Abstract
Epigenetic enzymes regulate higher-order chromatin architecture and cell-type specific gene expression. The ATPase BRG1 and the SWI/SNF chromatin remodeling complex are epigenetic enzymes that regulate chromatin accessibility during steady and transitional cell states. Experiments in mice show that the loss of BRG1 inhibits cellular reprogramming, while studies using human cells demonstrate that the overexpression of BRG1 enhances reprogramming. We hypothesized that the variation of SWI/SNF subunit expression in the human population would contribute to variability in the efficiency of induced pluripotent stem cells (iPSC) reprogramming. To examine the impact of an individuals, sex, ancestry, and age on iPSC reprogramming, we created a novel sex and ancestry balanced cohort of 240 iPSC lines derived from human dermal fibroblasts (DF) from 80 heathy donors. We methodically assessed the reprogramming efficiency of each DF line and then quantified the individual and demographic-specific variations in SWI/SNF chromatin remodeling proteins and mRNA expression. We identified that BRG1, BAF155, and BAF60a expression as strongly correlating with iPSC reprogramming efficiency. Additionally, we discovered that high efficiency iPSC reprograming is negatively correlated with donor age, positively correlated with African American descent, and uncorrelated with donor sex. These results show the variations in chromatin remodeling protein expression have a strong impact on iPSC reprogramming. Additionally, our cohort is unique in its large size, diversity, and focus on healthy donors. Consequently, this cohort can be a vital tool for researchers seeking to validate observational results from human population studies and perform detailed mechanistic studies in a controlled cell culture environment.
MeSH Terms: Human, Chromatin, Chromatin Assembly and Disassembly, Cellular Reprogramming, Induced Pluripotent Stem Cells, Demography, Cohort Studies
Keywords: Epigenetics, Reprogramming, BRG1, iPSC, Chromatin remodeling, Induced pluripotent stem cells, Chromatin insulator
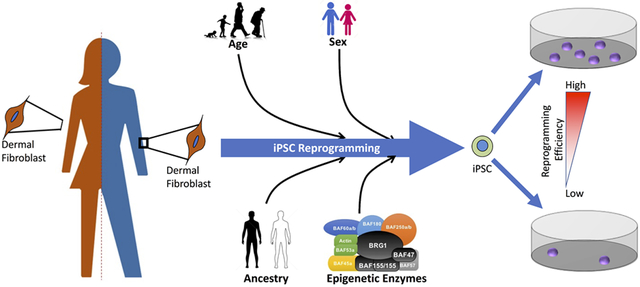
Grahical Abstract
We examined if human age, sex, ancestry, and/or expression of the SWI/SNF family of epigenetic enzyme influenced the ability to efficiently generate induced pluripotent stem cells (iPSC). Using a large sex and ancestry matched cohort of human dermal fibroblasts and iPSCs, we discovered that components of the SWI/SNF complex, donor age, and donor ancestry all correlated with proficient iPSC generation.
Introduction:
Epigenetic enzymes play a critical role in establishing higher-order chromatin architecture, modifying local nucleosome positioning, and finely regulating gene transcription in mammalian cells [1-3]. These enzymes identify patterns of post-translational modifications on histone tails (acetylation, phosphorylation, methylation, etc.) and DNA while initiating a complex series of events involving binding of DNA, recruiting cofactors, sliding and/or ejecting of nucleosomes resulting in the compaction of DNA to inhibit gene accessibility, or the opening of chromatin regions to facilitate rapid gene transcription [4]. Of the several families of chromatin remodeling epigenetic enzymes, the SWI/SNF family plays a critical role regulating ATP-dependent chromatin remodeling in response to hormone stimulation and during cellular reprograming [5]. SWI/SNF complexes can utilize either BRG1 or BRM as the catalytic ATPase [6]. The key to the specificity exhibited by the SWI/SNF complex is its combinatorial assembly with various BRG1 -associated factors (BAFs). The canonical SWI/SNF complex consists of BRG1, BAF170, BAF155, BAF250a /b, BAF45a/b, BAF60, BAF47, BAF57, and BAF53a /b [7]. Differences in the composition of the complex are seen in different cell types. For example, in embryonic stem cells, the most common SWI/SNF complex (esBAF) utilizes BRG1 and preferentially utilizes BAF180, Baf45b, Baf200 / BAF250a in lieu of BAF170, BAF45a, BAF250b respectively [8, 9]. While the mechanisms regulating the global changes in BAF subunit usage are still poorly defined, microRNAs have been shown to involved [3, 10, 11]. For instance, miR-302 has been shown to directly suppress BAF170 expression in human ES cells, and disruption of miR-302 dependent BAF170 suppression was shown to impair ES cell proliferation, block endodermal differentiation, and promote ectodermal differentiation [12].
Mouse models have shown that loss of SWI/SNF family members can have severe consequences specifically, deletion of BRG1, BAF180, BAF47, or BAF155 causes embryonic lethality [9, 13-18]. By contrast, global knockout of BRM is tolerated with little effect on the mouse [19]. While global knockout of BAF180 in mice is embryonic lethal, targeted deletion in mouse cell lines shows BAF180s role in stabilizing mitotic chromosomes as well as lymphocyte development [14]. Deletion of BRG1 or BAF250a in mouse ES cells resulted in a loss of self-renewal and defects in differentiation. Deletion of BAF155 and BAF170 resulted in several defects during neural differentiation and brain development [13]. While the canonical SWI/SNF complexes have been well characterized, the role and composition of chromatin remodeling complexes during transitional cell states are less clear [20].
During cellular reprogramming, terminally differentiated somatic cells (dermal fibroblast, urinary epithelial cell, circulating leukocytes, etc.) are induced into a pluripotent cell state by the transient activation of pluripotency-associated factors [21, 22]. The traditional method of producing these induced pluripotent stem cells (iPSC) is by inducing the expression of OCT4, SOX2, KLF4, and MYC (OSKM) [23]. The transient expression of OSKM proteins primes enhancer regions for pluripotency associated genes while cMyc associates with gene promoters to help activate endogenous expression of OCT4, SOX2, KLF4, and NANOG, thereby establishing long term pluripotency [24-28]. Several studies have shown that the method of introducing OSKM transgenes may slightly affect the efficiency of reprogramming without altering iPSC cell physiology, pluripotency, differentiation potential, or gene expression [29-34]. The activation of additional factors such as LIN28 and NANOG has been shown to greatly enhance reprogramming efficiency [34].
Murine reprogramming models provide evidence of an association between the age and genetic strain of the animal and the efficiency of iPSC generation during reprogramming. Tissues from young mice are more efficient at generating high quality iPSCs with fewer mutations or abnormalities then iPSC lines derived from older mice [33, 35]. Interestingly, studies using human somatic cells have not found a compelling link between donor age and iPSC reprogramming efficiency [36, 37]. This is evidenced by the ability to efficiently generate iPSC lines from fibroblasts of both neonates and donors over 100 years of age [38]. While many studies have utilized human iPSC reprogramming and observed no correlation between donor age and reprogramming efficiency, the majority of these reports have been performed with small sample sizes, different parental somatic cells, and samples from diverse genetic backgrounds [39-41]. These limitations decrease the statistical ability to detect correlations between donor demographics (age, ancestry, sex, etc.) and the efficiency of iPSC reprogramming and/or iPSC physiology. Overcoming these barriers will dramatically improve pharmaceutical development, personalized medicine, and regenerative therapies [26, 42, 43].
To elucidate the effect of donor demographics on iPSC physiology and reprogramming efficiency, we created a large novel cohort of human dermal fibroblasts and iPSCs from healthy human donors. We hypothesized that differences in reprogramming efficiency between demographic groups might be linked to steady state SWI/SNF subunit expression. To test our hypothesis, we analyzed the gene and protein expression of SWI/SNF components and pluripotency-associated genes in both dermal fibroblasts and iPSCs from our cohort of 80 healthy sex- and ancestry-matched donors. From our analysis, we determined that iPSC reprogramming efficiency is significantly enhanced in donors of African American (AA) descent compared to those of European American (EA) descent and that reprogramming efficiency is inversely correlated with donor age. Additionally, we identified several key SWI/SNF components whose expression levels were correlated with donor age, sex, ancestry, and reprogramming efficiency.
Material and Methods:
Cell lines, Cell culture and Reprogramming Experiments
Primary dermal fibroblast lines were derived from skin biopsies obtained from adult individuals at the NIEHS under institutional review board approved protocol human subjects 10-E-0063, “Sample Collection Registry for Quality Control of Biological and Environmental Specimens and Assay Development and Testing protocol” (ClinicalTrials.gov #NCT01087307). All participants gave written informed consent for tissue donation. Donor sex, age, and ancestry were voluntarily self-reported. Donor samples were excluded if sex-specific gene expression markers contradicted donor self-identified sex (n=2). Samples from several donors were excluded due to failure to yield high quality DF lines. 4mm in diameter skin punch biopsies were cut into small pieces and allowed to dry on a tissue culture dish for 5 minutes to form a physical attachment before adding DMEM (Gibco 11965–092) containing 10% FBS (BenchMark Gemini Bio-Products) and 1% Pen/Strep (Sigma). Media was changed daily for several weeks until a dermal layer of fibroblasts grew out and could be frozen. Most of the fibroblast lines were used for reprogramming between passage 1 and 5, with a small number of exceptions, but all were under passage 10. Human embryonic stem cells (hES), H1 and H9 (WiCell), were used as controls in the teratoma tumor assay.
Fibroblast cells were reprogrammed via lentiviral transduction using six transcription factors contained in three plasmids (ADDGENE/PSIN4-EF2-N2L, ADDGENE/PSIN4-EF2-O2S and ADDGENE/PSIN4-CMV-K2M) [34]. Viral supernatants at an approximate titer of 107 were added in combination with complete DMEM media and 8ug/ml polybrene to fibroblasts plated the day before in 6-well plates at 2.5 × 105 cells per well. Virus was removed after 12 hours and cells were refreshed with DMEM complete media for 48 hours. Cells were then harvested and transferred to 10cm dishes coated with Matrigel (Corning cat # 354234 diluted in DMEM/F-12, Gibco). The next day cells were fed with a 1:1 mixture of complete DMEM and E8 media (TeSR™-E8™ Stem Cell Technologies). Culture dishes were subsequently maintained in only E8 on a daily basis for 21 days until colonies appeared. The iPSC colonies were picked using a 20µl pipet and grown up individually in E8 media using ReLeSR (Stem Cell Technologies) for harvesting and splitting. All iPSC and hES cells were frozen in mFreSR (Stem Cell Technologies).
Reprogramming efficiency was determined by alkaline phosphatase staining (Stemgent AP Staining Kit II) of triplicate 10cm reprogrammed dishes containing colonies, which were then scanned. The saved images were analyzed with ImageJ 1.51h (Wayne Rasband, National Institutes of Health, USA) to count colonies, using color threshold adjusted and binary converted images of each dish. Triplicate plates were averaged and reported as colony counts or percent reprogramming efficiency ((# of colonies/250,000) x100).
Analysis of mRNA Gene Expression
RNA was isolated from frozen cell pellets using RNeasy Mini Kits (Qiagen) combined with QIAshredder Mini Spin Columns (Qiagen) and a 15 minutes on column treatment of RNase-Free DNase Set (Qiagen). After another 15-minute treatment at room temperature with DNase1 (Invitrogen), RNA was then converted to cDNA using SuperScript First Strand kits (Invitrogen) with SuperScript II (10,000 units). qPCR reactions were set up using either Brilliant II or Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies).
NanoString nCounter gene expression assay
Total RNA was isolated from 4 independent lines per donor, 3 iPSC sister clones and the parental dermal fibroblasts for total of 320 RNA samples. Utilizing a custom codeset the Nanostring nCounter gene expression system was used to assess the expression of 75 genes associated with pluripotency, ectoderm, mesoderm, and endoderm lineage commitment markers along with 6 housekeeping genes. RNA quality was examined by Advanced Analytics Fragment Analyzer and was loaded at 50 ng per sample. NanoString nCounter data were imported into R (version 3.4.3), and the log2 counts were normalized using the geometric mean of housekeeper genes: CLTC, GAPDH, GUSB, HPRT1, PGK1, and TUBB. Sister clone expression values were collapsed to single values using the mean of log2 counts. Genes were divided into pre-defined classes, and each class was hierarchically clustered using Euclidean distance and Ward linkage prior to visualization in a heatmap or box-whiskers plot.
Analysis of Protein Expression
Whole lysate protein was isolated from frozen cell pellets using Buffer X (100 mM Tris-HCL pH 8.5, 250 mM NaCl, 1% (v/v) NP-40, 1 mM EDTA) plus the addition of fresh protease inhibitors (cocktail mix, Sigma) and Phosphatase Inhibitor (PMSF). Protein concentrations were determined using the colorimetric Bradford Protein Analysis method with BioRad reagents. Protein Electrophoresis and Western blotting were performed on precast 18-well 7.5% Tris-HCL gels (Criterion cat# 345–0006) using 30μg of whole lysate protein with 2X SDS loading buffer. The transfer of proteins to nitrocellulose membranes was performed in Tris/Glycine buffer containing 20% methanol. Blotting of membranes was performed in 5% nonfat milk for one hour before overnight incubations with primary antibody(s). The antibodies used were as follows: BRG1 H88 (Santa Cruz sc-10768), BAF155 H-76 (Santa Cruz sc-10756), BAF170 H-116 (Santa Cruz sc-10757), BAF60a (Becton Dickson cat# 611728), β-Actin AC-15 (Sigma cat# A1978) and GAPDH FL-335 (Santa Cruz sc-25778). After incubation with appropriate secondary antibody, blots were quantified using digital fluorescence with LI-COR imaging reagents and the Odyssey™ CLx Imaging System.
Teratoma Tumor Assay and Histology
Human embryonic stem cell (hES) lines H1 and H9 and iPSC lines were first tested for mycoplasma contamination prior to use in our teratoma assay. Cells were grown on Matrigel, harvested using ReLeSR and collected in E8 media, spun down, resuspended in 1:1 E8 media and Matrigel (Matrigel Matrix hESC-qualified Corning cat# 354277) and then injected subcutaneously into Beige-SCID mice at 4 × 106 cells in 0.1ml total volume per injection site. Injections were done under sedation with isoflurane. Mice were monitored biweekly until tumors appeared, and tumors were removed when their size reached ≈1 cm3. After the mice were euthanized, the tumors were removed, weighed and placed into formalin for approximately 48 hours before being embedded into paraffin blocks for sectioning. Slides were prepared and stained with Hematoxylin and Eosin (H&E) and then examined for the presence of all three primal germ layers.
Statistical Analysis
Power analyses based on preliminary data indicated that n=30 subjects per demographic group were needed to achieve ≥ 90% power to detect reprogramming efficiency differences between demographic groups within the study at a significance level of 0.05. Reprogramming efficiency and qPCR expression of 14 genes measured in fibroblasts and measured in iPSCs were compared between men and women and between African-Americans and European-Americans using Mann-Whitney tests. In addition, Mann-Whitney tests were used to compare men and women within each ethnicity and to compare ethnic groups within each sex. Age was dichotomized at below 33 years old or above 33 years old, and both reprogramming efficiency and qPCR gene expression were compared between the two age groups using Mann-Whitney tests. Correlations between reprogramming efficiency and age and between reprogramming efficiency and qPCR gene expression were tested using Spearman’s rank order correlation coefficients. These correlations were also determined within each demographic group. P-values are two-sided and considered statistically significant if less than 0.05.
RESULTS
The Demographics of the Dermal Fibroblast Cohort
Healthy individuals were recruited through the Clinical Research Unit at NIEHS, where dermal biopsies were collected and primary dermal fibroblast lines were established in culture. The demographics, age, sex, and ancestry, for the cohort are listed in Table 1. The study enlisted 20 African American (AA) women, 19 European American (EA) women, 21 AA men and 20 EA men. Of the 80 donor individuals, age ranged from 21 to 64 years of age (mean of 34.9). Thirty-nine of the individuals were women (mean age of 32.2 years) and forty-one were male (mean age of 37.5 years). Forty-one of the individuals in the cohort identified their ancestry as AA (mean age of 35.1 years) and 39 as EA (mean age of 34.6 years). There was a natural break in the ages of the cohort so we divided donors into two groups, those ≤33 years of age (n=41) and those >33 years of age (n=39). To our knowledge, none of the individuals were consanguineous. Sample IDs for dermal fibroblasts (DF) were generated by including the donor ancestry as the first letter (A or E) and sex as the second letter (M or F) and numbered in the order the sample was received (Supplemental Table 1). iPSC lines are denoted by an “i” before the sample ID and the clone number is shown as decimal following the sample number. As examples, EF1 is an EA women, and iEF1.1 is the clone 1 iPSC line derived from EF1.
Table 1.
Summary of Donor Demographics
| Archer Cohort | N | Percent of Cohort |
Mean Age (Years) |
Mean Reprogramming Colony Counts |
|
|---|---|---|---|---|---|
| Donors | 80 | 100 | 34 ± 1.2 | 1416 ± 91 | |
| Sex | Men | 41 | 51.2 | 37 ± 1.6 | 1387 ± 131 |
| Women | 39 | 48.8 | 32 ± 1.6 | 1446 ± 128 | |
| Ancestry | African American | 42 | 52.5 | 35 ± 1.4 | 1634 ± 125 |
| European American | 38 | 47.5 | 35 ± 1.9 | 1175 ± 123 | |
| Age | Over 33 years old | 39 | 48.8 | 44 ± 1.0 | 1221 ± 122 |
| Under 33 years old | 41 | 51.2 | 26 ± 0.6 | 1601 ± 128 |
All the primary fibroblast lines were used at a low passage number (<10) to generate iPSC colonies and all fibroblast lines produced multiple pluripotent iPSC colonies. A minimum of three (3) iPSC lines were isolated, expanded, and characterized for each donor. To determine reprogramming efficiency each DF line was reprogrammed in triplicate. At the end of the experiment the plates were subjected to an alkaline phosphatase stain, scanned, and the images analyzed using ImageJ. Despite reprograming being considered a stochastic process, the number of colonies produced by each DF line was remarkably consistent with less than 10% variation between replicates of most lines. The average number of colonies produced by each line ranged from 47 to 3429 colonies (0.01% - 1.37% efficiency) with a cohort mean of 1416 colonies (Table 1).
Validating iPSC Pluripotency
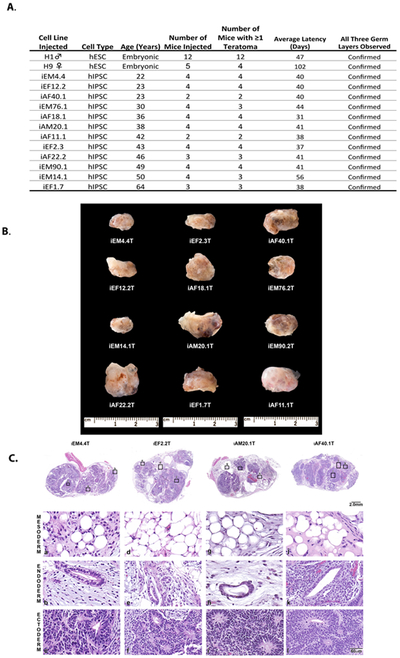
After isolation, all iPSC lines displayed traditional pluripotent cell morphology, growth rates, and alkaline phosphatase staining. The current gold standard assay to demonstrate true pluripotency is the teratoma assay. This assay demonstrates that a single cell has the potential ability to form all differentiated cell types from the 3 primary germ layers: mesoderm, endoderm and ectoderm. Due to the time, expense, and vast number of mice required, it was not feasible to generate teratomas for each iPSC line in our cohort. Therefore, we utilized a hybrid approach to verify pluripotency; twelve iPSC lines from different donors were picked at random and tested for their ability to form teratomas. Pluripotency-associated gene expression levels were assessed in all iPSC lines and compared to the expression levels of the H1 and H9 hES lines as well as iPSC lines confirmed pluripotent by teratoma formation. Figure 1A, shows that all 12 iPSC lines efficiently formed teratomas. Additionally, the average latency to iPSC teratoma formation (range 31–56 days) was similar to that for H1 hES teratomas (47 days) and significantly less then H9 hES teratomas (102 days). After tumors reached ≈1cm in size, they were removed (Figure 1B), fixed in formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E). Slides were scored for the presence of all three germ layers by a certified histopathologist. All teratomas had evidence of all three germ layers. Figure 1C, shows representative examples of the histologic findings confirming the presence of endoderm, mesoderm, and ectoderm-derived tissues. Gene expression analysis by qPCR of pluripotency-associated genes in iPSC lines showed a clear induction in pluripotency-associated (OCT4, SOX2, LIN28, PRI-miR302, and MYC) gene expression and a suppression of fibroblast markers such as Vimentin (Supplemental Figure 3). As a further check on the quality of our reprograming protocol we subsequently examined the cohort by Nanostring analysis for the expression levels of roughly 70 genes, comprising of markers of pluripotency and differentiation, (Supplemental Figure 4). The iPSC clones appeared to have equivalent expression levels of pluripotency genes as the hES clones H1 and H9 that clearly differed from their dermal fibroblast parental lines. Equally important the differentiation markers, Mesoderm, Mesendoderm, Endoderm and Ectoderm were all similar between the iPSCs and the ES cells but distinct form the dermal fibroblasts.
Figure 1. Gross and Histologic Confirmation of iPS Pluripotency.
A) Efficiency of hESC and iPSC teratomas formation. B) Gross Representative images of teratomas formed by injecting iPSC lines into SCID female mice. C) Hematoxylin and Eosin Stained sections of FFPE teratomas. Presence of all three germ layers were confirmed by a certified histopathologist.
Donor Demographics Influence SWI/SNF Subunit Variability
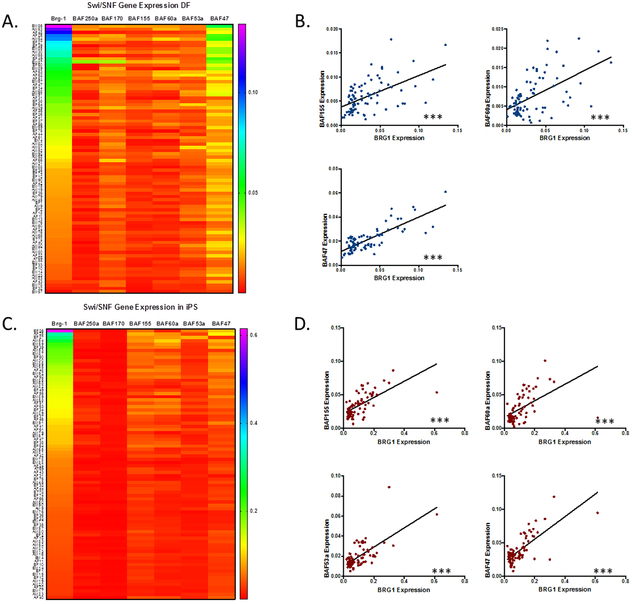
Having validated that the iPSC lines in our cohort are pluripotent, we next sought to assess the variability of SWI/SNF expression between donors and how it relates to reprogramming efficiency. The gene expression levels of BRG1, BAF250a, BAF170, BAF155, BAF60a, BAF53a and BAF47 were assessed by qPCR in both DF and iPSC. All gene targets were expressed at appreciable levels in both cell types. As shown in Figure 2, gene expression levels of SWI/SNF subunits was highly variable between individuals. BRG1 expression levels were the most variable compared to other SWI/SNF subunits tested. As shown in Figure 2B, BRG1 and BAF47 were expressed at the highest levels and exhibited the most variability in DF. Additionally, DF BRG1 expression strongly correlates with the expression of BAF155, BAF60a, and BAF47. In contrast, BAF170 and BAF53a have a distinct expression pattern with moderate gene expression and variability. The heterogeneity of SWI/SNF gene expression levels in the iPSCs was dramatically reduced following reprogramming (Figure 2C). Similarly to the DFs, BRG1 and BAF47 were the most highly expressed and heterogeneous SWI/SNF components in iPSCs. BAF155 and BAF60a were expressed at intermediate levels with moderate heterogeneity. While, BAF250a, BAF170, and BAF53a were expressed at low levels with relatively little heterogeneity. BRG1 expression was correlated with the expression patterns of BAF155, BAF60a, BAF53a, and BAF47 in iPSCs (Figure 2D).
Figure 2. SWI/SNF Expression is Highly Variable Between Donors.
Heatmaps showing individual GAPDH-normalized gene expression levels of SWI/SNF subunits in DF (A) and iPS cells (C). In DF, BRG1 expression levels have a strong positive correlation with BAF155, BAF60a, and BAF47 expression (B). In iPS cells, BAF155, BAF60a, BAF53a, and BAF47 expression is positively correlated with BRG1 expression (D).
*=p≤0.05, **=p≤0.01, ***=p≤0.001
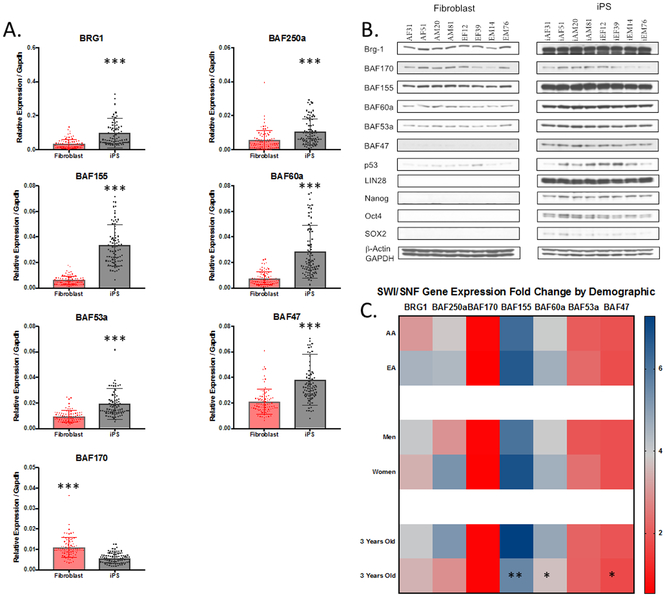
The SWI/SNF complex of epigenetic enzymes is known to be involved in cell state transitions, and subunit switching can alter chromatin architecture and contribute to cell-type specific gene expression. Comparing the gene expression values of SWI/SNF subunits between DFs and iPSCs, it becomes clear that there are several key reprogramming-dependent changes that occur during reprogramming (Figure 3A). iPSC reprogramming causes a 2.9-fold increase in BRG1, a 2-fold increase in BAF250a, a 5-fold increase in BAF155, a 3.5-fold increase in BAF60a, a 2-fold increase in BAF53a, a 1.8-fold increase in BAF47, and a 1.9-fold decrease in BAF170 iPSC mRNA expression compared to DFs (all p≤0.001). These changes highlight the dynamic nature of the SWI/SNF complex during iPSC reprogramming. To confirm that these mRNA expression changes had a functional impact on the protein levels, western immunoblots were performed on whole cell extracts from DF and iPSC lines. As shown in Figure 3B, for a representative sample of the DFs and iPSCs, the protein expression supported the gene expression data. While there is clearly some heterogeneity within the DF and iPSC sample groups, the reprogramming-dependent changes in SWI/SNF expression clearly overshadow individual variability. The increases in protein expression of BRG1, BAF155, BAF60a, BAF53a, and BAF47 are highly similar to the changes observed at the mRNA level. Also, consistent with the gene expression results, the protein expression of BAF170 is reduced following reprogramming. It is interesting that while BAF47 is detected in DF by qPCR, it is nearly undetectable by western blot, making for a stark contrast between DF and iPSC BAF47 protein expression.
Figure 3. Reprogramming-Dependent Changes in SWI/SNF Subunit Expression Dependent on Donor Age.
Reprogramming causes an elevation in gene (A) and protein (B) expression of all SWI/SNF subunits tested with the exception of BAF170. C. Heatmap showing the fold change in SWI/SNF gene expression between DF and iPS by donor demographics. Donor Ancestry and Sex had no impact on the gene expression fold change between DF and iPS (C). Donors over 33 years old had significantly reduced expression of BAF155, BAF60a,and BAF47. Error bars represent the standard deviation of the mean
*=p≤0.05,**=p≤0.01,***=p≤0.001
Having established the reprograming-dependent changes in SWI/SNF subunit expression, we next wanted to determine if these changes were correlated with donor ancestry, sex, and/or age. We used Mann-Whitney statistical tests to assess demographic specific changes in SWI/SNF gene expression and determined that ancestry and sex did not correlate with reprogramming-dependent changes. Age however, had a significant impact on BAF155, BAF60a, and BAF47 expression (Figure 3C). Donors over the age of 33 had significantly lower expression BAF155 (p=0.008), BAF60a (p=0.02), and BAF47 (p=0.04) than younger donors.
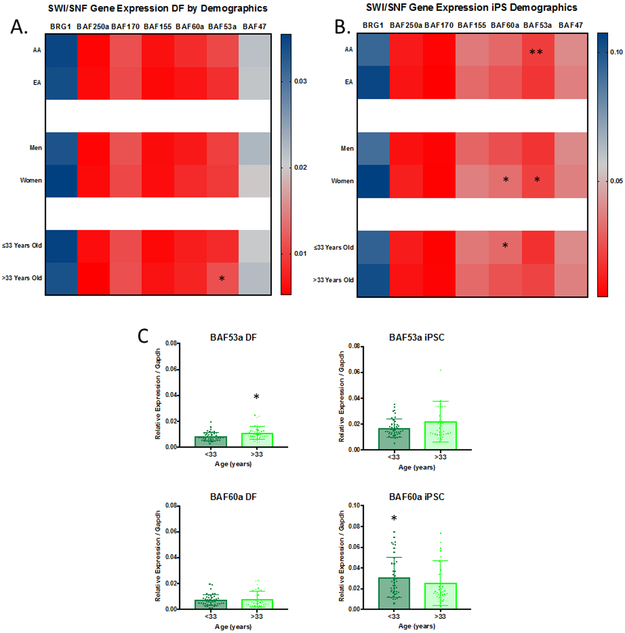
We next sought to determine if donor demographics influenced SWI/SNF expression level. As shown in Figure 4A, ancestry and sex had no significant effect on SWI/SNF expression levels. Donor age correlated with BAF53a expression, where DF from donors over 33 years of age had higher expressions levels of BAF53a (0.01 vs 0.008, p=0.002) compared to younger donors. In iPSCs, several subunits that correlated with donor demographics (Figure 4B); donors of AA descent had higher expression of BAF53a (0.02 vs 0.01, p=0.005) compared to EA samples, women had higher expression of BAF155 (0.04 vs 0.03, p=0.04) and BAF60a (0.03 vs 0.02, p=0.02) then men. While donors under 33 years of age had higher levels of BAF60a (0.03 vs 0.02, p=0.04) then samples from older donors (Figure 4C). Taken together these results show that donor age, sex, and ancestry can have a significant impact on SWI/SNF subunit expression.
Figure 4. SWI/SNF Subunit Gene Expression Profiles Differ Between Demographic Groups.
Heatmaps show the comparison of GAPDH-normalized gene expression for SWI/SNF subunits between demographic groups in DFs (A) and iPS (B). AA iPS had higher expression of BAF53a compared to EA iPS lines. Females have elevated expression of BAF60a and BAF53a compared to Males. Donor expression of BAF60a and BAF53a is dependent on age (C). Error shown as standard deviation of the mean.
*=p≤0.05,**=p≤0.01,***=p≤0.001
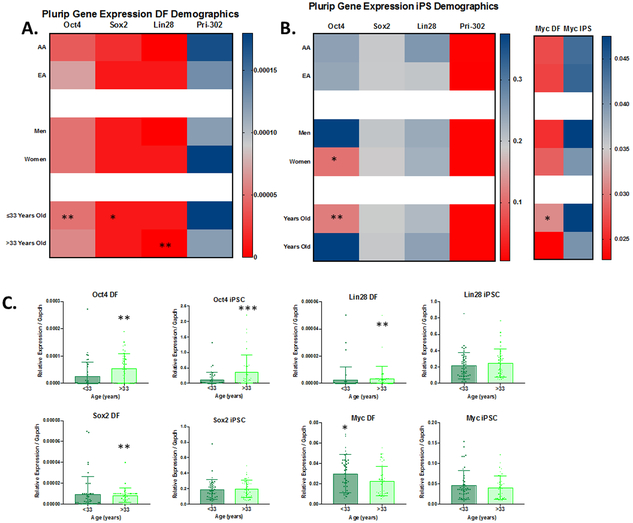
DF and iPSC Pluripotent-Associated Gene Expression Impacted by Age and Sex
Pluripotency associated genes such as OCT4 have been shown to help induce and orchestrate the pluripotent state, however their role in terminally differentiated cells is thought to be minimal. Despite this, we observed that in DF we could consistently detect low levels of expression of the pluripotency-associated genes OCT4, SOX2, LIN28, and Pri-302 (Figure 5A). While donor ancestry and sex did not associate with pluripotent gene expression in DF, donor age was correlated with OCT4, SOX2, LIN28 and MYC. Samples from individuals over 33 years of age had higher levels of OCT4 (p=0.002) and SOX2 (p=0.04) with significantly lower levels of LIN28 (p≤0.003) and MYC (p=0.04) compared to younger donors. We next assessed the iPSC expression of pluripotency associated genes (Figure 5B). We observed that OCT4 expression was elevated in men compared to women (0.372 vs 0.107, p=0.02) and donors over 33 years of age compared to those under 33 years of age (0.38 vs 0.11, p=0.001) (Figure 5C). While the function of pluripotent gene expression in DF needs further study, these findings make intriguing implications about a role for OCT4, SOX2, and LIN28 in priming or opposing iPSC preprogramming.
Figure 5. Pluripotency-Associated Gene Expression is Strongly Influenced by Donor Age.
Heatmaps show the comparison of GAPDH-normalized gene expression for pluripotency factors between demographic groups in DFs (A) and iPS (B). Females had lower expression of Oct4 compared to Males. Donor expression of Oct4, Sox2, Lin28 and Myc is dependent on age (C). Error shown and standard deviation of the mean.
*=p≤0.05,**=p≤0.01,***=p≤0.001
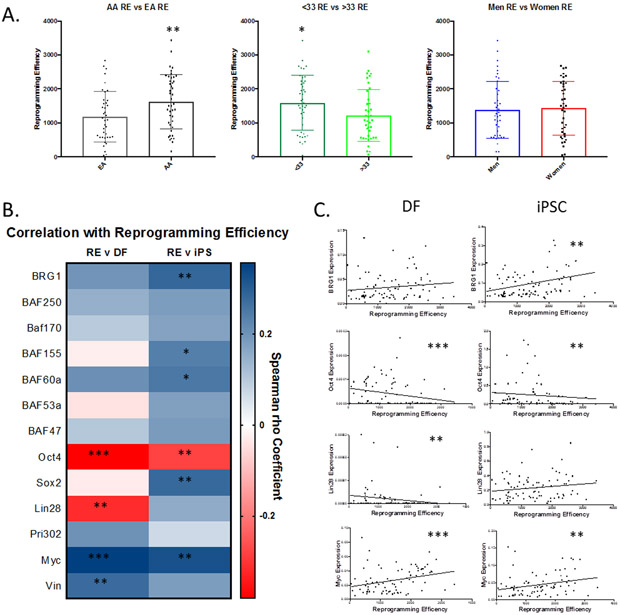
iPSC Reprogramming Efficiency is Correlated with Ancestry and Age
Consistent with our hypothesis, SWI/SNF expression correlates with donor demographics. We next sought to determine if both demographics and SWI/SNF expression influences reprogramming efficiency. Using Spearman Correlation Coefficient tests, we determined that ancestry does influence the DFs ability to reprogram. DFs from AA donors were significantly more efficient at reprogramming (p=0.01), producing on average 1634 iPSC colonies compared to an EA average of 1175 iPSC colonies (Figure 6A). We found no correlation between donor sex and reprogramming efficiency (Figure 6A). The results with age as a variable gave a more interesting result (Supplemental Figure 5), when examined as a continuum it misses significance with a p value for the correlation was approx. 0.08. However, when we took advantage of a natural break in our cohort and examined donors below or above age 33 age was found to correlate with reprograming efficiency (p=0.03) (Figure 6A). Donors under the age of 33 produced 1601 iPSC colonies while older donors produced 1221 iPSC colonies. These results establish that iPSC reprogramming efficiency correlated with the ancestry and age of the individual.
Figure 6. Efficiency of iPS Reprogramming is Correlated to Donor Demographics and Gene Expression Signature.
(A)AA DFs had higher iPS reprogramming efficiency then EA DFs (1634 vs. 1175, p=0.01). Donors under 33 consistently produced more iPS colonies then older donors (1601 vs. 1221, p=0.03). Error shown as standard deviation of the mean (B) Heat map showing Spearman Correlation Coefficients between gene expression and reprogramming efficiency. (C) Scatterplots showing correlations between gene expression of BRG1, Lin28, Oct4, or Myc with iPS reprogramming efficiency.
*=p≤0.05,**=p≤0.01,***=p≤0.001
In addition to the demographic associations, reprogramming efficiency is positively-associated with the expression of several SWI/SNF subunits in iPSCs (Figure 6B). BRG1, BAF155, and BAF60a expression levels were all positively correlated with iPSC reprogramming efficiency. BRG1 expression was the most significantly correlated with reprogramming efficiently with a coefficient of 0.28 (p=0.01), compared to that of BAF155 (0.23, p=0.03) and BAF60a (0.25, p=0.02). While DF SWI/SNF component gene expression was not significantly correlated with reprogramming efficiency, several genes had strong trends toward correlation. DF expression of BRG1 had a positive correlation coefficient of 0.199 with a p-value of 0.07 while BAF60a had a correlation coefficient of 0.206 and a p-value of 0.06.
Surprisingly, the weak pluripotency-associated gene expression in DF correlated with iPSC reprogramming efficiency (Figure 6C). In DFs, OCT4 and LIN28 had an inverse correlation with reprogramming efficiency with correlation coefficients of −0.38 (p=0.0005) and −0.31 (p=0.004) respectively. Higher levels of OCT4 and LIN28 in DFs expression reduced the number of iPSC colonies generated. In contrast, high levels of MYC and Vimentin in DF were strongly correlated with enhanced reprogramming efficiency, with correlation coefficients of 0.36 (p=0.001) and 0.27 (p=0.01) respectively. iPSC OCT4 expression was also negatively correlated with reprogramming efficiency in iPSCs with a correlation coefficient of −0.28 (p=0.01). In contrast, iPSC SOX2 and MYC expression, showed positive correlations of 0.28 (p=0.01) and 0.32 (p=0.003) with reprogramming efficiency (Figure 6C).
Discussion
We have tested iPSC reprogramming efficiency by reprogramming donor DF lines in triplicate, staining colonies with alkaline phosphatase, and utilizing a computer based counting method. Our findings show that samples from each donor had an intrinsic capacity to be reprogrammed to generate iPSC colonies, as evidenced by consistent colony counts (<10% variability) between replicates, even if the replicates were reprogrammed months apart. These results demonstrate that iPSC reprogramming is not a stochastic process and that the efficiency of iPSC conversion is governed by genetic and molecular mechanisms. Our results also show that donor age and ancestry can influence the efficacy of generating iPSCs.
It has long been appreciated in mice that age is inversely correlated with iPSC reprogramming efficiency [2, 33, 35, 39]. In humans, however, the impact of donor age on iPSC reprogramming has been controversial [36, 38, 44]. The results from our large cohort clearly show that samples from younger donors (<33 years of age) had a higher reprogramming efficiency than samples from older donors. On average, younger donors produced 400 more iPSC colonies per reprogramming experiment then did their older counter parts. Our findings support previous studies that found a correlation between donor age and reprogramming efficiency[44]. We believe that conflicting results stem from the use of small sample sizes (>10 donors on average) and attempts to assess a correlation with age on a continuum as opposed to distinct age groups [36, 38, 44].
To date, there have been no studies that have thoroughly examined the impact of ancestry on iPSC reprogramming efficiency. The results from this study show that samples from donors of AA descent had a significantly higher reprogramming efficiency. These findings have important implications for regenerative medicine and further highlight the need for advanced cellular models that incorporate complex demographic traits into mechanistic and pharmacologic studies.
While, further studies are needed to determine the specific underlying mechanisms regulating reprogramming efficiency, the results from our analyses show that the expression of SWI/SNF components can have a significant impact on efficiency of generating iPSC cells. BRG1 has been shown to be required for the initiation and maintenance of pluripotency in iPSC lines [3, 31]. Our results show that elevated levels of BRG1, BAF155, and BAF60a were correlated with higher reprogramming efficiency. This suggests that the BRG1 complex facilitates the resetting of the epigenetic landscape during iPSC reprogramming.
In addition to their role in reprogramming, BRG1, BAF155, BAF60a, BAF53a, and BAF47 were found to be expressed at higher levels in iPSCs compared to their parental DF lines. These findings coupled with the suppression of BAF170 in iPSC lines, confirms the role and composition of esBAF and somatic BAF complexes in human tissues and further supports that their function is closely conserved between humans and mice [8, 10, 13, 45]. We also determined that expression levels of BAF155, BAF60a, and BAF53a were associated with specific demographic traits. We identified unique age, sex, and ancestry-specific gene expression profiles for BRG1 complex subunits and pluripotency-associated transcription factors. For example, women on average had higher expression of BAF155 and BAF60a with lower expression of OCT4 than men. Additionally, younger donors had lower levels of BAF53a and OCT4 with higher levels of BAF60a, LIN28, SOX2, and MYC expression than older donors. Further studies are needed to elucidate the underlying mechanisms responsible for these demographic-specific signatures; however, our findings illustrate that there are functional and dynamic changes in epigenetic enzyme utilization across demographic groups.
The results from these studies clearly demonstrate the impact that individual diversity can have on cellular physiology and iPSC reprogramming. Given the pronounced impact of donor age, sex, and ancestry, it is vital that researchers take this diversity into account when designing human mechanistic and pharmacologic studies. Consisting of DF samples from 80 healthy donors, our cohort has been used to generate more than 240 iPSC lines. This large cohort is ancestry and sex balanced and is derived from donors around the Raleigh/Durham area of North Carolina. While several cohorts of human primary cells and derived iPSC lines exist [46, 47], our cohort is unique in its large size, diversity, and focus on healthy donors. These factors make this cohort a vital tool for researchers seeking to validate observational results from human population studies and to perform detailed mechanistic studies in a controlled cell culture environment. Recent clinical trials have shown that therapeutic compounds can be processed differently across individuals, requiring different dosing recommendations based on sex or ancestry[42, 43, 48-50]. Our cohort allows for the creation of sophisticated models to test a compound’s efficacy, metabolism, and toxicity while identifying demographic influences that may impact dosage, tolerance, and effectiveness.
Supplementary Material
Significance Statement:
We created a large novel, sex and ancestry balanced, cohort of primary dermal fibroblast and derived induced pluripotent stem cells (iPSC) from 80 healthy human donors. Creation of this cohort allowed us to determine key factors involved in the efficiency by which we were able to produce iPSCs. Specifically, components of the SWI/SNF family of epigenetic enzymes, donor age, and donor ancestry all correlated with proficient generation of iPSCs. The results from these studies will have broad implications in Stem Cell Biology and Regenerative Medicine. Additionally, our cohort will allow for the creation of sophisticated cell-based models for mechanism of action studies, pharmaceutical development, and toxicity assessments.
Acknowledgements
We would like to thank Drs. Guang Hu, Jack Tayor, Anton Jetton, and Erik Tokar for their critical evaluation of this manuscript. Drs. Harriet Kinyamu, Staton Wade, Lee Langer and Jackson Hoffman of the Archer lab for support. We would also like to thank Dr. Negin Martin and the Viral Vector Core Laboratory, Dr. Stavros Garantziotis and Rebecca Church of the Clinical Research Branch, the Comparative Medicine Branch, and the NTP Pathology Support Group.
Financial Support: This research was supported by the Intramural Research Program of the NIH – National Institute of Environmental Health Sciences [Z01 ES071006-17; nih.gov].
Footnotes
Conflicts of Interest
The authors identified no conflicts of interest.
References
- 1.Ferrari F, et al. , Rearranging the chromatin for pluripotency. Cell Cycle, 2014. 13(2): p. 167–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim K, et al. , Epigenetic memory in induced pluripotent stem cells. Nature, 2010. 467(7313): p. 285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singhal N, et al. , Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell, 2010. 141(6): p. 943–55. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi M, et al. , Chromatin remodelers: We are the drivers!! Nucleus, 2016. 7(4): p. 388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trotter KW, King HA, and Archer TK, Glucocorticoid Receptor Transcriptional Activation via the BRG1-Dependent Recruitment of TOP2beta and Ku70/86. Mol Cell Biol, 2015. 35(16): p. 2799–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trotter KW and Archer TK, The BRG1 transcriptional coregulator. Nucl Recept Signal, 2008. 6: p. e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hargreaves DC and Crabtree GR, ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res, 2011. 21(3): p. 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho L, et al. , An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A, 2009. 106(13): p. 5187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho L, et al. , An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A, 2009. 106(13): p. 5181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apostolou E and Hochedlinger K, Chromatin dynamics during cellular reprogramming. Nature, 2013. 502(7472): p. 462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koche RP, et al. , Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell, 2011. 8(1): p. 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade SL, et al. , MiRNA-Mediated Regulation of the SWI/SNF Chromatin Remodeling Complex Controls Pluripotency and Endodermal Differentiation in Human ESCs. Stem Cells, 2015. 33(10): p. 2925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachmann C, et al. , mSWI/SNF (BAF) Complexes Are Indispensable for the Neurogenesis and Development of Embryonic Olfactory Epithelium. PLoS Genet, 2016. 12(9): p. e1006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brownlee PM, Meisenberg C, and Downs JA, The SWI/SNF chromatin remodelling complex: Its role in maintaining genome stability and preventing tumourigenesis. DNA Repair (Amst), 2015. 32: p. 127–33. [DOI] [PubMed] [Google Scholar]
- 15.Huang X, et al. , Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol, 2008. 319(2): p. 258–66. [DOI] [PubMed] [Google Scholar]
- 16.Nargund AM, et al. , The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma. Cell Rep, 2017. 18(12): p. 2893–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, et al. , Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev, 2004. 18(24): p. 3106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. , Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports, 2014. 3(3): p. 460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Z, et al. , Knockdown of Brm and Baf170, Components of Chromatin Remodeling Complex, Facilitates Reprogramming of Somatic Cells. Stem Cells Dev, 2015. 24(19): p. 2328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Bi Y, and Gao S, Epigenetic regulation of somatic cell reprogramming. Curr Opin Genet Dev, 2017. 46: p. 156–163. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, et al. , Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov, 2017. 16(2): p. 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K and Yamanaka S, A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol, 2016. 17(3): p. 183–93. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K, et al. , Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 2007. 131(5): p. 861–72. [DOI] [PubMed] [Google Scholar]
- 24.Apostolou E, et al. , Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell, 2013. 12(6): p. 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayan S, et al. , OCT4 and SOX2 Work as Transcriptional Activators in Reprogramming Human Fibroblasts. Cell Rep, 2017. 20(7): p. 1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho R, Chronis C, and Plath K, Mechanistic insights into reprogramming to induced pluripotency. J Cell Physiol, 2011. 226(4): p. 868–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa M, et al. , Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol, 2008. 26(1): p. 101–6. [DOI] [PubMed] [Google Scholar]
- 28.Ohnuki M, et al. , Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci U S A, 2014. 111(34): p. 12426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu C and Li L, Current reprogramming systems in regenerative medicine: from somatic cells to induced pluripotent stem cells. Regen Med, 2016. 11(1): p. 105–32. [DOI] [PubMed] [Google Scholar]
- 30.Shaw A and Cornetta K, Design and Potential of Non-Integrating Lentiviral Vectors. Biomedicines, 2014. 2(1): p. 14–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh VK, et al. , Mechanism of Induction: Induced Pluripotent Stem Cells (iPSCs). J Stem Cells, 2015. 10(1): p. 43–62. [PubMed] [Google Scholar]
- 32.Trevisan M, et al. , Reprogramming Methods Do Not Affect Gene Expression Profile of Human Induced Pluripotent Stem Cells. Int J Mol Sci, 2017. 18(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, et al. , Reprogramming efficiency and quality of induced Pluripotent Stem Cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr, 2011. 3: p. RRN1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, et al. , Human induced pluripotent stem cells free of vector and transgene sequences. Science, 2009. 324(5928): p. 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Z, et al. , Establishment of induced pluripotent stem cells from aged mice using bone marrow-derived myeloid cells. J Mol Cell Biol, 2011. 3(2): p. 91–8. [DOI] [PubMed] [Google Scholar]
- 36.Ohmine S, et al. , Reprogrammed keratinocytes from elderly type 2 diabetes patients suppress senescence genes to acquire induced pluripotency. Aging (Albany NY), 2012. 4(1): p. 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somers A, et al. , Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells, 2010. 28(10): p. 1728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yagi T, et al. , Establishment of induced pluripotent stem cells from centenarians for neurodegenerative disease research. PLoS One, 2012. 7(7): p. e41572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. , The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature, 2009. 460(7259): p. 1136–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmoudi S and Brunet A, Aging and reprogramming: a two-way street. Curr Opin Cell Biol, 2012. 24(6): p. 744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phanthong P, et al. , Is aging a barrier to reprogramming? Lessons from induced pluripotent stem cells. Biogerontology, 2013. 14(6): p. 591–602. [DOI] [PubMed] [Google Scholar]
- 42.Avior Y, Sagi I, and Benvenisty N, Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol, 2016. 17(3): p. 170–82. [DOI] [PubMed] [Google Scholar]
- 43.Trounson A and DeWitt ND, Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol, 2016. 17(3): p. 194–200. [DOI] [PubMed] [Google Scholar]
- 44.Trokovic R, et al. , Combined negative effect of donor age and time in culture on the reprogramming efficiency into induced pluripotent stem cells. Stem Cell Res, 2015. 15(1): p. 254–62. [DOI] [PubMed] [Google Scholar]
- 45.Onder TT, et al. , Chromatin-modifying enzymes as modulators of reprogramming. Nature, 2012. 483(7391): p. 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panopoulos AD, et al. , iPSCORE: A Resource of 222 iPSC Lines Enabling Functional Characterization of Genetic Variation across a Variety of Cell Types. Stem Cell Reports, 2017. 8(4): p. 1086–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee G, et al. , Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature, 2009. 461(7262): p. 402–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu C and Li LJ, Improving the Reprogramming Efficiency of Somatic Cells to Induced Pluripotent Stem Cells. Crit Rev Eukaryot Gene Expr, 2015. 25(4): p. 323–34. [DOI] [PubMed] [Google Scholar]
- 49.Kwon D, et al. , Reprogramming Enhancers in Somatic Cell Nuclear Transfer, iPSC Technology, and Direct Conversion. Stem Cell Rev, 2017. 13(1): p. 24–34. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K and Yamanaka S, Induced pluripotent stem cells in medicine and biology. Development, 2013. 140(12): p. 2457–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.