ABSTRACT
Secondary metabolites can be viewed as a chemical language, facilitating communication between microorganisms. From an ecological point of view, this metabolite exchange is in constant flux due to evolutionary and environmental pressures. From a biomedical perspective, the chemistry is unsurpassed for its antibiotic properties. Genome sequencing of microorganisms has revealed a large reservoir of Biosynthetic Gene Clusters (BGCs); however, linking these to the secondary metabolites they encode is currently a major bottleneck to chemical discovery. This linking of genes to metabolites with experimental validation will aid the elicitation of silent or cryptic (not expressed under normal laboratory conditions) BGCs. As a result, this will accelerate chemical dereplication, our understanding of gene transcription and provide a comprehensive resource for synthetic biology. This will ultimately provide an improved understanding of both the biosynthetic and chemical space. In recent years, integrating these complex metabolomic and genomic data sets has been achieved using a spectrum of manual and automated approaches. In this review, we cover examples of these approaches, while addressing current challenges and future directions in linking these data sets.
Keywords: secondary metabolites, specialised metabolites, biosynthetic gene clusters, genome mining, comparative metabolomics
This review covers advances in methods used to link microbial genes to the chemistry they encode
INTRODUCTION
Recent improvements in the identification of BGCs has revolutionised our capacity to understand secondary metabolite production. Over the last few years, there has been a significant effort to link genomic data to secondary metabolite data for microorganisms, in particular bacteria. The first section of this review focuses on the creation of both biosynthetic and chemical data sets used for this purpose. The term linking in this review will cover all aspects of associating BGCs with the metabolite they encode. This linking process is divided into two main sections. The first is targeted approaches to linking, comprising the subsections, targeted genome mining linking, whereby strains are selected based on BGC information for further chemical analysis and multi-targeted linking, encompassing genome mining with bioactivity, metabolomics and proteomics approaches. The second is automated approaches, which is further divided into correlation-based and feature-based. Correlation-based approaches identify putative links via correlation of strain inclusion in clusters of spectra and BGCs. Feature-based approaches score individual spectra against individual BGCs based on shared properties. At the same time, we appreciate that studies rarely fall into these binary categories and that in reality, linking is often a spectrum using both approaches. In this review, we have selected recent studies to exemplify the discovery angle of each approach. We conclude with examples of experimental validation of these links through synthetic biology methods and a section on current challenges.
BIOSYNTHETIC AND CHEMICAL DATA SETS FOR LINKING
This section covers how data sets are made from the prediction of BGCs and detection of metabolites. This is a crucial step as the quality of the data sets will directly impact the success of linking across data sets. Predicting BGCs from bacterial genomes is a fairly mature discipline. Tools such as antiSMASH (Blin et al. 2017) and SMURF (Khaldi et al. 2010) provide BGC predictions by matching curated statistical models based on sequences of protein family domains (PFAM domains) to genomic sequences (Coggill, Finn and Bateman 2008). Such techniques typically exhibit high specificity (low numbers of false positives) at the expense of low sensitivity (high numbers of false negatives). For users wishing higher sensitivity, more speculative algorithms such as ClusterFinder (Cimermancic et al. 2014), MIDDAS-M (Umemura et al. 2013) or MIPS-GC (Umemura, Koike and Machida 2015) exist. An in-depth review of BGC detection is beyond the scope of this article – please refer to Chavali and Rhee 2018 (Chavali and Rhee 2018) for a detailed review – as are the community-driven comparative metabolomics platforms such as molecular networking, based on tandem-mass spectrometry data (Wang et al. 2016) and data bases such as NP Atlas (www.npatlas.org). While cross-referencing data bases has been the status quo for secondary metabolite identification, facilitating this through automation would greatly accelerate discovery.
TARGETED LINKING
Targeted genome mining approaches to linking
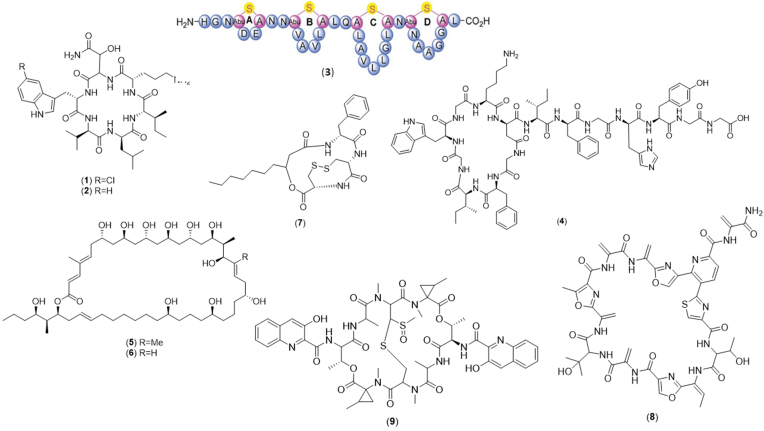
In this section, strains prioritised for chemical investigation based on genome mining information, for example the presence of specific BGCs, will be discussed. This linking is often a manual process and requires specialist biosynthetic and chemical knowledge. The first step involves the dereplication, or strain prioritisation (or strain elimination due to the presence of previously discovered metabolites), within complex biosynthetic and chemical data sets. Dereplication has been greatly aided by the data analysis platforms mentioned previously, resulting in new metabolite discovery (Fig.1) (Duncan et al. 2015; Kaweewan et al. 2017; Schneider et al. 2018; Son et al. 2018; Ueoka et al. 2018; Xu et al. 2018). For example, two new peptides, the halogenated curacomycin (1) and its dechlorinated derivative dechlorocuracomycin (2), produced by Streptomyces curacoi and Streptomyces noursei respectively, were discovered through a genome mining approach using antiSMASH to identify the presence of tryptophan halogenase genes in proximity to a nonribosomal peptide synthetase (NRPS) BGC (Kaweewan et al. 2017). Further genomic investigation of S. curacoi led to the discovery of an additional new cytotoxic peptide, curacozole, produced by a gene analogous to that of curacomycin (Kaweewan et al. 2019). Using a similar approach, the novel lanthipeptide tikitericin (3) was isolated from a thermophilic bacterium, Thermogemmatispora sp. by detecting a lanthionine synthetase, homologous to a class II lanthipeptide BGC (Xu et al. 2018). The putative BGC consisting of 10 genes encoding a new lasso peptide was observed through genome mining of the rare actinomycete Actinokineospora spheciospongiae and led to the isolation of actinokineosin, a new peptide with promising antibacterial activity (Takasaka et al. 2017). In another study, the product of the lasso peptide BGC uld from the genome of Streptomyces sp. KCB13F003 was targeted using a One Strain Many Compounds (OSMAC) approach, in which each strain was grown in multiple media in an attempt to elicit a wide range of BGC expression, resulting in the isolation of the new metabolite ulleungdin (4) (Son et al. 2018). Recently, a plant-associated Gynuella sunshinyii strain was prioritised based on the presence of several unassigned BGCs and six trans-AT PK clusters. Further investigation led to the isolation of four metabolites of which three, the polyketides lacunalide A and B (5, 6) and the cyclodepsitripeptide sunshinamide (7)-represented novel scaffolds. Further manual genome mining resulted in two ergoyne analogues, ergoyne A and B, revealing the importance of complementing automated genome mining with manual curation (Ueoka et al. 2018).
Figure 1.
Chemical structures discovered as a result of manual linking of gene clusters and metabolites, including curacomycin (1), dechlorocuracomycin (2), tikitericin (3), ulleungdin (4), lacunalide A and B (5, 6), sunshinamide (7), geninthiocin B (8) and retimycin A (9).
Multi-targeted linking
In the last five years, comparative metabolomics has been linked with comparative genome mining, proteomics and bioactivity to accelerate discovery. For example, genome mining of a lichen-associated Streptomyces sp. with metabolites bioactive against Bacillus subtilis revealed a BGC encoding a lantibiotic. By connecting the inactivation of the BGC to the loss of the observed bioactivity, a new 35-membered macrocyclic thiopeptide antibiotic geninthiocin B (8) was isolated (Schneider et al. 2018). Natural product proteomining – a quantitative proteomics platform – was introduced by Gubbens et al., for the identification of BGCs for targeted secondary metabolites by applying the OSMAC approach, metabolomics and quantitative proteomics. This approach allowed correlations between quantitative metabolomics or bioactivity data and protein expression profiles. A new juglomycin derivative was isolated from a soil-isolated Streptomyces sp. by applying this quantitative multi-omics approach (Gubbens et al. 2014).
Using pattern-based BGC genome mining combined with comparative metabolomics through molecular networking, the relationship between BGCs and the corresponding metabolites across 35 Salinispora strains was assessed. This resulted in an uncharacterised PKS BGC being linked to the previously reported metabolite arenicolide A, in addition to linking an uncharacterised NRPS BGC (NRPS40) to retimycin A (9), a new quinomycin-like depsipeptide (Duncan et al. 2015). The increasing complexity and scale of these combined data sets, often consisting of tens to thousands of both genomes and spectra, has resulted in the need to automate this process.
AUTOMATED LINKING
Two major tracks can be observed in the automated linking of BGCs to secondary metabolites. The first approach, feature-based linking, involves linking chemical features predicted from genomic information. For example, neutral losses indicative of amino acid residues, with the observed metabolomic data. The second approach, correlation-based linking, makes use of data sets where genomic and metabolomic data are available for a large number of strains. Related BGCs assembled into gene cluster families (GCFs) can be correlated with spectra belonging to molecular families (MFs) based on the occurrence of their source strains across the data set, with the assumption that true links would have high source strain correlations.
Feature-based linking
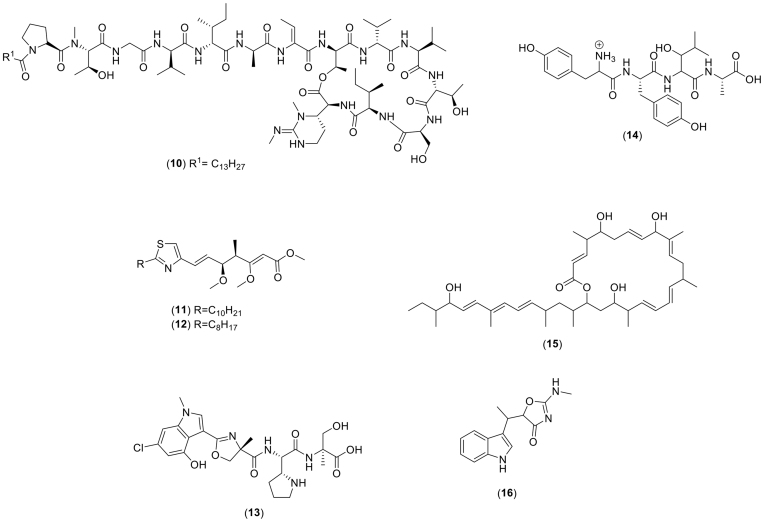
A fruitful approach for linking BGCs to secondary metabolites has been to predict the structural properties of the molecules based on genomic information to directly detect corresponding features in mass spectra (Fig.2). For example, tools such as SANDPUMA (Chevrette et al. 2017) can predict substrate specificity for adenylation domains in NRPS BGCs. These predictions can be matched to amino acid residue-derived MS/MS fragmentations. This approach, using molecular properties predicted from genomic information to guide the search in chemical space, termed peptidogenomics by Kersten et al. (Kersten et al. 2011), has been used to link peptidic natural product BGCs to metabolites. These include stendomycin I (10) and the ribosomal lantipeptide. A similar technique has been described by Panter et al. (Panter, Krug and Müller 2019) for polyketides, which facilitated the discovery and structure elucidation of fulvuthiacene A and B (11, 12). Other examples of tools intended to predict detectable features from genomic information include RODEO (Tietz et al. 2017), which is focused on RiPPs, although this has not yet been integrated with mass spectrometry data; PRISM (Skinnider et al., 2015, 2017), which focuses on NRPs and type I and II PKSs but is limited to LC-MS (but not LC-MS/MS data); GNP (Johnston et al. 2015), which links NRPS and PKS BGCs to LC-MS/MS spectra, Pep2path (Medema et al. 2014), which focuses on peptidic natural products, RippQUEST (Mohimani et al. 2014a) and NRPquest (Mohimani et al. 2014b), which detect RiPP and NRPS BGCs, respectively, and predict possible fragmentation patterns for their products.
Figure 2.
Chemical structures discovered as a result of automated linking of gene clusters and metabolites, including stendomycin I (10), fulvuthiacene A and B (11, 12), tambromycin (13), tyrobetaine (14), macrobrevin (15), and indolmycin (16).
Also relevant are tools that have been developed for dereplication. For example, DEREPLICATOR (Mohimani et al., 2017, 2018) predicts possible fragmentation patterns for peptidic natural products. By linking spectra to peptides, and identifying the BGCs responsible for the production of those peptides in databases such as MIBiG (Medema et al. 2015), similar BGCs from the organism can be tentatively linked to the spectra. This approach was used, for example, in Mohimani et al. 2018 (Mohimani et al. 2018), to link the polyketide antibiotic C35H56O13, which is structurally similar to chalcomycin, to its producing BGC.
Correlation-based linking
Another major approach is based on matching patterns of source strain occurrence between GCFs and MFs. The assumption that similar BGCs in different strains will produce similar molecules can be used to compute a score for the link between a GCF and a MF. This builds upon early work by Lin and co-workers (Lin, Zhu and Zhang 2006) on clustering homologous BGCs (structurally similar clusters that have a shared ancestry), which was then verified by comparison with known clusters of homologous genes. Later work on clustering BGCs (Cimermancic et al. 2014; Doroghazi et al. 2014; Navarro-Muñoz et al. 2018) has usually involved explicit verification of the GCF, i.e. that the BGCs being grouped together are, in fact, producing related metabolites, by heterologous expression or gene knockout, but follow a similar pattern of defining novel distance functions between BGCs and constructing a clustering based upon these distances. The distances are usually defined at least partly in terms of the composition of BGCs by protein family domains, (Lin, Zhu and Zhang 2006; Navarro-Muñoz et al. 2018) but are often composed of multiple factors which can include everything from direct sequence similarity (Navarro-Muñoz et al. 2018) to the output score of sequence alignment algorithms (Doroghazi et al. 2014).
The clustering of spectra into Molecular Families by molecular networking is incorporated into tools such as GNPS (Wang et al., 2016). The similarity between any two spectra in a data set is computed using a modified cosine similarity score, to account for certain structural modifications; as a result, two spectra are taken to belong to the same MF if their score exceeds a user-defined threshold.
Once the BGCs have been clustered based on the distance measurements, the shared source strains of GCFs and MFs can be used as a starting point to correlate between BGCs and products (Doroghazi et al. 2014; Goering et al. 2016) in an approach known as metabologenomics. A linking score between a GCF and a MF is computed, which is dependent on the degree of strain overlap, penalising strains being present in one side (GCF or MF) and not the other. Since the presence of a BGC in a strain does not guarantee that it will be active in all circumstances (cryptic BGCs), this penalty is often asymmetric, with a low penalty applied for strains that contribute to the GCF but not to the spectra, while strains that contribute to the spectra and not to the GCF are highly penalised. A new chlorinated antiproliferative compound named tambromycin (13) was discovered by applying this approach to a set of 178 actinomycete strains (Goering et al. 2016). Moreover, a new class of natural products and their BGC were discovered when metabologenomics was combined with molecular networking. As a result, six tyrobetaines (14) bearing an unusual N-terminal trimethylammonium were identified and their BGC was confirmed through heterologous expression (Parkinson et al. 2018). This approach was also used by McClure et al. to link the rimosamide family of natural products to their corresponding BGCs (McClure et al. 2016).
Hybrid approaches
Even though we have described two distinct approaches, they are not mutually exclusive. Clustering BGCs can be used in conjunction with previously established links to infer the product from the known link to the other BGCs in the cluster (Nguyen et al. 2013). This can also be done with databases of known BGCs, such as MIBiG, to determine which BGCs are likely to produce already known compounds (Helfrich et al. 2018). Clustering can therefore complement the matching of BGCs and metabolites based on predicted spectral features.
Similarly, the mutual strain information between a cluster of BGCs and a metabolite is not enough on its own to establish a correspondence between the two, especially for novel secondary metabolites. Instead, the strain content has been used to prioritise the potential links for further verification. This verification has, for instance, taken the form of predictions of common structural elements of the products. These have then been searched for in the metabolomic data (Parkinson et al. 2018). For example, the creation of knockout strains was used to verify the linking of macrobrevin (15) to its BGC, or the correspondence of parts of the BGCs with known parts of the pathway for the product from other organisms (Helfrich et al. 2018). An example of the last approach is the discovery of indolmycin (16) (Maansson et al. 2016), where publicly available databases and machine learning were used to create an integrated mining approach for linking gene clusters, biosynthetic pathways and secondary metabolites for 13 closely related strains of Pseudoaltreomonas luteoviolacea. In these strains, close to 10% of the total genes encode for secondary metabolites. This percentage is considerably higher compared to studies conducted on other Pseudoaltreomonas strains (Médigue et al. 2005; Thomas et al. 2008) and is corroborated by the high degree of chemical complexity reported in Maansson et al. 2016 as only 2% of the molecular features were shared between the investigated strains. Indeed, novel analogues of thiomarinols were detected in the molecular network of strains that were characterised as biosynthetically diverse.
VALIDATION OF LINKS
In this section, we focus on approaches to experimentally validate links between a BGC and a secondary metabolite using synthetic biology techniques, such as genetic manipulation of BGCs. One of the most common approaches to validate the link between BGCs and metabolite is heterologous expression, the experimental details of which are outside the scope of this review. The reader is referred to Huo et al. for a detailed description of heterologous expression of bacterial secondary metabolite pathways (Huo et al. 2019).
The advent of new methods such as CRISPR/Cas9-based editing (Tao et al. 2018), λ-red mediated recombination (Gust et al. 2003), overexpression of positive regulators (Bergmann et al. 2010) and promoter engineering (Myronovskyi and Luzhetskyy 2016) have recently been applied to GC-rich actinomycetes. For instance, Gomez-Escribano et al. engineered S. coelicolor M145 strains specifically for the heterologous expression of BGCs to simplify the metabolite profiles and eliminate antimicrobial activity. This was achieved by deleting the actinorhodin, prodiginine, CPK and CDA BGCs and adding point mutations into the rpoB and rpsL genes to increase the production of secondary metabolites. The point mutations in rpoB and rpsL increased the production of chloramphenicol and congocidine by 40- and 30-fold, respectively, therefore validating the BGC-metabolite link (Gomez-Escribano and Bibb 2011). In another example, genome mining was recently used to confirm the presence of a lasso peptide BGC in the genome of a marine Streptomyces sp. SCSIO ZS0098 strain that was known to produce the antimicrobial type I lasso peptide aborycin. In this study, the utility of strain engineering was used to validate this link through the heterologous expression of the candidate aborycin BGC in S. coelicolor M1152 (Shao et al. 2019).
Genetic manipulations can also be applied in combination with genome mining to induce metabolite production. For example, genome mining of a marine Streptomyces strain previously known to produce anthracenes and xiamycin A also revealed an ansamycin BGC. By removing the anthracenes and xiamycin A BGCs, the mutant strain was found to produce two new napthoquinone macrolides, olimyicn A and B (Maansson et al. 2016; Sun et al. 2018). A conserved set of five regulatory genes previously characterised by Sidda et al. were used as a query to both search and identify atypical BGCs in Streptomyces sclerotialus NRRL ISP-5269 (Sidda et al. 2014; Alberti et al. 2019). This approach was then used to identify an atypical scl BGC which was transferred and heterologously expressed in Streptomyces albus, resulting in the production of scleric acid, a secondary metabolite with moderate activity against Mycobacterium tuberculosis and inhibitory activity on the cancer-associated enzyme NNMT (Alberti et al. 2019). An additional example of genome mining combined with heterologous expression and chemical analysis, was a study of the fungal strains Arthrinium sp. NF2194 and Nectria sp. Z14-w, which resulted in the isolation of eight new meroterpenoids, two of which exhibited immunosuppressive bioactivity (Zhang et al. 2018).
CURRENT CHALLENGES
While genome mining and the application of genomic techniques have hugely benefited the genome-led secondary metabolite discovery pipeline, there are still important challenges that need to be addressed in order to maximise the potential that these approaches offer. Arguably, the bottleneck in discovery is our narrow understanding of the total biosynthetic and chemical space of microbial secondary metabolites. While prediction pipelines like SMURF (Khaldi et al. 2010) and antiSMASH (Medema et al. 2011; Blin et al. 2017) greatly facilitate the characterisation of BGCs, our knowledge of secondary metabolites is impeded as a result of up to 90% of BGCs being cryptic or silent (Abdelmohsen et al. 2015; Rutledge and Challis 2015; Baltz 2017; Machado, Tuttle and Jensen 2017). The lack of global transcriptome and translation data therefore makes it difficult to distinguish between BGCs that are transcriptionally silent and those that are actively transcribed but lack a link with their products (Jeong et al. 2016). A recent comparative transcriptomics study focused on four Salinispora strains to assess the effect of gene expression in the production of secondary metabolites. Only 13 out of the 49 BGCs were previously linked to their products, whereas the remaining were considered cryptic gene clusters. However, global transcriptome analyses at exponential and stationary phase revealed that more than half of the BGCs were in fact expressed (Amos et al. 2017). Further knowledge of protein translation will enable greater understanding of transcription levels and metabolite detection.
Another issue commonly encountered in secondary metabolites research is the lack of metabolite detection due to extraction constraints or the analytical technique limitations. For instance, studies have demonstrated the impact of extraction solvent on the detection of metabolites (Floros et al. 2016; Crüsemann et al. 2017). Although advances in instrument sensitivity (mass spectrometry and NMR spectroscopy) could arguably remedy problems of detection (Bouslimani et al. 2014), the use of limited experimental conditions can greatly impact the number and diversity of metabolites detected. These include, for example, culture conditions, media composition, growth stage and extraction solvent (Romano et al. 2018). If not taken into consideration, these variables could undermine the biosynthetic potential of the studied organism, complicating the linking process further.
The high rediscovery rate of molecules is another setback commonly encountered in secondary metabolite research. The efficient prioritisation of strains and extracts using combined comparative genomic and metabolomic approaches has proven to be a useful strategy to avoid this. For example, using an integrated approach of combining metabolomic and genomic techniques, Ong and co-workers identified novel metabolites with anti-quorum sensing activity from five bacterial strains isolated from subtidal marine samples (Ong et al. 2019). Their work is a good example of the application of molecular networking-based dereplication in the discovery of secondary metabolites. Effective dereplication greatly relies on the availability of comprehensive, curated, chemical databases and several commercially available databases to this effect are already in place, including AntiBase (Laatsch 2017), Dictionary of Natural Products (Buckingham 1994) and MarinLit (‘MarinLit’). However, data analysis using this approach is often complex and manual. The recent development of the Global Natural Products Social Molecular Networking (GNPS) platform represents a step-change that facilitates community-driven data curation, enabling open access analysis and sharing of MS/MS spectra (Wang et al. 2016). The expansion of such data sets will greatly facilitate our understanding of chemical space.
FUTURE DIRECTIONS AND CONCLUDING REMARKS
Currently, researchers are biased towards the study of putative BGCs that encode variants of already known compounds or biosynthetic pathways, consequently biasing discovery towards analogues of known natural products. Efforts to overcome this include a trend towards the creation of datasets built upon increasing numbers of strains. Recently, a dataset consisting of genomic and metabolomic data for 363 bacterial strains was published and it is likely that more datasets of increasing size will become available (Navarro-Muñoz et al. 2018). As they do, the performance of automated linking approaches will improve.
Data sets are also likely to increase in terms of the data modalities they cover. Already, large datasets connecting bioactivity with genomics exist. For example, a recently published study linked genomic data for 224 bacterial strains (found on the leaves of Arabidopsis plants) to bioactivity data for the same strains (Helfrich et al. 2018). As high-throughput bioactivity screening becomes standard (Pye et al. 2017), it is likely that data sets combining metabolomics, genomics and bioactivity will become available, in turn, motivating the development of new computational techniques capable of analysing them. Crypticity of BGCs will always be a challenge in this domain. Transcriptomic analysis can indicate activity of BGCs (Amos et al. 2017), and the coupling of transcriptomic data with metabolomics, genomics and bioactivity is now possible. This would be particularly powerful when coupled with data generated using the OSMAC approach.
In conclusion, as data sets increase in strain coverage and modalities, increasingly advanced bioinformatics tools are required for their analysis. We believe that modern computing techniques, such as machine learning and artificial intelligence, have a key role to play in elucidating the links between genomes, transcriptomes, metabolomes and phenotypes. Computational tools to date have largely focused on modular secondary metabolites (e.g. NRPS and RIPPs), reflecting the relatively repetitive nature of their biosynthesis. The creation and continued growth of ground-truth data sets such as MiBIG (Medema et al. 2015) provides the necessary infrastructure for the development of tools based upon recent advances in machine learning, that are able to learn mappings between genomic information and molecular structure (as observed in mass spectrometry data). Current research is biased towards areas of the BGC space for which much is known about biosynthesis. Machine learning tools may be able to uncover patterns that help us illuminate larger, unknown areas of both the biosynthetic and chemical space.
ACKNOWLEDGEMENTS
KRD and SR would like to acknowledge funding from the Carnegie Trust for the Universities of Scotland Collaborative Research Grant and the Biotechnology and Biological Sciences Research Council Transformative Research Technologies Tools and Research Development Fund BB/R022054/1. KRD acknowledges funding received from the Scottish University Life Science Alliance Early Career Researcher Exchange Scheme Fellowship.
Conflicts of Interests. None declared.
REFERENCES
- Abdelmohsen UR, Grkovic T, Balasubramanian S et al.. Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv. 2015;33:798–811. [DOI] [PubMed] [Google Scholar]
- Alberti F, Leng DJ, Wilkening I et al.. Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chem Sci. 2019;10:453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos GCA, Awakawa T, Tuttle RN et al.. Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc Natl Acad Sci. 2017;114:E11121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz RH. Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol. 2017;44:573–88. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Funk AN, Scherlach K et al.. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76:8143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Wolf T, Chevrette MG et al.. AntiSMASH 4.0 - improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A, Sanchez LM, Garg N et al.. Mass spectrometry of natural products: current{,} emerging and future technologies. Nat Prod Rep. 2014;31:718–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham J. Dictionary of Natural Products. Chapman and Hall/CRC, 1994. [Google Scholar]
- Chavali AK, Rhee SY. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief Bioinform. 2018;19:1022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrette MG, Aicheler F, Kohlbacher O et al.. SANDPUMA: Ensemble predictions of nonribosomal peptide chemistry reveal biosynthetic diversity across Actinobacteria. Bioinformatics. 2017;33:3202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P, Medema MH, Claesen J et al.. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database. Curr Protocols Bioinform. 2008, DOI: 10.1002/0471250953.bi0205s23. [DOI] [PubMed] [Google Scholar]
- Crüsemann M, O'Neill EC, Larson CB et al.. Prioritizing natural product diversity in a collection of 146 bacterial strains based on growth and extraction protocols. J Nat Prod. 2017;80:588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Albright JC, Goering AW et al.. Aroadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KR, Crüsemann M, Lechner A et al.. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from salinispora species. Chem Biol. 2015;22:460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floros DJ, Jensen PR, Dorrestein PC et al.. A metabolomics guided exploration of marine natural product chemical space. Metabolomics. 2016;12:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering AW, McClure RA, Doroghazi JR et al.. Metabologenomics: Correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent Sci. 2016;2:299–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4: DOI: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbens J, Zhu H, Girard G et al.. Natural product proteomining, a quantitative proteomics platform, allows rapid discovery of biosynthetic gene clusters for different classes of natural products. Chem Biol. 2014;21:707–18. [DOI] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K et al.. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci. 2003;100:1541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich EJN, Vogel CM, Ueoka R et al.. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat Microbiol. 2018;3:909–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L, Hug JJ, Fu C et al.. Heterologous expression of bacterial natural product biosynthetic pathways. Nat Prod Rep. 2019, DOI: 10.1039/c8np00091c. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Kim JN, Kim MW et al.. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat Commun. 2016;7:11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CW, Skinnider MA, Wyatt MA et al.. An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat Commun. 2015;6, DOI: 10.1038/ncomms9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaweewan I, Komaki H, Hemmi H et al.. Isolation and structure determination of new antibacterial peptide curacomycin based on genome mining. Asian J Org Chem. 2017;6:1838-44, DOI: 10.1002/ajoc.201700433. [DOI] [Google Scholar]
- Kaweewan I, Komaki H, Hemmi H et al.. Isolation and structure determination of a new cytotoxic peptide, curacozole, from Streptomyces curacoi based on genome mining. J Antibiot (Tokyo). 2019;72:1–7. [DOI] [PubMed] [Google Scholar]
- Kersten RD, Yang YL, Xu Y et al.. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G et al.. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laatsch H. AntiBase: The Natural Compound Identifier. Wiley-Vch. 2017. ISBN:978-3-527-33841-2 [Google Scholar]
- Lin K, Zhu L, Zhang DY. An initial strategy for comparing proteins at the domain architecture level. Bioinformatics. 2006;22:2081–6. [DOI] [PubMed] [Google Scholar]
- Maansson M, Vynne NG, Klitgaard A et al.. An integrated metabolomic and genomic mining workflow to uncover the biosynthetic potential of bacteria. mSystems. 2016;1:e00028–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado H, Tuttle RN, Jensen PR. Omics-based natural product discovery and the lexicon of genome mining. Curr Opin Microbiol. 2017;39:136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure RA, Goering AW, Ju K-S et al.. Elucidating the rimosamide-detoxin natural product families and their biosynthesis using metabolite/gene cluster correlations. ACS Chem Biol. 2016;11:3452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P et al.. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucl Acids Res. 2011;39:W339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P et al.. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Paalvast Y, Nguyen DD et al.. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLOS Comput Biol. 2014;10:e1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C, Krin E, Pascal G et al.. Coping with cold: The genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Gurevich A, Mikheenko A et al.. Dereplication of peptidic natural products through database search of mass spectra. Nat Chem Biol. 2017;13:30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Gurevich A, Shlemov A et al.. Dereplication of microbial metabolites through database search of mass spectra. Nat Commun. 2018;9:4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Kersten RD, Liu WT et al.. Automated genome mining of ribosomal peptide natural products. ACS Chem Biol. 2014a;9:1545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Liu WT, Kersten RD et al.. NRPquest: Coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J Nat Prod. 2014b;77:1902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myronovskyi M, Luzhetskyy A. Native and engineered promoters in natural product discovery. Nat Prod Rep. 2016;33:1006–19. [DOI] [PubMed] [Google Scholar]
- Navarro-Muñoz J, Selem-Mojica N, Mullowney M et al.. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv. 2018, DOI: 10.1101/445270. [DOI] [Google Scholar]
- Nguyen DD, Wu C-H, Moree WJ et al.. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci. 2013;110:E2611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong JFM, Goh HC, Lim SC et al.. Integrated genomic and metabolomic approach to the discovery of potential anti-quorum sensing natural products from microbes associated with marine samples from Singapore. Mar Drugs. 2019;17:E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panter F, Krug D, Müller R. Novel methoxymethacrylate natural products uncovered by statistics-based mining of the myxococcus fulvus secondary metabolome. ACS Chem Biol. 2019;14:88–98. [DOI] [PubMed] [Google Scholar]
- Parkinson EI, Tryon JH, Goering AW et al.. Discovery of the tyrobetaine natural products and their biosynthetic gene cluster via metabologenomics. ACS Chem Biol. 2018;13:1029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye CR, Bertin MJ, Lokey RS et al.. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci. 2017;114:5601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano S, Jackson SA, Patry S et al.. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar Drugs. 2018;16:E244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–23. [DOI] [PubMed] [Google Scholar]
- Schneider O, Simic N, Aachmann FL et al.. Genome Mining of Streptomyces sp. YIM 130001 Isolated From Lichen Affords New Thiopeptide Antibiotic. Front Microbiol. 2018;9:3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Ma J, Li Q et al.. Identification of the anti-infective aborycin biosynthetic gene cluster from deep-sea-derived streptomyces sp. SCSIO ZS0098 enables production in a heterologous host. Mar Drugs. 2019;17:E127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidda JD, Song L, Poon V et al.. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem Sci. 2014;5:86–9., DOI: 10.1039/c3sc52536h. [DOI] [Google Scholar]
- Skinnider MA, Dejong CA, Rees PN et al.. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucl Acids Res. 2015;43:9645–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider MA, Merwin NJ, Johnston CW et al.. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucl Acids Res. 2017;45:W49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Jang M, Lee B et al.. Ulleungdin, a Lasso Peptide with Cancer Cell Migration Inhibitory Activity Discovered by the Genome Mining Approach. J Nat Prod. 2018;81:2205–11., DOI: 10.1021/acs.jnatprod.8b00449. [DOI] [PubMed] [Google Scholar]
- Sun C, Zhang C, Qin X et al.. Genome mining of Streptomyces olivaceus SCSIO T05: Discovery of olimycins A and B and assignment of absolute configurations. Tetrahedron. 2018;74:199–203. [Google Scholar]
- Takasaka N, Kaweewan I, Ohnishi-Kameyama M et al.. Isolation of a new antibacterial peptide actinokineosin from Actinokineospora spheciospongiae based on genome mining. Lett Appl Microbiol. 2017;64:150–7. [DOI] [PubMed] [Google Scholar]
- Tao W, Yang A, Deng Z et al.. CRISPR/Cas9-based editing of streptomyces for discovery, characterization, and production of natural products. Front Microbiol. 2018;9:1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Evans FF, Schleheck D et al.. Analysis of the Pseudoalteromonas tunicata Genome Reveals Properties of a Surface-Associated Life Style in the Marine Environment. PLoS One. 2008;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz JI, Schwalen CJ, Patel PS et al.. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol. 2017;13:470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueoka R, Bhushan A, Probst SI et al.. Genome-based identification of a plant-associated marine bacterium as a rich natural product source. Angew Chemie - Int Ed. 2018;57:14519–23. [DOI] [PubMed] [Google Scholar]
- Umemura M, Koike H, Machida M. Motif-independent de novo detection of secondary metabolite gene clusters-toward identification from filamentous fungi. Front Microbiol. 2015;6:371, DOI: 10.3389/fmicb.2015.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura M, Koike H, Nagano N et al.. MIDDAS-M: Motif-independent de novo detection of secondary metabolite gene clusters through the integration of genome sequencing and transcriptome data. PLoS One. 2013;8:e84028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan V V et al.. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Aitken EJ, Baker BP et al.. Genome mining, isolation, chemical synthesis and biological evaluation of a novel lanthipeptide, tikitericin, from the extremophilic microorganism: Thermogemmatispora strain T81. Chem Sci. 2018;9:7311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang TT, Xu QL et al.. Genome Mining and Comparative Biosynthesis of Meroterpenoids from Two Phylogenetically Distinct Fungi. Angew Chemie - Int Ed. 2018;57:8184–8. [DOI] [PubMed] [Google Scholar]