Acinetobacter baumannii is an important nosocomial pathogen resistant to many, and sometimes all, antibiotics. The A. baumannii K2 capsular type has been associated with elevated antibiotic resistance. The capsular depolymerase characterized here fits the new trend of alternative antibacterial agents needed against multidrug-resistant pathogens. They are highly specific, stable, and refractory to resistance, as they do not kill bacteria per se; instead, they remove bacterial surface polysaccharides, which diminish the bacterial virulence and expose them to the host immune system.
KEYWORDS: A. baumannii, antivirulence, bacteriophage, capsular depolymerase
ABSTRACT
Acinetobacter baumannii is emerging as a major nosocomial pathogen in intensive care units. The bacterial capsules are considered major virulence factors, and the particular A. baumannii capsular type K2 has been associated with high antibiotic resistance. In this study, we identified a K2 capsule-specific depolymerase in a bacteriophage tail spike C terminus, a fragment that was heterologously expressed, and its antivirulence properties were assessed by in vivo experiments. The K2 depolymerase is active under a broad range of environmental conditions and is highly thermostable, with a melting point (Tm) at 67°C. In the caterpillar larva model, the K2 depolymerase protects larvae from bacterial infections, using either pretreatments or with single-enzyme injection after bacterial challenge, in a dose-dependent manner. In a mouse sepsis model, a single K2 depolymerase intraperitoneal injection of 50 μg is able to protect 60% of mice from an otherwise deadly infection, with a significant reduction in the proinflammatory cytokine profile. We showed that the enzyme makes bacterial cells fully susceptible to the host complement system killing effect. Moreover, the K2 depolymerase is highly refractory to resistance development, which makes these bacteriophage-derived capsular depolymerases useful antivirulence agents against multidrug-resistant A. baumannii infections.
IMPORTANCE Acinetobacter baumannii is an important nosocomial pathogen resistant to many, and sometimes all, antibiotics. The A. baumannii K2 capsular type has been associated with elevated antibiotic resistance. The capsular depolymerase characterized here fits the new trend of alternative antibacterial agents needed against multidrug-resistant pathogens. They are highly specific, stable, and refractory to resistance, as they do not kill bacteria per se; instead, they remove bacterial surface polysaccharides, which diminish the bacterial virulence and expose them to the host immune system.
INTRODUCTION
Acinetobacter baumannii is an important nosocomial pathogen able to cause wound, skin, lung, and bloodstream infections, which are especially problematic in intensive care units (1). Current treatments are becoming less effective, as many isolates are resistant to most and sometimes all available antibiotics (2, 3). The international surveillance Healthcare Safety Network and Eurofins programs have already demonstrated that >50% of A. baumannii isolates in patients admitted to intensive care units have acquired resistance to carbapenem and that A. baumannii is by far the leading antibiotic-resistant pathogen surveyed (4, 5). Reports show that case fatality rates associated with bloodstream infections range between 50% and 60% (6, 7).
The presence of capsule polysaccharide-based structures (K types) at the A. baumannii surface represents an important virulence factor. These polysaccharide coats modulate growth within soft-tissue infection sites (8), confer intrinsic resistance to peptide antibiotics (9), and protect bacteria from host immune defenses (8, 10). Currently, more than 125 capsule synthesis loci have been found in A. baumannii, with 40 different structures having been determined (11, 12). Nevertheless, the lack of serotyping or genotyping schemes have limited the information available about the virulence and prevalence of A. baumannii K types.
Capsular depolymerases are emerging as a new line of antivirulence agents. These enzymes are typically displayed at the tips of bacteriophage (phage) tails or baseplates to “shave” bacterial capsules, exposing their receptors for phage binding and further cell infection (12). Phage-derived capsular depolymerases have been shown to reduce the virulence of Gram-negative bacteria, namely, Escherichia coli (K1, K2, K5, and K30) (13, 14), Klebsiella pneumoniae (K1, K5, K64, and KN2) (15–18), and A. baumannii (undefined capsule type) (19), in murine models of sepsis by enhancing killing by complement systems, neutrophils, and macrophages. Our research group has also recently shown that capsular depolymerases are widespread in Acinetobacter phage genomes, being responsible for the activity and binding to specific Acinetobacter bacterial capsules (20), and that they can be heterologously expressed and digest A. baumannii capsules, making bacteria susceptible to serum killing (21). However, the in vivo therapeutic potential of phage depolymerases remains poorly explored against A. baumannii infections.
In this study, we tested the antivirulence properties of a new phage-derived capsular depolymerase against a prevalent bacterial capsular type (K2), which has been associated with high antibiotic resistance (e.g., encoding oxacillinase and cephalosporinases genes) (22, 23) and reported to be one of the most common K type in patients admitted to hospitals of Portugal (24). The enzyme was tested in Galleria mellonella, murine models, and human serum.
RESULTS
Depolymerase functional characterization.
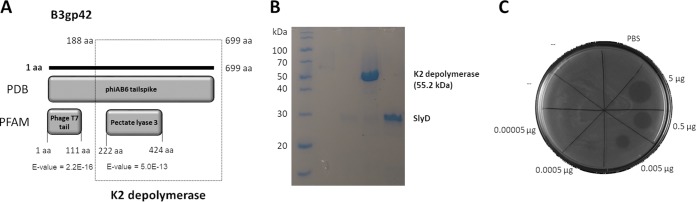
We previously isolated phage vB_AbaP_B3 (B3 for short), which infects A. baumannii K2 capsule type (named NIPH 2061 strain) (12). This virus makes clear plaques with large sorrowing haloes, which is indicative of capsule depolymerization activity (25). The analysis of the phage B3 genome predicted the existence of a tail spike gene (B3gp42) with a high level of identity to five other Acinetobacter phage tail-associated proteins (>97% overall amino acid identity). This gene harbors a conserved N-terminal phage_T7 domain (Pfam entry PF03906.14) and a less conserved C-terminal pectate lyase domain (PF12708.7), which has been previously linked to enzymes able to degrade bacterial capsules (20) (Fig. 1A). Therefore, we further cloned, recombinantly expressed, and purified the C-terminal fragment, named K2 depolymerase, which contained the pectate lyase domain (Fig. 1A). The heterologous production yielded a protein with >95% purity and with a molecular mass of 55.2 kDa, matching the theoretical value (Fig. 1B).
FIG 1.
K2 depolymerase identification. (A) In silico analysis of the A. baumannii phage vB_AbaP_B3 tail spike (B3gp42). The PBD database shows homology to the phiAB6 tail spike (5JS4_A), and PFAM identifies an N-terminal tail domain and a C-terminal depolymerase domain (pectate lyase 3, named K2 depolymerase) that was cloned (genetic region of bp 567 to 2100 of the B3_42 open reading frame). (B) SDS-PAGE gel demonstrating the expression and purification of hybrid protein expression (SlyD-K2 depolymerase) after TEV protease digestion to separate the K2 depolymerase from the SlyD fusion partner. (C) Drop tests of the K2 depolymerase onto NIPH 2061 strain.
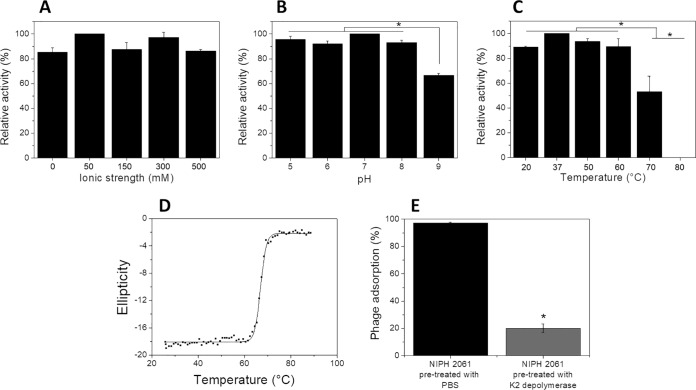
To assess the K2 depolymerase ability to degrade bacterial capsules, we performed drop tests. The enzyme was only active on K2 out of 22 different K types of A. baumannii (Table 1). It remained active from a 0.0005- to 5-μg range (Fig. 1C). To determine the protein hydrolytic activity, we digested extracted capsular polysaccharides and quantified the sugar-reducing ends with the 3,5-dinitrosalicylic acid (DNS) method. K2 depolymerase exhibited high and similar hydrolytic activity in all ranges of ionic strength (0 to 500 mM) and pH values (pH 5 to 9) (Fig. 2A and B). The enzyme was also highly active between 20 and 60°C, retained 50% of activity at 70°C, and was inactivated at higher temperatures (Fig. 2C). These results are in agreement with the heat-induced changes in the K2 depolymerase secondary structure, where thermal unfolding curves were achieved by following the circular dichroism (CD) signal at 222 nm as a function of temperature. Thermal unfolding curves showed a sigmoidal transition with a melting temperature (Tm) of 67°C (Fig. 2D), temperatures above which no enzymatic activity was observed.
TABLE 1.
Activity spectrum of the K2 depolymerasea
| Acinetobacter baumannii strain (n = 28) | ST | K type | GenBank accession no. | K2 depolymerase spot test |
|---|---|---|---|---|
| NIPH 290 (CIP 110431) | ST1 | 1 | KB849940.1 (bp 126651–104645) | − |
| NIPH 2061 (CIP 110467) | ST2 | 2 | KB849309.1 (bp 77375–101575) | + |
| H466 | ST2 | 2 | NA | + |
| H580 | ST2 | 2 | NA | + |
| H603 | ST2 | 2 | NA | + |
| H678 | ST2 | 2 | NA | + |
| NIPH 501T (ATCC 19606T) | ST52 | 3 | KB849970.1 (bp 174731–119233) | − |
| NIPH 528 (CIP 110436; RUH 134) | ST2 | 9 | KB849906.1 (bp 78004–102899) | − |
| NIPH 80 (CIP 110427) | ST37 | 9 | KB849944.1 (bp 156383–131489) | − |
| J9 | ST49 | 11 | KF002790 | − |
| A85 | ST1 | 15 | KC118540 (bp 8456–36738) | − |
| RBH2 | ST111 | 19 | KU165787 | − |
| NIPH 190 (CIP 110429) | ST9 | 30 | KB849477.1 (bp 592918–572137) | − |
| NIPH 67 (CIP 110425) | ST35 | 33 | KB849903.1 (bp 1301423–1278652) | − |
| LUH5535 | NA | 35 | KC526896 | − |
| NIPH 146 (CIP 110428) | ST25 | 37 | KB849308.1 (bp 572444–592959) | − |
| ANC 4097 (CIP 110499) | ST1 | 40 | KB849962.1 (bp 39134–62621) | − |
| NIPH 60 (CIP 110424) | ST34 | 43 | KB849508.1 (bp 140959–120981) | − |
| NIPH 70 (CIP 110426) | ST36 | 44 | KB849923.1 (bp 574942–546765) | − |
| NIPH 201 (CIP 110430) | ST38 | 45 | KB849844.1 (bp 365379–344305) | − |
| NIPH 329 (CIP 110432) | ST11 | 46 | KB849871.1 (bp 2591085–2567472) | − |
| NIPH 601 (CIP 110437) | ST40 | 47 | KB849894.1 (bp 3226845–3205785) | − |
| NIPH 615 (CIP 110438) | ST12 | 48 | KB849301.1 (bp 114143–93442) | − |
| NIPH 1734 (CIP 110466) | ST15 | 49 | KB849325.1 (bp 2998512–2965610) | − |
| NIPH 335 (CIP 110433) | ST10 | 49 | KB849886.1 (bp 1318556–1286966) | − |
| BAL_212 | ST52 | 57 | KY434631 | − |
| SGH0703 | ST2 | 73 | MF362178 | − |
| LUH5538 | NA | 83 | KC526898 | − |
Drop test of the enzyme was made in bacterial lawns of a collection of A. baumannii strains with known capsular types. For all strains, the sequence types (ST) according the multilocus sequence analysis using the Pasteur scheme, capsular type (K), and respective accession numbers are given. K type, determined capsule structure; NA, not available.
FIG 2.
In vitro functional analysis. Hydrolytic activity of the K2 depolymerase on extracted exopolysaccharides from NIPH 2061 (K2 capsule type) host at different ionic strengths (pH 6, 0 to 500 mM NaCl, 37°C) (A), pH values (5 to 9, 0 mM NaCl, 37°C) (B), and temperatures (pH 6, 0 mM NaCl, 37 to 80°C) (C). The results are expressed as relative activity compared with the best activity value obtained at pH 6.0, 37°C. Significance was determined by Student t test (*, P < 0.05). (D) Circular dichroism analysis of the K2 depolymerase. Melting curve was acquired by measuring ellipticity at 222 nm from 25 to 90°C. (E) A. baumannii phage vB_AbaP_B3 adsorption onto NIPH 2061 cells pretreated with K2 depolymerase. Results are expressed as residual PFU percentages compared with results from adsorption assays with NIPH 2061 cells pretreated with PBS. *, statistically different (P < 0.01).
To further illuminate the role of the K2 depolymerase in phage B3 adsorption onto the host bacterium, experiments were conducted using hosts with and without enzymatic treatments. Results showed that phage adsorbs 97% versus 20% for phosphate-buffered saline (PBS)- and enzyme-treated cells (P < 0.01), respectively (Fig. 2E). Therefore, by stripping the NIPH 2061 cells from their capsules, the K2 depolymerase is removing the receptors that are no longer available for phage adsorption. This demonstrates an important role of the K2 depolymerase, which is located at the tail spike of the virion particle, in the initial step of the phage B3 infection.
Resistance development.
The emergence of resistant isolates toward K2 depolymerase under selective pressure was also analyzed by challenging the NIPH 2061 strain with PBS or with K2 depolymerase at 3 different enzyme concentrations (at 0.1, 1, or 5 μM) and three subcultures (of 24 h each), as described in Materials and Methods. There were no differences in the growth rate of NIPH 2061 strain in the presence of either PBS or K2 depolymerase at 0.1, 1, or 5 μM, which presented a steady increase reaching an OD from 0.1 to 1.0 in a few hours. As expected, all 20 isolated colonies from PBS-challenged cultures remained sensitive to enzyme drops. Interestingly, similar observations were made with all 20 isolated colonies from enzyme-challenged cultures and for all three different enzyme concentrations used. This demonstrates that resistance development of A. baumannii cells toward K2 depolymerase under selective pressure is not easily observed.
Capsular depolymerase treatment in A. baumannii-infected larvae.
To validate the K2 depolymerase antivirulence properties in vivo, we used the G. mellonella infection model (Fig. 3). Two types of experiments were performed. Larva survival rates were measured by either (i) injecting K2 depolymerase-pretreated NIPH 2061 cells for 2 h or (ii) injecting the K2 depolymerase 30 min after NIPH 2061 inoculation without pretreatment. In all experiments, control groups with either HEPES or enzyme alone resulted in a 100% survival rate, which seems to demonstrate the absence of toxicity (data not shown).
FIG 3.
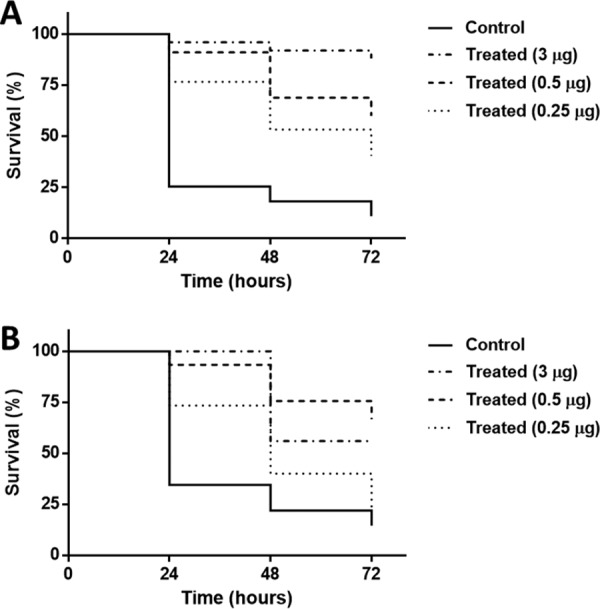
In vivo Galleria mellonella model. The larva survival rates of larvae infected with NIPH 2061 cells (K2 capsule type) (106 CFU inoculum) were measured by injecting K2 depolymerase-pretreated NIPH 2061 cells for 2 h (A) or K2 depolymerase 30 min after NIPH 2061 bacterial inoculation (B). Three different enzyme doses of 0.25 μg, 0.5 μg, and 3 μg were used. For clarity, control groups (HEPES-treated and enzyme-treated larvae) resulted in 100% survival rate and were omitted from the figures. Significance was determined by Mantel-Cox test (*, P < 0.05).
In the first experiment (Fig. 3A), bacterially infected larvae pretreated with HEPES resulted in a quick and abrupt death, with only 25%, 20%, and 10% survival rates after 24, 48, and 72 h, respectively. In contrast, bacterially infected larvae pretreated with K2 depolymerase significantly prevented worm death in a time- and dose-dependent manner. After 72 h, 53%, 69%, and 88% of larvae survived using 0.25 μg, 0.5 μg, and 3 μg of K2 depolymerase pretreatments, respectively (P < 0.01, P < 0.0001, and P < 0.0001). We also confirmed that K2 depolymerase did not affect bacterial viability given that no differences were observed in the number of the NIPH 2061 CFU before and after K2 depolymerase treatments.
In the second set of experiments using larvae with an established bacterial infection (Fig. 3B), in the first 24 h, HEPES injections resulted in 35% survival of larvae, whereas increasing doses of the K2 depolymerase increased the survival rate from 73% up to 100% of larvae. At 48 h, only 22% of bacterially infected and HEPES-treated larvae survived, being significantly increased between 40% and 76% when treated with increasing doses of the K2 depolymerase. At 72 h, bacterially infected and untreated larvae displayed less than a 15% survival rate. K2 depolymerase treatments with 0.25 μg did not show any improvement. Nevertheless, larvae injected with 0.5 μg and 3 μg of K2 depolymerase still exhibited 56% and 70% worm survival, respectively (P < 0.0001 and P < 0.0001). Thus, the NIPH 2061 bacterial capsule (K2 type) influences bacterial virulence, and higher doses of K2 depolymerase improve the therapeutic effect.
Capsular depolymerase treatment in A. baumannii-infected mice.
To further validate the K2 depolymerase antivirulence properties in vertebrate animals, we used mice infected with the NIPH 2061 strain (107 CFU) and treated them with a single 50-μg dose of K2 depolymerase 1 h postinfection (Fig. 4A). In the control group, infection in all PBS-treated mice quickly progressed by 20 h postinfection. Mice exhibited severe signs of septicemia, with abnormal posture behavior and more than 10% weight loss, at which point they were sacrificed for ethical reasons. In contrast, at 20 h postinfection, 90% of enzyme-treated mice had survived, decreasing to 60% at 42 h postinfection, after which mice remained healthy until the end of the experimental period. Statistical analysis of survival rates corroborated that the K2 depolymerase therapy was successful against A. baumannii infection (P = 0.004). The fact that in treated mice the CFU counts were lower in the spleen, but not in the liver, than in the control group also attest to the success of depolymerase treatment (Fig. 4B).
FIG 4.
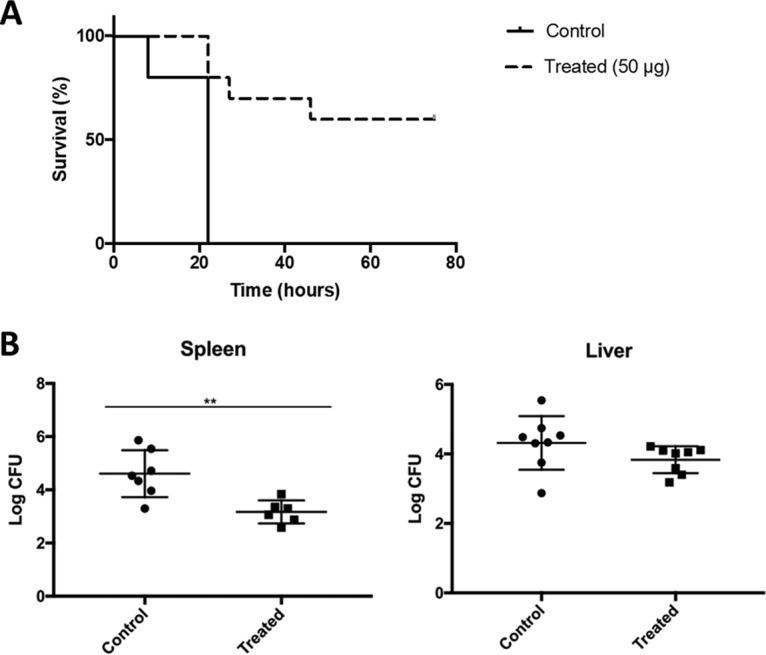
In vivo murine model. (A) The survival rates of mice infected with NIPH 2061 (K2 capsule type) cells (107 CFU inoculum) were measured after intraperitoneal injection of PBS or the K2 depolymerase (50 μg/mouse) 1 h after challenge. (B) CFU counts of the spleen and liver of mice 12 h after challenge. Significance was determined by Mantel-Cox test (*, P < 0.05).
Capsular depolymerase treatment biochemical effect.
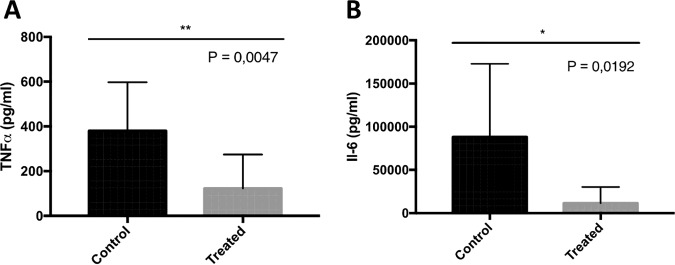
Levels of tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) were analyzed from serum of mock- and enzyme-treated mice, and the results showed the induction of this proinflammatory cytokine production upon infection. As shown in Fig. 5, the amounts of both TNF-α and IL-6 were significantly higher in nontreated mice than in enzyme-treated mice.
FIG 5.
Biochemical analysis. The biochemical levels of TNF-α (A) and IL-6 (B) were measured in blood of PBS-treated mice and mice treated with 50 μg K2 depolymerase. Significance was determined by two-tailed Student's t test (*, P < 0.05).
Serum sensitivity of depolymerase-treated A. baumannii.
To further validate the antivirulence effect of K2 depolymerase, we used human serum mixed with the NIPH 2061 strain (Fig. 6). After adding K2 depolymerase to the contaminated serum, NIPH 2061 CFU were reduced below the detection limit (<10 CFU/ml). As expected, addition of heat-inactivated K2 depolymerase did not sensitize the NIPH 2061 strain to serum killing. Thus, enzymatic capsule removal via K2 depolymerase is crucial to help the host complement system to control the infection. As an additional study, we also tested the susceptibility to serum killing of the previously isolated bacterial colonies (n = 20) plated after being challenged with K2 depolymerase. These strains survived the serum complement system but, after being in contact with K2 depolymerase, were reduced below the detection limit (<10 CFU/ml) (data not shown).
FIG 6.
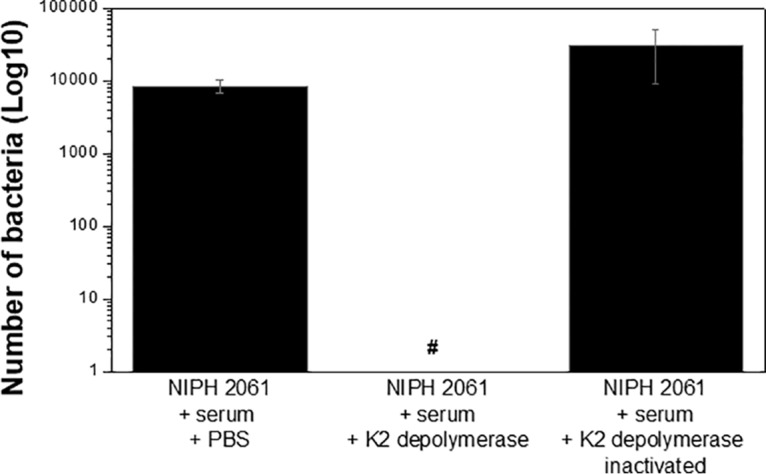
Host serum complement system effect. Human serum was infected with NIPH 2061 (K2 capsule type) cells at a 3:1 ratio and incubated with PBS, K2 depolymerase, or heat-inactivated (15 min at 100°C) enzyme for 1 h at 37°C. Enzyme final concentration of 1 μM was used. #, Below detection limit (10 CFU/ml).
DISCUSSION
Capsular polysaccharides, also termed K types, are major virulence factors of bacteria that are involved in protecting cells from a range of environmental pressures, mostly against host immunity (26, 27). As documented for E. coli (28) and K. pneumoniae (29), capsules may also be the primary virulence determinant of A. baumannii. However, so far, only a few K types of A. baumannii (K1 and K45) have been proven to invade or overwhelm mammalian defenses (9, 21, 30). The existence of at least 125 K types in A. baumannii might be related to different degrees of clinical manifestation of infections and antibiotic resistance (21). However, this knowledge is limited due to the lack of robust typing schemes available to determine the most predominant and virulent K types.
To the best of our knowledge, the first epidemic capsule typing study demonstrated that the A. baumannii bacterial capsule type K2 has been frequently associated with infections in Portugal (24). Strains of this capsular type, such as NIPH 2061 used in this study, also have been linked to high antibiotic resistance (encoding oxacillinase and cephalosporinase genes), thereby demanding the development of new therapeutic options (22, 23). Therefore, the present study aimed at isolating a new phage-derived capsular depolymerase that degrades the A. baumannii K2 and evaluating its antivirulence potential and efficacy in controlling A. baumannii systemic infections using in vivo models.
We found that the phage vB_AbaP_B3 genome (B3 for short) contains a tail spike (B3gp42) with a pectate lyase domain at the C-terminal fragment. We further showed that this C-terminal fragment (named K2 depolymerase) has a specific depolymerase activity on K2 out of 22 different capsular types tested, with activity between micro- and millimolar ranges. A similar extremely narrow host range has been found in capsular depolymerases from phages infecting A. baumannii (21), E. coli (13), and K. pneumoniae (16, 18, 31), often restricted to one or two K types. We also showed that the K2 depolymerase binds to the host receptor, as NIPH 2061 cells pretreated with the enzyme heavily affected phage B3 adsorption. As phage tail spikes function as host recognition elements (32), phage tail spike proteins probably have high diversity, having been evolved to encode several depolymerase domains to recognize a wide range of bacterial K antigen possibilities.
K2 depolymerase was also demonstrated to be a versatile and thermostable enzyme, as it is active across a range of different environmental conditions of pH (5 to 9), ionic strengths (0 to 500 mM), and temperatures (20 to 70°C). We noted that the loss of enzymatic activity at temperatures of ≥70°C related well with the loss of protein structure observed during CD spectroscopy measurements (Tm of 67°C). These impressive characteristics have been shown for the few capsular depolymerases also tested from K. pneumoniae-infecting phages (31, 33). They are likely a reflection of their structural nature, i.e., being part of the virion particle, all structural proteins are evolved to endure broad and harsh conditions to maintain phage infectivity and survival.
To assess if A. baumannii capsule type K2 is an important virulence factor, we first used the G. mellonella larva model for its simplicity. These insects are easy to use and have host mechanisms of resistance similar of those of vertebrates against A. baumannii infections (34). We demonstrated that K2 capsule could overcome the immune system of larvae, causing significant death. We also showed that the K2 depolymerase could significantly rescue larvae either by pretreating K2 cells with K2 depolymerase before inoculation or by injecting the enzyme 2 h postinfection. As expected, these results were dose dependent. Overall, since capsule removal by the K2 depolymerase attenuated the pathogenicity of the cells and prolonged the larval life span, it was demonstrated that the capsule K2 type is a major virulence factor of A. baumannii. Generally, other researchers have also shown success in preventing and treating the lethal effects of encapsulated K. pneumoniae (K3, K21, and K36) (31, 35) and A. baumannii (undefined capsule type) (19) with single injections of capsular-specific depolymerases. However, these models have administered enzymes immediately after infection (5 min or less), which may raise the question of whether they are only providing a protective effect rather than a therapeutic one.
To further investigate the antivirulence efficacy in a model that better mimics the human immune systems, we tested the K2 depolymerase in a murine model of sepsis. Our results indicated that a single administration of the K2 depolymerase (50 μg/mouse) had a significant therapeutic effect by rescuing 60% of mice, controlling bacterial proliferation, and reducing local inflammation. Other studies have also demonstrated that a single administration of capsular depolymerases could significantly protect mice infected with E. coli (K1, K2, K5, and K30) (13, 14) and K. pneumoniae (K1, K5, K64, and KN2) (15–18). A recent study also showed that a capsule depolymerase administered intraperitoneally could rescue 100% of mice with A. baumannii systemic infections (19). In that study, the A. baumannii strain used has an undefined capsule type, which enabled us to correlate the activity with our K2 depolymerase. Furthermore, the authors used the whole tail spike protein (N-terminal tail domain plus the C-terminal pectate lyase 3 domain), while we cloned and used only the pectate lyase fragment that confers capsule depolymerase activity, which makes a direct comparison more difficult. Still, these previous works prove that these phage-derived proteins are able to reproductively reduce bacterial virulence in vivo. Nevertheless, particular attention must be given to the time and route of administration, as well as the serotype strains used, which have been shown to influence the therapeutic outcomes (36). Overall, the in vivo efficacy of capsular depolymerases suggests that a therapeutic approach based on capsular depolymerases represents a promising alternative to treat bacterial infections and, in particular, those of A. baumannii associated with extended drug resistance.
Aiming to elucidate the mechanism of cooperation between the capsular depolymerase and host immune system to control bacterial infections, we performed additional tests. We hypothesized that a complement system-dependent mechanism should be responsible for the clearance of enzyme-treated bacteria and protection against infection, since cyclophosphamide, used to immunocompromise mice, reduces the level of leukocytes but not those associated with the host complement system (37). To prove this, we isolated human serum and mixed it with the NIPH 2061 strain (K2 type) cells in the presence and absence of the K2 depolymerase. The experiments indicated that the enzyme could efficiently sensitize the bacteria in serum. The fact that the bacteria could not be sensitized to the host complement system in the presence of heat-inactivated enzyme showed that the killing effect was solely attributed to the lytic effect of the complement system.
The K2 depolymerase characterized here fit the new trend of alternative antibacterial agents needed against multidrug-resistant A. baumannii. Besides being highly specific, stable, and able to reduce the virulence of A. baumannii in in vivo models, the enzyme also seems to be refractory to resistance development. This was shown by challenging the NIPH 2061 strain with the enzyme for 24 h, where the isolated colonies remained sensitive to the K2 depolymerase in drop tests and to the serum complement only after stripping the cells from their polymeric coats with the K2 depolymerase. This can be explained by the unique mode of action of the capsular depolymerases that do not kill bacteria but instead degrade the extracellular polymers from the bacteria that protect them from the environment, which makes the development of bacterium-resistant variants unlikely.
In conclusion, the antivirulence properties, together with the high stability, versatility of the K2 depolymerase toward extreme conditions of pH, ionic strength, and temperatures, and the low probability of resistance development, make this enzyme a potential therapeutic agent against multidrug-resistant A. baumannii infections. In an era where multidrug-resistant infections are increasing, capsular depolymerases may play a vital role as surrogate antibiotics.
MATERIALS AND METHODS
Capsular depolymerase cloning and expression.
The depolymerase coding sequence was amplified (forward primer, 5′-GGATCCGATCCGAATATTGATATGACTGG; reverse primer, 5′- CTCGAGTTAACTCGTTGCTGTAAATGC; with restriction sites for BamHI and XhoI) from the C-terminal region of the B3_42 open reading frame (genetic region of bp 567 to 2100) of the previously isolated A. baumannii phage vB_AbaP_B3 (GenBank accession number MF033348), further mentioned as phage B3 (20). This fragment, named K2 depolymerase, was cloned into a pTSL vector (GenBank accession no. KU314761) containing an N-terminal SlyD leader protein, as well as solubility enhancer, with a His tag as a leader (38). The K2 depolymerase was expressed and purified exactly as previously described (21). Briefly, recombinant hybrid protein (SlyDK2 depolymerase) was expressed in BL21 cells at 21°C for 16 h with agitation, pelleted, suspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl), disrupted by sonication, and purified in immobilized metal affinity chromatography using nickel resins. The hybrid protein (SlyDK2 depolymerase) was digested with tobacco etch virus protease (protease/protein ratio of 1:100 [wt/wt]) to separate the His-SlyD expression tag from the K2 depolymerase and repurified. The K2 depolymerase eluted fraction was dialyzed in 20 mM PBS and quantified by a Pierce bicinchoninic acid protein assay kit (Thermo Scientific).
Capsular depolymerase functional analysis.
K2 depolymerase was characterized in vitro in terms of (i) activity spectrum, (ii) degradation of extracted exopolysaccharides, (iii) structural thermostability, and (iv) ability to remove the phage B3 receptor. First, the enzyme activity spectrum was determined using drop tests. Enzyme serial dilution drops between 0.0005 and 5 μg (equivalent to 10 μl drops of 0.001 to 10 μM solutions) were spotted onto 28 A. baumannii strains (having 22 different capsular types, namely, K1 to K3, K9, K11, K15, K30, K33, K35, K37, K40, K43 to K49, K57, K73, and K83, described in reference 21 and listed in Table 1) overlaid in tryptic soy broth-agar plates (TSB with 0.6% [wt/vol] agar). The presence of a hazy spot indicates capsule-degrading activity. Second, the ability of the K2 depolymerase to hydrolyze extracted exopolysaccharides under different environmental conditions was performed exactly as described elsewhere (21). Briefly, 5 mg/ml of extracted exopolysaccharides was dissolved into different buffer systems to simulate different pH (5 to 9), ionic strength (0 to 500 mM), and temperature (20 to 80°C) conditions and incubated with the K2 depolymerase or with PBS at 37°C for 1 h. A final concentration of 0.05 μg/μl was used (equivalent to 1 μM). The hydrolytic activity was assessed with the 3,5-dinitrosalicylic acid (DNS) method. Circular dichroism (CD) spectroscopy then was employed to measure the melting temperature (Tm) exactly as described previously (21). Briefly, thermal denaturation was monitored through changes in the ellipticity recorded at 222 nm from 25°C to 90°C with a heating rate of 1°C/min. Measurements were performed using the K2 depolymerase at 10 μM in 10 mM potassium phosphate buffer (pH 7). The melting curves were plotted as a function of temperature and fitted to the Boltzmann sigmoidal curve. Finally, we further illuminated the role of K2 depolymerase in phage adsorption onto the host bacterium as described previously (21). Briefly, NIPH 2061 strain was incubated with PBS or K2 depolymerase for 2 h, spun down, washed twice, and added to the phage B3 at a multiplicity of infection of 0.001. After 5 min of incubation, phage B3 was quantified in the supernatants and the adsorptions were calculated as percentages by the difference between total phage titer and the phage that did not adsorb.
Resistance development.
To evaluate possible resistance development, the frequency of bacterial variants insensitive to K2 depolymerase under selective pressure was determined as follows. An overnight culture of NIPH 2061 strain was 100× diluted in TSB and incubated with PBS or K2 depolymerase for 24 h with agitation, pelleted, washed twice with TSB, and subcultured two more times in the presence of PBS or K2 depolymerase. Three different K2 depolymerase concentrations of 0.005 μg/μl, 0.05 μg/μl, and 0.25 μg/μl (equivalent to 0.1, 1, and 5 μM) were used. The cultures were plated to isolate colonies, and 20 were subcultured three times in Trypticase soy agar (TSA) plates before testing the sensitivity toward K2 depolymerase using drop tests.
Galleria mellonella model.
G. mellonella wax moth larvae were reared in our insectarium at 25°C in the dark, from egg to last-instar larvae, on a natural diet (beeswax and pollen grains). Worms of the final-instar larval stage, weighing 250 ± 25 mg, were selected to be used in the experiments. The G. mellonella survival experiment was adapted from previous studies with small changes (31, 39). Briefly, NIPH 2061 strain overnight cultures were grown in fresh TSB at 37°C and 200 rpm to exponential phase, and cells were harvested, washed, and suspended in 20 mM HEPES (pH 7) to 106 CFU per volume of injection (5.5 μl). Using a hypodermic microsyringe, the larvae were injected with 106-CFU suspensions via the hindmost left proleg, previously surface sanitized with 70% (vol/vol) alcohol. To assess the antivirulence effect of the K2 depolymerase, the wax moth larvae were injected either with bacteria pretreated with K2 depolymerase for 2 h at 37°C prior to inoculation or with K2 depolymerase administered 30 min after bacterial infection via the penultimate right proleg. Three different enzyme doses (0.25 μg, 0.5 μg, and 3 μg/larvae) were used. Two group controls were used: larvae injected with HEPES to monitor the killing due to injection trauma and larvae injected with K2 depolymerase to evaluate the toxicity of the enzyme. After inoculation, larvae were kept in petri dishes and maintained in the dark at 37°C for 72 h. The larval survival was assessed daily during that period, and caterpillars were considered dead based on the lack of mobility in response to touch.
Mouse model.
Animal experimentation was performed at the Life and Health Sciences Research Institute at the University of Minho, where a license in accordance with European guidelines for the care and use of animals for research purposes is granted. Animals were handled in accordance with Directive 2010/63/EU of the European Parliament and of the Council on the Protection of Animals Used for Scientific Purposes (transposed to Portuguese law Decreto-Lei 2013/113, 7 August 2013). The study was approved by the Portuguese national authority for animal experimentation, Direção Geral de Alimentação e Veterinária (DGAV 8421 from 2018).
In vivo experimentation was carried out with 6- to 8-week-old BALB/c mice (n = 20). To avoid gender bias, both sexes were used, and mice were caged separately. After a period of acclimatization, mice were immunosuppressed with two intraperitoneal injections of cyclophosphamide at a dose of 100 mg/kg of body weight 4 days and 1 day before infection. Mice were challenged with a 1 × 107-CFU suspension of NIPH 2061 strain via intraperitoneal injections. One hour after challenge, mice were treated with PBS (n = 10) or K2 depolymerase (n = 10) with a dose of 50 μg/mouse via intraperitoneal injections. Mock- and enzyme-treated groups were monitored up to 30 days to assess survival rate.
Pathological examinations.
In an independent experiment, cyclophosphamide-pretreated mice (n = 24) were challenged with 1 × 107 CFU of NIPH 2061 strain and treated 1 h later with PBS (n = 12) or K2 depolymerase (n = 12) at 50 μg/mouse via intraperitoneal injections. Before organ harvesting, mice were euthanized via CO2 inhalation, starting with a CO2 flow rate that displaced 10 to 30% of the cage volume per minute. Spleen and livers were aseptically removed from mice 12 h postinfection, and homogenates were 10-fold serially diluted in sterile saline and cultured on LB agar plates at 37°C for 18 h. Quantification of viable bacteria was assessed by CFU counts and expressed as log10 CFU per organ.
Blood biochemical assays.
Serum was collected from whole blood by centrifugation at 3,000 × g for 15 min. TNF-α and IL-6 were measured in the mocked- and enzyme-treated groups by enzyme-linked immunosorbent assay (ELISA) using commercially available ELISA kits (mouse TNF-α max standard set and mouse IL-6 max standard set; BioLegend) according to the manufacturer’s instructions.
Human serum assay.
The serum killing assay was adapted from a previous protocol (40). The NIPH 2061 strain was grown overnight in TBS, diluted in TBS to ≈5 × 104 CFU/ml, and mixed with fresh human serum at a 1:3 (serum/cells) ratio. The serum was isolated from donations by healthy volunteers. The samples were mixed with (i) PBS, (ii) K2 depolymerase, or (iii) heat-inactivated (100°C at 15 min) enzyme for 1 h at 37°C. A final concentration of 0.05 μg/μl of enzyme was used (equivalent to 1 μM). Percent survival of enzyme-treated cells was determined based on viable counts relative to controls without enzyme. A similar experimental was also performed using ten isolated colonies from NIPH 2061 challenge cultures with the K2 depolymerase to evaluate their susceptibility to serum killing effect.
Statistical analysis.
Differences between two experimental groups were analyzed with the two-tailed Student's t test. Survival curves were plotted using the Kaplan-Meier method, and the differences in survival were calculated by using the log-rank Mantel-Cox statistical test, all performed with GraphPad Prism6 (GraphPad Software, Inc., La Jolla, CA). A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Filipa Grosso for providing the A. baumannii K2 isolates H466, H580, H603, and H678. This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of the UID/BIO/04469/2019 unit, Project PTDC/BBB-BSS/6471/2014, and COMPETE 20202 (POCI-01-0145-FEDER-016678, POCI-01-0145-FEDER-007038, and POCI-01-0145-FEDER-006684). This work was also supported by a BioTecNorte operation (NORTE-01-0145-FEDER-000004, NORTE-01-0145-FEDER-000013, and NORTE-01-0145-FEDER-000023), funded by the European Regional Development Fund under the scope of Norte2020–Programa Operacional Regional do Norte. This study was additionally supported by Infect-ERA grant Infect-ERA/0002/2015: BU_SPONT_HEAL. H.O., A.G.F., and D.M.-H. acknowledge the FCT grants SFRH/BPD/111653/2015, FRH/BPD/112903/2015, and SFRH/BDP/91831/2012. A.M. acknowledges the NORTE-08-5369-FSE-000041 project. Funding received by the Institute for Bioengineering and Biosciences (iBB) from FCT (UID/BIO/04565/2013) and from Programa Operacional Regional de Lisboa 2020 (Project N. 007317) is acknowledged. We have no conflicts of interest to declare.
REFERENCES
- 1.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tal-Jasper R, Katz DE, Amrami N, Ravid D, Avivi D, Zaidenstein R, Lazarovitch T, Dadon M, Kaye KS, Marchaim D. 2016. Clinical and epidemiological significance of carbapenem resistance in Acinetobacter baumannii infections. Antimicrob Agents Chemother 60:3127–3131. doi: 10.1128/AAC.02656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 5.Zilberberg MD, Kollef MH, Shorr AF. 2016. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: a survey study. J Hosp Med 11:21–26. doi: 10.1002/jhm.2477. [DOI] [PubMed] [Google Scholar]
- 6.Spellberg B, Rex JH. 2013. The value of single-pathogen antibacterial agents. Nat Rev Drug Discov 12:963. doi: 10.1038/nrd3957-c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esterly JS, Griffith M, Qi C, Malczynski M, Postelnick MJ, Scheetz MH. 2011. Impact of carbapenem resistance and receipt of active antimicrobial therapy on clinical outcomes of Acinetobacter baumannii bloodstream infections. Antimicrob Agents Chemother 55:4844–4849. doi: 10.1128/AAC.01728-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun 78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lees-Miller RG, Iwashkiw JA, Scott NE, Seper A, Vinogradov E, Schild S, Feldman MF. 2013. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol 89:816–830. doi: 10.1111/mmi.12300. [DOI] [PubMed] [Google Scholar]
- 11.Kenyon JJ, Kasimova AA, Shneider MM, Shashkov AS, Arbatsky NP, Popova AV, Miroshnikov KA, Hall RM, Knirel YA. 2017. The KL24 gene cluster and a genomic island encoding a Wzy polymerase contribute genes needed for synthesis of the K24 capsular polysaccharide by the multiply antibiotic resistant Acinetobacter baumannii isolate RCH51. Microbiology 163:355–363. doi: 10.1099/mic.0.000430. [DOI] [PubMed] [Google Scholar]
- 12.Arbatsky NP, Shneider MM, Dmitrenok AS, Popova AV, Shagin DA, Shelenkov AA, Mikhailova YV, Edelstein MV, Knirel YA. 2018. Structure and gene cluster of the K125 capsular polysaccharide from Acinetobacter baumannii MAR13-1452. Int J Biol Macromol 117:1195–1199. doi: 10.1016/j.ijbiomac.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Lin H, Paff ML, Molineux IJ, Bull JJ. 2017. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front Microbiol 8:2257. doi: 10.3389/fmicb.2017.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. 2005. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J Antimicrob Chemother 56:160–165. doi: 10.1093/jac/dki177. [DOI] [PubMed] [Google Scholar]
- 15.Pan YJ, Lin TL, Lin YT, Su PA, Chen CT, Hsieh PF, Hsu CR, Chen CC, Hsieh YC, Wang JT. 2015. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother 59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin TL, Hsieh PF, Huang YT, Lee WC, Tsai YT, Su PA, Pan YJ, Hsu CR, Wu MC, Wang JT. 2014. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J Infect Dis 210:1734–1744. doi: 10.1093/infdis/jiu332. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh PF, Lin HH, Lin TL, Chen YY, Wang JT. 2017. Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci Rep 7:4624. doi: 10.1038/s41598-017-04644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu CR, Lin TL, Pan YJ, Hsieh PF, Wang JT. 2013. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS One 8:e70092. doi: 10.1371/journal.pone.0070092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Leung SSY, Guo Y, Zhao L, Jiang N, Mi L, Li P, Wang C, Qin Y, Mi Z, Bai C, Gao Z. 2019. The capsule depolymerase Dpo48 rescues Galleria mellonella and mice from Acinetobacter baumannii systemic infections. Front Microbiol 10:545. doi: 10.3389/fmicb.2019.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira H, Costa AR, Konstantinides N, Ferreira A, Akturk E, Sillankorva S, Nemec A, Shneider M, Dotsch A, Azeredo J. 2017. Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ Microbiol 19:5060–5077. doi: 10.1111/1462-2920.13970. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira H, Costa AR, Ferreira A, Konstantinides N, Santos SB, Boon M, Noben JP, Lavigne R, Azeredo J. 2018. Functional analysis and anti-virulent properties of a new depolymerase from a myovirus that infects Acinetobacter baumannii capsule K45. J Virol 93:e01163-18. doi: 10.1128/JVI.01163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perichon B, Goussard S, Walewski V, Krizova L, Cerqueira G, Murphy C, Feldgarden M, Wortman J, Clermont D, Nemec A, Courvalin P. 2014. Identification of 50 class D beta-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob Agents Chemother 58:936–949. doi: 10.1128/AAC.01261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenyon JJ, Marzaioli AM, Hall RM, De Castro C. 2014. Structure of the K2 capsule associated with the KL2 gene cluster of Acinetobacter baumannii. Glycobiology 24:554–563. doi: 10.1093/glycob/cwu024. [DOI] [PubMed] [Google Scholar]
- 24.Liliana Silva CR, Filipa G, Luisa M, Vieira P. 2018. The secret is on sugar: capsular type explains the discrimination of Acinetobacter baumannii clones by Fourier-transform infrared (FT-IR) spectroscopy and multilocus sequence typing. 28th ECCMID–European Congress of Clinical Microbiology, Madrid, Spain. [Google Scholar]
- 25.Pires DP, Oliveira H, Melo LD, Sillankorva S, Azeredo J. 2016. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol 100:2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- 26.Taylor CM, Roberts IS. 2005. Capsular polysaccharides and their role in virulence. Contrib Microbiol 12:55–66. doi: 10.1159/000081689. [DOI] [PubMed] [Google Scholar]
- 27.Zhensong Wen J-R. 2015. Bacterial capsules, p 33–53. In Tang Y-W, Schwartzman J, Sussman M, Poxton I (ed), Molecular medical microbiology, 2nd ed, vol 1 Academic Press, New York, NY. [Google Scholar]
- 28.Jann K, Jann B. 1992. Capsules of Escherichia coli, expression and biological significance. Can J Microbiol 38:705–710. doi: 10.1139/m92-116. [DOI] [PubMed] [Google Scholar]
- 29.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang-Lin SX, Olson R, Beanan JM, MacDonald U, Balthasar JP, Russo TA. 2017. The capsular polysaccharide of Acinetobacter baumannii is an obstacle for therapeutic passive immunization strategies. Infect Immun 85:e00591-17. doi: 10.1128/IAI.00591-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majkowska-Skrobek G, Łątka A, Berisio R, Maciejewska B, Squeglia F, Romano M, Lavigne R, Struve C, Drulis-Kawa Z. 2016. Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 8:E324. doi: 10.3390/v8120324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, Dutilh BE, Brouns S. 2018. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16:760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Li GY, Mo ZL, Chai ZH, Shang AQ, Mou HJ. 2014. Properties of Klebsiella phage P13 and associated exopolysaccharide depolymerase. J Ocean Univ China 13:163–168. doi: 10.1007/s11802-014-2042-6. [DOI] [Google Scholar]
- 34.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majkowska-Skrobek G, Latka A, Berisio R, Squeglia F, Maciejewska B, Briers Y, Drulis-Kawa Z. 2018. Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front Microbiol 9:2517. doi: 10.3389/fmicb.2018.02517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin H, Paff ML, Molineux IJ, Bull JJ. 2018. Antibiotic therapy using phage depolymerases: robustness across a range of conditions. Viruses 10:622. doi: 10.3390/v10110622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XY, Lin ZY, Zhu PP, Jin YF. 1987. Effect of cyclophosphamide on serum complement in mice. Zhongguo Yao Li Xue Bao 8:79–83. [PubMed] [Google Scholar]
- 38.Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG. 2016. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533:346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- 39.Mil-Homens D, Rocha EPC, Fialho AM. 2010. Genome-wide analysis of DNA repeats in Burkholderia cenocepacia J2315 identifies a novel adhesin-like gene unique to epidemic-associated strains of the ET-12 lineage. Microbiology 156:1084–1096. doi: 10.1099/mic.0.032623-0. [DOI] [PubMed] [Google Scholar]
- 40.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]