SUMMARY
In the past four decades, tremendous progress has been made in understanding how plants respond to microbial colonization and how microbial pathogens and symbionts reprogram plant cellular processes. In contrast, our knowledge of how environmental conditions impact plant-microbe interactions is less understood at the mechanistic level, as most molecular studies are performed under simple and static laboratory conditions. In this review, we highlight research that begins to shed light on the mechanisms by which environmental conditions influence diverse plant-pathogen, plant-symbiont and plant-microbiota interactions. There is a great need to increase efforts in this important area of research in order to reach a systems-level understanding of plant-microbe interactions that are more reflective of what occurs in nature.
Keywords: Plant pathogen, symbiosis, abiotic stress, temperature, light, circadian clock, humidity, nutrient, innate immunity, climate change
INTRODUCTION
In nature, plants live in a microbe-rich environment and must interact with a myriad of pathogenic, commensal and beneficial microbes. How plants harness the beneficial functions provided by microbes and, at the same time, combat microbial pathogens has attracted the attention of generations of plant and microbial scientists. Impressive molecular work conducted since the early 1980s has revealed a number of basic principles underlying plant-microbe interactions. Among them are discovery of (i) signals from microbes that are perceived by cognate plant immune receptors to initiate defense or symbiotic responses (Jones et al., 2016); (ii) microbial DNA and/or protein secretion systems that transport effector molecules into the plant cell to modulate host cell functions (Buttner and He, 2009; Hwang et al., 2017); (iii) microbial and plant developmental programs that orchestrate the formation of specialized nutrient-exchanging/producing organs (e.g. nodules and galls) during symbiotic and pathogenic interactions (Zipfel and Oldroyd, 2017); and (iv) binary and community-level antagonistic warfare in plant-microbiota interactions (Hacquard et al., 2017).
In addition, it has been long noted that many, if not all, plant-microbe interactions are profoundly affected by external environmental conditions (Figure 1A). In fact, “disease triangle”, a famous concept in describing plant-pathogen interactions, states that, for a plant disease to occur, the plant needs to be genetically susceptible, the pathogen genetically virulent and environmental conditions conducive to pathogen virulence and plant susceptibility (Stevens, 1960). Similarly, symbiotic and commensal plant-microbe interactions can be altered by external conditions, including temperature, moisture and nutrient status. As such, understanding how environmental conditions influence plant-microbe interactions is crucial in predicting disease outbreaks, engineering effective symbiotic and biocontrol agents, and designing “dream” crop plants with increased resilience to current and future climate change.
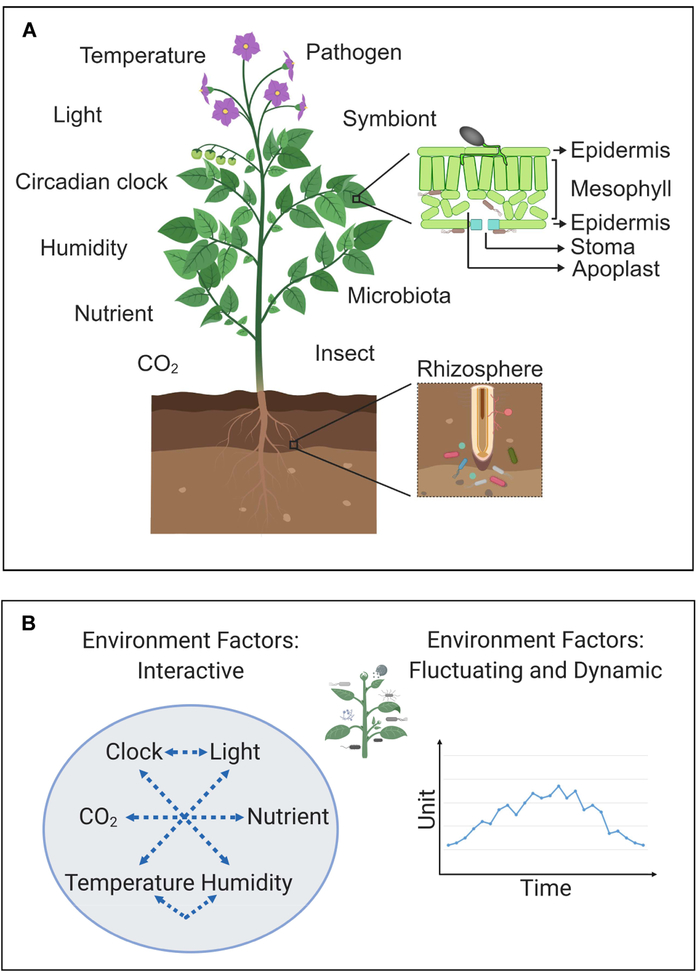
Figure 1.
(A) An overview diagram depicting environmental conditions that are known to affect plant-microbe interactions in plants. (B) The dynamic nature of environmental conditions that fluctuate and influence one another.
In this review, we will highlight selected environmental conditions for which there is a substantial body of knowledge that begins to explain how they modulate plant-microbe interactions at the molecular level. We will attempt to infer general principles, when possible. However, as it will become clear, in many cases, environmental influences are complex; the current knowledge has not reached a stage where one can pinpoint general principles, illustrating a great need for increased efforts in this important area of research.
SECTION 1: TEMPERATURE
1.1. Impact of elevated temperature on the plant immune system
The plant immune system consists of a complex web of signaling modules, transcriptional networks and hormonal crosstalk; it can be activated by at least two types of microbial signals. The first type includes broadly conserved microbe/pathogen-associated molecular patterns (PAMPs hereinafter), such as flagellin from bacteria or chitin from fungi. Recognition of PAMPs lead to PAMP-triggered immunity (PTI) that is believed to be a principal component of basal defense against all microbes. However, PTI is often suppressed by evolved pathogens, mostly via virulence “effector” proteins that are delivered into the plant cell by pathogens. To counter pathogen virulence, plants have evolved an ability to recognize the second type of microbial signals (individual effector proteins) through nucleotide-binding leucine-rich repeat (NLR) immune receptors, resulting in the activation of a more violent form of immunity called effector-triggered immunity (ETI). Despite different modes of signaling perception, PTI and ETI share a number of downstream responses (Peng et al., 2018).
Elevated temperature (i.e., usually a few degrees warmer than the optimal temperature range for growth; Balasubramanian et al., 2006) has long been known to suppress ETI in plants and has emerged as a great concern as major crops across the globe rely on ETI for protection against many devastating plant pathogens. In response to even a brief exposure to elevated temperature, expression of an ETI-responsive gene (WRKY46) is dampened (Cheng et al., 2013; Figure 2A). Exactly how elevated temperature inhibits ETI is not fully understood. Study on two NLR proteins, N in tobacco and SUPPRESSOR OF NPR1–1, CONSTITUTIVE1 (SNC1) in Arabidopsis, showed that nuclear localization of these two proteins is reduced at elevated temperature (Zhu et al., 2010). Elevated temperature also induces expression and nuclear localization of transcription factors TEOSINTE BRANCHED1, CYCLOIDEA AND PROLIFERATING CELL FACTORS (TCPs) as well as the accumulation of HOPZ-ETI-DEFICIENT1 (ZED1) and ZED1-RELATED KINASES (ZRKs), resulting in suppression of SNC1 expression (Wang et al., 2019; Zhang et al., 2018). Another Arabidopsis NLR, RPS4, together with EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1, an essential immune regulator downstream of many NLRs), also exhibit nucleo-cytoplasmic partitioning changes (Bhattacharjee et al., 2011; Heidrich et al., 2011). Interestingly, nuclear localization of SNC1 and RPS4 proteins at elevated temperature could be reversed either in the abscisic acid (ABA) biosynthesis mutant or by application of an ABA biosynthesis inhibitor (Mang et al., 2012; Figure 2A). Taken together, these results suggest that differential nucleo-cytoplasmic localization of NLRs in response to temperature changes may underlie the negative impact of elevated temperature on ETI.
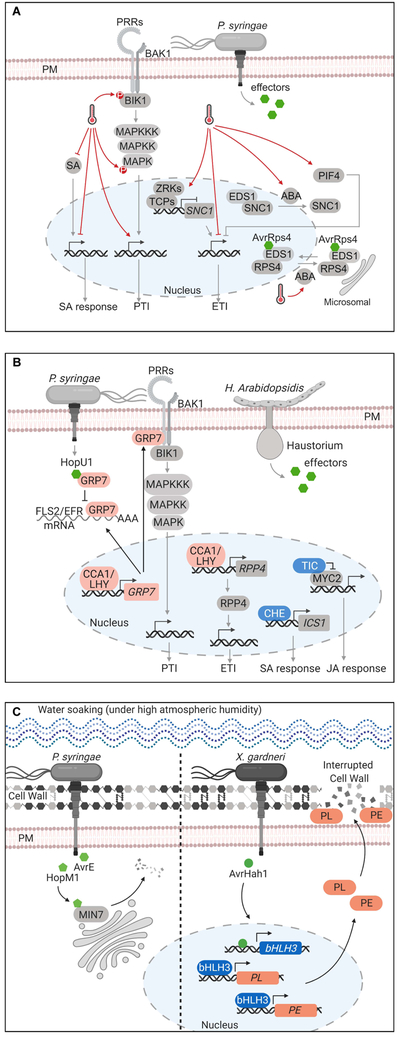
Figure 2.
Schematic diagram of temperature-, circadian- and humidity-mediated effects on plant immunity.
(A) Effect of elevated temperature on immune signaling in plants. At elevated temperature, PTI responsive genes and phosphorylation of MPKs and BIK1 are activated more robustly. Elevated temperature suppresses ETI through (i) dampening expression of ETI-responsive genes, (ii) disruption of nuclear localization of NLR proteins, including SNC1 and RPS4, possibly through an ABA-dependent mechanism, (iii) reduced transcripts of SNC1 by the action of the ZRK-TCP module, and (iv) PIF4-mediated growth-defense tradeoff. Elevated temperature inhibits SA accumulation and defense gene expression via an unknown mechanism.
(B) Effects of circadian clock on immune signaling in plants. The slave oscillator, GRP7, acting downstream of CCA1 and LHY, binds to the transcripts of FLS2 and EFR. Bacterial effector protein HopU1 blocks this interaction and reduces FLS2 protein level. GRP7 could also interact with FLS2 or EFR protein and translational machinery components. RPP4-mediated plant defense against avirulent Hpa isolates is modulated by CCA1 or LHY via transcriptional regulation. Clock protein TIC interacts with and contributes to the reduction of MYC2 protein accumulation. Clock protein CHE either directly regulates the expression of ICS1 gene or through CBP60g and SARD1.
(C) Effects of humidity on immune signaling in plants. Left panel, under high atmospheric humidity, water-soaked lesions can be caused by two Pst DC3000 effectors, AvrE and HopM1. While how AvrE induces water-soaking remains unknown, HopM1 causes water-soaking in part through mediating the removal of HopM1-interactor7 (MIN7) in the host (Nomura et al., 2006). Right panel, AvrHah1, effector secreted by X. gardneri, transcriptionally activates bHLH3 and bHLH6. bHLH3 and bHLH6 regulate the induction of pectinesterase (PE) and pectate lyase (PL) that likely changes/loosens plant cell wall structure to create water soaking lesions.
However, temperature-mediated nucleo-cytoplasmic partitioning of NLR immune receptors may not be generalized as an universal mechanism because not all NLRs show nuclear localization and some NLRs seem to function better in warmer temperature (Webb et al., 2010). Other possible mechanisms could include differential growth-defense tradeoff. For example, Arabidopsis pif4 mutant, defective in a growth-promoting transcription factor, PHYTOCHROME-INTERACTING FACTOR 4, could partially alleviate elevated temperature-mediated suppression of ETI caused by an auto-active mutant of SNC1 via an unknown mechanism (Gangappa et al., 2017; Figure 2A).
The effect of elevated temperature on plant immunity is beyond ETI. Elevated temperature increases basal Arabidopsis susceptibility to a compatible pathogen (Pseudomonas syringae pv. tomato (Pst) DC3000) in the absence of ETI activation (Gangappa et al., 2017; Huot et al., 2017). This increased basal disease development is associated with a marked reduction of plant defense hormone salicylic acid (SA) and SA-associated defense gene expression (Huot et al., 2017; Figure 2A). The molecular mechanism by which elevated temperature suppresses SA accumulation and enhances disease susceptibility remains to be elucidated. Recently, Janda and colleagues (2019) showed that temporary heat stress also suppresses PTI signaling and resistance to Pst DC3000 in Arabidopsis. Conversely, Cheng and colleagues (2013) showed that PTI-responsive genes and MAPK and BOTRYTIS-INDUCED KINASE1 (BIK1) phosphorylation were activated more robustly in response to a brief exposure of elevated temperature (Figure 2A). Further research is needed to resolve these contrasting results. Finally, recent work by Liu and colleagues (2019) showed that heat stress affects immunity of unstressed progeny and that this transgenerational memory is achieved by epigenetic machinery.
A fundamental question that remains to be answered is the identity of the thermosensor(s) involved in regulating plant immunity at elevated temperature. Recent studies showed that the red and far-red light receptor, phytochrome B (phyB) is a thermosensor; together with the transcription factor PIF4, it regulates temperature-dependent growth in Arabidopsis (Jung et al., 2016; Legris et al., 2016). As mentioned above, the Arabidopsis pif4 mutant could partially alleviate elevated temperature-mediated suppression of ETI caused by an auto-active mutant of SNC1 (Gangappa et al., 2017) and was proposed to act as a signal integration hub underlying temperature/circadian-modulated growth-defense tradeoff (Gangappa and Kumar, 2018). However, it is not clear whether PIF4 plays a broad role in temperature-mediated suppression of ETI beyond the auto-active mutant of SNC1. Furthermore, beyond ETI, Huot and colleagues (2017) found that the phyB/PIF pathway was not responsible for the enhanced basal susceptibility to virulent P. syringae at elevated temperature. Future research is needed to rigorously examine the role of phyB and PIF4 as thermosensors in immune responses and to possibly discover new thermosensors that are important for immune modulation.
1.2. Impact of temperature on microbial mechanisms
In a given plant-microbe interaction, it is expected that the effect of an environmental condition would be imposed on both the plant and the microbe. Velásquez and colleagues (2018) recently reviewed how environmental conditions affect pathogens, emphasizing that each pathogen has an optimal temperature range for growth and virulence. Due to space limitation, we will only highlight a few examples here (see Compant et al. (2010) and Velásquez et al. (2018) for more comprehensive examples). In the case of Agrobacterium infection, elevated temperature has been shown to inhibit type IV secretion-associated pilus formation and expression of virulence (vir) genes (Baron et al., 2001; Jin et al., 1993). Conversely, increased virulence was detected in the soft-rot bacterium Pectobacterium atrosepticum at elevated temperature, which is associated with increased production of plant cell wall-degrading enzymes, quorum-sensing signals and accelerated disease development (Hasegawa et al., 2005). Elevated temperature also affects beneficial plant-microbe interactions. In most cases, it positively affects hyphal growth and plant colonization of arbuscular mycorrhizal fungi (AMF), probably due to faster plant carbon allocation to the rhizosphere where AMF lives (Compant et al., 2010). Environmental conditions (especially heat, moisture and UV radiation) also directly affect the survival of microbes (Fahimipour et al., 2018).
However, it is often not clear whether conclusions based on in vitro data on temperature effects on microbes always reflect what occur during an active in planta interaction. This was especially evident in the study of the effect of temperature on type III secretion of P. syringae. While it has been well documented that elevated temperature negatively affects type III secretion in vitro (Smirnova et al., 2001), increased type III translocation of effectors into host plants was detected during Pst DC3000 infection in Arabidopsis at elevated temperature (Huot et al., 2017). Therefore, it would be desirable if future research to assess environmental effects on microbes includes more experiments performed in planta and uses new techniques (e.g., dual RNA-seq) to reveal both host and microbe changes (Nobori et al., 2018).
1.3. Plant-microbe interaction helps partners cope with temperature challenges
Some rhizosphere bacteria and endophytes could alleviate the negative impact of temperature stress on plants and expand the ability of host plants to grow at different temperatures. An interesting example is the symbiosis between tropical panic grass Dichanthelium lanuginosum and the fungus Curvularia protuberate, which allows both organisms to grow at high soil temperatures, whereas, separately, neither the plant nor the fungus can survive at this condition (Marquez et al., 2007). Moreover, the ability of C. protuberate to confer heat tolerance to the host plant requires infection of the fungus by Curvularia thermal tolerance virus (Marquez et al., 2007). In addition to panic grass, C. protuberate-mediated heat tolerance could be observed in tomato (Rodriguez et al., 2008), suggesting that the underlying mechanism may be broadly applicable to help diverse plants to cope with elevated temperature.
Some microbes can even help plants to cope with multiple stresses. An intriguing example is Burkholderia phytofirmans strain PsJN, which improves plant tolerance to cold in grapevine, heat in tomato, drought in wheat, and salt and freezing in Arabidopsis (Issa et al., 2018; Miotto-Vilanova et al., 2016). This bacterium also has direct antifungal effects, can prime plant defense and makes better resource mobilization in plants (Miotto-Vilanova et al., 2016; Timmermann et al., 2017). The mechanism(s) by which PsJN confers multi-stress tolerance remains to be elucidated; its elucidation may be of special interest for microbe-mediated crop improvement.
SECTION 2: CIRCADIAN CLOCK
2.1. Circadian clock and plant immunity
Many aspects of plant biology are linked to external light and internal circadian clock. The influence of circadian clock on plant immunity is an emerging topic (Karapetyan and Dong, 2018; Lu et al., 2017). Circadian clock is regulated through interlocked transcription-translation feedback loops. Relevant to this review are two morning-phased transcription factors, CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). Although the circadian clock is a self-sustaining system, recent studies showed that, in addition to light, certain aspects of the clock function may vary in response to changes in environmental conditions, including temperature and humidity (Lu et al., 2017; Mwimba et al., 2018). Mwimba and colleagues (2018) reported that ETI at night is strengthened by humidity oscillation, suggesting possible host anticipation of increased pathogen infection under high humidity at night. This is consistent with an earlier study showing the effect of circadian clock on ETI against the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) of Arabidopsis. Hpa normally disperses the spores at dawn to initiate infection. Circadian clock was shown to regulate RPP4 (NLR)-mediated immunity against avirulent Hpa isolates through CCA1-modulated peak expression of RPP4 and RPP4-dependent genes at dawn (Wang et al., 2011; Figure 2B). Moreover, LHY acts synergistically with CCA1 not only in RPP4-mediated immunity but also in RPS2 (NLR)-mediated defense against P. syringae, indicating that circadian clock potentially controls multiple NLR-mediated pathways (Zhang et al., 2013).
In addition to the effect of clock on ETI, several features of PTI signaling are also under clock regulation. Bhardwaj et al. (2011) infected Arabidopsis plant with P. syringae at different times of a day under constant light and found that clock controls temporal regulation of PTI against P. syringae infection. It has also been shown that GLYCINE-RICH RNA-BINDING PROTEIN7 (GRP7, a RNA-binding protein), acting downstream of CCA1 and LHY, binds specifically to the transcripts of pattern-recognition receptor (PRR) genes FLAGELLIN SENSITIVE2 (FLS2) and EF-Tu RECEPTOR (EFR), and associates with translational machinery components as well as with FLS2 and EFR proteins (Nicaise et al., 2013; Zhang et al., 2013). PRRs regulate a plethora of downstream immune outputs including stomatal closure in response to pathogen invasion (Melotto et al., 2017). Many pathogenic and nonpathogenic microbes enter plant leaves through stomata. By regulating the production of PRRs via GRP7, CCA1 and LHY may affect the responsiveness of stomata to pathogen invasion in a diurnal cycle (Zhang et al., 2013). Remarkably, as a virulence counter strategy, P. syringae produces an effector protein, HopU1, to block the interaction between GRP7 and FLS2/EFR transcripts, reduce FLS2 protein level, and promote plant susceptibility, including pathogen invasion through stomata (Nicaise et al., 2013; Figure 2B). The ability of pathogen to modulate the plant clock activity has been observed in plants infected with different pathogens, such as P. syringae, Hpa and B. cinerea, indicating that influencing clock activity may be a common strategy in pathogen infection (Lu et al., 2017).
In addition to PTI and ETI, the clock affects the biosynthesis and signaling of defense hormones SA and jasmonate (JA). SA levels show daily oscillations, with peak in the middle of the night, implying that plants may activate defenses in anticipation of attacks from biotrophic pathogens at dawn (Goodspeed et al., 2012). Besides SA accumulation, expression of SA biosynthetic genes and accumulation of NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) monomer, which activates SA signaling, show circadian oscillations (Lu et al., 2017). Furthermore, the clock protein CCA1 HIKING EXPEDITION (CHE) directly regulates the transcription level of SA biosynthetic gene ICS1 during daily oscillations (Zheng et al., 2015; Figure 2B). CHE also contributes to the induction of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT1 (SARD1) and CALMODULIN BINDING PROTEIN 60g (CBP60g), which encode transcription factors that regulates ICS1 expression (Zheng et al., 2015). Recently, it was shown that transient treatment of SA triggers a significant clock phase delay and amplitude reduction (Li et al., 2018). In addition, the cca1 lhy double mutant showed lower levels of local defense to P. syringae, while maintaining SA biosynthesis (Zhang et al., 2013), suggesting that clock genes also regulate plant defense through mechanisms independent of SA biosynthesis.
In contrast to SA levels, JA levels peak in the middle of the day (Goodspeed et al., 2012). Some of JA biosynthetic and signaling genes are direct targets of clock proteins CCA1 or TIME FOR COFFEE (TIC) (Nagel et al., 2015; Shin et al., 2012). For example, TIC interacts with the JA transcription factor MYC2 protein and acts as a negative regulator of JA signaling (Shin et al., 2012; Figure 2B).
2.2. Effect of light/circadian clock on pathogens
Not only plants, microbes also show time-of-day behavior or response. Fungal and oomycete pathogens develop hyphae and spores as well as disseminate spores often at specific times of a day. However, circadian-modulated pathogenicity is much less investigated. Hevia and colleagues (2015) provided the first evidence of microbial clock-mediated plant-pathogen interaction. Experiments involving genetic disruption of a circadian oscillator in B. cinerea, treatment of constant light to suppress fungal rhythmicity, and application of out-of-phase light:dark cycles, revealed that the outcome of susceptible Arabidopsis-B. cinerea interaction is mainly influenced by the fungal clock. Pathogens also use light exposure as a cue to initiate infection. For example, in the maize fungal pathogen Cercospora zeae-maydis, the blue light receptor Cercospora Regulator of Pathogenesis1 (Crp1) is required for sensing plant stomata and could mediate the biosynthesis of the light-activated toxin cercosporin to disrupt stomatal guard cell membranes, thereby facilitating fungal infection through stomata (Kim et al., 2011). Light could also affect the fitness and virulence of P. syringae. Red light, for example, down-regulates the expression of coronatine toxin biosynthetic genes (Santamaria-Hernando et al., 2018). Because coronatine is required for P. syringae to open stomata and facilitate bacterial entry (Melotto et al., 2006), red light may reduce bacterial entry through stomata. Taken together, similar to temperature, light and circadian clock affect both host plant and microbe during the establishment of a plant-microbe interaction.
SECTION 3: MOISTURE
3.1. Drought and plant-pathogen interactions
Water is vital to life on earth. Too little water (under water deficit or osmotic stress) or too much water (during flooding) can greatly impact many aspects of plant and microbe biology. Plants react to water deficit by regulating the level of the phytohormone ABA. ABA increase triggers a signaling cascade, resulting in large-scale transcriptional reprogramming and physiological changes, including closure of stomata to reduce transpiration (Zhu, 2016). Studies in Arabidopsis showed that bacterial pathogens, such as P. syringae, or PAMPs, such as flg22 (a 22-amino-acid epitope of Pseudomonas flagellum), can be perceived by FLS2, resulting in stomatal closure to reduce pathogen entry (Melotto et al., 2006). Thus, during drought stress, ABA-induced stomatal closure may reduce bacterial entry through stomata. However, increased ABA can lead to suppression of SA signaling pathway in the mesophyll cells inside the leaf, thus compromising post-invasion, SA-mediated resistance (Jiang et al., 2010).
3.2. Drought and plant-root microbiome interactions
Drought also affects plant-microbiome interactions. In particular, Santos-Medellin and colleagues (2017) found that, while drought affected microbial community composition in all sampled compartments (bulk soil, rhizosphere and root endosphere), the more intimate the community is associated with the root, the greater the shift of composition in drought-stressed rice plants. Similarly, in a study to examine the effects of soil moisture on sorghum root microbiome, Xu et al. (2018) found that while bacterial community diversity in surrounding soil is mostly unchanged, drought significantly reduces diversity in the rhizosphere and the root endosphere. At the phylum level, drought increases the abundance of Actinobacteria and Firmicutes. Decrease in community diversity and increase in Actinobacteria and Firmicutes transcript abundance with specific enrichment in amino acid and carbohydrate transport functions are most pronounced in the root endosphere. On the host side, drought stress causes a shift in root metabolites. Whether and how these drought-enriched metabolites “configure” root microbiome composition to promote plant stress responses remains to be determined. Nevertheless, this interesting correlation suggests that, under drought, there may be molecular dialogues between plants and associated microbiome to reshape root microbiota in order to cope with drought stress. Deciphering this molecular dialogue should advance our fundamental knowledge necessary to employ microbiota to enhance drought-tolerance in crop plants.
3.3. High air humidity and foliar pathogenesis
Rain and/or high air humidity have long been recognized as a prerequisite for many disease outbreaks in plants. During ETI, plants often undergo localized cell death at the site of pathogen infection, a phenomenon called the hypersensitive response (HR). The HR is thought to prevent the growth and spread of biotrophic pathogens and to activate secondary immune responses. High atmospheric humidity suppresses HR cell death in a number of plant-pathogen interactions and might be one of the reasons for increased plant susceptibility and disease outbreaks under high humidity (Wang et al., 2005; Wright and Beattie, 2004).
In contrast to host immune suppression, high humidity generally favors pathogen virulence. In addition to the well-recognized role of water and high humidity in promoting spore germination and bacterial motility prior to entry into the plant (Dechesne et al., 2010), recent studies showed that high humidity is critical for promoting post-invasion bacterial virulence and/or survival (i.e., inside the plant). Water-soaked lesions are a common early symptom of foliar diseases; they are formed due to abnormal accumulation of liquid inside the leaf apoplast. Water-soaking creates a disease-favorable micro-environment that could potentially increase the flow of nutrients to bacteria, diluting plant-derived defense molecules and/or facilitate bacterial spread beyond initial infection sites. Xin et al. (2016) found that P. syringae uses the type III secretion system to deliver two “water-soaking” effector proteins, AvrE and HopM1 (two widely conserved effectors in P. syringae and/or other bacterial pathogens; Degrave et al., 2015), into the plant cell to cause water soaking as an integral part of bacterial pathogenesis (Figure 2C). Notably, the ability of AvrE and HopM1 to cause water soaking requires high atmospheric humidity, as low air humidity might promote evaporation of excess apoplastic water through open stomata, countering the virulence function of AvrE and HopM1. This study illustrates the interesting phenomenon of environment-dependent function of pathogen virulence factors.
Other pathogens may use different virulence factors to cause water-soaked lesions. Schwartz et al. (2017) showed that the foliar bacterial pathogen Xanthomonas gardneri uses the type III secretion system to deliver a transcription activator-like effector, AvrHah1, into the plant cell to cause water-soaked lesions in N. benthamiana and tomato. Two tomato basic helix-loop-helix (bHLH) transcription factor genes, bHLH3 and bHLH6, were found to be the direct targets of AvrHah1. bHLH3 and bHLH6 regulate the induction of plant cell wall-modifying enzymes pectinesterase (PE) and pectate lyase (PL). Schwartz and colleagues hypothesized that AvrHah1 hijacks host bHLH3 and bHLH6 to changes/loosen plant cell wall structure to create water-soaked lesions (Figure 2C).
3.4. High soil moisture and root pathogens
Moisture also affects root disease development. Bacterial wilt in ginger plants caused by soil-borne Ralstonia solanacearum is more severe when the soil moisture is high. Jiang et al. (2018) found that expression of two wall-associated kinase (WAK) genes, WAK16 and WAK3–2, are high under low soil moisture. WAK1 is important for monitoring cell-wall integrity and acts as a receptor for oligogalacturonides, a damage-associated molecular pattern (DAMP) in Arabidopsis (Brutus et al., 2010). High soil moisture dampens WAK16 and WAK3–2 expression and weakens plant immunity toward R. solanacearum, suggesting WAK16 and WAK3–2 may play important roles in sensing soil moisture and mediate cell wall-based plant immunity against root pathogens (Jiang et al., 2018).
SECTION 4: NUTRITIONAL STATUS
One of the ultimate forces that drive plant-microbe interactions is nutrient acquisition. It is of no surprise that plant nutritional status and nutrient availability in the environment have significant effects on plant-microbe interactions.
4.1. Phosphate
It is well known that the intricate symbiotic relationship between land plants and phosphate-acquiring AMF is tightly regulated by phosphate status in the soil and in the plant (Muller and Harrison, 2019). However, the effect of phosphate on plant-microbe interactions is beyond plant-AMF interactions. Arabidopsis thaliana is a non-host for phosphorus-acquiring AMF (Fernandez et al., 2019). Hiruma et al. (2016) identified a natural endophytic fungus, Colletotrichum tofieldiae (Ct), in wild Arabidopsis in central Spain. Ct can transfer phosphate to Arabidopsis and promotes plant growth and fertility; however, Ct-mediated growth promotion can only be observed when plants were grown under phosphate-deficient conditions. Further investigation found that an intact plant phosphate starvation response (PSR) system (Chiou and Lin, 2011) is required for Ct-dependent plant growth promotion (Figure 3A). Hacquard and colleagues (2016) later found that host defense responses are transcriptionally suppressed in Ct-colonized plants under phosphate-starving state, presumably to facilitate the symbiotic relation. These two studies illustrate that host nutritional status determines the outcome of Arabidopsis-Ct interaction: partners form a mutualistic interaction if the host experiences phosphate starvation and the interaction becomes commensal (i.e., not mutualistic) if the host is grown under phosphate-sufficient conditions.
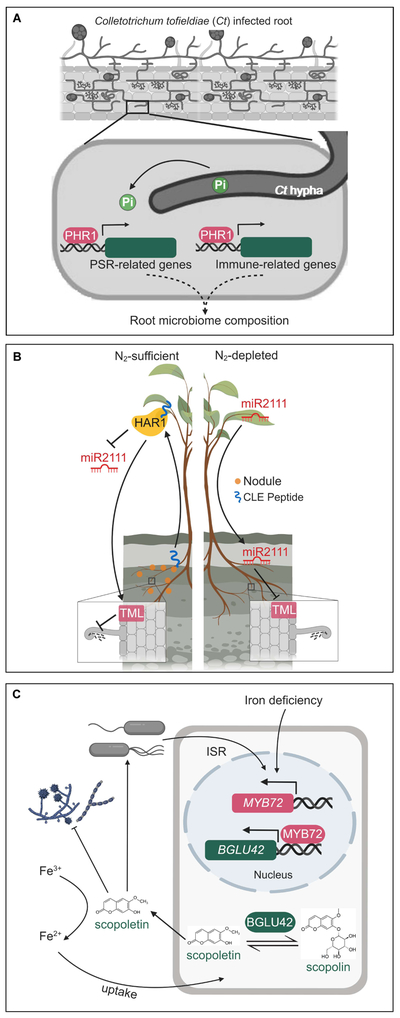
Figure 3.
Nutrient status and plant-microbe interactions. (A) Phosphate status and Arabidopsis-root microbiome interaction. Under phosphate-limiting state, C. tofieldiae (Ct) colonization activates phosphate starvation response (PSR) genes and promotes phosphate uptake, which requires a functional PHR1. Additionally, PHR1 represses immune-related gene expression and is required for assembling root endophytic microbiome.
(B) Nitrogen status and legume-Rhizobium interaction. Under nitrogen-sufficient condition, nodule formation is repressed through autoregulation of nodulation (AON; left panel). When plant is nitrogen-depleted (right panel), shoot-produced miR2111 post-transcriptionally downregulates TML (a positive regulator of AON) in the root to maintains host susceptibility to rhizobium for nodulation.
(C) Iron status and Arabidopsis-root microbiome interaction. MYB72- and BGLU42-mediated production and exudation of scopoletin is induced by iron deficiency and during induced systemic resistance (ISR). Scopoletin is hypothesized to solubilize iron for uptake and to select plant-associated microbes.
Castrillo et al. (2017) investigated the effects of host phosphate status and immunity on root-associated bacterial microbiome composition. It was found that PSR-defective mutants assemble an anomalous root endophytic microbiota and identified PHOSPHATE STARVATION RESPONSE1 (PHR1), the master regulator of the PSR, to be an integrating hub for regulating plant immunity, PSR and root-microbiota. Specifically, under phosphate-deficient environment, PHR1 transcriptionally represses immune-related gene expression and shapes root microbiota structure presumably to optimize plant performance (Figure 3A).
Studies by Hiruma et al. (2016), Hacquard et al. (2016), and Castrillo et al. (2017) set a foundation for future investigations to identify specific microbial consortia that could improve plant fitness under phosphate-limiting soils.
4.2. Nitrogen
Interactions between legumes and Rhizobium spp. result in the formation of symbiotic nodules in the root, representing a remarkable biological process in which biologically inert atmospheric N2 is converted to biologically usable NH3 to facilitate plant growth and development. However, it is energetically costly to the legume host to form nodules and may not be cost-effective when plants are grown in a nitrogen-rich environment (Morgan et al., 2005). To avoid the situation where the energy cost to the host outweighs the benefits from the interaction, plants have developed autoregulation of nodulation (AON) to optimize the number of nodules formed in the root based on the need for nitrogen in the shoot. Key players in AON include a shoot CLAVATA1-like LRR receptor kinase HYPERNODULATION ABERRANT ROOT FORMATION1 (HAR1), which perceives rhizobium/nitrate-induced, root-producing CLV3/EMBRYO SURROUNDING REGION (CLE) peptides (Okamoto et al., 2013). After perception, shoot-derived inhibitory signals are transmitted through TOO MUCH LOVE (TML), a root-acting F-box protein, to impede nodule formation (Takahara et al., 2013; Figure 3B).
Under nitrogen-deficient condition, shoots communicate with roots to maintain a susceptible state to rhizobium symbiotic interaction. Using Lotus japonicus and Mesorhizobium loti as the model system, Tsikou and colleagues (2018) found that the miR2111 abundance in lotus is negatively correlated with M. loti infection and nitrogen availability. Further investigation revealed that shoot-produced miR2111 is translocated through the phloem to the root to post-transcriptionally silence TML, a positive regulator of AON (Figure 3B). Tsikou et al. (2018) proposed that shoots systemically regulate TML expression in roots via miR2111 to ensure un-infected tissue remain susceptible to rhizobium. If the plant has ample nitrogen or a symbiotic interaction with rhizobium has been established, miR2111 level is lowered in a HAR1-dependent manner and progression of nodulation is restricted (Figure 3B). Their findings highlight the important roles the legume hosts play in steering beneficial interactions with symbiotic partners in response to environmental changes.
4.3. Iron
Induced systemic resistance (ISR) is a form of plant immunity that is triggered by certain rhizosphere mutualistic microbes to prime the host against potential pathogens and herbivore attacks. Microarray and mutant analyses identified the Arabidopsis transcription factor MYB72 as a key regulator of ISR (Van der Ent et al., 2008). Interestingly, expression of MYB72 in roots is also highly induced by iron deficiency (Buckhout et al., 2009). BGLU42, a β-glucosidase, was identified as a key player downstream of MYB72 in both ISR and response to iron deficiency. MYB72 activates genes that encode enzymes producing iron-mobilizing phenolic metabolites and BGLU42, which is required for releasing these phenolic compounds into the rhizosphere under iron-deficient condition (Zamioudis et al., 2014). Stringlisa and colleagues (2018) found that scopoletin, a member of coumarins, is the most abundant phenolic compound produced and secreted into the Arabidopsis rhizosphere in iron deficiency in a MYB72- and BGLU42-dependent manner. Metagenomics analysis found that microbial community associated with the rhizosphere of the scopoletin biosynthesis mutant f6′h1 is very different from that of wild-type plants. In vitro antimicrobial activity assays showed that while scopoletin has no or a minimal effect on two ISR-inducing plant growth-promoting rhizobacteria (PGPR), it has a dose-dependent inhibitory effect on two known soil-borne pathogens of Arabidopsis, Fusarium oxysporum f. sp. raphani and Verticillium dahliae JR2. Thus, under iron-deficient conditions, MYB72, BGLU42 and scopoletin appear to constitute a regulatory module to increase iron solubility for acquisition and reconfigure rhizosphere microbiota to protect the host from potential pathogen and insect attack via ISR (Figure 3C).
SECTION 5: CONCLUSION AND OUTLOOK
One of the most challenging tasks of the 21st century is to find novel methods to increase global crop production for the growing human population. A major roadblock to global food security is persistent loss of crops due to plant diseases. In the past four decades, tremendous progress has been made in understanding how plant diseases occur and how the plant immune system works. In contrast, our knowledge of why environmental conditions have such strong impact on plant-microbe interactions is still at a novice stage in terms of molecular and mechanistic insights. This is, in part, because most contemporary investigations into molecular plant-pathogen interactions have been performed in plant growth chambers with simple, static environmental setups or in “test tubes”. While these studies are important in revealing the first principles underlying disease and immunity, they do not provide molecular insights into the influence of environmental conditions on pathogen virulence and host immunity. Unlike controlled and often static environmental conditions in the lab, in nature, changes in one environmental condition (e.g. temperature) is often associated with changes in multiple other environmental parameters (e.g. humidity and CO2; Figure 1B) and static environmental condition is exception rather than norm. We envision that future studies of plant-pathogen interactions will increasingly consider the multi-dimensional nature of “plant-microbe-environment” interactions by conducting experiments in next-generation growth facilities that could simulate natural dynamic and interactive abiotic conditions and incorporating plant-microbe systems in addition to Arabidopsis-microbe interactions, which have so far played a dominating role in generating first sights into basic mechanisms. In addition, plant natural accessions, a rich source of genetic variation, should be utilized to understand the influences of environmental conditions on plant-microbe interactions (Alcazar and Parker, 2011). Together, such studies are necessary to reach a systems-level understanding with a strong predictive power for forecasting the performance of plant-microbe interactions under dynamic climatic conditions in crop fields and natural ecosystems.
Basic research into how environmental conditions shape plant-microbe interactions will hopefully identify a set of environment-sensitive “switches” in the plant defense network, symbiosis biology and microbial community structure that are genetically or chemically modifiable. Identification of such switches should provide a platform for developing a new generation of plant varieties in which beneficial plant-microbe interactions can be made more resilient, and pathogenic plant-microbe interactions more vulnerable to a warming and harsher climate. Likewise, the emerging field of plant-microbiome interactions is a promising area of research for making crop plants more resilient to both abiotic and biotic stresses.
In concluding this review, we would like to point out that, although we have focused our discussion on how temperature, circadian, moisture and nutrients affect plant-microbe interactions, other environmental conditions, including, most notably, atmospheric CO2 concentration, have been receiving increasing attention (Zhou et al., 2017). Furthermore, there are numerous examples in which animal-microbe interactions are affected by their surrounding environment. These examples include: 1) the effects of sunlight (UV-R) on skin microbiome (Patra et al., 2016), 2) perturbation of circadian clock by gut microbiome (Wang et al., 2017), 3) global warming temperature on the prevalence and severity of marine animal infectious diseases and coral reef bleaching (Harvell et al., 2019; Hughes et al., 2017), and 4) the vital role of nutrition on animal immunity (Belkaid and Hand, 2014; Lazar et al., 2018). It is likely that there are significant cross-kingdom principles that await to be discovered. As such, study of environmental effects on plant-microbe interactions could have a broader impact on understanding how global climatic conditions shape current and future host-microbe in both plant and animal kingdoms.
In this review, we highlight studies that begin to shed light on how environmental conditions influence diverse plant-pathogen, plant-symbiont and plant-microbiota interactions. Study of environmental effects on plant-microbe interactions has significant ramifications in understanding how global climatic change might shape future host-microbe interactions.
ACKNOWLEDGEMENTS
We thank lab members Christian Danve Castroverde and André Velásquez for their critical comments during the preparation of this review. We apologize to any colleagues whose work was not cited due to space limitation. Authors are funded by US National Institute of General Medical Sciences (GM109928 to S.Y.H.); US National Science Foundation (IOS-1557437 and NSF MCB 011272–00001 to S.Y.H); US Department of Agriculture, NIFA (2015-67017-23360 and 2017-67017-26180 to S.Y.H); US Department of Energy (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science; DE–FG02–91ER20021 for infrastructural support to S.Y.H.); Michigan State University Plant Resilience Institute (to S.Y.H.) and NSERC Postdoctoral Fellowship (to Y.T.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alcazar R, and Parker JE (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16, 666–675. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, and Weigel D (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C, Domke N, Beinhofer M, and Hapfelmeier S (2001). Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol 183, 6852–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Meier S, Petersen LN, Ingle RA, and Roden LC (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One 6, e26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, and Gassmann W (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334, 1405–1408. [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, and De Lorenzo G (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U. S. A 107, 9452–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJW, and Schmidt W (2009). Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D, and He SY (2009). Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Gao X, Feng B, Sheen J, Shan L, and He P (2013). Plant immune response to pathogens differs with changing temperatures. Nat. Commun 4, 2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, and Lin S-I (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Compant S, van der Heijden MGA, and Sessitsch A (2010). Climate change effects on beneficial plant-microorganism interactions. FEMS Microbiol. Ecol 73, 197–214. [DOI] [PubMed] [Google Scholar]
- Dechesne A, Wang G, Gulez G, Or D, and Smets BF (2010). Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl. Acad. Sci. U. S. A 107, 14369–14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave A, Siamer S, Boureau T, and Barny M-A (2015). The AvrE superfamily: ancestral type III effectors involved in suppression of pathogen-associated molecular pattern-triggered immunity. Mol. Plant Pathol 16, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, Verhagen BWM, Van Doorn R, Bakker D, Verlaan MG, Pel MJC, Joosten RG, Proveniers MCG, Van Loon LC, Ton J, et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahimipour AK, Hartmann EM, Siemens A, Kline J, Levin DA, Wilson H, Betancourt-Roman CM, Brown GZ, Fretz M, Northcutt D, et al. (2018). Daylight exposure modulates bacterial communities associated with household dust. Microbiome 6, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I, Cosme M, Stringlis IA, Yu K, de Jonge R, van Wees SM, Pozo MJ, Pieterse CMJ, and van der Heijden MGA (2019). Molecular dialogue between arbuscular mycorrhizal fungi and the nonhost plant Arabidopsis thaliana switches from initial detection to antagonism. New Phytol. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, and Kumar SV (2018). DET1 and COP1 Modulate the Coordination of Growth and Immunity in Response to Key Seasonal Signals in Arabidopsis. Cell Rep. 25, 29–37.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Berriri S, and Kumar SV (2017). PIF4 Coordinates Thermosensory Growth and Immunity in Arabidopsis. Curr. Biol 27, 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, and Covington MF (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U. S. A 109, 4674–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard S, Kracher B, Hiruma K, Munch PC, Garrido-Oter R, Thon MR, Weimann A, Damm U, Dallery J-F, Hainaut M, et al. (2016). Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun 7, 11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard S, Spaepen S, Garrido-Oter R, and Schulze-Lefert P (2017). Interplay Between Innate Immunity and the Plant Microbiota. Annu. Rev. Phytopathol 55, 565–589. [DOI] [PubMed] [Google Scholar]
- Harvell CD, Montecino-Latorre D, Caldwell JM, Burt JM, Bosley K, Keller A, Heron SF, Salomon AK, Lee L, Pontier O, et al. (2019). Disease epidemic and a marine heat wave are associated with the continental-scale collapse of a pivotal predator (Pycnopodia helianthoides). Sci. Adv 5, eaau7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Chatterjee A, Cui Y, and Chatterjee AK (2005). Elevated temperature enhances virulence of Erwinia carotovora subsp. carotovora strain EC153 to plants and stimulates production of the quorum sensing signal, N-acyl homoserine lactone, and extracellular proteins. Appl. Environ. Microbiol 71, 4655–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, and Parker JE (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334, 1401–1404. [DOI] [PubMed] [Google Scholar]
- Hevia MA, Canessa P, Muller-Esparza H, and Larrondo LF (2015). A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A 112, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K, Gerlach N, Sacristan S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramirez D, Bucher M, O’Connell RJ, et al. (2016). Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell 165, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Alvarez-Noriega M, Alvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. [DOI] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velásquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, and He SY (2017). Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat. Commun 8, 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H-H, Yu M, and Lai E-M (2017). Agrobacterium-mediated plant transformation: biology and applications. Arab. B 15, e0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa A, Esmaeel Q, Sanchez L, Courteaux B, Guise J-F, Gibon Y, Ballias P, Clement C, Jacquard C, Vaillant-Gaveau N, et al. (2018). Impacts of Paraburkholderia phytofirmans Strain PsJN on Tomato (Lycopersicon esculentum L.) Under High Temperature. Front. Plant Sci 9, 1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Lamparova L, Zubikova A, Burketova L, Martinec J, and Krckova Z (2019). Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol. Plant Pathol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C-J, Shimono M, Sugano S, Kojima M, Yazawa K, Yoshida R, Inoue H, Hayashi N, Sakakibara H, and Takatsuji H (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant. Microbe. Interact 23, 791–798. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Huang M, Zhang M, Lan J, Wang W, Tao X, and Liu Y (2018). Transcriptome analysis provides novel insights into high-soil-moisture-elevated susceptibility to Ralstonia solanacearum infection in ginger (Zingiber officinale Roscoe cv. Southwest). Plant Physiol. Biochem. PPB 132, 547–556. [DOI] [PubMed] [Google Scholar]
- Jin S, Song YN, Deng WY, Gordon MP, and Nester EW (1993). The regulatory VirA protein of Agrobacterium tumefaciens does not function at elevated temperatures. J. Bacteriol 175, 6830–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Vance RE, and Dangl JL (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354. [DOI] [PubMed] [Google Scholar]
- Jung J-H, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889. [DOI] [PubMed] [Google Scholar]
- Karapetyan S, and Dong X (2018). Redox and the circadian clock in plant immunity: A balancing act. Free Radic. Biol. Med 119, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ridenour JB, Dunkle LD, and Bluhm BH (2011). Regulation of stomatal tropism and infection by light in Cercospora zeae-maydis: evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathog. 7, e1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar V, Ditu L-M, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L, and Chifiriuc MC (2018). Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front. Immunol. 9, 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CCR, Neme M, Hiltbrunner A, Wigge PA, Schafer E, Vierstra RD, and Casal JJ (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900. [DOI] [PubMed] [Google Scholar]
- Li Z, Bonaldi K, Uribe F, and Pruneda-Paz JL (2018). A Localized Pseudomonas syringae Infection Triggers Systemic Clock Responses in Arabidopsis. Curr. Biol 28, 630–639.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Feng L, Gu X, Deng X, Qiu Q, Li Q, Zhang Y, Wang M, Deng Y, Wang E, et al. (2019). An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 29, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, McClung CR, and Zhang C (2017). Tick Tock: Circadian Regulation of Plant Innate Immunity. Annu. Rev. Phytopathol 55, 287–311. [DOI] [PubMed] [Google Scholar]
- Mang H-G, Qian W, Zhu Y, Qian J, Kang H-G, Klessig DF, and Hua J (2012). Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24, 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez LM, Redman RS, Rodriguez RJ, and Roossinck MJ (2007). A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315, 513–515. [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, and He SY (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, and He SY (2017). Stomatal Defense a Decade Later. Plant Physiol. 174, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto-Vilanova L, Jacquard C, Courteaux B, Wortham L, Michel J, Clement C, Barka EA, and Sanchez L (2016). Burkholderia phytofirmans PsJN Confers Grapevine Resistance against Botrytis cinerea via a Direct Antimicrobial Effect Combined with a Better Resource Mobilization. Front. Plant Sci. 7, 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JAW, Bending GD, and White PJ (2005). Biological costs and benefits to plant-microbe interactions in the rhizosphere. J. Exp. Bot 56, 1729–1739. [DOI] [PubMed] [Google Scholar]
- Muller LM, and Harrison MJ (2019). Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol 50, 132–139. [DOI] [PubMed] [Google Scholar]
- Mwimba M, Karapetyan S, Liu L, Marques J, McGinnis EM, Buchler NE, and Dong X (2018). Daily humidity oscillation regulates the circadian clock to influence plant physiology. Nat. Commun 9, 4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, and Kay SA (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 112, E4802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, Joe A, Jeong B, Korneli C, Boutrot F, Westedt I, Staiger D, Alfano JR, and Zipfel C (2013). Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J 32, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori T, Velásquez AC, Wu J, Kvitko BH, Kremer JM, Wang Y, He SY, and Tsuda K (2018). Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. U. S. A 115, E3055–E3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, and He SY (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313, 220–223. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, and Kawaguchi M (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun 4, 2191. [DOI] [PubMed] [Google Scholar]
- Patra V, Byrne SN, and Wolf P (2016). The Skin Microbiome: Is It Affected by UV-induced Immune Suppression? Front. Microbiol 7, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, van Wersch R, and Zhang Y (2018). Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol. Plant. Microbe. Interact 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim Y-O, and Redman RS (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. [DOI] [PubMed] [Google Scholar]
- Santamaria-Hernando S, Rodriguez-Herva JJ, Martinez-Garcia PM, Rio-Alvarez I, Gonzalez-Melendi P, Zamorano J, Tapia C, Rodriguez-Palenzuela P, and Lopez-Solanilla E (2018). Pseudomonas syringae pv. tomato exploits light signals to optimize virulence and colonization of leaves. Environ. Microbiol 20, 4261–4280. [DOI] [PubMed] [Google Scholar]
- Santos-Medellin C, Edwards J, Liechty Z, Nguyen B, and Sundaresan V (2017). Drought Stress Results in a Compartment-Specific Restructuring of the Rice Root-Associated Microbiomes. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AR, Morbitzer R, Lahaye T, and Staskawicz BJ (2017). TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc. Natl. Acad. Sci. U. S. A 114, E897–E903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, and Davis SJ (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24, 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova A, Li H, Weingart H, Aufhammer S, Burse A, Finis K, Schenk A, and Ullrich MS (2001). Thermoregulated expression of virulence factors in plant-associated bacteria. Arch. Microbiol 176, 393–399. [DOI] [PubMed] [Google Scholar]
- Stevens RB in Plant Pathology, An Advanced Treatise (ed. Horsfall JG) 357–429 (Academic Press, New York, 1960). [Google Scholar]
- Stringlis IA, Yu K, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, and Pieterse CMJ (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U. S. A 115, E5213–E5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M, Magori S, Soyano T, Okamoto S, Yoshida C, Yano K, Sato S, Tabata S, Yamaguchi K, Shigenobu S, et al. (2013). Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 54, 433–447. [DOI] [PubMed] [Google Scholar]
- Timmermann T, Armijo G, Donoso R, Seguel A, Holuigue L, and Gonzalez B (2017). Paraburkholderia phytofirmans PsJN Protects Arabidopsis thaliana Against a Virulent Strain of Pseudomonas syringae Through the Activation of Induced Resistance. Mol. Plant. Microbe. Interact 30, 215–230. [DOI] [PubMed] [Google Scholar]
- Tsikou D, Yan Z, Holt DB, Abel NB, Reid DE, Madsen LH, Bhasin H, Sexauer M, Stougaard J, and Markmann K (2018). Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236. [DOI] [PubMed] [Google Scholar]
- Velásquez AC, Castroverde CDM, and He SY (2018). Plant-Pathogen Warfare under Changing Climate Conditions. Curr. Biol 28, R619–R634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cai X, and Zheng Z (2005). High humidity represses Cf-4/Avr4- and Cf-9/Avr9-dependent hypersensitive cell death and defense gene expression. Planta 222, 947–956. [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, Lee D, Fu X-D, and Dong X (2011). Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, and Hooper LV (2017). The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cui D, Liu C, Zhao J, Liu J, Liu N, Tang D, and Hu Y (2019). TCP transcription factors interact with ZED1-related kinases as components of the temperature-regulated immunity. Plant. Cell Environ 42, 2045–2056. [DOI] [PubMed] [Google Scholar]
- Webb KM, Ona I, Bai J, Garrett KA, Mew T, Vera Cruz CM, and Leach JE (2010). A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 185, 568–576. [DOI] [PubMed] [Google Scholar]
- Wright CA, and Beattie GA (2004). Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A 101, 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X-F, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, and He SY (2016). Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Naylor D, Dong Z, Simmons T, Pierroz G, Hixson KK, Kim Y-M, Zink EM, Engbrecht KM, Wang Y, et al. (2018). Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U. S. A 115, E4284–E4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C, Hanson J, and Pieterse CMJ (2014). beta-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204, 368–379. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xie Q, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong D, Kwak JM, et al. (2013). Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 9, e1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang Z, Bao Z, Yang L, Wu D, Shu X, and Hua J (2018). MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. Plant J. 93, 66–78. [DOI] [PubMed] [Google Scholar]
- Zheng X-Y, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, and Dong X (2015). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. U. S. A 112, 9166–9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Vroegop-Vos I, Schuurink RC, Pieterse CMJ, and Van Wees SCM (2017). Atmospheric CO2 Alters Resistance of Arabidopsis to Pseudomonas syringae by Affecting Abscisic Acid Accumulation and Stomatal Responsiveness to Coronatine. Front. Plant Sci 8, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K (2016). Abiotic Stress Signaling and Responses in Plants. Cell 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qian W, and Hua J (2010). Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6, e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, and Oldroyd GED (2017). Plant signalling in symbiosis and immunity. Nature 543, 328–336. [DOI] [PubMed] [Google Scholar]