We successfully achieved efficient secretory production and improved the catalytic efficiency of diacetylchitobiose deacetylase in Bacillus subtilis, and this provides a good foundation for the application of diacetylchitobiose deacetylase in the production of chitosan oligosaccharides and monosaccharides.
KEYWORDS: 5′-untranslated-region mutations, Bacillus subtilis, diacetylchitobiose deacetylase, secretory expression, site-directed saturation mutagenesis
ABSTRACT
Diacetylchitobiose deacetylase has great application potential in the production of chitosan oligosaccharides and monosaccharides. This work aimed to achieve high-level secretory production of diacetylchitobiose deacetylase by Bacillus subtilis and perform molecular engineering to improve catalytic performance. First, we screened 12 signal peptides for diacetylchitobiose deacetylase secretion in B. subtilis, and the signal peptide YncM achieved the highest extracellular diacetylchitobiose deacetylase activity of 13.5 U/ml. Second, by replacing the HpaII promoter with a strong promoter, the P43 promoter, the activity was increased to 18.9 U/ml. An unexpected mutation occurred at the 5′ untranslated region of plasmid, and the extracellular activity reached 1,548.1 U/ml, which is 82 times higher than that of the original strain. Finally, site-directed saturation mutagenesis was performed for the molecular engineering of diacetylchitobiose deacetylase to further improve the catalytic efficiency. The extracellular activity of mutant diacetylchitobiose deacetylase R157T reached 2,042.8 U/ml in shake flasks. Mutant R157T exhibited much higher specific activity (3,112.2 U/mg) than the wild type (2,047.3 U/mg). The Km decreased from 7.04 mM in the wild type to 5.19 mM in the mutant R157T, and the Vmax increased from 5.11 μM s−1 in the wild type to 7.56 μM s−1 in the mutant R157T.
IMPORTANCE We successfully achieved efficient secretory production and improved the catalytic efficiency of diacetylchitobiose deacetylase in Bacillus subtilis, and this provides a good foundation for the application of diacetylchitobiose deacetylase in the production of chitosan oligosaccharides and monosaccharides.
INTRODUCTION
Diacetylchitobiose deacetylase (Dac) derives from Archaea and plays an important role in a unique chitin degradation pathway in Archaea (1). It belongs to the carbohydrate esterase family 14 and works in combination with glucosaminidase to hydrolyze diacetylchitobiose (GlcNAc2) to glucosamine (GlcN). First, the N-acetyl group of GlcNAc2 is catalyzed by Dac from the nonreducing end residue N-acetylglucosamine (GlcNAc) of GlcNAc2 and generates the product GlcN-GlcNAc; the product (GlcN-GlcNAc) is then hydrolyzed by glucosaminidase following degradation into GlcN and GlcNAc. Finally, the resulting monomer GlcNAc is catalyzed by Dac to generate GlcN (2). The pathway of chitin degradation involving deacetylation by Dac is shown in Fig. S1 in the supplemental material. Since Dac is derived from hyperthermophilic archaea, it has excellent resistance and stability to high temperature, providing great advantages in industrial applications. Currently, Dac is a key enzyme used in the biodegradation of chitin and chitosan to produce chitosan oligosaccharides and monosaccharides. For example, it can be used for the production of GlcN (3, 4), which is utilized as a medicine and functional food to treat osteoarthritis and preserve cartilage around the joints (5).
To date, Dacs derived from diverse sources can be expressed in Escherichia coli to investigate the degradation mechanism of chitin (1, 2, 4, 6); however, all the Dacs overexpressed by the plasmids were insoluble and accumulated in inclusion bodies. The gene encoding Dac was inserted into E. coli’s chromosome to produce recombinant protein which is soluble in a suitable buffer solution, while the yield of Dac is extremely low and the amount of soluble protein was lower than 40 mg/liter (7). A recombinant Bacillus subtilis strain expressing Dac without using signal peptide for whole-cell biocatalytic production of GlcN was constructed in our previous work, while the yield of intracellular Dac is still low, around 80 mg/liter (4). To date, there are no reports about the secretory production of Dac. After decades of investigation, the secretion system of genus Bacillus is now very evident (8). In addition, B. subtilis is one of the most identifiable Gram-positive bacteria. In contrast to Gram-negative bacteria, such as E. coli, Gram-positive bacteria can secrete proteins directly into the medium because they have only one layer of membrane (9, 10). Besides, misfolded synthesized proteins and inclusion bodies can be removed by the cellular control systems, which could provide a high-quality protein production experiment (11, 12). Therefore, it has unique advantages as an industrial host strain to massively produce recombinant proteins (13, 14). Thus, we chose B. subtilis as the host to achieve secretory production of Dac.
There are many strategies to enhance the yield of heterologous proteins. Signal peptides are key factors for protein expression and secretion. To achieve high-level secretion of enzymes, it is necessary to fuse a suitable signal peptide at its N terminus, as a guided functional fraction (15). The prediction and verification of many signal peptides have been performed at the genome and proteome levels (16). In B. subtilis, there are several secretion pathways, of which the general secretory (Sec) pathway and the twin-arginine translocation (Tat) pathway are the most typical and have been studied in depth. Compared with that of the Tat pathway, the mechanism of secretion by signal peptides has been studied more thoroughly (17). Many studies have indicated that the Sec pathway achieves high-level secretion of different enzymes (14, 18, 19). In a heterologous protein expression system, the promoter is also a critical regulatory element to control the strength of gene expression. Altering the protein expression in engineered cells by changing promoters is an effective strategy to construct industrial strains. An inducible promoter can control the expression timing and levels of different target genes. However, it needs real-time monitoring of the fermentation process, and the addition of inducers increases the cost of industrialization. In this work, for the high-intensity expression of a single gene in the plasmid, we choose the stronger constitutive promoters to regulate the expression of Dac (20).
The 5′ untranslated region (5′ UTR) exists both in eukaryotes and in prokaryotes. The 5′ UTR initiates at the transcription start site and finishes at the first nucleotide of the initiation codon, containing numerous regulatory components (21). The length of 5′ UTR varies greatly, and it can possibly range from a few to several thousand nucleotides. Generally, the 5′ UTR of prokaryotic mRNA is usually much shorter than that of eukaryotic mRNA (22). It has been proved that the secondary structures of 5′ UTRs in mRNAs can stabilize mRNAs of many bacterial genes (23, 24). The stabilizing effect of 5′ structures is also important in controlling the expression of genes (25). Therefore, the changes in the secondary structure of mRNA may have an enormous impact on increasing the production of heterologous protein in B. subtilis.
In this study, we constructed a recombinant B. subtilis to produce Dac efficiently by overexpressing the gene encoding Dac (PH0499), which comes from Pyrococcus horikoshii. First, the appropriate signal peptide and strong promoter were selected to achieve successful secretory expression of Dac. Then, a mutation occurred at the 5′ UTR that significantly enhanced the expression of Dac. Further determination of the transcriptional level of the gene encoding Dac in recombinant strains by quantitative real-time PCR (qRT-PCR) showed that the 5′ UTR mutation greatly increased the transcription level of the Dac gene at different fermentation phases. Besides, the mRNA stability and the translation initiation rate (TIR) of 5′ UTR sequences were predicted by RBS Calculator v2.0. The results indicated that the TIR was increased by the 5′ UTR mutation. Then, the catalytic efficiency of Dac was increased by molecular engineering. The kinetic parameters of Dac and its positive mutants were analyzed. The final engineered B. subtilis strain can efficiently produce extracellular Dac, which is meaningful for the use of Dac to produce chitosan oligosaccharides and monosaccharides in the future.
RESULTS
Signal peptide and promoter engineering for achieving high-level secretion of Dac.
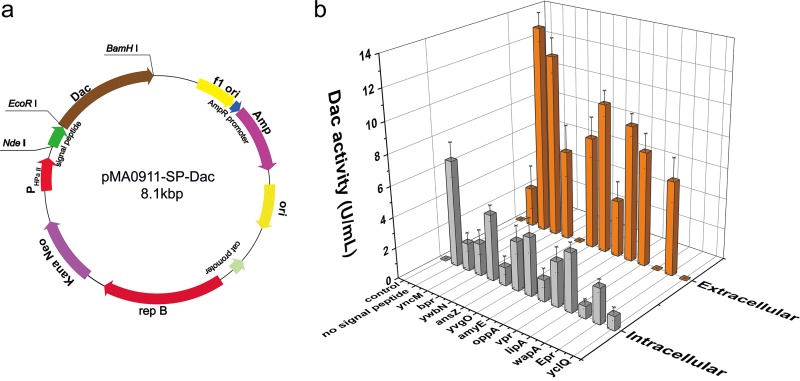
To screen for a suitable signal peptide, 12 signal peptides (Table 1) from the Sec pathway which were reported to achieve high-level secretion of different endogenous proteins and heterologous recombinant enzymes in B. subtilis were selected (17, 18, 26). According to the preference of codons in a different host, the complete open reading frame (ORF) of P. horikoshii Dac (PH0499) was optimized for expression in B. subtilis (the nucleotide sequence is shown in Fig. S2 in the supplemental material). The sequence encoding the mature Dac (272 residues) was cloned into the expression vector pMA0911. Next, 12 signal peptides was inserted in front of the N terminus of the mature Dac. The Dac gene fused with the corresponding signal peptide was expressed in B. subtilis WB600 under the HpaII promoter. We successfully constructed a series of plasmids carrying the Dac gene and different signal peptides. The structure of the expression plasmid is shown in Fig. 1a. Then, the recombinant plasmids were transformed into B. subtilis WB600. Figure 1b reveals that neither intracellular nor extracellular Dac activity was found in the control group (B. subtilis harboring pMA0911 plasmid), while some intracellular Dac activity appeared in the strain expressing Dac without the signal peptide. The intracellular activity of this strain was the highest among all strains. As shown in Fig. 1b, the result suggests that 9 signal peptides (YncM, Bpr, AmyE, Vpr, LipA, Epr, YwbN, YvgO, and OppA) result in the secretion of mature Dac. Among them, the signal peptide YncM achieved the highest extracellular Dac activity of 13.5 U/ml, while the lowest extracellular activity of 3.7 U/ml was obtained with the signal peptide OppA. The other three signal peptides (AnsZ, WapA, and YclQ) did not result in secretion of mature Dac; extracellular Dac activities were not detected in their fermentation supernatants. The SDS-PAGE analysis (see Fig. S3) revealed the presence of 32-kDa bands from the the culture supernatants of cells which expressed Dac with signal peptides YncM, Vpr, Bpr, and AmyE, while the others have no distinct Dac band. Among them, the 32-kDa band of Dac guided by YncM had the thickest stripe. The above results indicate that YncM is the optimal signal peptide to produce Dac in B. subtilis. Furthermore, the recombinant strain harboring pMA0911J (Dac gene fused with a sequence encoding YncM) and a control strain were collected and disrupted with an ultrasonic cell disruption system, as manifested in the SDS-PAGE analysis (see Fig. S4); there were no insoluble fractions in the recombinant strain. By comparing the activities of Dac in and out of cells, it was found that majority of the Dac was conveyed by signal peptide YncM.
TABLE 1.
Signal peptides used for Dac secretion in B. subtilis WB600
| Name | Amino acid sequencea | Charge in N regionb | Hydrophobic amino acidc |
|---|---|---|---|
| YncM | MAKPLSKGGILVKKVLIAGAVGTAVLFGTLSSGIPGLPAADAQVAKA | 4 | 35 |
| Bpr | MRKKTKNRLISSVLSTVVISSLLFPGAAGA | 3 | 17 |
| YwbN | MSDEQKKPEQIHRRDILKWGAMAGAAVA | 0 | 15 |
| AnsZ | MKKQRMLVLFTALLFVFTGCSHS | 2 | 13 |
| YvgO | MKRIRIPMTLALGAALTIAPLSFASA | 3 | 19 |
| AmyE | MFAKRFKTSLLPLFAGFLLLFHLVLAGPAAASA | 3 | 26 |
| OppA | MKKRWSIVTLMLIFTLVLSA | 3 | 13 |
| Vpr | MKKGIIRFLLVSFVLFFALSTGITGVQA | 2 | 20 |
| LipA | MKFVKRRIIALVTILMLSVTSLFALQPSAKA | 4 | 20 |
| WapA | MKKRKRRNFKRFIAAFLVLALMISLVPADVLA | 7 | 21 |
| Epr | MKNMSCKLVVSVTLFFSFLTIGPLAHA | 1 | 17 |
| YclQ | MKKFALLFIALVTAVVISACGNQSTSSKG | 2 | 17 |
Boldface letters present the hydrophobic H domain. The residues at positions −3 to −1 relative to the predicted signal peptidase (SPase) cleavage sites are underlined.
Calculated with aspartate and glutamate defined as −1, arginine and lysine defined as +1, and any other amino acid defined as 0.
Amino acids G, A, V, L, I, M, F, W, and P were defined as hydrophobic, and the other residues were characterized as hydrophilic.
FIG 1.
Dac production in the recombinant B. subtilis WB600 using different signal peptides. (a) Construction of recombinant plasmid pMA0911-SP-Dac with different signal peptides. (b) The intracellular (light gray) and extracellular (dark orange) Dac activities of the recombinant strains. Control, B. subtilis WB600 harboring pMA0911 plasmid; no signal peptide, B. subtilis WB600 harboring pMA0911NS plasmid. The recombinant strains were grown on a fermentation medium (see Materials and Methods) containing 10 mg/liter of kanamycin in 500-ml flask and incubated at 37°C and 220 rpm for 48 h. Error bars indicate the average values from three parallel experiments.
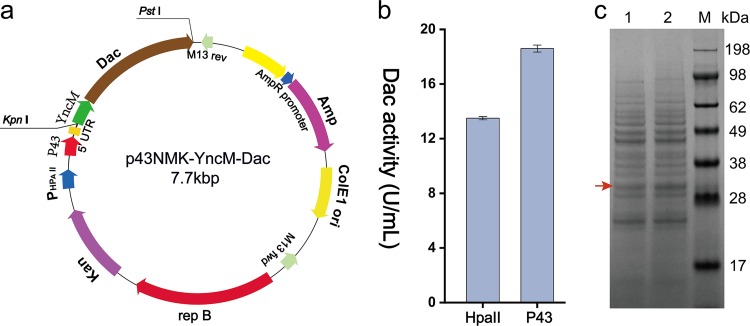
To enhance Dac production further, the HpaII promoter was replaced with a stronger promoter, P43. The Dac gene with the sequence encoding signal peptide YncM was cloned into the vector pP43NMK to produce pP43NMK-YncM-Dac, in which the expression system is controlled by the P43 promoter. The structure of the expression plasmid is shown in Fig. 2a. Then, the plasmid pP43NMK-YncM-Dac was transferred into B. subtilis WB600. The results shown in Fig. 2b and c indicate that Dac was expressed successfully, and the activity of extracellular Dac in the culture supernatant increased from 13.5 U/ml with the HpaII promoter to 18.6 U/ml with the P43 promoter.
FIG 2.
Dac production in the recombinant B. subtilis WB600 using different promoters. (a) Construction of recombinant plasmid p43NMK-YncM-Dac using p43NMK vector with P43 promoter. (b) The extracellular Dac activities of the recombinant strains using different promoters. (c) SDS-PAGE analysis of extracellular Dac of the recombinant strains. M, protein marker; 1, B. subtilis WB600 strains expressing Dac using HpaII promoter; 2, B. subtilis WB600 strains expressing Dac using P43 promoter. The recombinant strains were grown on fermentation medium (see Materials and Methods) containing 10 mg/liter of kanamycin in 500-ml flask and incubated at 37°C and 220 rpm for 48 h. Error bars indicate the average values from three parallel experiments.
5′ UTR mutation significantly enhanced the expression of Dac.
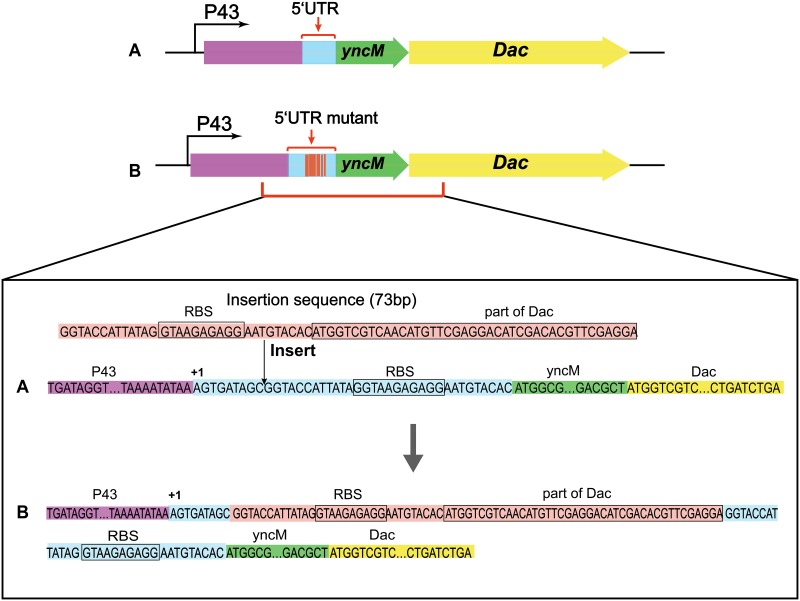
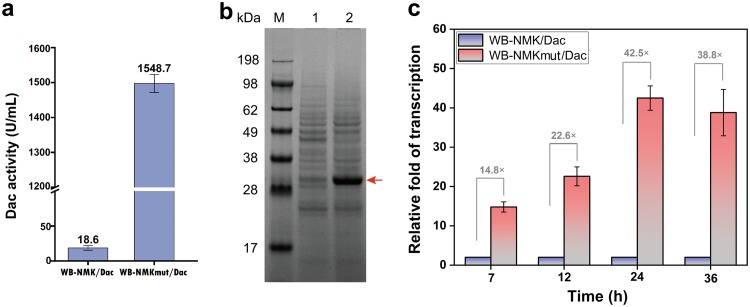
In the process of plasmid construction, we unexpectedly obtained a plasmid mutant named p43NMKmut-YncM-Dac. The nucleotide sequencing results showed that the mutation was a 73-bp insertion including part of gene encoding Dac and the original ribosome binding site (RBS) in the 5′ UTR; the specific sequences are shown in Fig. 3 We transformed the plasmid mutant p43NMKmut-YncM-Dac into B. subtilis WB600, yielding the recombinant strain WB-NMKmut/Dac which produced an extracellular Dac activity of 1,548.1 U/ml, 82 times that from the strain that expressed the plasmid pP43NMK-YncM-Dac (Fig. 4a). Furthermore, SDS-PAGE analysis shows that WB-NMKmut/Dac exhibited a 32-kDa Dac band of higher intensity in the culture supernatant (Fig. 4b).
FIG 3.
The 5′ UTR sequence of the plasmids p43NMK-YncM-Dac and p43NMKmut-YncM-Dac. (A) 5′ UTR sequence of recombinant plasmid p43NMK-YncM-Dac. (B) 5′ UTR sequence of the plasmid p43NMKmut-YncM-Dac with a 73-bp insertion.
FIG 4.
The effect of 5′ UTR mutation on the production of Dac by the recombinant B. subtilis WB600. (a) The extracellular Dac activities of the recombinant strains. (b) SDS-PAGE analysis of extracellular Dac of the recombinant strains. M, protein marker; 1, WB-NMK/Dac (mature Dac expressed in B. subtilis WB600 strains harboring p43NMK/Dac plasmid); 2, WB-NMKmut/Dac (mature Dac expressed in B. subtilis WB600 strains harboring p43NMKmut/Dac plasmid). (c) qRT-PCR analysis of the Dac mRNA in the recombinant strains WB-NMK/Dac and WB-NMKmut/Dac at different culture phases. The recombinant strains were grown in fermentation medium (see Materials and Methods) containing 10 mg/liter of kanamycin in 250-ml flask and incubated at 37°C and 220 rpm for 60 h. Error bars indicate the average values from three parallel experiments.
To further determine the role of the 5′ UTR mutation in the increase of Dac expression, the transcriptional level of Dac mRNA was measured through qRT-PCR. For the qRT-PCR analysis, the total RNA was derived from the WB-NMK/Dac and WB-NMKmut/Dac strains at culture times of 7, 12, 24, and 36 h, using the WB-NMK/Dac strain as a control. The B. subtilis 16S rRNA gene and hbsU gene served as the internal control genes to normalize the results. Compared with the control strain WB-NMK/Dac (without the 5′ UTR mutation), the transcription level of the Dac gene in the strain WB-NMKmut/Dac at 7, 12, 24, and 36 h was increased by 14.8-, 22.6-, 42.5-, and 38.8-fold, respectively. The results indicate that the 5′ UTR mutation contributed to the significant increase in the transcription of the Dac gene (Fig. 4c).
Additionally, the transcription start sites of the Dac gene in the recombinant strains were determined by rapid amplification of cDNA ends (RACE) using terminal deoxynucleotidyl transferase. The 5′-terminal sequencing maps after RACE experiments in different strains are shown in Fig. S5. The result of RACE suggests that the transcriptional initiation site of Dac in the recombinant strain WB-NMK/Dac is the same as that in strain WB-NMKmut/Dac. Then, the sequences of the 5′ UTRs were analyzed using an equilibrium statistical thermodynamic model named RBS Calculator v2.0. The model calculated the free energies of key molecular interactions, including energetic changes of translation, energetic changes of initiation, and other sequence-dependent energetic variations occurring in the RNA folding and hybridization. The intensity of the molecular interaction between the mRNA transcript and the 30S ribosome was quantified to anticipate the final TIR (25). The analysis results are shown in Table 2 and indicate that the TIR of the 5′ UTR mutant is 4.5 times that the original 5′ UTR sequence. The amount of protein expressed in strain is proportionate to the TIR. As shown in Table 3, TIR is decided by diverse molecular interactions. The Gibbs free energy (ΔGtotal) is a key parameter in the characterization of TIR. ΔGtotal, which could be anticipated by the energy model (equation), depends on the mRNA sequence that surrounds a specific initiation codon (25):
TABLE 2.
Free energy calculated by equilibrium statistical thermodynamic model
| Name of sequence | TIR (au)a | ΔG (kcal/mol) |
|||||
|---|---|---|---|---|---|---|---|
| Total | mRNA-rRNA | Spacing | Standby | Start | mRNA | ||
| Original 5′ UTR | 7,164.11 | −3.91 | −10.42 | 2.94 | −3.51 | −2.76 | −9.8 |
| 5′ UTR mutant | 31,774.77 | −7.22 | −22.15 | 0.92 | −2.48 | −2.76 | −19.6 |
au, artificial units (manually defined relative value obtained through calculation).
TABLE 3.
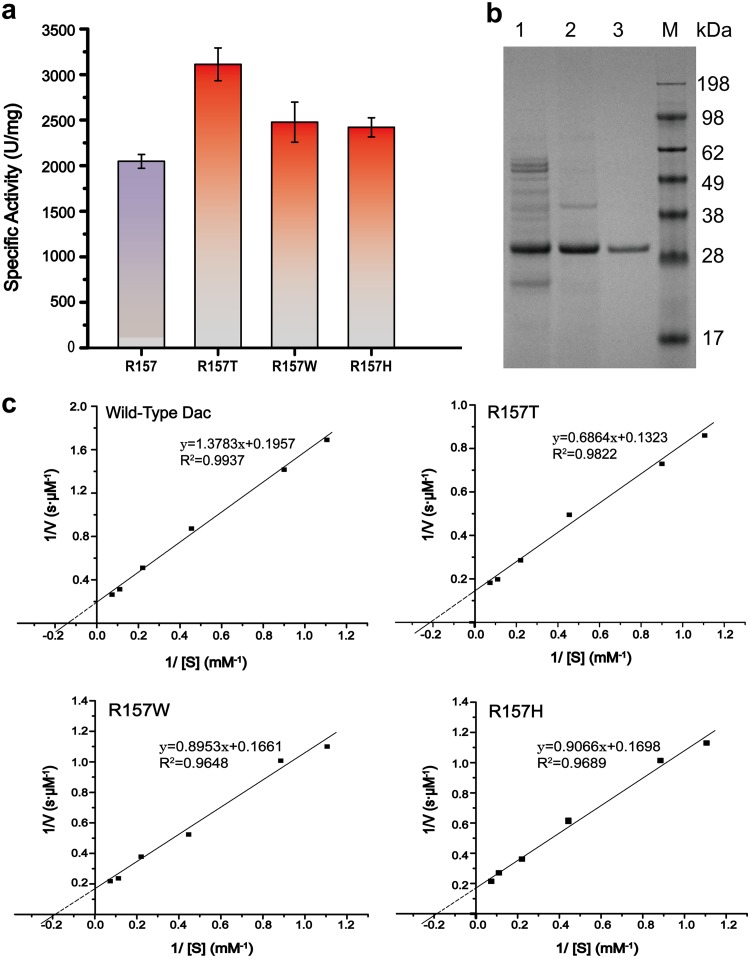
Kinetic parameters of Dac and its mutantsa
| Protein name | Kmb (mM) | Vmaxc (μM s−1) |
|---|---|---|
| Wild-type Dac | 7.04 ± 0.27 | 5.11 ± 0.14 |
| R157T | 5.19 ± 0.16 | 7.56 ± 0.21 |
| R157W | 5.39 ± 0.16 | 6.02 ± 0.19 |
| R157H | 5.34 ± 0.16 | 5.89 ± 0.26 |
Each value was calculated from three independent experiments. The enzyme concentration of the reaction system was 6.16 mg/liter, and the total volume of each reaction mixture was 0.2 ml (50 mM sodium phosphate buffer, pH 8.0).
Km, Michaelis constant.
Vmax, the maximum rate of glucosamine (GlcN) production.
The ΔGmRNA-rRNA represents the released energy resulting from the hybridization and cofolding between the last nine nucleotides of 16S rRNA and the mRNA subsequence; ΔGspacing is one kind of free energy penalty that exists when the spatial distance between the 16S rRNA binding site and the initiation codon is not optimum; ΔGstandby is the work needed to unfold any category of secondary structures isolating the standby site after the 30S complex assembly; ΔGstart is the energy that is released due to the hybridization between the initiation codon and the initiating tRNA anticodon loop (3′-UAC-5′), and ΔGmRNA is the work to fully unfold the minimum free energy structure of the mRNA subsequence, which is the most stable secondary structure of mRNA.
There are some differences in free energy values between the two strains that are apparent by comparing the predicted results of the two strains, WB-NMK/Dac and WB-NMKmut/Dac. The ΔGmRNA-rRNA values of the strains WB-NMK/Dac and WB-NMKmut/Dac are −10.42 and −22.15 kcal/mol, respectively. Therefore, it is reasonable to deduce that the sequence in the WB-NMKmut/Dac strain contained a stronger 16S rRNA binding site than the original 5′ UTR sequence. The ΔGspacing values of the strains WB-NMK/Dac and WB-NMKmut/Dac are 2.94 kcal/mol and 0.92 kcal/mol, respectively. The penalty for nonoptimal spacing of the binding site in the WB-NMKmut/Dac strain is lessened, and the spatial distance between the 16S rRNA binding site and the initiation codon is more suitable, resulting in an increased TIR. The values of ΔGstandby and ΔGstart in the two strains WB-NMK/Dac and WB-NMKmut/Dac were similar, indicating that the interactions, including the binding of tRNA to the initiation codon and the existence of RNA secondary structures that obstruct the standby site, have little difference between the two strains. The ΔGmRNA values of the strains WB-NMK/Dac and WB-NMKmut/Dac differed significantly. The values of the two strains were −9.8 kcal/mol and −19.6 kcal/mol, respectively. The results indicate that in strain WB-NMKmut/Dac, the work required to unfold the subsequence of mRNA, when it folds into its most steady secondary structure, was much more than in strain WB-NMK/Dac. That is to say, the mRNA secondary structure of the WB-NMKmut/Dac strain is stronger than that of strain WB-NMK/Dac. The change in the target gene mRNA secondary structure can make the mRNA more stabilized and thus not effortless to degrade. The predicted secondary structure of mRNA by RBS Calculator v2.0 is shown in Fig. S6.
Molecular engineering of Dac for improved catalytic efficiency.
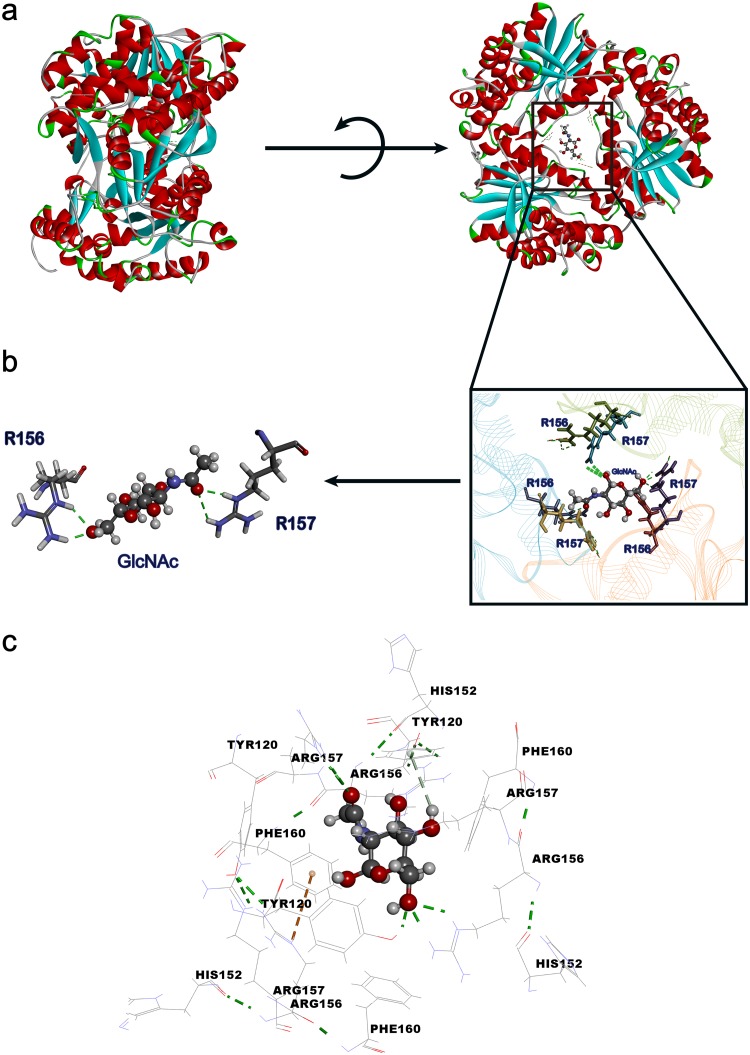
To enhance the catalytic efficiency of Dac, the molecular docking between the substrate GlcNAc and protein Dac was carried out to select the GlcNAc binding sites and determine the molecular engineering targets. As indicated in the three-dimensional (3D) structure of Dac (27), the symmetric structure, a hexamer, is made up of two trimers consisting of three identical molecules (2). According to the result of the molecular docking (Fig. 5a), the active pocket lies in the center of the hexamer. Compared with the hydrogen-bonding capability, we found that the pivotal sites for GlcNAc binding are R156 and R157 (Fig. 5b). The results of the computational analysis of simulated site-saturation mutation indicate that F160 is an important site for the interaction with GlcNAc and has a great influence on the stability of GlcNAc binding to Dac. Moreover, as predicted in a previous study, the bond angles and spatial distances between the Tyr120 and His152 side chains support hydrogen-bonding interactions (27). Thus, we selected these five sites, R156, R157, F160, Tyr120, and His152, for saturation mutation to enhance the catalytic efficiency. Molecular modeling showed that these residues are located at the center, and amorphous loop regions of Dac are commonly reckoned to be of high flexibility (Fig. 5c). According to the site-directed saturation mutagenesis results of these five sites, the saturation mutations of four sites, R156, F160, Tyr120, and His152, have no positive mutants (data not shown). However, we obtained 3 positive R157 mutants, R157W, R157T, and R157H, with higher specific activity (Fig. 6a).
FIG 5.
Protein structure of substrate binding to Dac and the key site of interaction with substrate and substrate entrance sites in Dac. (a) The structure of protein (Dac) and the substrate (GlcNAc) binding to Dac of the inside active pocket. The substrate entrance was located in the center of Dac. (b) The interaction between critical residues and substrate. Green sticks show the hydrogen bonds between R157/R156 residues and GlcNAc. (c) The substrate entrance sites in Dac and distance of key residues from substrate.
FIG 6.
Analysis results of Dac and its mutants. (a) The specific activities of the R157 mutants. (b) SDS-PAGE analysis of the purified Dac. M, protein marker; 1, culture supernatant of recombinant strain WB-NMKmut/Dac after fermentation for 60 h; 2, culture supernatant was heated at 85°C for 30 min; 3, Dac purified by using an anion-exchange chromatography column. (c) Calculation of kinetic parameters of Dac and its mutants according to Lineweaver-Burk plot. To test the specific activity and kinetic parameters of the R157 mutants, the experiments were performed using the purified enzymes at 40°C and pH 8.0. Error bars indicate the average values from three parallel experiments.
The purified recombinant enzyme was the refined product of the culture supernatant of recombinant B. subtilis. As shown in Fig. 6b, SDS-PAGE indicated that the molecular weight of Dac is approximately 32 kDa, consistent with the previously reported result (2). The majority of the thermally unstable proteins were removed through incubation at 85°C for 30 min (compare lane 1 and lane 2 in Fig. 6b), and Dac was still stable. The supernatant was then purified by using an anion-exchange chromatography column, and the pure Dac was obtained (lane 3 in Fig. 6b).
The enzymatic properties of Dac mutants R157W, R157T, and R157H were analyzed after purification. The mutant R157T exhibited the highest specific activity (3,112.2 U/mg) compared with that of wild-type Dac (2,047.3 U/mg) (Fig. 6a). In addition, when GlcNAc was used as the substrate, according to the kinetic analysis, compared with those of R157 mutants, the reaction rate and substrate specificity were increased in the R157T mutant (Table 3). The kinetic parameters were estimated by plotting the data on the double reciprocal graph of Michaelis-Menten equation. As shown in Fig. 6c, the Km decreased from 7.04 mM in wild-type Dac to 5.19 mM in mutant R157T, and the Vmax of 5.11 μM s−1 in wild-type Dac increased to 7.56 μM s−1 in mutant R157T.
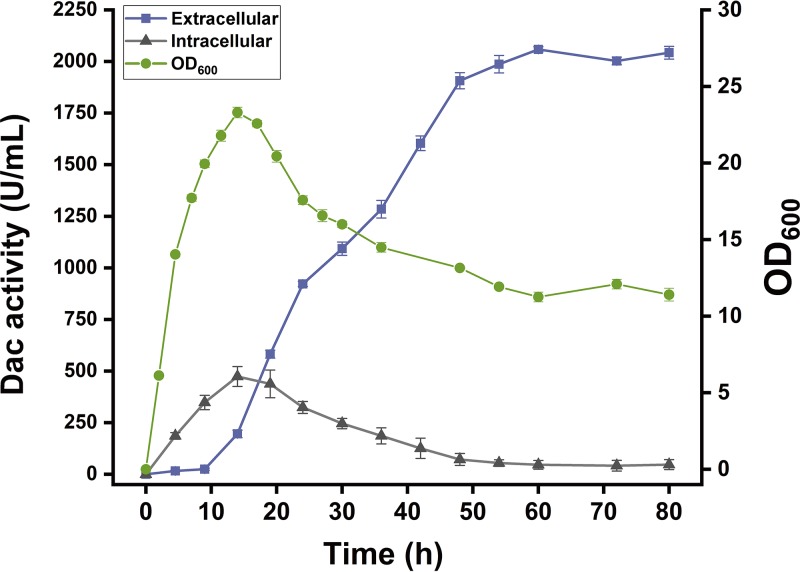
Figure 7 reveals the time curve of Dac production by batch culture in a 500-ml flask. During batch culture, the recombinant strain continuously grew from 0 h to 14 h. After 14 h, the cells entered the stationary phase; the biomass reached the maximum (optical density at 600 nm [OD600] of 23.3) at 14 h. The biomass began to decline at around 16 h when the carbon source was depleted. After the stationary phase, the extracellular Dac activity continuously increased; after 48 h, the Dac activity increased slowly, reaching the highest level of 2,042.8 U/ml at 60 h.
FIG 7.
Time profile of Dac activity of the recombinant strain WB-NMKmut/Dac. ●, cell growth; ■, extracellular activity of Dac; ▲, intracellular activity of Dac. Error bars indicate the average values from three parallel experiments.
DISCUSSION
Appropriate signal peptides and promoters are vital to realize efficient secretory expression of a desired protein in a prokaryotic system. Signal peptides share certain common features, such as containing a positively charged N domain, hydrophobic H domain, and conserved C domain (28). Based on its compositions, mainly the differences of charge distribution in the N domain and H domain, 12 signal peptides of B. subtilis from the Sec pathway, which had been proved to lead to the massive secretion of different enzymes successfully, were compared in this work (Table 1). The YncM signal peptide, which possesses higher positive charge in the N-terminal region and higher hydrophobicity in the core H domain, achieved the highest extracellular Dac activity. Some studies have proved that the hydrophobicity of the signal peptide has a widespread and profound impact on the early stage of the recognition of signal peptide in B. subtilis (29, 30), which means that suitable signal peptide structures, such as a higher positive charge in the N-terminal region and higher hydrophobicity in the core H domain, can improve the efficiency of protein secretion. At the same time, by using the constitutive promoter P43, the activity of Dac was improved by 1.38 times. In B. subtilis, the promoter P43 is a typical promoter that δ55 and rδ37 RNA polymerase holoenzymes can recognize and transcribe (31). It is an efficient way to use an artificial promoter with high activity to improve the yield of heterologous proteins.
The transcription level and translation level determine the efficiency of heterologous gene expression in prokaryotes. The 5′ UTR sequence is a necessary region regulating the expression of target genes. The elements in the region at the 5′ end of a mature transcript (preceding the initiation codon), including the 5′ stem loop, the RBS, and the spacer region between the RBS and the 5′ stem loop, influence the expression of heterologous proteins both at the transcription and translation levels (23). Altering the sequence of target gene mRNA transcripts can significantly change the gene expression, which directly determines the amount of translation products by affecting the effective extension of mRNA, termination of mRNA, and stability of mRNA (32, 33). At present, secondary structure reconstruction by adding repetitive extragenic palindromic sequences in the 3′ UTR of a target gene provides stability to mRNA in some studies. It makes the mRNA of target gene more stable and not readily degradable. The change of the UTR protects the mature mRNA from being digested by exonuclease and then improves the activity of cyclodextrin glucosyltransferase and enhances the product titers (34). In this study, the mutation in the 5′ UTR significantly enhanced the expression of Dac by 82 times, indicating that the 5′ UTR of mRNA is highly important to the control of gene expression. The results presented in Fig. 4c indicate the transcription level of target gene Dac in the WB-NMKmut/Dac strain was increased by 14.8- to 42.5-fold during the different fermentation phases. The analysis results from the equilibrium statistical thermodynamic model conform to the results of the qRT-PCR transcriptional analysis. The mutation in the 5′ UTR generates a far more stable secondary structure, which provides stability to mRNA and protects the mRNA from degradation.
Directed evolution is one type of efficient tool to improve the catalytic efficiency of enzymes. Site-saturation mutagenesis in the substrate-binding pocket has been shown to be efficient for enhancing enzyme catalytic performance (30). In this study, the mutant R157T exhibited higher specific activity and catalytic efficiency than wild-type Dac. A complete Dac is a hexamer consisting of two trimers as shown in the molecular model, which is made up of three molecules, and the boundary of the two polypeptides in assembled Dac forms a cleft that allows the substrate to access the catalytic site. The docking result shows that the residue R157 is located at the center of Dac, and substrate GlcNAc is bound to the center of the enzyme. There are hydrogen bond interactions between the R157 residue and GlcNAc (Fig. 5b). We first demonstrated that the R157 site is a key site for the GlcNAc deacetylation, which was verified by the fact that the R157G mutant completely lost catalytic activity (data not shown).
Overall, we report on the high-level secretory production of Dac, and an unexpected insertion of a 73-bp sequence in 5′ UTR resulted in a significant increase of Dac expression due to the increase of TIR. In addition, the site R157 of Dac is identified as a key catalytic site by molecular docking, and the mutant R157T exhibits much higher specific activity and catalytic efficiency. The results obtained here will expand the applications of Dac in the production of chitosan oligosaccharides and monosaccharides in the future.
MATERIALS AND METHODS
Materials, plasmids, and strains.
Bacterial strains and plasmids used in this study are listed in Table 4, and primers utilized in this study are listed in Table 5. E. coli JM109 was used for gene cloning and plasmid construction. The plasmid pMA0911 derivative was a gift from Yu Xia (Jiangnan University, China), and plasmid pP43NMK was a generous donation from X. Z. Zhang (Department of Biological Systems Engineering, Virginia Tech, USA) (31). The materials used in this study are listed in Table 6. DNA sequencing and primer synthesis were performed by Genewiz (Suzhou, China). Seed cultures of E. coli and B. subtilis were cultivated in Luria-Bertani (LB) medium (5 g/liter yeast extract, 10 g/liter tryptone, and 10 g/liter NaCl), and the cells were cultured in Terrific broth (TB) (24 g/liter yeast extract, 12 g/liter tryptone, 4 ml/liter glycerol, 2.31 g/liter KH2PO4, and 12.54 g/liter K2HPO4) with 10 mg/liter kanamycin.
TABLE 4.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | Cloning strain | Takara, Japan |
| E. coli BL21(DE3) | Expression strain | Invitrogen, CA |
| B. subtilis WB600 | Deficient in NprE AprE Epr Bpr Mpr NprB | Lab stock |
| WB-MA-DS | B. subtilis WB600 containing pMA0911NS | This work |
| WB-MA-J | B. subtilis WB600 containing pMA0911J | This work |
| WB-MA-C | B. subtilis WB600 containing pMA0911C | This work |
| WB-MA-M | B. subtilis WB600 containing pMA0911M | This work |
| WB-MA-Z | B. subtilis WB600 containing pMA0911Z | This work |
| WB-MA-O | B. subtilis WB600 containing pMA0911O | This work |
| WB-MA-A | B. subtilis WB600 containing pMA0911A | This work |
| WB-MA-F | B. subtilis WB600 containing pMA0911F | This work |
| WB-MA-G | B. subtilis WB600 containing pMA0911G | This work |
| WB-MA-L | B. subtilis WB600 containing pMA0911L | This work |
| WB-MA-H | B. subtilis WB600 containing pMA0911H | This work |
| WB-MA-E | B. subtilis WB600 containing pMA0911E | This work |
| WB-MA-I | B. subtilis WB600 containing pMA0911I | This work |
| WB-NMK/Dac | B. subtilis WB600 containing pP43NMK-YncM-Dac | This work |
| WB-NMKmut/Dac | B. subtilis WB600 containing pP43NMKmut-YncM-Dac | This work |
| Plasmids | ||
| pET-28a-Dac | pET-28a(+) containing Dac gene | Lab stock |
| pMA0911 | pMA5 derivative, PHpaII, Kanr, Ampr | Lab stock |
| pP43NMK | PHpaII, P43, Kanr, Ampr | 31 |
| pMA0911J | pMA0911 derivative, Dac gene, YncM | This work |
| pMA0911C | pMA0911 derivative, Dac gene, Bpr | This work |
| pMA0911M | pMA0911 derivative, Dac gene, YwbN | This work |
| pMA0911Z | pMA0911 derivative, Dac gene, AnsZ | This work |
| pMA0911O | pMA0911 derivative, Dac gene, YvgO | This work |
| pMA0911A | pMA0911 derivative, Dac gene, AmyE | This work |
| pMA0911F | pMA0911 derivative, Dac gene, OppA | This work |
| pMA0911G | pMA0911 derivative, Dac gene, Vpr | This work |
| pMA0911L | pMA0911 derivative, Dac gene, LipA | This work |
| pMA0911H | pMA0911 derivative, Dac gene, WapA | This work |
| pMA0911E | pMA0911 derivative, Dac gene, Epr | This work |
| pMA0911I | pMA0911 derivative, Dac gene, YclQ | This work |
| pMA0911NS | pMA0911 derivative, Dac gene, without signal peptide | This work |
| pP43NMK-YncM-Dac | pP43NMK containing Dac gene, YncM | This work |
| pP43NMKmut-YncM-Dac | pP43NMK/Dac derivative, 5′ UTR mutation | This work |
| pP43NMKmut-YncM-Dac-R157T | pP43NMK/Dac derivative, 5′ UTR mutation, R157T | This work |
Ampr, ampicillin resistance; Kanr, kanamycin resistance.
TABLE 5.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| Dac-F1 | CCGGAATTCATGGTCGTCAACATGTTCGAG |
| Dac-R1 | CGCGGATCCTCAGATCAGGTCCGTAAACGGT |
| Dac-F2 | GTTTACCCGCGGCAGACGCTATGGTCGTCAACATGTTCGAG |
| Dac-R2 | CTGCAGATTACGCCAAGCTTCTGCAGTCAGATCAGGTCCGTAAACGGT |
| yncM-F | GGTACCATTATAGGTAAGAGAGGAATGTACACATGGCGAAACCACTATCAAAAG |
| yncM-R | AGCGTCTGCCGCGGGTAAAC |
| qDac-F | GGACGATTGCGTTATTGGAATG |
| qDac-R | GCTTTCTTCTTCTTCTTTGCGG |
| qPCR16S-F | CCACGTGTAGCGGTGAAATG |
| qPCR16S-R | GGCGGAGTGCTTAATGCG |
| qPCRhbsU-F | ATCGGTTTTGGTAACTTCGAGG |
| qPCRhbsU-F | GCAGGTACTTTGCTTGCTGGA |
| 28a-R156-F | TCACATCCGGATCATNNKCGCACAGGCTTTCTG |
| 28a-R156-R | CAGAAAGCCTGTGCGMNNATGATCCGGATGTGA |
| 28a-R157-F | ATCCGGATCATCGTNNKACCGGTTTTTTAGCC |
| 28a-R157-R | GGCTAAAAAACCGGTMNNACGATGATCCGGAT |
| 28a-F160-F | CATAGACGCACAGGCNNKCTGGCGATTGAATC |
| 28a-F160-R | GATTCAATCGCCAGMNNGCCTGTGCGTCTATG |
| 28a-R92-R | GAATTAGCAGCAATCNNKCGCAAAGAAGAAGAAG |
| 28a-R92-R | CTTCTTCTTCTTTGCGMNNGATTGCTGCTAATTC |
| 28a-H152-F | CTTCCGTACGAATCANNKCCGGATCATAGACGC |
| 28a-H152-R | GCGTCTATGATCCGGMNNTGATTCGTACGGAAG |
| NMK-R157W-F | ATCCGGATCATCGTTGGACCGGTTTTTTAGCC |
| NMK-R157W-R | GGCTAAAAAACCGGTCCAACGATGATCCGGAT |
| NMK-R157T-F | ATCCGGATCATCGTACGACCGGTTTTTTAGCC |
| NMK-R157T-R | GGCTAAAAAACCGGTCGTACGATGATCCGGAT |
| NMK-R157H-F | ATCCGGATCATCGTCATACCGGTTTTTTAGCC |
| NMK-R157H-R | GGCTAAAAAACCGGTATGACGATGATCCGGAT |
| 5′RACE1 | GGCCACGCGTCGACTAGTAGGGGGGGGGGGGG |
| Dac1 | TCTTCTTCTTCTTTGCGGCGGATTG |
| 5′RACE2 | GGCCACGCGTCGACTAGTAG |
| Dac2 | AGCCGTCTGTCATGCAAACGTAGAT |
Italic letters represent the restriction enzyme sites, underlined letters represent homologous sequences for cloning, and boldface letters represent degenerate base.
TABLE 6.
Materials used in this study
| Name | Source |
|---|---|
| SanPrep Column Plasmid Mini-Preps kit | Sangon (Shanghai, China) |
| DNA purification kit | TaKaRa (Dalian, China) |
| Restriction enzymes | TaKaRa (Dalian, China) |
| ClonExpress MultiS One Step Cloning kit | Vazyme (Nanjing, China) |
| PrimeScriptRT reagent kit | TaKaRa (Dalian, China) |
| RevertAid First Strand cDNA synthesis kit | Thermo Scientific (USA) |
| PrimeSTAR HS DNA polymerase | TaKaRa (Otsu, Japan) |
| Glucosamine (GlcN) | Sigma-Aldrich (St. Louis, MO, USA) |
| ortho-Phthalaldehyde (OPA) | Sigma-Aldrich (St. Louis, MO, USA) |
| Dithiothreitol (DTT) | Sigma-Aldrich (St. Louis, MO, USA) |
| Ampicillin (Amp) | Amresco (Solon, OH, USA) |
| Isopropyl-β-d-1-thiogalactopyranoside (IPTG) | Merck (Darmstadt, Germany) |
| Other chemicals | Sangon (Shanghai, China) |
Construction of plasmids expressing the Dac with different signal peptides.
The primers used in this work are listed in Table 5. The signal peptides used in this work are described in Table 1. The chromosome of B. subtilis 168 was used as the template to amplify the gene encoding signal peptides by PCR, and the signal peptide fragments were inserted into pMA0911 between the NedI and EcoRI sites. The gene encoding Dac (PH0499) was codon optimized, according to the preference of codons in different hosts, to achieve suitable expression in B. subtilis, and the Dac gene from the strain that we constructed in a previous work was amplified with primers Dac-F1 and Dac-R1 (4). By inserting the target gene encoding Dac into the EcoR I-BamH I sites of pMA0911 derivatives which harbor sequences encoding signal peptides YncM, Bpr, Vpr, WapA, LipA, Epr, AmyE, YclQ, YwbN, AnsZ, YvgO, and OppA, the plasmids pMA0911J, pMA0911C, pMA0911A, pMA0911G, pMA0911L, pMA0911H, pMA0911E, pMA0911I, pMA0911M, pMA0911Z, pMA0911O, and pMA0911F, respectively, were obtained. The structure of recombinant plasmid is shown in Fig. 1a. The plasmid pMA0911NS was obtained by inserting target gene encoding Dac into pMA0911 without signal peptides.
Construction of plasmids expressing Dac with the P43 promoter.
To express Dac under the strong promoter P43, plasmid pP43NMK was digested with KpnI and PstI and subjected to column purification. The Dac gene with the sequence encoding signal peptide YncM was amplified from plasmid pMA0911J by PCR with gene-specific primers Dac-F2, Dac-R2, yncM-F, and yncM-R. The gene fragments were ligated into vector pP43NMK between the KpnI and PstI sites by using ClonExpress MultiS One Step Cloning kit. By cloning the gene encoding Dac and the YncM signal peptide downstream of the P43 promoter, the plasmid pP43NMK-YncM-Dac was finally obtained. The structure of expression plasmid pP43NMK-YncM-Dac is shown in Fig. 2a.
Expression and purification of Dac in B. subtilis.
The recombinant plasmids that had been constructed in the above-described procedures were transformed into host strain B. subtilis WB600 to obtain the recombinant strains producing Dac. The plasmid pMA0911 without the gene insertion was transformed into B. subtilis WB600 and used as the control. All the recombinant plasmids were confirmed by DNA sequencing and restriction analysis. Transformation of B. subtilis was performed according to the manufacturer’s specification (MoBiTec). The recombination B. subtilis strain was cultivated in LB medium containing kanamycin at a final concentration of 10 mg/liter. The bacteria were cultured at 37°C overnight (220 rpm) to prepare the seed cultures; 4% of the seed culture was inoculated into the TB medium containing 10 mg/liter kanamycin and was then cultured at 37°C and 220 rpm for 48 to 72 h. OD600 was determined on an UVmini-1240 spectrophotometer (Shimadzu, Kyoto, Japan). The culture supernatant was used to determine the extracellular Dac activity.
The mature Dac was harvested from the culture supernatant of recombinant B. subtilis. The culture supernatant was incubated at 85°C for 30 min and then centrifuged at 10,000 × g for 10 min to remove the thermally unstable proteins to yield the crude enzyme. The crude enzyme solution was filtered by using a 0.45-μm-pore-size membrane and purified from the supernatant by using a HiTrap Q HP column with AKTA explorer (GE Healthcare, Piscataway, NJ, USA). Binding buffer (50 mM sodium phosphate buffer, pH 7.0) was used to equilibrate the column. After that, the recombinant enzyme was eluted with gradient elution buffer (50 mM sodium phosphate buffer with 500 mM NaCl, pH 7.0). The fractions obtained from every step were stored for SDS-PAGE and enzymatic property analyses.
Transcriptional analysis of Dac in recombinant strains.
Cells were harvested at different culture phases (7 h, 12 h, 24 h, and 36 h) in shake flasks. According to the manufacturer’s directions (TIANGEN, China), the total RNA was extracted from 5-ml cultures. The cDNA was synthesized using the PrimeScriptRT reagent kit. Then, the cDNA was used for qRT-PCR with primers qDac-F and qDac-R to analyze the Dac transcriptional level (Table 5). qRT-PCR was performed by using iQ SYBR green Supermix (Bio-Rad, Hercules, CA, USA) and CFX Connect (Bio-Rad). During that, 0.5 ng of cDNA served as the template, and the 16S rRNA gene and hbsU gene served as the internal standard using primer pairs of qPCR16S-F, qPCR16S-R, qPCRhbsU-F, and qPCRhbsU-R. At the same time, the negative control was the reaction without template (34, 35), and the conditions were 95°C for 3 min followed by 50 three-step cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Rapid amplification of cDNA ends by terminal deoxynucleotidyl transferase.
The method of total RNA extraction was as mentioned above. The synthesis of first-strand cDNA was performed with a RevertAid First Strand cDNA synthesis kit (Table 6). To prepare the template of rapid amplification of cDNA ends (RACE), homopolymeric tailing processes were performed by using terminal deoxynucleotidyl transferase (TDT). The reaction system included 5 μl 5× TDT buffer, 2.5 μl 0.1% (wt/vol) bovine serum albumin (BSA), 2.5 μl 10 mM dCTP, 15 U TDT, and the synthesis product of the first-strand cDNA. Then, double distilled-water (ddH2O) was added up to 25 μl and was incubated at 37°C for 30 min. Then, the first PCR was performed using primers 5′RACE1 and Dac1 (Table 5), and the tailing product as the template. PCR conditions were 95°C for 5 min followed by 35 three-step cycles of 95°C for 40 s, 60°C for 30 s, and 72°C for 90 s. Next, nest PCR was performed using primers 5′RACE2 and Dac2 (Table 5), and the product of the first PCR served as the template. PCR conditions were 95°C for 5 min followed by 20 three-step cycles of 95°C for 40 s, 60°C for 30 s, and 72°C for 90 s.
Docking calculations and site-directed saturation mutagenesis.
The Discovery Studio (DS CDOCKER) software was used to recognize underlying binding sites for the substrate GlcNAc. The 3D structural data were obtained from the Protein Data Bank database, and the accession number is 3WE7; one of the catalytic activity regions was located in the center of the 3D structure (7). In this work, substrate GlcNAc was the ligand, and Dac was the protein accepter. The protein file and ligand file were prepared to fit the docked state. The series of procedures mentioned above were according to the manufacturer’s protocol with the default settings. After calculations were performed, the binding modes within 1 kcal/mol, which is the most favorable binding free energy conformation, were chosen.
The method of targeted whole-plasmid mutagenesis with PrimeSTAR HS DNA polymerase was used to carry out the site-directed saturation mutagenesis, and the pET-28a-Dac plasmid we constructed in a previous work was used as the template DNA (4). The specific sequences of primers are given in Table 5. The DpnI restriction enzyme, which is specific for methylated DNA, was used to degrade the template DNA. The products were transformed into E. coli BL21(DE3) to establish a mutant library for expression and further screening. According to the results of docking calculations, we chose several specific residues of Dac to analyze the active site and to perform the saturation mutagenesis of each site.
High-throughput screening of Dac mutants.
To screen high-enzyme-activity mutants rapidly, we established a high-throughput screening method by using derivatization reagent ortho-phthalaldehyde (OPA). The screening process was performed by the QPix 420 system (Molecular Devices, Sunnyvale, CA). Single colonies from the mutant library obtained in the above-described step were selected and inoculated in 96-deep-well microtiter plates (MTPs); each well contained 600 μl LB medium and 50 mg/liter kanamycin. After an overnight incubation at 37°C, 200 μl seed culture was transferred to new 96-deep-well MTPs containing 600 μl of LB medium per well where the final concentrations of kanamycin and IPTG (isopropyl-β-d-thiogalactopyranoside) were 50 mg/liter and 0.05 mM, respectively. The MTPs were incubated for 6 h at 37°C and 900 rpm on a Titramax 1000 multiplate shaker (Heidolph, Schwabach, Germany). After that, the MTPs were centrifuged in a 96-well MTP centrifuge (Beckman, Brea, CA) at 8,000 × g for 5 min to collect whole cells, and 100 μl of 50 g/liter GlcNAc in 50 mM sodium phosphate buffer (pH 8.0) was added to each well to start the reaction under the conditions of 40°C and 900 rpm for 20 min. The reaction was stopped by adding 50 μl of 0.5 M HCl to each well. Then, the mixtures were diluted by adding 350 μl distilled water to every well. Subsequently, the mixtures were centrifuged at 3,500 × g for 10 min, and 10 μl supernatant from every well was transferred to another well on a new 96-well MTP which contained 100 μl OPA indicator (5 mg OPA, 10 μl 1.0 M dithiothreitol [DTT], and 100 μl alcohol in 10 ml [pH 10.5] sodium carbonate buffer) in each well. Finally, the mixture was used to measure the absorbance at 330 nm with a Multiskan Spectrum spectrophotometer (Thermo Scientific; Vantaa, Finland). According to the rule that higher quantity of GlcN corresponds to higher absorbance, high-enzyme-activity mutants with higher absorbance values were recognized. The second screening in 500-ml flasks was performed to obtain the best mutants. The plasmid in the best mutant was sequenced to confirm the mutations. Then, site-directed mutagenesis was carried out by using the method of targeted whole-plasmid mutagenesis with the pP43NMK-YncM-Dac plasmid serving as the template. The primers used in this part are given in Table 5. DNA sequencing was carried out to determine the specific sequences of these plasmids, and correct clones were transformed into B. subtilis WB600 for expression and measurement of catalytic efficiency.
Measurement of Dac activity.
The measurement of Dac activity was performed in 1.5-ml centrifuge tubes at 40°C. A 50 mM sodium phosphate buffer (pH 8.0) served as the buffer solution in this study. Diluted Dac samples and GlcNAc solutions (452 mM) were prewarmed at 40°C for 10 min. To initiate the reaction, 0.1 ml of dilute Dac sample was added to 0.1 ml GlcNAc solution. The reaction mixture was incubated at 40°C. After precisely 5 min, 0.1 ml of 0.5 M HCl was added to stop the reaction immediately. The supernatant was recovered for quantification of GlcN. The GlcN produced through enzymatic reaction was measured by the OPA indicator with a Multiskan Spectrum spectrophotometer. One unit of Dac activity was defined as the amount of the enzyme liberating 1 μM GlcN in 1 h at 40°C.
Determination of kinetic parameters of the evolved mutants.
The recombinant enzyme and the evolved mutants were purified as described above. Apparent kinetic parameters (Km and Vmax) were measured by varied GlcNAc concentrations ranging from 0.9 to 13.36 mM (0.9 mM, 1.13 mM, 2.26 mM, 4.52 mM, 9.04 mM, and 13.36 mM) at 40°C. A 50 mM sodium phosphate buffer (pH 8.0) served as the buffer solution. Each value was calculated via three mutually independent experiments. The enzyme concentration of the reaction system was 6.16 mg/liter, and the total volume of each reaction mixture was 0.2 ml (50 mM sodium phosphate buffer, pH 8.0). The assays were performed in a linear range of the reaction and less than 10% substrate to convert into product.
The kinetic parameters were evaluated and estimated by plotting the data on the double reciprocal graph according to the following equation:
where V is the reaction rate, Km is the Michaelis constant (mM), Vmax is the maximum reaction rate (micromoles GlcN per minute per milligram protein), and [S] is the GlcNAc concentration (mM).
The double reciprocal graphs are shown in Fig. 6c.
Analytical methods.
The GlcN concentration was measured with Multiskan Spectrum spectrophotometer (Thermo Scientific; Vantaa, Finland). Five-microliter aliquots of samples were added to 100 μl OPA indicator (5 mg OPA, 10 μl 1.0 M DTT, and 100 μl alcohol in 10 ml sodium carbonate buffer [pH 10.5]). The mixture was used to detect the absorbance at 330 nm. Pure substances were used as the standards to get the standard curve, and the concentration of standard GlcN ranged from 0.1 to 5.0 g/liter.
To analyze the protein by SDS-PAGE, the experiment was performed on a 10% running gel in MES (morpholineethanesulfonic acid) SDS running buffer (Bio-Rad Laboratories, Hercules, CA, USA). The gel was dyed by Coomassie brilliant blue G250 to visualize the proteins.
The Bradford method was used to determine the concentration of protein (36). Bovine serum albumin served as the standard.
Statistical analysis.
All the experiments were independently conducted at least three times, and the results were expressed as means ± standard deviations (n = 3).
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation (31622001, 31671845, 21808084, 21676119, and 31870069), the 111 project (111-2-06), and the China Postdoctoral Science Foundation (2018M630523).
Z.J. designed and performed the experiments; X.L., Y.L., T.N, L.L., J.L., and G.D. conceived the project and analyzed the data; Z.J., Y.L., and L.L. wrote the paper.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01076-19.
REFERENCES
- 1.Tanaka T, Fukui T, Fujiwara S, Atomi H, Imanaka T. 2004. Concerted action of diacetylchitobiose deacetylase and exo-beta-d-glucosaminidase in a novel chitinolytic pathway in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Biol Chem 279:30021–30027. doi: 10.1074/jbc.M314187200. [DOI] [PubMed] [Google Scholar]
- 2.Mine S, Ikegami T, Kawasaki K, Nakamura T, Uegaki K. 2012. Expression, refolding, and purification of active diacetylchitobiose deacetylase from Pyrococcus horikoshii. Protein Expr Purif 84:265–269. doi: 10.1016/j.pep.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Park J, Lee SY, Ooshima A, Yang KM, Kang JM, Kim YW, Kim SJ. 2013. Glucosamine hydrochloride exerts a protective effect against unilateral ureteral obstruction-induced renal fibrosis by attenuating TGF-beta signaling. J Mol Med (Berl) 91:1273–1284. doi: 10.1007/s00109-013-1086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Z, Lv X, Liu Y, Shin HD, Li J, Du G, Liu L. 2018. Biocatalytic production of glucosamine from N-acetylglucosamine by diacetylchitobiose deacetylase. J Microbiol Biotechnol 28:1850–1858. doi: 10.4014/jmb.1805.05061. [DOI] [PubMed] [Google Scholar]
- 5.Chen JK, Shen CR, Liu CL. 2010. N-Acetylglucosamine: production and applications. Mar Drugs 8:2493–2516. doi: 10.3390/md8092493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Niiyama M, Hashimoto W, Ida K, Abe M, Morita J, Uegaki K. 2015. Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus. Acta Crystallogr F Struct Biol Commun 71:657–662. doi: 10.1107/S2053230X15005695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mine S, Niiyama M, Hashimoto W, Ikegami T, Koma D, Ohmoto T, Fukuda Y, Inoue T, Abe Y, Ueda T, Morita J, Uegaki K, Nakamura T. 2014. Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase. FEBS J 281:2584–2596. doi: 10.1111/febs.12805. [DOI] [PubMed] [Google Scholar]
- 8.Harwood CR, Cranenburgh R. 2008. Bacillus protein secretion: an unfolding story. Trends Microbiol 16:73–79. doi: 10.1016/j.tim.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Westers L, Westers H, Quax WJ. 2004. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim Biophys Acta 1694:299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Zweers JC, Barak I, Becher D, Driessen AJ, Hecker M, Kontinen VP, Saller MJ, Vavrova L, van Dijl JM. 2008. Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb Cell Fact 7:10. doi: 10.1186/1475-2859-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darmon E, Noone D, Masson A, Bron S, Kuipers OP, Devine KM, van Dijl JM. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J Bacteriol 184:5661–5671. doi: 10.1128/JB.184.20.5661-5671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyyryläinen HL, Bolhuis A, Darmon E, Muukkonen L, Koski P, Vitikainen M, Sarvas M, Prágai Z, Bron S, van Dijl JM, Kontinen VP. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol Microbiol 41:1159–1172. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 14.Promchai R, Promdonkoy B, Tanapongpipat S, Visessanguan W, Eurwilaichitr L, Luxananil P. 2016. A novel salt-inducible vector for efficient expression and secretion of heterologous proteins in Bacillus subtilis. J Biotechnol 222:86–93. doi: 10.1016/j.jbiotec.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji S, Tanaka K, Takenaka S, Yoshida K. 2015. Enhanced secretion of natto phytase by Bacillus subtilis. Biosci Biotechnol Biochem 79:1906–1914. doi: 10.1080/09168451.2015.1046366. [DOI] [PubMed] [Google Scholar]
- 16.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. 2002. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, Hecker M. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res 11:1484–1502. doi: 10.1101/gr.182801. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Liu S, Jiao Y, Gao H, Wang M, Du G, Chen J. 2017. Enhanced extracellular production of l-asparaginase from Bacillus subtilis 168 by B. subtilis WB600 through a combined strategy. Appl Microbiol Biotechnol 101:1509–1520. doi: 10.1007/s00253-016-7816-x. [DOI] [PubMed] [Google Scholar]
- 19.Kang Z, Yang S, Du G, Chen J. 2014. Molecular engineering of secretory machinery components for high-level secretion of proteins in Bacillus species. J Ind Microbiol Biotechnol 41:1599–1607. doi: 10.1007/s10295-014-1506-4. [DOI] [PubMed] [Google Scholar]
- 20.Song Y, Nikoloff JM, Fu G, Chen J, Li Q, Xie N, Zheng P, Sun J, Zhang D. 2016. Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. PLoS One 11:e0158447. doi: 10.1371/journal.pone.0158447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emory SA, Bouvet P, Belasco JG. 1992. A 5'-terminal stem loop structure can stabilize messenger-RNA in Escherichia coli. Genes Dev 6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 22.Tian T, Salis HM. 2015. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res 43:7137–7151. doi: 10.1093/nar/gkv635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan TT, Nguyen HD, Schumann W. 2013. Construction of a 5'-controllable stabilizing element (CoSE) for over-production of heterologous proteins at high levels in Bacillus subtilis. J Biotechnol 168:32–39. doi: 10.1016/j.jbiotec.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Carrier TA, Keasling JD. 1999. Library of synthetic 5' secondary structures to manipulate mRNA stability in Escherichia coli. Biotechnol Prog 15:58–64. doi: 10.1021/bp9801143. [DOI] [PubMed] [Google Scholar]
- 25.Salis HM, Mirsky EA, Voigt CA. 2009. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y, Fu G, Dong H, Li J, Du Y, Zhang D. 2017. High-efficiency secretion of beta-mannanase in Bacillus subtilis through protein synthesis and secretion optimization. J Agric Food Chem 65:2540–2548. doi: 10.1021/acs.jafc.6b05528. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T, Yonezawa Y, Tsuchiya Y, Niiyama M, Ida K, Oshima M, Morita J, Uegaki K. 2016. Substrate recognition of N, N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii. J Struct Biol 195:286–293. doi: 10.1016/j.jsb.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Brockmeier U, Caspers M, Freudl R, Jockwer A, Noll T, Eggert T. 2006. Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria. J Mol Biol 362:393–402. doi: 10.1016/j.jmb.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Fu LL, Xu ZR, Li WF, Shuai JB, Lu P, Hu CX. 2007. Protein secretion pathways in Bacillus subtilis: implication for optimization of heterologous protein secretion. Biotechnol Adv 25:1–12. doi: 10.1016/j.biotechadv.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Zanen G, Houben EN, Meima R, Tjalsma H, Jongbloed JD, Westers H, Oudega B, Luirink J, van Dijl JM, Quax WJ. 2005. Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J 272:4617–4630. doi: 10.1111/j.1742-4658.2005.04777.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XZ, Cui ZL, Hong Q, Li SP. 2005. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl Environ Microbiol 71:4101–4103. doi: 10.1128/AEM.71.7.4101-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brocato J, Fang L, Chervona Y, Chen D, Kiok K, Sun H, Tseng H-C, Xu D, Shamy M, Jin C, Costa M. 2014. Arsenic induces polyadenylation of canonical histone mRNA by down-regulating stem-loop-binding protein gene expression. J Biol Chem 289:31751–31764. doi: 10.1074/jbc.M114.591883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YS, Beyer A, Aebersold R. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Deng C, Lv XQ, Li JH, Liu YF, Du GC, Amaro RL, Liu L. 2019. Synthetic repetitive extragenic palindromic (REP) sequence as an efficient mRNA stabilizer for protein production and metabolic engineering in prokaryotic cells. Biotechnol Bioeng 116:5–18. doi: 10.1002/bit.26841. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Chen T, Liu Y, Lv X, Li J, Du G, Ledesma-Amaro R, Liu L. 2018. CRISPRi allows optimal temporal control of N-acetylglucosamine bioproduction by a dynamic coordination of glucose and xylose metabolism in Bacillus subtilis. Metab Eng 49:232–241. doi: 10.1016/j.ymben.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.