Abstract
Respiratory exposure to either multiwalled carbon nanotubes (MWCNT) or asbestos results in fibrosis; however, the mechanisms to reach this endpoint may be different. A previous study by our group identified pulmonary effects and significantly altered mRNA signaling pathways following exposure to 1, 10, 40, and 80 μg MWCNT and 120 μg crocidolite asbestos on mouse lungs over time at 1-month, 6-months, and 1-year postexposure following pulmonary aspiration. As a continuation to the above study, this current study took an in depth look at the signaling pathways involved in fibrosis development at a single time point, 1 year, and exposure, 40 mg MWCNT, which was the lowest exposure at which fibrosis was pathologically evident. The 120 μg asbestos exposure was included to compare MWCNT-induced fibrosis with asbestos-induced fibrosis. A previously validated computational model was used to identify mRNAs with expression profiles matching the fibrosis pathology patterns from exposed mouse lungs. mRNAs that matched the pathology patterns were then input into Ingenuity Pathway Analysis to determine potential signaling pathways and physiological disease functions inherent to MWCNT and asbestos exposure. Both MWCNT and asbestos exposure induced changes in mouse lungs regarding gene expression, cell proliferation, and survival, while MWCNT uniquely induced alterations in pathways involved in oxidative phosphorylation, mitochondrial dysfunction, and transcription. Asbestos exposure produced unique alterations in pathways involved in sustained inflammation. Although typically considered similar due to scale and fiber-like appearance, the different compositional properties inherent to either MWCNT or asbestos may play a role in their ability to induce fibrosis after pulmonary exposure.
Keywords: asbestos, multiwalled carbon nanotubes, lung fibrosis, toxicogenomics, pathway analysis, non-negative matrix factorizations
Introduction
Nanotechnology is the understanding and control of matter at dimensions between 1 and 100 nanometers that represents a multibillion dollar worldwide industry.1,2 Manipulating matter at this scale increases the potential for engineered nanomaterials and nanoscale processes to store energy, reinforce materials, sense contaminants, enable life-saving drugs, and shrink and accelerate computational devices in incremental and paradigm-shifting ways.2 With the wide application of nanomaterials comes the possibility of nanoparticle exposure to workers and potential wide-spread exposure due to release of nanoparticles from consumer products, particularly during mechanical operations, such as dry and wet cutting of materials, which have been shown to release particles of different sizes.3 While nanotechnology focuses on the creation and advancement of novel nanotechnology products, nanotoxicology must closely follow to assess toxicological risk assessment of nanomaterial design.1,4
Multiwalled carbon nanotubes (MWCNT), multiple concentric rolled up sheets of hexagonally-ordered carbon atoms, are appealing for industrial and medical purposes due to their unique electrical, thermal, and vibrational properties.5,6 The high aspect ratio and surface characteristics of MWCNT contribute to their technological appeal but may also contribute to their bioactivity upon exposure during product synthesis and disposal.7,8
MWCNT exposure induces inflammation as early as 1 day postexposure, and fibrosis has been noted following MWCNT exposure as early as 1 month postexposure and up to 1 year.9,10,11,12,13,14 Gene expression in mouse lungs following MWCNT exposure has also been correlated with human lung cancer risk.15 A previous study by our group (Snyder-Talkington et al., 2016), exposed mice by aspiration to 1, 10, 40, or 80 μg MWCNT or 120 μg crocidolite asbestos and noted increased bronchioalveolar lavage fluid lactate dehydrogenase (LDA) activity and polymorphonuclear (PMN) lymphocyte infiltration at 1-month, 6-months, and 1-year postexposure compared with mice who were exposed to dispersion media (DM) control. Elevated LDA activity and PMN infiltration rose with increasing MWCNT dose and were also significantly higher than control following exposure to asbestos, suggesting that both MWCNT and asbestos induce lung inflammation up to 1-year postexposure.14 PMN infiltration was higher in mouse lungs exposed to 80 μg at 1-year postexposure than in lungs exposed to 120 μg asbestos.14 Fibrosis scores of pathologically evident fibrosis were also elevated with increasing doses of MWCNT up to 1-year postexposure, with the 40 μg exposure of MWCNT being the lowest amount at which fibrosis was significantly pathologically evident.14 Analysis of gene expression in mouse lungs exposed to 1, 10, 40, or 80 μg MWCNT or 120 μg crocidolite asbestos showed that differential gene expression in mouse lungs exposed to MWCNT tended to favor pathways involved in protein ubiquitination, cell proliferation and survival, and immune response, particularly immune responses mediated by T cells.14
The previous study by Snyder-Talkington et al (2016) showed that asbestos-induced fibrosis was associated with high levels of persistent inflammation, while this disease progression did not appear to correlate with MWCNT-induced fibrogenic responses. While Snyder-Talkington et al. (2016) focused on potential mechanisms underlying fibrosis development over a period of 1 year following exposure to either MWCNT or asbestos over multiple doses, this current study was formed as a continuation of data analysis using the same dataset to determine key players involved in biological processes underlying the lung fibrosis induced by either MWCNT or asbestos. This current study took a more comprehensive look at the biological processes induced at a single dose of MWCNT (40 μg), the lowest exposure at which fibrosis was pathologically significantly evident at 1-year postexposure to MWCNT, and compared these with the biological processes induced by asbestos. This study hypothesizes that systematic analysis using a sophisticated computational matching of time-series gene expression profiles and MWCNT- or asbestos-induced fibrosis pathological patterns during 1-year postexposure will identify important genes, as well as similar or different signaling pathways, perturbed by these 2 substances. A previously validated computational model was used to identify genes with expression profiles matching fibrosis pathology patterns from mouse lungs exposed to 40 μg MWCNT or 120 μg crocidolite asbestos 1-year postexposure.16 This approach enables the identification of genes uniquely relevant to either MWCNT- or asbestos-induced lung fibrosis or those matching both MWCNT- and asbestos-induced fibrosis pathological patterns. Significantly up- or downregulated genes involved in similar or different pathways perturbed by MWCNT or asbestos were pinpointed in this study. These identified genes were then analyzed with Ingenuity Pathway Analysis (IPA) to determine differential and similar canonical pathways associated with MWCNT- or asbestos-induced lung fibrosis.
Materials and Methods
MWCNT and Crocidolite Asbestos
The MWCNT and asbestos used in this study have been previous characterized. For full characterization, please see Snyder-Talkington et al. 2016.14
Animals
All animals were housed in an AAALAC International-accredited, specific-pathogen-free, and environmentally controlled facility. All animal studies and procedures were approved by the National Institute for Occupational Safety and Health Animal Care and Use Committee. For full characterization of animal husbandry, please see Snyder-Talkington et al. 2016.14
MWCNT and Crocidolite Asbestos Exposure and Lung Collection
All methods for MWCNT and asbestos pulmonary exposure and RNA collection from exposed lungs have been previously described.14
Pathology Methods
Lungs were excised from mice exposed to DM; 1, 10, 40, or 80 μg MWCNT; or asbestos at 1-year postexposure, fixed, and analyzed as previously described.14 This current study only concerned lungs from mice exposed to 40 μg MWCNT.
mRNA Microarray Processing
Global mRNA expression profiles were generated with Mouse Gene ST 2.1 plates at the University of Michigan Microarray Facility using an Affymetrix Plus kit. cDNA was synthesized from 500 ng total RNA, amplified, fragmented, and biotinylated using a GeneChip WT PLUS Reagent kit (Affymetrix; Santa Clara, CA) according to manufacturer’s instructions. cDNA was then prepared for hybridization with reagents from the Affymetrix GeneTitan Hybridization, Wash, and Stain Kit for WT Array Plates. For hybridization, 2.76 μg labeled cDNA were hybridized to the Affymetrix Mouse Gene ST 2.1 Arrays, which were then washed, stained, and scanned using the GeneTitan Multi-Channel Instrument according to Affymetrix’s User Guide for Expression Array Plates (P/N 702933 Rev. 2, 2013). Data were analyzed using the Limma, Oligo, and Affy Bioconductor packages implemented in the R statistical environment. The robust multi-array average was used to normalize the data and fit log2 transformed expression values. The raw microarray data were processed using a robust multi-array average method (Irizarry, et al. 2003), and expression values were log2 transformed, with a principal component analysis utilized as the final quality control step to visualize mRNA expression values. The analysis was performed with the oligo package of Bioconductor in the R statistical environment. The microarray data is publically available at NCBI Gene Expression Omnibus with the accession number GSE112780 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112780).
mRNA Microarray Data Analysis
The mRNA microarray consisted of 41,345 probes. Variance was calculated for each probe across all samples to remove any probe with variance >0.2. All probes resulted in variance <0.2; therefore, no probes were removed due to variance. Unknowns were then removed from the data, leaving 25,616 probes. Data analysis was performed with 26,191 probes. ANOVA was used to compare each treatment group at each postexposure time point to its respective DM control to determine significantly up- and downregulated mRNAs with a p-value <0.05, false discovery rate of 10%, and fold change (FC) ≥1.5 (Supplemental File 1).
Identifying genes associated with MWCNT and Crocidolite Asbestos-induced fibrosis pathological patterns
The MEGPath system was employed to identify mRNAs with expression transcriptionally concordant with fibrotic pathologies in mice exposed to crocidolite asbestos or 40 μg MWCNT by aspiration at 1-month, 6-month, or 1-year postexposure as previously described.14,16,17 Fibrotic pathology was defined by the analysis of trichrome and Sirius Red staining of collagen in mouse lungs at 1 month, 6 months, and 1 year, MWCNT dose 40 μg and crocidolite asbestos as previously reported.14 Using constraint-based non-negative matrix factorization, underlying biological patterns were identified, where fibrotic pathology was used as the constraint.18 Identified patterns were then related back to gene expression profiles producing genome-wide coefficients that related each gene to varying pathological patterns. Sets of genes with expressions matching the fibrotic pathological patterns were identified from the annotated C2 and C5 datasets of the curated Molecular Signatures Database (MSigDB).19 Genes may be contained in multiple biological processes and pathways. The gene sets were functionally related by annotations and belonged to the same biological process, but were not required to be coexpressed.16 This method was used to identify genes with expression concordant with time-series fibrosis pathological patterns induced by MWCNT dose 40 μg and crocidolite asbestos, genes concordant uniquely with MWCNT dose 40 μg, and genes concordant uniquely with crocidolite asbestos.
IPA analysis to identify pathways, diseases and physiological functions
IPA (QIAGEN, Redwood City, CA, www.qiagen.com/ingenuity) was used to determine mRNAs associated with canonical pathways and diseases and functions. All associations are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Knowledge Base.
Results
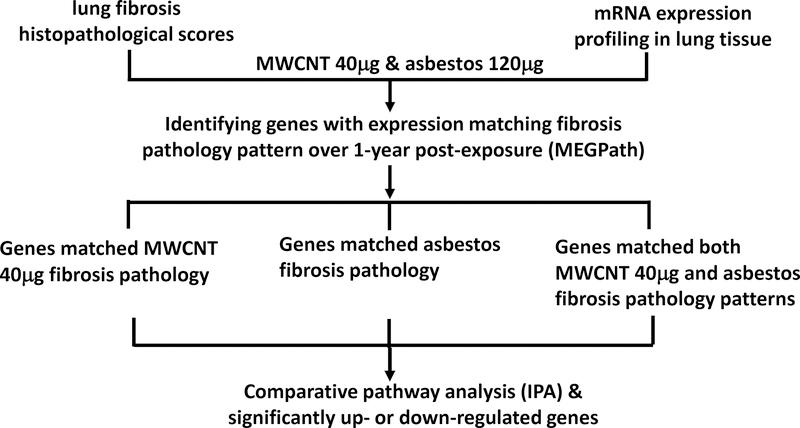
In this study, similar and different pathways and biological processes underlying lung fibrosis induced by MWCNT or asbestos were identified using mRNA data and histopathological quantification of lung fibrosis from mice exposed to 40 μg MWCNT or 120 μg crocidolite asbestos (Figure 1). Asbestos and MWCNT are often thought to similarly result in inflammation and fibrosis following exposure due to their similar fibrous nature; however, MWCNT and asbestos have differing lengths and compositions that may contribute to different signaling pathways leading to these outcomes following exposure (Figure 2). The 40 μg MWCNT treatment was selected as this is an occupationally-relevant exposure to MWCNT and is the lowest quantity at which pathologically evident fibrosis was induced.14 A novel computational model (MEGPath) was used to identify mRNAs that either matched the MWCNT 40 μg fibrosis pathology pattern, asbestos pathology pattern, or both the MWCNT 40 μg and asbestos pathology patterns over the 1-year postexposure time course. These identified genes were then input into IPA (Fall Release 2016) to determine potential signaling pathways and physiological disease functions inherent to MWCNT and asbestos. A total of 2,462 mRNAs matched the MWCNT 40 μg pathology pattern, 3,669 mRNAs matched the asbestos pathology pattern, and 3,551 mRNAs matched both 40 μg and asbestos patterns (Supplemental File 1). Among the identified genes involved in similar or different pathways underlying MWCNT and asbestos-induced lung fibrosis, those significantly up- or downregulated genes are summarized in Table 1.
Figure 1.
Study overview.
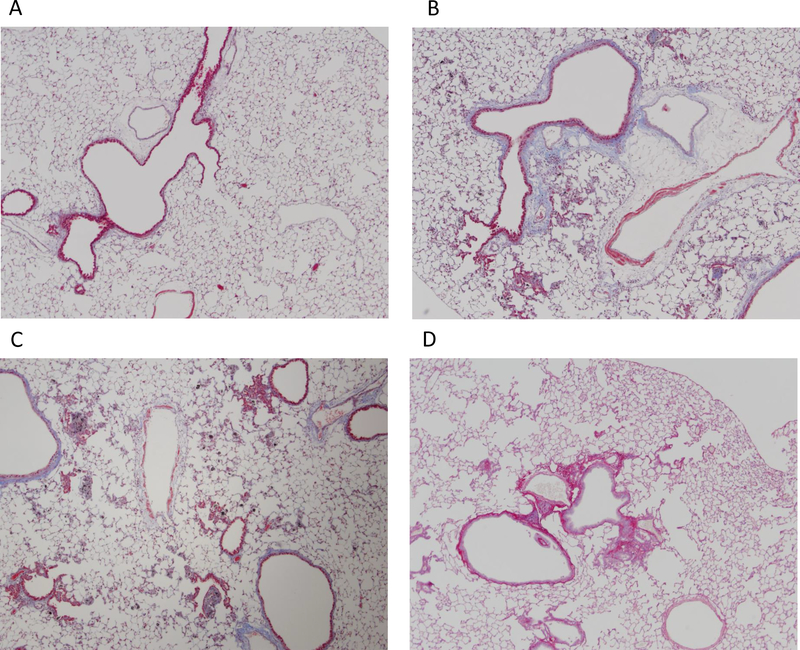
Figure 2.
Left lung trichrome and Sirius red pathology staining at 1 year. Lung sections were evaluated by visualization of collagen at 4× magnification. (A) DM (negative control) trichrome staining; (B) mild fibrosis with focal emphysema, 40 μg MWCNT, trichrome staining; (C) mild fibrosis with focal emphysema, 80 μg MWCNT, trichrome staining; (D) mild fibrosis, 120 μg asbestos, Sirius red staining.
Table 1:
Significantly up- and downregulated genes at 1-month, 6-month, and 1-year postexposure to MWCNT 40 μg and asbestos. Genes that were uniquely associated with pathology of MWCNT 40 μg, but not asbestos, are listed in the column “MWCNT 40 μg”; genes that were uniquely associated with pathology of asbestos, but not MWCNT 40 μg, are listed in column asbestos; genes that are associated with both MWCNT 40 μg and asbestos pathology are listed in column “MWCNT 40 μg AND asbestos”.
| MWCNT 40 μg | Asbestos | MWCNT 40 μg AND asbestos | ||||
|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | |
| 1 month | CHEK2 | BMPR2 | BMPR2 | NFKB1, CHEK2, CYCS, TAF2, ICAM1, MYD88 | ||
| 6 months | COX7B | RPL7 | CCL3, JUN, KRAS, MAP2K1, NFKBIA, PIK3CD, PIK3RA, PIK3R5, PIK3R4, APAF1, LRP1, MAP3K2, SIRPA | APAF1, LRP1, PIK3R5, PRKCH, NFKBIA, PIK3CD, KRAS, ARHGEF1, PIK3R4, MAP2K1, CCL3, FPR2, PTGER3 | TGFA | |
| 1-year | EIF3F, RPL5, RPL29, RPL36AL, RPS8, RPS13, RPS16, RPS19, PARK7, SDHB | CCL5, IL1B, CYBA, CD79B | PTGER3 | HDAC10, TRAF5, ARAF, CCL5, CD3G, CYCS, RHOH | ||
Identifying pathways associated with the MWCNT 40 μg or asbestos pathology patterns
The 2,462 genes that were concordant with MWCNT 40 μg pathology pattern and the 3,669 genes that were concordant with asbestos were input into IPA. A Core Analysis was run on each set of mRNAs, followed by a Comparison Analysis to determine signaling pathways in which both the MWCNT 40 μg and asbestos genes were involved. A review of the Comparison Analysis was completed to determine the top 5 canonical pathways in which more MWCNT 40 μg mRNAs were involved than asbestos mRNAs and the top 5 canonical pathways in which more asbestos mRNAs were involved than MWCNT 40 μg mRNAs. Genes involved in each of the top 5 canonical pathways were then analyzed to determine those genes that were both involved in the identified pathways and significantly up- or downregulated.
MWCNT 40 μg
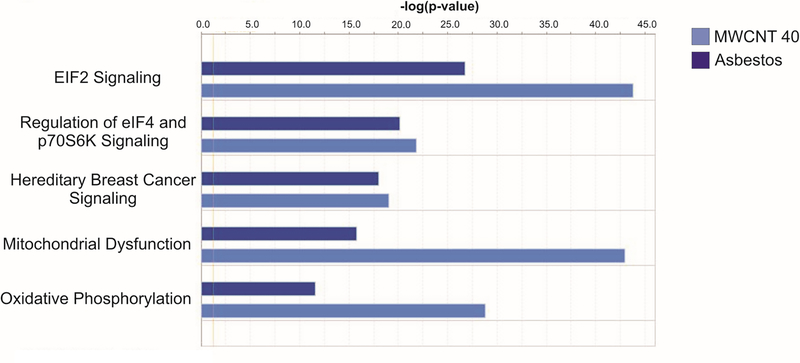
The top 5 pathways associated with mRNA expression that was concordant with the MWCNT 40 μg pathology pattern versus the asbestos pathology pattern were EIF2 signaling, regulation of eIF4 and p70S6K signaling, hereditary breast cancer signaling, mitochondrial dysfunction, and oxidative phosphorylation (Figure 3). Of the genes involved in these pathways, CHECK2 was significantly downregulated at 1-month postexposure; COX7B was significantly upregulated and RPL7 was significantly downregulated at 6-months postexposure. EIF3F, RPL5, RPL29, RPL36AL, RPS8, RPS13, RPS16, RPS19, PARK7, and SDHB were significantly downregulated at 1-year postexposure (Table 1).
Figure 3.
Top 5 IPA pathways associated with mRNA expression concordant with the MWCNT 40 μg pathology pattern versus the asbestos pathology pattern.
Asbestos
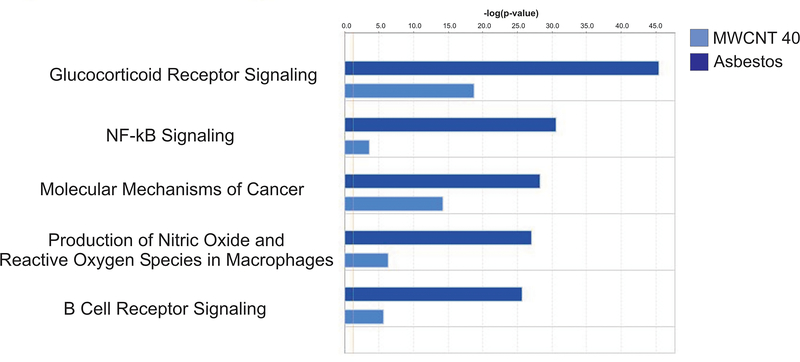
The top 5 pathways associated with mRNA expression that was concordant with the asbestos pathology pattern versus the MWCNT 40 μg pathology pattern were glucocorticoid receptor signaling, NF-κB signaling, Molecular Mechanisms of Cancer, production of nitric oxide and reactive oxygen species in macrophages, and B cell receptor signaling (Figure 4). Of the genes involved in these pathways, BMPR2 was significantly upregulated at 1-month postexposure; CCL3, JUN, KRAS, MAP2K1, NFKBIA, PIK3CD, PIK3RA, PIK3R5, PIK3R4, APAF1, LRP1, MAP3K2, SIRPA were significantly upregulated at 6-months postexposure; and CCL5, IL1B, CYBA, and CD79B were significantly downregulated at 1-year postexposure (Table 1).
Figure 4.
Top 5 IPA pathways associated with mRNA expression concordant with the asbestos pathology pattern versus the MWCNT 40 μg pathology pattern.
Identifying pathways associated with both the MWCNT 40 μg and asbestos patterns
The 3,551 genes that were concordant with MWCNT 40 μg pathology pattern and asbestos pathology were inputted into IPA. A Core Analysis was run on the set of mRNAs to determine the top 5 canonical pathways associated with both MWCNT 40 μg mRNA pathology pattern expression and asbestos mRNA pathology pattern expression.
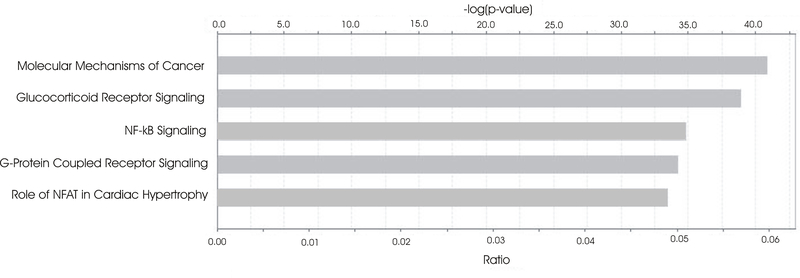
The top 5 canonical pathways associated with mRNA expression that were concordant with both the MWCNT 40 mg and asbestos pathology patterns were Molecular Mechanisms of Cancer, Glucocorticoid Receptor Signaling, NF-κB Signaling, G-Protein Coupled Receptor Signaling, and Role of NFAT in Cardiac Hypertrophy (Figure 5). Of the genes involved in these pathways, BMPR2 was significantly upregulated and NFKB1, CHEK2, CYCS, TAF2, ICAM1, and MYD88 were significant downregulated at 1-month postexposure; APAF1, LRP1, PIK3R5, PRKCH, NFKBIA, PIK3CD, KRAS, ARHGEF1, PIK3R4, MAP2K1, CCL3, FPR2, and PTGER3 were upregulated and TGFA was downregulated at 6-months postexposure; and PTGER3 was upregulated and HDAC10, TRAF5, ARAF, CCL5, CD3G, CYCS, and RHOH were downregulated at 1-year postexposure (Table 1).
Figure 5.
Top 5 canonical pathways associated with mRNA expression concordant with both the MWCNT 40 μg and asbestos pathology patterns.
Discussion
Inflammation and fibrosis are hallmarks of both MWCNT and asbestos lung exposure, potentially due to their high aspect ratio and biopersistence.11,20,21 While the endpoint of exposure to both MWCNT and asbestos may be similar, the mechanisms to reach this endpoint may be different. Previously, Snyder-Talkington et al (2016) showed that asbestos-induced fibrosis was associated with high levels of persistent inflammation, while this disease manifestation did not appear to correlate with MWCNT-induced fibrogenic responses. Increasing doses of MWCNT-induced genomic profiles of fibrosis were associated with numerous diseases, including lung cancer and canonical pathways involved in the immune response.15 Lung carcinogenicity of MWCNT exposure was later confirmed in animal studies.22,23 In this study, a systematic approach was used to determine similar and differing pathways and biological processes associated with lung fibrosis induced by MWCNT and asbestos at a single MWCNT exposure at which lung fibrosis was first significantly pathologically evident.
This study utilized a sophisticated computational model (MEGPath) to discover genes whose transcriptional activities uniquely matched lung fibrosis pathology patterns of either MWCNT or asbestos over the 1-year postexposure time course with three time points: 1 month, 6 months, and 1 year. These genes revealed different biological processes and pathways underlying exposure to these two substances. Genes whose expression profiles matched both MWCNT and asbestos pathology patterns were also identified; the canonical pathways represented by these genes might be relevant to similar disease mechanisms underlying MWCNT- and asbestos-mediated lung fibrosis. The novel computational analysis of matching time-course gene expression profiles with pathology patterns using non-negative matrix factorization identified genes functionally involved in similar and differing processes of fibrotic pathogenesis induced by MWCNT and asbestos, offering important insight into the toxicity assessment of these two substances.
The major advantages of the time-series method compared with the previous single time-point analysis are: 1) pathological patterns of lung fibrosis over the 1-year post-exposure time course were used as constraints to identify genes that had transcriptional activities similar to the pathological patterns. Such computational ability is not possessed by the earlier single time-point analysis. 2) The current time-series analysis could also identify genes that are more similar to MWCNT-induced fibrotic pathological patterns than asbestos- induced ones. The identified genes are considered to be more relevant to MWCNT-induced lung fibrosis pathogenesis. Similarly, genes more relevant to asbestos-induced lung fibrosis could also be identified by using the time-series method. When both MWCNT and asbestos-induced fibrotic pathological patterns are used as constraints in the time series method, genes that had transcriptional profiles matching both pathology patterns were identified and considered to be relevant to both MWCNT and asbestos-induced fibrotic pathogenesis. Only the time-series method has the capacity to identify unique and similar genes and biological pathways perturbed by MWCNT and asbestos exposures, whereas the earlier single time-point analysis does not have all these features. Many of our identified genes were not previously reported as relevant to lung fibrosis induced by MWCNT or asbestos in the current Ingenuity Knowledge Base. These findings provide evidence that the transcriptional activities of the genes identified in this study were associated with MWCNT- or asbestos-induced fibrosis in mouse lungs.
It is noteworthy that BMPR2 appears in both the asbestos-only analysis and the MWCNT 40 μg and asbestos analysis at 1 month, similarly for CCL5 at 1 year (Table 1). The MWCNT 40 μg and asbestos analysis was run with a single constraint that found gene sets which were significantly associated with the single combined fibrosis pattern in both MWCNT 40 μg exposed mice and the asbestos treated mice. A gene could have associated so strongly with asbestos and almost significantly associated with MWCNT that it would be associated with asbestos and MWCNT when looking at the constraint which included both pathological patterns. The other two columns in Table 1 were found from a separate analysis which ran with two constraints, asbestos and MWCNT 40 μg, respectively. This was to find genes that were very strongly related to one of the constraints. Details of the computational analysis of matching time-course genomic profiles with pathology patterns were provided in our previous studies of time-series of MWCNT exposure at 1 day, 28 days, and 56 days postexposure.16
The top five signaling pathways uniquely associated with the MWCNT 40 μg pathology pattern were most often involved in protein synthesis, DNA replication and repair, cell cycle and survival, and metabolic diseases. Significantly up- and downregulated genes within these pathways suggest a role for increased oxidative phosphorylation and mitochondrial dysfunction and decreased transcriptional ability. The tumor suppressor CHEK2 was significantly downregulated at 1-month postexposure to MWCNT 40 μg, suggesting potential for increased growth and proliferation following MWCNT exposure. Several ribosomal proteins (RPL7, RPL5, RPL29, RPL36AL, RPS8, RPS13, RPS16, and RPS19) were found to be downregulated at 1-year postexposure. These proteins belong to the small and large subunits of the larger ribosomal subunits, suggesting that protein translation may be affected following MWCNT exposure, an effect that has overarching effects on cell growth, proliferation, differentiation, and development.24 Disturbances in ribosomal biogenesis can result in ribosomal stress and the release of ribosome-free forms of ribosomal proteins, which can have numerous extraribosomal functions, such as activation of tumor suppressor p53 or oncogenic processes.24 In particular, RPL5 has been shown to activate tumor suppressor p53, and this activation may be lost by the loss of RPL5 following MWCNT exposure.24 The abundance of alterations in ribosomal proteins following MWCNT exposure that is not evident following asbestos exposure may suggest that MWCNT have greater effects on ribosomal stress. An interesting finding that correlated with our previous study was downregulation of oxidative stress sensor PARK7. PARK7 downregulation may play a role in loss of oxidative stress protection and also a role for mitochondrial dysfunction following MWCNT exposure.14
The top five signaling pathways uniquely associated with the asbestos pathology pattern were more often involved in cellular development, cellular growth and proliferation, gene expression, and free radical scavenging. Significantly up- and downregulated genes within these pathways suggest increased sustained inflammation over time after asbestos exposure with enhanced cellular growth. Asbestos exposure has been linked to diseases such as pulmonary fibrosis (asbestosis), pleural effusion and plaques, and malignancies, such as bronchogenic carcinoma and mesothelioma.25,26 Inflammation and reactive oxygen species-induced DNA damage resulting in apoptosis are well-known effects attributed to asbestos exposure.25 Genes involved in inflammation (CCL3, CCL5, NFKBIA, and IL1B) and apoptosis (APAF1 and LRP1) were significantly up- and downregulated in our study. Further, asbestos exposure has been consistently linked to development of asbestos-associated lung cancer.25 Numerous genes involved in growth (PIK3RA, PIK3R5, PIK3R4, SIRPA), proliferation (MAP2K1), and malignancy (JUN, KRAS) were up- and downregulated in our study of genes significantly associated with asbestos pathology patterns.
The top five signaling pathways associated with both MWCNT 40 μg and asbestos pathology patterns were most often involved in cellular development and survival, cellular growth and proliferation, gene expression, and posttranslational modification. Significantly up- and downregulated genes in pathways associated with both MWCNT and asbestos suggest increases in growth and proliferation over time with changes in mitochondrial function and transcription. Both MWCNT and asbestos exposure were found to result in lung fibrosis over time; however, previous research suggested that this endpoint is reached through differing mechanisms.14 In this study, several significantly up- and downregulated genes were identified by IPA to be involved in the overall process of fibrosis. Following asbestos exposure, the upregulated genes BMPR2, JUN, KRAS, and NFKIA were found to be involved in the fibrosis process. In signaling pathways that were representative of both MWCNT and asbestos pathology, the upregulated genes NFKBIA and KRAS and the downregulated genes NFKB1, ICAM1, MYD88, and TGFA were shown to be involved in the fibrosis process. No specific fibrosis-related genes were found to be up- or downregulated in the top signaling pathways representative of only MWCNT exposure. These findings may further validate varying fibrotic patterns of MWCNT versus asbestos exposure.
The computational analysis used in this study discovered genes with time-series expression profiles matching pathological patterns induced by MWNCT and/or asbestos over the course of three time points: 1-month, 6-months, and 1-year postexposure. The identified significantly up- and downregulated genes associated with MWCNT- and asbestos-induced pathology patterns were significantly represented in different signaling pathways. Although MWCNT and asbestos may result in similar outcomes, the pathways to these endpoints may be different due to different sizes and compositions of the different particles. This study, a follow-up to the previous study by Snyder-Talkington et al. (2016), identified significant similar and differential signaling pathways associated with MWCNT and asbestos exposure at increasing doses over time. This study presented results on one MWCNT exposure, 40 μg, over time to identify signaling pathways associated with the lowest exposure to MWCNT at which significant pathologically evident fibrosis was identified (results on the comparison of MWCNT 80 μg and asbestos are included in Supplementary File 2). In particular, an alteration in ribosomal proteins was identified following MWCNT exposure that was not identified when asbestos exposure was analyzed. This finding suggests that MWCNT may induce ribosomal stress, in addition to mitochondrial stress identified previously, which can have long-term detrimental effects on the lung.14 In contrast, numerous genes involved in inflammation, growth, proliferation, apoptosis, and malignancy were perturbed and uniquely associated with asbestos pathology patterns. It is noteworthy that among the genes matching both MWCNT- and asbestos-induced fibrotic pathology patterns, NFKBIA and NFKB1 had significant differential expression in mouse lungs, which is consistent with the previous prediction of NF-κB regulation of MWCNT-induced gene expression changes in qRT-PCR assays of a panel of lung cancer signaling hallmarks and biomarkers in human small airway epithelial cells (SAEC).27 Moreover, our previous systematic analysis of MWCNT-induced cellular signaling and gene expression in SAEC showed that MWCNT exposure induced a concordant increase of both mRNAs and proteins of CCL2 and VEGFA, two prognostic biomarkers of lung inflammation, fibrosis, and carcinogenesis.27 Although adenocarcinoma formation was not identified in this current study, there were several changes in genes involved in oncogenic processes in this study which may set the stage for tumor development in the long term. Gene expression signatures in mouse lungs following MWCNT exposure were previously identified to predict human lung cancer risk.15 MWCNT was later confirmed as a promoter of lung adenocarcinoma formation as well as a tumor initiator.22,23,28
Both MWCNT and asbestos exposure produces changes in mouse lungs regarding gene expression, cell proliferation, and survival. MWCNT uniquely produce alterations in pathways involved in oxidative phosphorylation, mitochondrial dysfunction, and transcription, while asbestos exposure produces unique alterations in pathways involved in sustained inflammation. These results suggest that asbestos exposure may lead to fibrosis through a continuous inflammatory process, while MWCNT exposure leads to fibrosis through different means. Although similar in scale, the differential compositional properties of MWCNT versus asbestos may play into their fibrotic potential. Limitations of this study include the use of whole lungs for genomic analysis in place of subpopulations of specific cell types. The validation of the signaling pathways and overall cellular changes identified in this study are an ongoing endeavor.
Supplementary Material
Supplementary File 1: Significantly up- and downregulated mRNAs concordant with the MWCNT 40 μg pathology pattern, asbestos pathology pattern, or both MWCNT 40 μg and asbestos pathology patterns.
Supplementary File 2: Genes matching the MWCNT 80 μg pathology pattern, asbestos pathology pattern, or both MWCNT 80 μg and asbestos pathology patterns in the MSigDB C2 and C5 databases.
Acknowledgments
Funding
Lan Guo is supported by an R01 from NIEHS (ES021764; Guo). The WV-INBRE (IPA license) is supported by NIH Grant P20GM103434.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
References
- 1.Bakand S, Hayes A. Toxicological Considerations, Toxicity Assessment, and Risk Management of Inhaled Nanoparticles. Int J Mol Sci. 2016;17(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NNI. National Nanotechnology Initiative Strategic Plan. 2014.
- 3.Wang J, Schlagenhauf L, Setyan A . Transformation of the released asbestos, carbon fibers and carbon nanotubes from composite materials and the changes of their potential health impacts. J Nanobiotechnology. 2017;15(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kermanizadeh A, Gosens I, MacCalman L, et al. A Multilaboratory Toxicological Assessment of a Panel of 10 Engineered Nanomaterials to Human Health--ENPRA Project--The Highlights, Limitations, and Current and Future Challenges. J Toxicol Environ Health B Crit Rev. 2016;19(1):1–28. [DOI] [PubMed] [Google Scholar]
- 5.Iijima S Helical microtubules of graphitic carbon. Nature. 1991;354:56–58. [Google Scholar]
- 6.Serpell CJ, Kostarelos K, Davis BG. Can Carbon Nanotubes Deliver on Their Promise in Biology? Harnessing Unique Properties for Unparalleled Applications. ACS Cent Sci. 2016;2(4):190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NIOSH. Occupational Exposure to Carbon Nanotubes and Nanofibers. Current Intelligence Bulletin 652013. [Google Scholar]
- 8.Oberdorster G, Castranova V, Asgharian B, Sayre P. Inhalation Exposure to Carbon Nanotubes (CNT) and Carbon Nanofibers (CNF): Methodology and Dosimetry. J Toxicol Environ Health B Crit Rev. 2015;18(3–4):121–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer RR, Hubbs AF, Scabilloni JF, et al. Pulmonary fibrotic response to aspiration of multi-walled carbon nanotubes. Part Fibre Toxicol. 2011;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer RR, Hubbs AF, Scabilloni JF, et al. Distribution and persistence of pleural penetrations by multi-walled carbon nanotubes. Part Fibre Toxicol. 2010;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer RR, Scabilloni JF, Hubbs AF, et al. Distribution and fibrotic response following inhalation exposure to multi-walled carbon nanotubes. Part Fibre Toxicol. 2013;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer RR, Scabilloni JF, Hubbs AF, et al. Extrapulmonary transport of MWCNT following inhalation exposure. Part Fibre Toxicol. 2013;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter DW, Hubbs AF, Mercer RR, et al. Mouse pulmonary dose- and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology. 2010;269(2–3):136–147. [DOI] [PubMed] [Google Scholar]
- 14.Snyder-Talkington BN, Dong C, Porter DW, et al. Multiwalled carbon nanotube-induced pulmonary inflammatory and fibrotic responses and genomic changes following aspiration exposure in mice: A 1-year postexposure study. J Toxicol Environ Health A. 2016;79(8):352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo NL, Wan YW, Denvir J, et al. Multiwalled carbon nanotube-induced gene signatures in the mouse lung: potential predictive value for human lung cancer risk and prognosis. J Toxicol Environ Health A. 2012;75(18):1129–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder-Talkington BN, Dymacek J, Porter DW, et al. System-based identification of toxicity pathways associated with multi-walled carbon nanotube-induced pathological responses. Toxicol Appl Pharmacol. 2013;272(2):476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dymacek J, Guo NL. Systems Approach to Identifying Relevant Pathways from Phenotype Information in Dose-Dependent Time Series Microarray Data. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2011;2011:290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401(6755):788–791. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cyphert JM, McGee MA, Nyska A, Schladweiler MC, Kodavanti UP, Gavett SH. Long-term toxicity of naturally occurring asbestos in male Fischer 344 rats. J Toxicol Environ Health A. 2016;79(2):49–60. [DOI] [PubMed] [Google Scholar]
- 21.Donaldson K, Murphy FA, Duffin R, Poland CA. Asbestos, carbon nanotubes and the pleural mesothelium: a review of the hypothesis regarding the role of long fibre retention in the parietal pleura, inflammation and mesothelioma. Part Fibre Toxicol. 2010;7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukushima S, Kasai T, Umeda Y, Ohnishi M, Sasaki T, Matsumoto M. Carcinogenicity of multi-walled carbon nanotubes: challenging issue on hazard assessment. J Occup Health. 2018;60(1):10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasai T, Umeda Y, Ohnishi M, et al. Lung carcinogenicity of inhaled multi-walled carbon nanotube in rats. Part Fibre Toxicol. 2016;13(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7(2):92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, Cheresh P, Kamp DW. Molecular basis of asbestos-induced lung disease. Annu Rev Pathol. 2013;8:161–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mossman BT, Lippmann M, Hesterberg TW, Kelsey KT, Barchowsky A, Bonner JC. Pulmonary endpoints (lung carcinomas and asbestosis) following inhalation exposure to asbestos. J Toxicol Environ Health B Crit Rev. 2011;14(1–4):76–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder-Talkington BN, Pacurari M, Dong C, et al. Systematic analysis of multiwalled carbon nanotube-induced cellular signaling and gene expression in human small airway epithelial cells. Toxicol Sci. 2013;133(1):79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sargent LM, Porter DW, Staska LM, et al. Promotion of lung adenocarcinoma following inhalation exposure to multi-walled carbon nanotubes. Part Fibre Toxicol. 2014;11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Significantly up- and downregulated mRNAs concordant with the MWCNT 40 μg pathology pattern, asbestos pathology pattern, or both MWCNT 40 μg and asbestos pathology patterns.
Supplementary File 2: Genes matching the MWCNT 80 μg pathology pattern, asbestos pathology pattern, or both MWCNT 80 μg and asbestos pathology patterns in the MSigDB C2 and C5 databases.