice, primed NOD-LIRKO mice showed significantly lower Tet+ cell counts compared to primed NOD-Lox animals (Supplementary Fig 10a). Furthermore, although the number of Tet+CD44hi cells in untreated animals was similar, we observed that NOD-LIRKO mice have significantly less Tet+CD44hi cell counts following immunization (Supplementary Fig 10b) suggesting that autoreactive immune cell subsets in NOD-LIRKO mice are less active compared to NOD-lox mice.
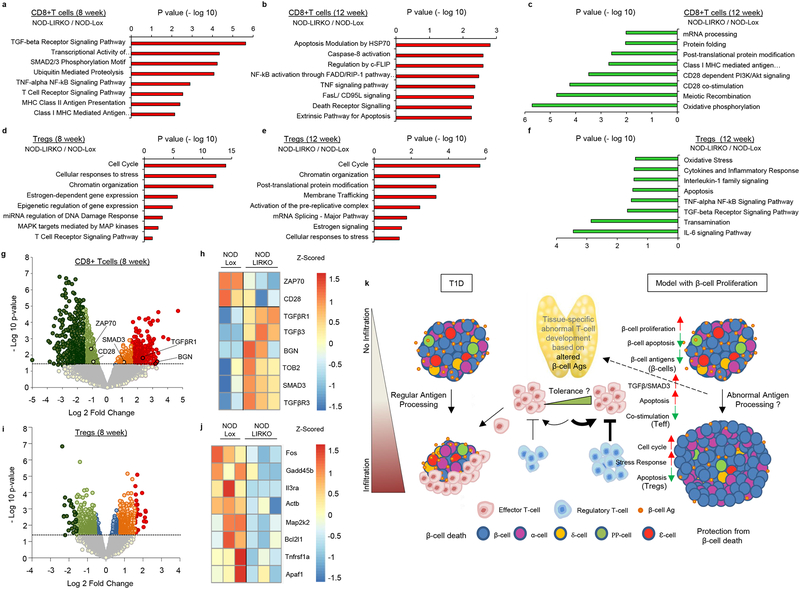
The TGF-β/Smad3 signaling axis, a key regulator in T-cell homeostasis23, has been reported to inhibit CD28-dependent cell growth and proliferation in T-cells24 and to enhance co-inhibitory receptor signaling that down-regulates the activity of tumor-infiltrating lymphocytes in cancer25. Consistent with the adoptive transfer and islet transplantation data, analyses of microarray data revealed an increased regulation of TGF-β receptor signaling and apoptosis in CD8+ T-cells from NOD-LIRKO mice compared to control NOD-Lox mice at both 8 and 12 weeks of age (Fig. 4a,b and Supplementary Table 1,2). The key mechanisms in gene and protein regulation such as mRNA processing, protein folding and post-translational modifications, and vital T-cell activation signals such as CD28 co-stimulation signaling were significantly downregulated in CD8+ T-cells from the NOD-LIRKO mice (Fig. 4c and Supplementary Table 3). On the other hand, pathway analysis in Tregs pointed to the cell cycle as the most upregulated pathway in NOD-LIRKOs at both 8 and 12 weeks of age as well as gene and protein regulation and response to stress (Fig. 4d,e, Supplementary Fig 11a–c and Supplementary Table 4,5). Furthermore, IL-6, TNF-α, and apoptosis pathways were significantly downregulated in NOD-LIRKO Tregs (Supplementary Fig 11d–g, and Supplementary Table 6). Detailed analysis of the differentially expressed transcripts (defined as ≥ 1.5-fold difference in expression and a P-value of < 0.05) in NOD-Lox and NOD-LIRKO CD8+ T-cells showed enrichment of key mediators of TGF-β signaling such as Tgfβr1 (~3.5-fold; 8wks) and Smad3 (~2.2-fold; 8wks and ~2-fold; 12wks) and repression of genes implicated in T-cell activation such as Cd28 (~1.9-fold; 8wks and ~1.7-fold; 12wks) and Zap70 (~2.4-fold; 8wks and ~1.7-fold; 12wks), an essential kinase in transduction of signals from the T-cell receptor (TCR)26 (Fig. 4g,h). Since Biglycan (BGN), forms a complex with TGFβ1 and has been recently reported as a positive regulatory molecule of the TGFβR1-SMAD2/3 TGF-β1 signaling27, we examined its expression profile and observed a significant change (~7.5-fold; 8wks and ~9.5-fold; 12wks) in the protected mice (Fig. 4g). Of note, TOB (transducer ErbB-2) a well-known negative regulator of IL-2 transcription and T-cell proliferation is expressed in anergic and quiescent T-cells28 and enhances binding of SMADs to the negative regulator element of the IL-2 promoter and represses IL-2 expression29. Real-time qPCR analyses on an independent cohort of 8 week-old NOD-Lox and NOD-LIRKO CD8+ T-cells, revealed a significant (p=0.006) increase in Tob expression along with an increase in several mediators of the TGF-β/SMAD3 pathways in the latter group (Supplementary Fig 12a–d). Further analyses of the microarray data revealed estrogen signaling as one of the most up-regulated pathways in Tregs both at 8 and 12 weeks, (Fig. 4d,e). This finding gains significance because oestradiol (E2) signals via the estrogen receptor alpha (ERα) and binds to the FoxP3 promoter to drive Treg expansion to induce their suppressive capacity30,31. Indeed, analyses of plasma revealed significantly higher levels in the NOD-LIRKO compared to NOD-Lox (age 2 mo; NOD-Lox; 6.9±0.6 pg/ml vs NOD-LIRKO; 14.9±0.8 pg/ml; n=3; p<0.001; age 4 mo; NOD-Lox; 7.2±0.4 pg/ml vs NOD-LIRKO; 16.5±2.6 pg/ml; n=3–4; p<0.05) and may be linked to hepatic insulin resistance in the former. Further work is necessary to directly examine the relevance of E2 in the expansion and function of Tregs in the protection from diabetes.
Figure 4|. Key mechanisms in cell homeostasis are upregulated in Tregs in NOD-LIRKO mice.
a,b,c, Selected pathways of differentially expressed genes in CD8+ T cells upregulated at 8 weeks (NOD-Lox: n=2; NOD-LIRKO: n=3) (a), and 12 weeks (n=3 per genotype) (b) of age and downregulated at 12 weeks (n=3 per genotype) (c) in NOD-LIRKO mice. d,e,f, Selected pathways in Tregs upregulated at 8 weeks (n=3 per genotype) (d), and 12 weeks (n=3 per genotype) (e) of age and downregulated at 12 weeks (n=3 per genotype) (f) in NOD-LIRKO mice. g,i, Volcano plots showing the distribution of differential transcript expression defined as a function of fold change (NOD-LIRKO/NOD-Lox) and P value for CD8+ T-cells (NOD-Lox: n=2; NOD-LIRKO: n=3) (g) and Tregs (n=3 per genotype) (i) at the age of 8 weeks. Colored circles (dark green; FC<−3, blue −1.5<FC<1.5 and red FC>3) above the dashed line delineate the area corresponding to fold change of at least 1.5 and a P value <0.05. h,j, Heat-map representation of differentially expressed genes in CD8+ T-cells related to TGFβ/SMAD3 signaling and T-cell activation (h) and downregulated genes in Tregs related to apoptosis (j). All samples in each panel are biologically independent. k, Cartoon depicting changes in β-cell identity before immune cell infiltration. Enhanced β-cell proliferation leads to altered antigen processing and antigen presentation which leads to immune cell alterations early in life. A modified immune repertoire with increased regulatory T cells and poorly effective cytotoxic CD8 T cells allow protection from T1D.
It will be important to explore the translational significance of the findings in the mouse models to human T1D32. In this context, it is interesting to note that parental history of type 2 diabetes (T2D) is associated with a late onset of T1D, the metabolic syndrome, and a metabolic profile related to insulin resistance33,34. In addition, it has been reported that offspring of males or females with insulin-dependent diabetes mellitus (IDDM) have diabetes by age 20 and was higher percentage in the offspring from the males35. Whether alterations secondary to obesity, T2D or gender in the parents have an impact on the identity of β-cells to influence the onset of T1D in the offspring requires careful examination.
In sum these data provide novel evidence for a model (Fig. 4k) that enhanced β-cell proliferation starting early in life alters the identity of β-cells, leading to an improved pathologic self-reactivity of effector cells and a beneficial increase in Tregs to prevent progression of T1D.
METHODS
Mice:
Mice were housed on a 12-h light/12-h dark cycle with water and food ad libitum. Female mice were used for all experiments throughout the study. Alb-Cre;InsRflox/flox (LIRKO) mice5 on the C57BL/6 background were obtained from the colony maintained at the Joslin (obtained from C.R Kahn MD) were backcrossed onto the NOD background to >99.9% purity by speed congenic and T1D susceptible loci were ensured to be expressed in experimental animals by using 150 SNP markers (Jackson Laboratory). NOD.RAG1−/−, NOD-Lox, and NOD-LIRKO mice were bred at Joslin Diabetes Center. NOD and NOD-scid IL2Rgnull (NSG) mice were purchased from Jackson Laboratory. 16–20 week-old NOD, NOD-Lox and NOD-LIRKO mice were used as splenocytes donors. 16–20 week-old NOD.RAG1−/− and NOD-LIRKO mice were used as islet donors. 6–8 week-old NOD.RAG1−/− and NSG mice were used as either islet or splenocyte recipients. 10–12 week-old NOD-Lox mice were used as pre-diabetic recipient animals. Bone marrow chimeras were generated by transferring 5 × 105 T-cell-depleted NOD-Lox (6–8 week-old) and NOD.BDC2.5 (6–8 week-old) mice bone marrow cells into sub-lethally irradiated 6–8 week-old NOD-LIRKO mice, followed by adoptive transfer experiments detailed below. Blood glucose was measured weekly for follow-up studies and mice were considered diabetic when two consecutive measurements of blood glucose exceed 200 mg/dl. All mice were kept in a specific pathogen-free facility in the Animal Facility at Joslin Diabetes Center, and animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC).
Adoptive transfer of diabetes.
Total splenocytes were purified from freshly harvested spleens of 16–20 weeks-old NOD or NOD-Lox mice with diabetes or age-matched normoglycemic NOD-LIRKO mice by filtering through a nylon mesh followed by lysis of the red blood cells with ACK Lysing buffer (Lonza). A total of or 5×106 or 1×107 splenocytes were injected intravenously into a healthy NOD.RAG1−/−, pre-diabetic NOD or normoglycemic NOD-LIRKO mice22.
Treg depletion.
NSG, NOD-Lox, and NOD-LIRKO mice were treated with 400 μg of anti-CD25 Ab (PC61.5.3, BioXcell) intraperitoneally on three occasions within fifteen days at the age of 10 to 12 weeks. Alternatively, 10 to 12 weeks old NOD-Lox and NOD-LIRKO mice were treated intraperitoneally with 200 mg/kg/bw Cyclophosphamide (CY2) (Sigma) followed by a second injection two weeks after the first treatment. All mice were followed-up for diabetes development.
Islet isolation and transplantation.
Islets were isolated either from 16–20 week-old NOD.RAG1−/− or from age-matched NOD-LIRKO mice using the intraductal collagenase technique followed by a density gradient separation with Histopaque-1077 (Sigma-Aldrich)36. Freshly isolated 150 islets were handpicked, concentrated in a pellet in PE50 polyethylenetubing (Beckton Dickinson). A Hamilton syringe (Fisher) was used to transplant the islet pellet underneath the kidney capsule of 6–8 week-old immunodeficient NOD.RAG1−/− mice37. In parallel, diabetogenic splenocyte were prepared by accelerating diabetes progression in 12 week-old pre-diabetic NOD-Lox mice by using multiple low-dose streptozotocin (STZ) (MLDS) (Sigma-Aldrich). One-week post-islet engraftment, a total of 107 total splenocytes were injected intravenously and followed for diabetes development.
Immunohistochemistry and insulitis scoring.
At the end of the follow-up period, mice were sacrificed, pancreata were harvested, fixed, and embedded in paraffin 6 h post-injection with BrdU (100 mg/kg/bw). Sections were stained using antibodies to BrdU (Dako, M0744, 1:100), Ki67 (Dako, M7240, 1:100), phosphohistone H3 (Millipore, 06–570, 1:250), or insulin (Abcam, ab7842, 1:500), glucagon (Sigma, G2654, 1:8000), somatostatin (Abcam, ab30788, 1:1000), β2M (Abcam, ab87483, 1:80), serum anti-ZnT8 (c-term), 1:500, serum anti-phogrin (C-term), 1:250 (gift from H. Davidson, Univ Colorado) and appropriate secondary antibodies and counterstained with DAPI (Sigma, D9564, 1:6600). Cell death was detected by TUNEL assay (ApopTag S7100; Chemicon). At least 1,000–2,000 β-cell nuclei were counted per animal, and data were expressed as a percentage of BrdU+, Ki67+, pHH3+ or TUNEL+ β-cells. Insulitis was evaluated as reported previously38. β-cell mass was evaluated by point-counting morphometry on immunofluorescence-stained sections of the pancreas5.
Western blotting.
Total proteins were harvested from the liver using RIPA buffer (Thermo Fisher) supplemented with proteinase and phosphatase inhibitors (Sigma) according to standard protocols. Protein concentrations were determined using BCA followed by western immunoblotting for proteins. The blots were developed using chemiluminescent substrate ECL (Thermo Fisher).
Islet dispersion, cell sorting and proteomics.
Overnight cultured islets were dispersed by treating with a solution of 1 mg/ml trypsin and 30 μg/ml DNase followed by incubation for 15 min in a 37°C incubator. During the digestion, the islets were vortexed every 5 min for 10 s. Cold media including serum was added to stop the digestion, and the cells were washed two times in PBS. Before sorting islet cells were filtered through a 35 μm filter and sorted using MoFlo Cytometer (Dako), where cells were gated according to forward scatter, and then sorted on the basis of endogenous flourescence39. Sorted cell pellets were analyzed using the simplified nanoproteomic platform (SNaPP)40. Briefly, cells were lysed by adding 10 μL of 8M urea, 10 mM dithiothreitol (DTT), in 50 mM ammonium bicarbonate at pH 8, followed by sonication in a bath sonicator. After incubation at 37 °C, cell lysate was quantitatively transferred to an ALS vial with a 20 μL aliquot of 50 mM ammonium bicarbonate pH 8. A 25 μL injection was used on the SNaPP system41, and separated using a 300 min gradient. The separation gradient started at 5% mobile phase increasing to 8% B at 6 minutes, 12% at 60 minutes, 35% at 225 minutes, 60% at 291 minutes, and 75% at 300 minutes. Carryover on the analytical column is addressed by running a “washing” gradient which ramps to 35% B twice, followed by a ramp to 95% B and then 2 more ramps to 35% B over 25 minutes. The SNaPP system was coupled to a QExactive Plus mass spectrometer (Thermo Scientific) using a custom ESI interface. Mass spectra were collected from 400–2,000 m/z at a resolution of 70 k followed by data dependent HCD MS/MS at a resolution of 17.5 K for the ten most abundant ions. All raw files were processed using Maxquant (version 1.5.3.30) for feature detection, database searching, and protein/peptide quantification42.
Generation of bone-marrow chimeras.
For the generation of bone-marrow chimeras, irradiated NOD-LIRKO mice (900 rad) were injected intravenously with lineage-depleted bone marrow cells (5×105 / mouse) isolated from NOD-Lox or NOD.BDC2.5 mice. Lineage depletion was performed using magnetic separation with MACS lineage-depletion kit (Miltenyi Biotech) according to the manufacturer’s instructions. Lineage depletion efficiency was measured by Flow Cytometry.
ELISA Assay.
Serum insulin was measured using the Ultra Sensitive Mouse Insulin ELISA kit (Crystal Chem) and serum C-peptide was measured using the Mouse C-peptide ELISA kit (Crystal Chem) according to manufacturer’s instructions. Plasma estradiol (E2) was measured using the mouse/rat ELISA kit (Calbiotech).
Antigen assay.
The antigenicity was assessed through the IFN-γ responses of T-cell clones43. Forms of antigens used included islet cell suspensions (0.5 or 2×104 cells) from either NOD/Lox mice or NOD/LIRKO or a membrane fraction obtained from β-cell tumors as a source of antigen, referred to as β-membrane (bMem). After 18 h, IFN-γ concentrations were determined in culture supernatants by ELISA.
Tetramer assay.
NOD/Lox and NOD/LIRKO mice were immunized subcutaneously in the flank with 25 μg insulin B10–23 peptides (p8E, p8G) in Complete Freund’s Adjuvant, and harvested 10 days after immunization, and compared with non-immunized age-matched controls. To detect the two major populations of insulin B10–23-specific CD4+ T cells, we used I-Ag7 tetramer reagents covalently linked to HLVERLYLVCGEEG (p8E) or HLVERLYLVCGGEG (p8G)44,45. Briefly, spleen and lymph node cell suspensions were stained separately from immunized and non-immunized mice with 10nM APC- and PE-conjugated tetramers for 1 hour at room temperature in Fc block (2.4G2). Tetramer+ cells were subsequently magnetically enriched using anti-APC and anti-PE EasySep selection kit (STEMCELL Technologies), stained with fixable live/dead dye, and antibodies targeting surface markers44,45. Insulin-specific CD4+ T cells were defined as lineageneg (B220−, CD8−, CD11b−, CD11c−), CD4+, PE- and APC-tetramer+ cells. A 5-μL aliquot of the bound fraction was removed and added to 200 μL AccuCheck counting beads to determine total CD4+ T-cell number.
Glucose and insulin tolerance test.
For glucose tolerance test (GTT) animals were fasted O/N and injected intraperitoneally with 20% (v/v) dextrose (Hospira) (2g/kg/bw). Blood glucose levels were measured with an automated glucose monitor (Contour, Bayer) by tail punch immediately before and at 15, 30, 60, and 120 minutes after injection36. For insulin tolerance test (ITT) animals were fasted for 3 hours (7 a.m.- 10 a.m.) and intraperitoneally injected with 1U/kg insulin (Humalog). Glucose levels were measured with an automated gucose monitor (Contour, Bayer) by tail punch before and 15, 30, 45, and 60 minutes after insulin injection and plotted as % of basal values.
Microarray.
Microarray was performed by the Molecular Genetic Core Facility at Children’s Hospital Boston. Three biological replicates of CD8+ T-cells and Tregs from both NOD-Lox and NOD-LIRKO at eight and twelve weeks of age were analyzed. Briefly, the microarray data was analyzed by removing probes that were not expressed in any sample (at a p-value below 0.01), correcting for background and normalizing46, and estimating empirical array quality weights47. One sample was removed, because iys estimated weight was 1% of the average weight. We performed differential expression of the microarray data using the R package limma48. Analysis was performed in R/Bioconductor49.
Flow cytometry.
For analyses of surface markers, cells were stained in PBS containing 2% (wt/vol) BSA and pre-incubated with the CD16/32 Fc-receptor blocking antibody prior to labeling with fluorescently-conjugated antibodies; anti-B220 (RA3–6B2) anti-CD4 (GK1.5), anti-CD8α (53–6.7), anti-CD3Ɛ (145–2C11), anti-CD49b (HMα2) anti-TCRβ (H57–597), anti-CD25 (PC61.5); all antibodies were from BioLegend. Intracellular FoxP3 (FJK-16s) staining was performed with the anti-mouse/Rat FoxP3 Staining Set (eBioscience). Flow cytometry measurements were performed using LSRII instrument (BD Biosciences). Data were analyzed with FlowJo software (TreeStar Inc.)
RNA isolation and RT-PCRs.
For real-time experiments, total RNAs from T-cells were extracted using standart Trizol reagent (Invitrogen) according to manufacturer’s instructions and resultant aqueous phase was mixed (1:1) with 70% RNA-free ethanol and added to Qiagen Rneasy Mini Kit columns (QIAGEN) followed by the manufacturer’s protocol. RNA quality and quantity were analyzed by Nanodrop 1000 and used for reverse transcription step. Reverse Transcription was performed by using high-capacity cDNA Archive Kit (Applied Biosystems). cDNA was analyzed using the ABI 7900HT system (Applied Bioscinces) and gene expression was calculated using the double delta Ct (ΔΔCt) method. TBP was used as an internal control. Primers for TOB: 5’-CACAGGATCTTAGTGTTTGGATCGA-3’ (forward) and 5’-TTCTTCATTTTGGTAGAGCCGAACT-3’ (reverse), for TBP; 5’-ACCCTTCACCAATGACTCCTATG-3’ (forward) and 5’-ATGATGACTGCAGCAAATCGC-3’ (reverse), for TGFβR2; 5’-CCGGAAGTTCTAGAATCCAG-3’ (forward) and 5’-TAATCCTTCACTTCTCCCAC-3’ (reverse), for SMAD2; 5’-GGAAAGGGTTGCCACATGTT-3’ (forward) and 5’-AGAATCTCCGTGTGCCGAGG-3’ (reverse) and for SMAD3; 5’-GTTGGACGAGCTGGAGAAGG-3’ (forward) and 5’-TGCTGTGGTTCATCTGGTGG-3’ (reverse).
Statistical analysis.
Data presented are shown of mean ± s.e.m. Graphpad Prism software (v.7) was used for graphing and statistical analysis. Survival curves were analyzed by log-rank (Mantel-cox) test. Statistics were performed using unpaired two-tailed Student’s t-test with statistical significance defined as P < 0.05. For microarray analyses, Moderated t-test with LIMMA (linear models for microarray data) was used to analyze P-values. For the pathway analyzes performed via ConsensusPathDB, the P-value was calculated according to the hypergeometric test based on the number of physical entites present in both the predefined set and user-specified list of physical entities50.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Transcriptomics data have been deposited on the National Center for Biotechnology Information (NCBI) gene expression omnibus (GEO) under accession code GSE128315 for murine microarray analysis. Mass spectrometry proteomics data reported in this paper have been deposited in ProteomeXchange (accession code: PXD013100) and MassIVE (accession code: (MSV000083576). Further information on statistical parameter, software, study design and so forth is in the Nature Reporting Summary. The data that supports the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request. Correspondence and requests for material should be addressed to R.N.K.
Supplementary Material
Acknowledgments
We thank the late A. Rossini MD (Joslin) for discussions during the early stages of this project. We thank C.R. Kahn MD (Joslin) for discussions and sharing the LIRKO model. We thank D. Mathis, Ph.D. (Harvard), J. Gaglia, MD Ph.D. (Joslin), C. Mathews PhD (Univ. Gainesville, Florida), and G.C. Weir MD (Joslin) and F. Bosch PhD (UAB, Spain) for discussions regarding various aspects of the studies and the NOD.RAG1−/− mice. We thank H. Davidson (Univ Colorado) for kindly providing serum anti-ZnT8 (C-term) and serum anti-Phogrin (C-term) and F.M. Jarvis MD PhD (Tulane Univ. Health Sciences, New Orleans) for assistance with E2 assays. We thank C. Cahill (Joslin Advanced Microscopy Core) for assistance with confocal microscopy, to L. Kannan, Z. Fu and G. Sankaranarayanan (Joslin Assay Core) for ELISA assays, J. Hollister-Lock (Joslin) for assistance with mouse islet isolations and J. Dreyfuss and H. Pan (Joslin Bioinformatics Core) for assistance in data analyses. Proteomics experiments were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by Department of Energy and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for DOE under Contract DE-AC05–76RL0 1830. E.D. was supported by a Senior Juvenile Diabetes Research Foundation Fellowship (JDRF-3-APF-2014–220-A-N). This project was funded by JDRF Grant 1-SRA-355-Q-R (R.N.K.) and in part from NIH RO1 067536 (R.N.K.), NIH RO1 103215 (R.N.K.), UC4 DK104167 (W.J.Q. and R.N.K.), R.N.K. acknowledges support from the Margaret A. Congleton Chair, the Joslin DRC (P30 386836), P41 GM103493 (R.D.S), R01 AI106791 (B.T.F), R21 AI133059 (R.L.B.), ADA Junior Faculty Award 1–15-JF-04 (R.L.B.) and R01 DK081166 (K.H.).
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Competing interests
The authors declare no competing interests.
References
- 1.Mathis D, Vence L & Benoist C beta-Cell death during progression to diabetes. Nature 414, 792–798, doi: 10.1038/414792a (2001). [DOI] [PubMed] [Google Scholar]
- 2.Rezania A et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature biotechnology 32, 1121–1133, doi: 10.1038/nbt.3033 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Tooley JE, Waldron-Lynch F & Herold KC New and future immunomodulatory therapy in type 1 diabetes. Trends in molecular medicine 18, 173–181, doi: 10.1016/j.molmed.2012.01.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannesson B, Sui L, Freytes DO, Creusot RJ & Egli D Toward beta cell replacement for diabetes. The EMBO journal 34, 841–855, doi: 10.15252/embj.201490685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michael MD et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Molecular cell 6, 87–97 (2000). [PubMed] [Google Scholar]
- 6.Chen YG, Mathews CE & Driver JP The Role of NOD Mice in Type 1 Diabetes Research: Lessons from the Past and Recommendations for the Future. Frontiers in endocrinology 9, 51, doi: 10.3389/fendo.2018.00051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchwald P et al. Comprehensive Metabolomics Study To Assess Longitudinal Biochemical Changes and Potential Early Biomarkers in Nonobese Diabetic Mice That Progress to Diabetes. Journal of proteome research 16, 3873–3890, doi: 10.1021/acs.jproteome.7b00512 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Wilcox NS, Rui J, Hebrok M & Herold KC Life and death of beta cells in Type 1 diabetes: A comprehensive review. Journal of autoimmunity 71, 51–58, doi: 10.1016/j.jaut.2016.02.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Ouaamari A et al. SerpinB1 Promotes Pancreatic beta Cell Proliferation. Cell metabolism 23, 194–205, doi: 10.1016/j.cmet.2015.12.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stadinski BD et al. Chromogranin A is an autoantigen in type 1 diabetes. Nature immunology 11, 225–231, doi: 10.1038/ni.1844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delong T et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714, doi: 10.1126/science.aad2791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster C et al. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity 42, 942–952, doi: 10.1016/j.immuni.2015.04.011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhal G et al. Fibroblast Growth Factor 21 (FGF21) Protects against High Fat Diet Induced Inflammation and Islet Hyperplasia in Pancreas. PloS one 11, e0148252, doi: 10.1371/journal.pone.0148252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun MY et al. Autofluorescence imaging of living pancreatic islets reveals fibroblast growth factor-21 (FGF21)-induced metabolism. Biophysical journal 103, 2379–2388, doi: 10.1016/j.bpj.2012.10.028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wicker LS et al. Beta 2-microglobulin-deficient NOD mice do not develop insulitis or diabetes. Diabetes 43, 500–504 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Kay TW, Parker JL, Stephens LA, Thomas HE & Allison J RIP-beta 2-microglobulin transgene expression restores insulitis, but not diabetes, in beta 2-microglobulin null nonobese diabetic mice. Journal of immunology 157, 3688–3693 (1996). [PubMed] [Google Scholar]
- 17.Tang Q & Bluestone JA The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nature immunology 9, 239–244, doi: 10.1038/ni1572 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Bandala-Sanchez E & Harrison LC Revisiting regulatory T cells in type 1 diabetes. Current opinion in endocrinology, diabetes, and obesity 19, 271–278, doi: 10.1097/MED.0b013e328355a2d5 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Tang J et al. IL-25 promotes the function of CD4+CD25+ T regulatory cells and prolongs skin-graft survival in murine models. International immunopharmacology 28, 931–937, doi: 10.1016/j.intimp.2015.03.036 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Shultz LD et al. Humanized NOD/LtSz-scid IL2 receptor common gamma chain knockout mice in diabetes research. Annals of the New York Academy of Sciences 1103, 77–89, doi: 10.1196/annals.1394.002 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Tsai S, Shameli A & Santamaria P CD8+ T cells in type 1 diabetes. Advances in immunology 100, 79–124, doi: 10.1016/S0065-2776(08)00804-3 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Dirice E et al. Soluble factors secreted by T cells promote beta-cell proliferation. Diabetes 63, 188–202, doi: 10.2337/db13-0204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKarns SC & Schwartz RH Distinct effects of TGF-beta 1 on CD4+ and CD8+ T cell survival, division, and IL-2 production: a role for T cell intrinsic Smad3. Journal of immunology 174, 2071–2083 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Delisle JS et al. The TGF-beta-Smad3 pathway inhibits CD28-dependent cell growth and proliferation of CD4 T cells. Genes and immunity 14, 115–126, doi: 10.1038/gene.2012.63 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Park BV et al. TGFbeta1-Mediated SMAD3 Enhances PD-1 Expression on Antigen-Specific T Cells in Cancer. Cancer discovery 6, 1366–1381, doi: 10.1158/2159-8290.CD-15-1347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H et al. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harbor perspectives in biology 2, a002279, doi: 10.1101/cshperspect.a002279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara T et al. Biglycan Intensifies ALK5-Smad2/3 Signaling by TGF-beta1 and Downregulates Syndecan-4 in Cultured Vascular Endothelial Cells. Journal of cellular biochemistry 118, 1087–1096, doi: 10.1002/jcb.25721 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzachanis D et al. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nature immunology 2, 1174–1182, doi: 10.1038/ni730 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Tzachanis D & Boussiotis VA Tob, a member of the APRO family, regulates immunological quiescence and tumor suppression. Cell Cycle 8, 1019–1025, doi: 10.4161/cc.8.7.8033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adurthi S et al. Oestrogen Receptor-alpha binds the FOXP3 promoter and modulates regulatory T-cell function in human cervical cancer. Scientific reports 7, 17289, doi: 10.1038/s41598-017-17102-w (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polanczyk MJ et al. Cutting edge: estrogen drives expansion of the CD4+CD25+ regulatory T cell compartment. Journal of immunology 173, 2227–2230 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Skyler JS Hope vs hype: where are we in type 1 diabetes? Diabetologia 61, 509–516, doi: 10.1007/s00125-017-4530-x (2018). [DOI] [PubMed] [Google Scholar]
- 33.Thorn LM et al. Effect of parental type 2 diabetes on offspring with type 1 diabetes. Diabetes care 32, 63–68, doi: 10.2337/dc08-0472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zalloua PA et al. Type-2 diabetes family history delays the onset of type-1 diabetes. The Journal of clinical endocrinology and metabolism 87, 3192–3196, doi: 10.1210/jcem.87.7.8649 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Warram JH, Krolewski AS, Gottlieb MS & Kahn CR Differences in risk of insulin-dependent diabetes in offspring of diabetic mothers and diabetic fathers. The New England journal of medicine 311, 149–152, doi: 10.1056/NEJM198407193110304 (1984). [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni RN et al. Altered function of insulin receptor substrate-1-deficient mouse islets and cultured beta-cell lines. The Journal of clinical investigation 104, R69–75, doi: 10.1172/JCI8339 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dirice E et al. Inhibition of DYRK1A Stimulates Human beta-Cell Proliferation. Diabetes 65, 1660–1671, doi: 10.2337/db15-1127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flodstrom-Tullberg M et al. Target cell expression of suppressor of cytokine signaling-1 prevents diabetes in the NOD mouse. Diabetes 52, 2696–2700 (2003). [DOI] [PubMed] [Google Scholar]
- 39.King AJ et al. Normal relationship of beta- and non-beta-cells not needed for successful islet transplantation. Diabetes 56, 2312–2318, doi: 10.2337/db07-0191 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Huang EL et al. SNaPP: Simplified Nanoproteomics Platform for Reproducible Global Proteomic Analysis of Nanogram Protein Quantities. Endocrinology 157, 1307–1314, doi: 10.1210/en.2015-1821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clair G et al. Spatially-Resolved Proteomics: Rapid Quantitative Analysis of Laser Capture Microdissected Alveolar Tissue Samples. Scientific reports 6, 39223, doi: 10.1038/srep39223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyanova S, Temu T & Cox J The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature protocols 11, 2301–2319, doi: 10.1038/nprot.2016.136 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Delong T et al. Islet amyloid polypeptide is a target antigen for diabetogenic CD4+ T cells. Diabetes 60, 2325–2330, doi: 10.2337/db11-0288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pauken KE et al. Cutting edge: type 1 diabetes occurs despite robust anergy among endogenous insulin-specific CD4 T cells in NOD mice. Journal of immunology 191, 4913–4917, doi: 10.4049/jimmunol.1301927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pauken KE et al. Cutting edge: identification of autoreactive CD4+ and CD8+ T cell subsets resistant to PD-1 pathway blockade. Journal of immunology 194, 3551–3555, doi: 10.4049/jimmunol.1402262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi W, Oshlack A & Smyth GK Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic acids research 38, e204, doi: 10.1093/nar/gkq871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie ME et al. Empirical array quality weights in the analysis of microarray data. BMC bioinformatics 7, 261, doi: 10.1186/1471-2105-7-261 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentleman RC et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology 5, R80, doi: 10.1186/gb-2004-5-10-r80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herwig R, Hardt C, Lienhard M & Kamburov A Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nature protocols 11, 1889–1907, doi: 10.1038/nprot.2016.117 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomics data have been deposited on the National Center for Biotechnology Information (NCBI) gene expression omnibus (GEO) under accession code GSE128315 for murine microarray analysis. Mass spectrometry proteomics data reported in this paper have been deposited in ProteomeXchange (accession code: PXD013100) and MassIVE (accession code: (MSV000083576). Further information on statistical parameter, software, study design and so forth is in the Nature Reporting Summary. The data that supports the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request. Correspondence and requests for material should be addressed to R.N.K.