Abstract
Background
Lower-grade gliomas (LGGs, defined as WHO grades II and III) with 1p19q codeletion have increased chemosensitivity when compared to LGGs without 1p19q codeletion, but the mechanism is currently unknown.
Methods
RNAseq data from 515 LGG patients in the Cancer Genome Atlas (TCGA) were analyzed to compare the effect of expression of the 9 DNA repair genes located on chromosome arms 1p and 19q on progression free survival (PFS) and overall survival (OS) between patients who received chemotherapy and those who did not. Chemosensitivity of cells with DNA repair genes knocked down was tested using MTS cell proliferation assay in HS683 cell line and U251 cell line.
Results
The expression of 9 DNA repair genes on 1p and 19q was significantly lower in 1p19q-codeleted tumors (n = 175) than in tumors without the codeletion (n = 337) (p < 0.001). In LGG patients who received chemotherapy, lower expression of LIG1, POLD1, PNKP, RAD54L and MUTYH was associated with longer PFS and OS. This difference between chemotherapy and non-chemotherapy groups in the association of gene expression with survival was not observed in non-DNA repair genes located on chromosome arms 1p and 19q. MTS assays showed that knockdown of DNA repair genes LIG1, POLD1, PNKP, RAD54L and MUTYH significantly inhibited recovery in response to temozolomide when compared with control group (p < 0.001).
Conclusions
Our results suggest that reduced expression of DNA repair genes on chromosome arms 1p and 19q may account for the increased chemosensitivity of LGGs with 1p19q codeletion.
Keywords: Chemosensitivity, DNA repair genes, Lower-grade gliomas, 1p19q codeletion
Introduction
Gliomas are central nervous system tumors arising from glial cells that comprise approximately 30% of all primary brain and CNS tumors [1]. The lower-grade gliomas (LGG) include diffuse low-grade and intermediate-grade gliomas (World Health Organization grades II and III) [2]. The prognosis of LGGs varies, and their infiltrative nature makes recurrence common [3]. Treatment for these tumors involves resection which may be followed by adjuvant radiotherapy and chemotherapy [3]. The results of the Radiation Therapy Oncology Group (RTOG) 9802 trial suggested that progression-free survival (PFS), but not overall survival (OS), was improved for patients with LGG who received radiotherapy (RT) and chemotherapy compared to those who received RT alone [4]. Recently, a study by Buckner et al. found that in certain populations with grade II gliomas, adjuvant chemotherapy in addition to RT improves both PFS and OS compared to RT alone [5].
A study including 615 grade II and III gliomas from the Cancer Genome Atlas (TCGA) suggested that IDH mutation and 1p19q codeletion status can better predict prognosis than histological grading [6]. Furthermore, there is evidence to suggest that 1p19q codeletion is associated with increased chemosensitivity [7]. In a randomized controlled trial comparing RT alone to RT followed by chemotherapy for the treatment of anaplastic oligodendroglioma, adjuvant chemotherapy was more beneficial for tumors with 1p19q-codeletion [8]. The mechanism conferring this increased susceptibility to chemotherapy is poorly understood, but it has been suggested that DNA repair genes on chromosome 1p and 19q may play a role [9]. In this study, we investigated relationship between the expression of 9 DNA repair gene located on chromosome arms 1p and 19q and chemosensitivity/survival outcomes using data from TCGA and conducted experiments in vitro to validate our results.
Materials and methods
Statistical analyses
Wood et al. published one hundred and fifty DNA repair genes on human chromosomes [10]. Nine of the 150 genes were located on chromosome arms 1p and 19q (ERCC1, LIG1, ERCC2, POLD1, RUVBL2, PNKP, RAD54L, MUTYH and MAD2L2). In this study, the expression level of these nine DNA repair genes was extracted from the RNAseq data of 515 TCGA LGG patients along with age, KPS, WHO grade, histological type, extent of resection, IDH mutation status, TERT mutation status, TP53, H3F3A, 1p19q codeletion status, receipt of RT and CT, PFS, and OS. To demonstrate specificity, the expression level of five DNA repair genes (CHAF1A, CLK2, EXO1, PARP1 and XAB2) located on chromosome arms 1q and 19p was also extracted.
1p19q codeleted status was assigned by using Gistic2 results by chromosome arm as found on the TCGA data portal [11]. Student’s t-test was performed to compare gene expression in patients with and without 1p19q codeletion. The TCGA cohort was divided into two groups according to whether or not the patients received chemotherapy. Univariate Cox proportional hazard regression was performed to evaluate the association of WHO tumor grade, IDH1 mutation, TERT mutation, TP53, H3F3A, 1p19q status, histological type, extent of resection, receipt of RT (as categorical variables), and age, KPS and gene expression (as continuous variables) with PFS and OS in each group. The significant variables on univariate Cox proportional hazard regression (significance threshold set to be p < 0.1) were taken into multivariate Cox proportional hazard regression (significance threshold set to be p < 0.05). PFS and OS were also evaluated by Kaplan–Meier analysis. All comparisons between high-and low-expressing genes groups using log-rank tests were made by separating genes into two equal-sized groups with the median expression levels as cut-off values. To demonstrate specificity, univariate followed by multivariate Cox proportional hazard regression was performed to evaluate the association of the five most upregulated (ID3, APOC1, RPS19, GNAI3, RPS9) and five most downregulated non-DNA repair genes (PRKCZ, SPINT2, EXTL1, RIMS3, FUT1) located on chromosome arms 1p and 19q in LGG compared to non-glioma tissue with PFS and OS [12].
Cell line
Human oligodendrogliomas cell line HS683 and glioblastomas cell line U251 were purchased from American Type Culture Collection (ATCC). Human Embryo Kidney cell line 293T used for the creation of lentiviral particles was a gift from the Cancer Research Institute of Central South University. Cells were cultured in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), maintained at 37 °C and 5% CO2 in a humidified incubator.
shRNA
shRNA oligo pairs corresponding to LIG1, POLD1, PNKP, RAD54L, MUTYH and GFP were designed (Sangon Biotech, China) and inserted into lentiviral vectors, pLVX-shRNA1 (Clontech). The shRNA sequences used are shown in Supplementary Table 1. The two most effective shRNAs were selected for each DNA repair gene based on downregulation demonstrated by quantitative reverse transcription polymerase chain reaction (RT-qPCR) and western blot experiments. shRNA targeting GFP was used as control and empty vector (pLVX-shRNA1) for judgment of specificity. The lentiviral particles were harvested from the HEK 293T cell lines 3 days after transfection with vectors (pLV-cDNA 1.5 μg, Rev 0.3 μg, VSV-G 0.45 μg, pMDLg 0.75 μg) using the jet PRIME transfection reagent (Polyplus). HS683 cells and U251 cells were infected with lentiviral particles in 6-well plates with puromycin used for selection.
Real-time quantitative reverse transcription-PCR
Total RNA isolation was carried out according to the standard RNA extraction protocol. cDNA was synthesized from 1000 ng of total RNA using RT reagent Kit with gDNA Eraser (Takara). Primers designed for Quantitative real-time PCR are listed in Supplementary Table 2. Quantitative real-time PCR was conducted on 10 ng of cDNA template using SYBR green mix (Roche) in a final volume of 20 μl.
Western blot
Cells were prepared and lysed in RIPA buffer for total protein extraction. Protein concentrations were determined using the BCA reagent. 50 μg of total cellular protein was added to each lane, separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gels, transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore). Then, bands were blocked with 5% non-fat milk. Blots were incubated at 4 °C overnight with antibodies against β-Actin (Cell Signaling Technology), LIG1 (Abnova), POLD1 (Abcam), PNKP (Abcam), RAD54L (Novus) and MUTYH (Novus). Horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) were used for blots detection at room temperature. Western blot band was evaluated using a chemo-luminescence detection system.
MTS
MTS is a cytotoxicity assay which uses a novel tetrazolium compound and colorimetric method to determine the number of viable cells in proliferation. Cells were seeded at 5000 cells per well in a set of 96-well plates, and cultured in humidified incubator for 24 h. After 24 h of seeding, medium was removed and temozolomide (TMZ) (Sigma) of 2, 5, and 10 mg/L was added to the cells separately, representing the estimated human plasma, the maximum in mouse plasma and estimated human cerebrospinal fluid (CSF) TMZ concentrations [13]. The absorbance was measured at 0, 24, 48, and 72 h using a plate reader at a wavelength of 490 nm. Finally, the absorbance at 24, 48, and 72 h was normalized by the absorbance at 0 h, one-way ANOVA test and Student’s t-test performed (significance threshold set to be p < 0.05), and cell proliferation curves plotted. The cells with DNA repair genes knocked down were in the experimental groups, while cells with GFP knocked down were in the control groups.
All statistical analyses were performed using Stata IC 14.2 (StataCorp, College Station, TX) and all figures were made using GraphPad Prism 6.01 (GraphPad Software Inc., La Jolla, CA).
Results
Lower expression of DNA repair genes on chromosome 1p and 19q is specific for tumors with 1p19q codeletion
Among the 512 patients in the TCGA LGG cohort with information on 1p19q codeletion status, 175 patients had 1p19q codeleted tumors, while 337 patients had tumors without the codeletion. The median age was 41 (range 14–87). The expression of all 9 DNA repair genes on chromosome arms 1p and 19q was significantly lower in patients with 1p19q codeletion than in those without the codeletion (p < 0.001) (Fig. 1). For the 5 DNA repair genes on chromosome arms 1q and 19p, expression was significantly higher in the 1p19q codeleted group for XAB2 (p < 0.001), significantly lower for CHAF1A and EXO1 (p < 0.05), and not significantly different for CLK2 and PARP1 (p > 0.05) (Supplementary Fig. 1). The expression of the five most upregulated non-DNA repair genes (ID3, APOC1, RPS19, GNAI3, RPS9) on chromosome arms 1p and 19q in LGG compared to non-glioma tissue was significantly lower in patients with 1p19q codeletion than in those without the codeletion (p < 0.001) (Supplementary Fig. 2). For the most downregulated non-DNA repair genes on chromosome arms 1p and 19q, expression was significantly higher in the 1p19q codeleted group for FUT1 (p = 0.02) and EXTL1 (p = 0.02), and not significantly different for PRKCZ, SPINT2 and RIMS3 (p > 0.05) (Supplementary Fig. 2).
Fig. 1.
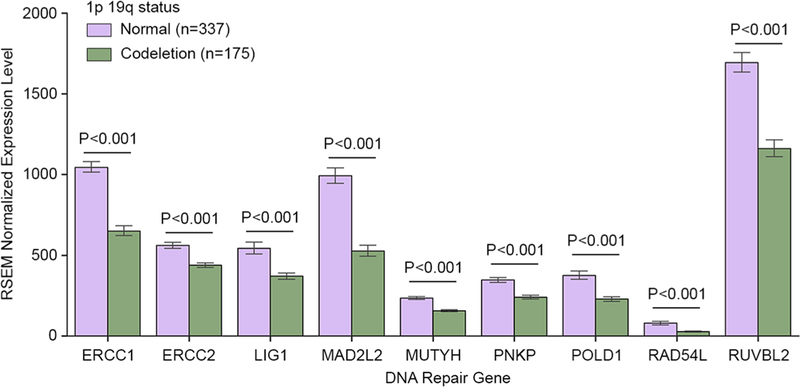
The expression of 9 DNA repair genes on chromosome arms 1p and 19q in patients with versus without 1p19q codeletion
The effect of gene expression on survival outcomes in patients who received chemotherapy
A total of 281 patients received chemotherapy with 238 patients receiving TMZ, two patients receiving PCV (procarbazine, lomustine and vincristine) and 41 patients receiving other agents. The results of the univariate Cox analysis for patients who received chemotherapy are shown in Supplementary Table 3 and Supplementary Table 4 for PFS and OS, respectively. In the univariate Cox regression analysis, age, KPS, IDH1 mutant status, 1p19q codeletion status, and histological type were significant predictors of PFS. Higher expression of LIG1 (p = 0.009), POLD1 (p = 0.045), PNKP (p = 0.005), RAD54L (p = 0.017) and MUTYH (p = 0.001) were associated with shorter PFS (n = 268) on multivariate analysis (Table 1 and Supplementary Table 5). The Kaplan–Meier plots for PFS are shown in Supplementary Fig. 3.
Table 1.
Summary of univariate Cox and multivariate Cox regression analyses for progression free survival and overall survival in the Cancer Genome Atlas lower-grade gliomas patients
| Repair gene | PFS |
OS |
||||||
|---|---|---|---|---|---|---|---|---|
| Chemotherapy |
No chemotherapy |
Chemotherapy |
No chemotherapy |
|||||
| Univariate (n = 268) p < 0.10 |
Multivariate (n = 268) p < 0.05 |
Univariate (n = 218) p < 0.10 |
Multivariate (n = 218) p < 0.05 |
Univariate (n = 280) p < 0.10 |
Multivariate (n = 280) p < 0.05 |
Univariate (n = 231) p < 0.10 |
Multi- variate (n = 231) p < 0.05 |
|
| ERCC1 | X | |||||||
| LIG1 | X | X | X | X | X | X | ||
| ERCC2 | X | X | ||||||
| POLD1 | X | X | X | X | X | X | ||
| RUVBL2 | X | |||||||
| PNKP | X | X | X | X | ||||
| RAD54L | X | X | X | X | X | |||
| MUTYH | X | X | X | X | X | |||
| MAD2L2 | X | X | ||||||
X means the p value is below the significance threshold (0.1 for univariate cox and 0.05 for multivariate-cox)
PFS progression free survival, OS overall survival
In the univariate Cox regression analysis, age, KPS, IDH1 mutant status, TP53, 1p19q codeletion, status, histological type and tumor grade were significant predictors of OS. Higher expression of LIG1 (p = 0.024), POLD1 (p = 0.013), PNKP (p = 0.047), RAD54L (p = 0.001) and MUTYH (p = 0.001) was associated with shorter OS (n = 280) on multivariate analysis (Table 1 and Supplementary Table 5). The Kaplan–Meier plots for OS are shown in Supplementary Fig. 4.
The effect of gene expression on survival outcomes in patients who did not receive chemotherapy
A total of 234 patients did not receive chemotherapy and the results of the univariate Cox regression analysis for patients who did not receive chemotherapy are shown in Supplementary Table 6 and Supplementary Table 7. In patients who did not receive chemotherapy, no significant association between the expression of the nine DNA repair genes and PFS (n = 218) or OS (n = 231) was found on multivariate analysis after accounting for confounding variables (Table 1).
The effect of gene expression on survival outcomes for non-DNA repair genes located on chromosomes 1p and 19q
For the five most upregulated genes and five most down-regulated non-DNA repair genes on chromosome arms 1q and 19p in LGG compared to non-glioma tissue, only the expression of GNAI3 was significantly associated with PFS and OS in the chemotherapy group (Supplementary Table 10). However, GNAI3 expression was also significantly associated with OS in patients who did not receive chemotherapy (Supplementary Table 11). None of the other genes demonstrated significant association between expression and survival (Supplementary Tables 8–11).
Cells with DNA repair genes on chromosome 1p and 19q knockdown demonstrated increased sensitivity to TMZ
Cell lines with DNA repair genes LIG1, POLD1, PNKP, RAD54L and MUTYH knockdown were successfully established after selection with puromycin. The results of RT-qPCR (Supplementary Figs. 5, 6) and western blot (Figs. 2f, 3f) demonstrated good knockdown efficiency for at least one shRNA oligo. Knockdown sequence of GFP did not increase chemosensitivity (Supplementary Fig. 7). Cells with DNA repair gene RAD54L, MUTYH, LIG1, PNKP and POLD1 knocked down had significantly lower proliferation than control group at 72 h after adding TMZ of 2 mg/L in HS683 cell line (Fig. 2) and U251 cell line (Fig. 3), as well as after adding 5 mg/L (Supplementary Figs. 8, 9) and 10 mg/L (Supplementary Figs. 10, 11). There was no significant association between MTS results and TMZ concentration for either HS683 cell line or U251 cell line (Fig. 4).
Fig. 2.
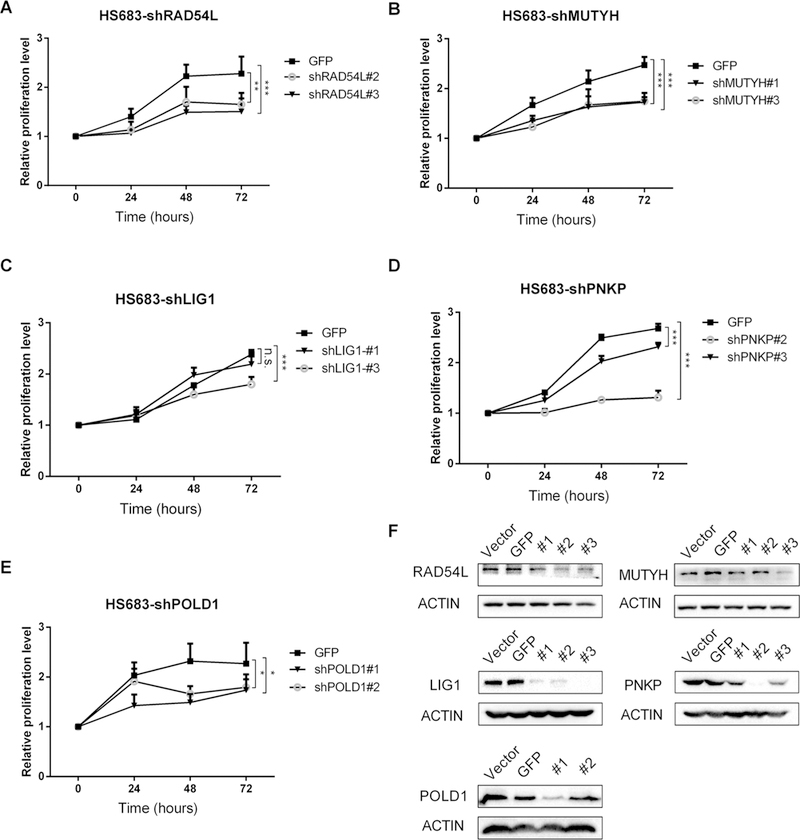
In HS683 cell line: MTS curve at 2 mg/L for: a RAD54L, b MUTYH, c LIG1, d PNKP, e POLD1, and f western blot bands demonstrate good knockdown efficiency. *p < 0.05; **p < 0.01; ***p < 0.001
Fig. 3.
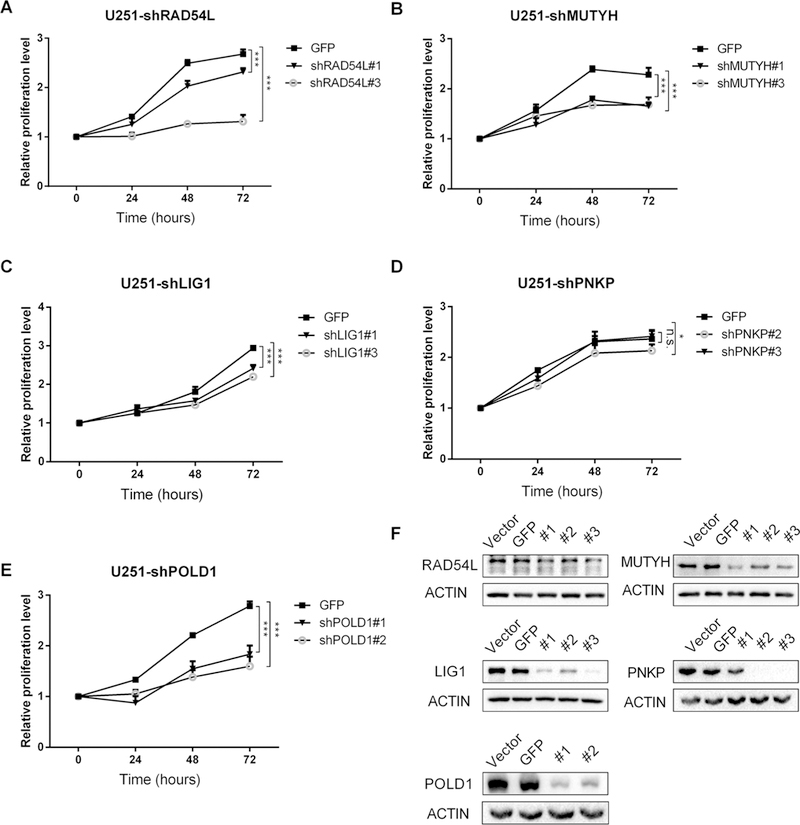
In U251 cell line: MTS curve at 2 mg/L for: a RAD54L, b MUTYH, c LIG1, d PNKP, e POLD1, and f western blot bands demonstrate good knockdown efficiency. *p < 0.05; **p < 0.01; ***p < 0.001
Fig. 4.
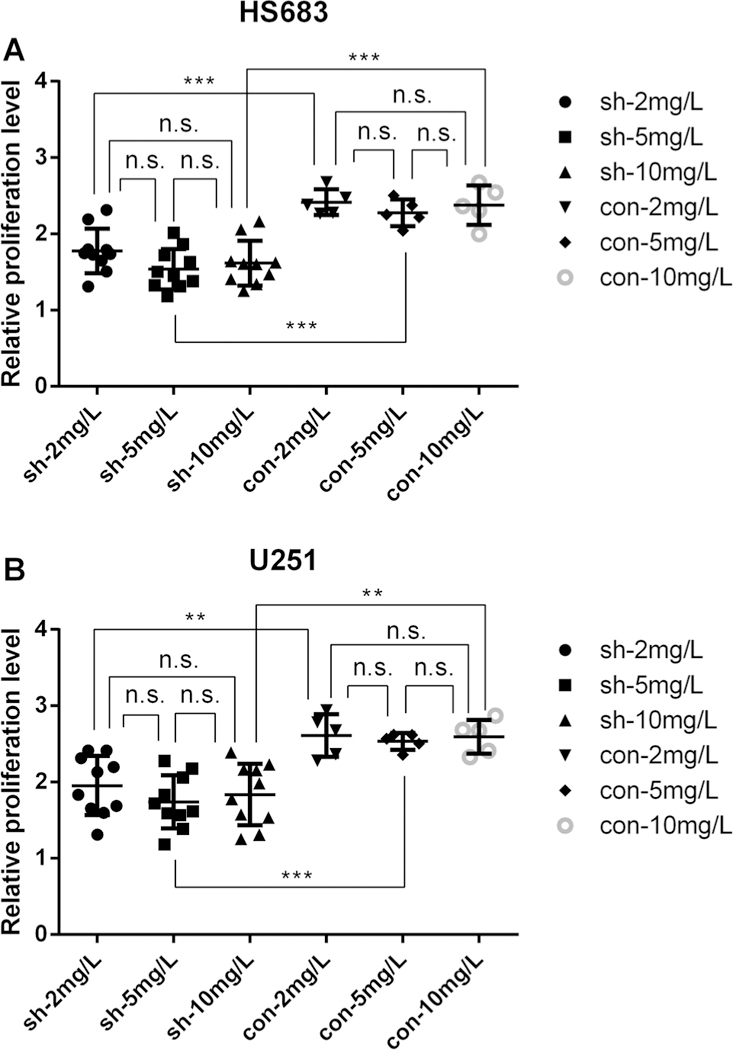
The association between MTS results and TMZ concentration for: a HS683 cell line, b U251 cell line. Under TMZ treatment at 72 h, sh DNA repair gene knockdown, con GFP knockdown. *p < 0.05; **p < 0.01; ***p < 0.001
Discussion
The deletion of chromosomes 1p and 19q is found in 70% of oligodendrogliomas and 50% of mixed oligoastrocytomas [14]. LGG with this codeletion have been demonstrated to have increased chemosensitivity. The mechanism behind this is not fully elucidated, but evidence suggests that DNA repair genes may play a role. Deregulation of DNA repair system plays an important role in cancer therapy, and many chemotherapy drugs work through disruption of DNA repair pathways [15]. Sensitivity of tumors to alkylating agents can be enhanced by impaired DNA repair. For example, in a study of 206 glioblastoma patients, those with silenced O-6-methylgua-nine–DNA methyltransferase (MGMT), which encodes a DNA repair protein, benefited from alkylating agents while those without silenced MGMT did not [16]. Furthermore, MGMT silencing via shRNA in vitro in combination with alkylating agents has been demonstrated to reduce tumor size when compared with alkylating agents alone [17]. Other genes involved in DNA repair such as RAD51, RBBP4 and MSH2 may also increase the sensitivity of gliomas to alkylating agents [18–20]. In addition to these DNA repair genes, the onco-metabolite 2-hydroxyglutarate (2HG) can play a role in the chemosensitivity of 1p19q codeleted tumors. In 2009, a study by Dang et al. found that IDH1 mutation results in a gain-of-function mutation leading to accumulation of the onco-metabolite 2-hydroxy-glutarate (2HG) [21]. Sulkowski et al. demonstrated that 2HG impairs DNA double-strand break repair and thereby increases sensitivity to poly (adenosine 5′-diphosphate-ribose) polymerase (PARP) inhibitors [22]. In our study, we showed that lower expression of specific DNA repair genes in 1p19q codeletion (LIG1, POLD1, PNKP, RAD54L and MUTYH) only prolonged PFS and OS in LGG patients who received TMZ, but not in patients who did not receive TMZ. The results of vitro experiments in HS683 and U251 supported our hypothesis that these genes may account for chemosensitivity to TMZ in 1p19q codeleted lower-grade glioma patients.
Boccard et al. demonstrated that inhibition of certain DNA repair genes (ERCC1, ERCC2, MUTYH, and PNKP) located on chromosome 1p and 19q significantly increased astrocytoma cell chemosensitivity to TMZ [9]. Our results agree with those of Boccard et al. on the effect of MUTYH and PNKP downregulation in increasing glioma’s chemosensitivity. MUTYH and PNKP are involved in base excision repair [10, 23], which has a role in the repair of damage induced by TMZ [24]. However, in contrast to the findings of Boccard et al., we did not find ERCC1 or ERCC2 to be associated with longer survival in LGG patients who received chemotherapy. Some previous research has suggested that abnormalities in copy number of ERCC1 or ERCC2 is not associated with response to therapy or survival in patients with gliomas [25]. However, another study of 32 gliomas showed that ERCC1 DNA methylation levels differ significantly between cisplatin-sensitive samples and cisplatin-resistant samples, suggesting that this gene does play a role in chemosensitivity [26]. Further studies with larger cohort size and repeat in vitro experiments are needed to resolve these differences.
LIG1 encodes DNA ligase I which is involved in base excision repair [27]. Human pancreatic cancer cells demonstrated increased levels of DNA ligase I when exposed to cytostatic concentrations of cisplatin [28]. Mutations in this gene have been associated with increased sensitivity to DNA damaging agents [29]. POLD1 plays several roles in different aspects of DNA repair [30]. There is evidence to suggest that POLD1 knockdown increases sensitivity to ATR inhibitors in colorectal cancer cells [31]. RAD54L encodes protein Rad54 which functions in homologous recombination [32]. In mice, RAD54L deficiency may be associated with sensitivity to clastogens [33], and the loss of RAD54L can result in increased sensitivity to DNA-damaging agents [34]. In summary, there is evidence to suggest that either mutation or downregulation of these DNA repair proteins may influence sensitivity to chemotherapy, which is consistent with our findings.
There are several limitations of our current study. First, response to chemotherapy is difficult to assess in LGG patients since chemotherapy is only administered after surgery and commonly used in combination with radiation. Consequently, we used survival outcomes as surrogate for response to chemotherapy while accounting for confounding variables. The specificity of the DNA repair genes in mediating chemosensitivity was confirmed by the lack of similar effects of non-DNA repair genes located on chromosome 1p and 19q as well as in vitro experiments. Second, we did not account for the chemotherapy agents used in the TCGA cohort, since the majority of the cohort received TMZ (238/281). However, previous studies have suggested no difference in survival between TMZ and PCV [35]. Finally, the TMZ adopted in this study has a maximum recommended concentration at 10 mg/mL in DMSO according to the specification, and the concentration of DSMO in culture should < 0.1%. Therefore 10 mg/L is the highest concentration we could achieve in our MTS.
Conclusions
Reduced expression of DNA repair genes on chromosome arms 1p and 19q, particularly LIG1, POLD1, PNKP, RAD54L and MUTYH, may account for the increased chemosensitivity of LGGs with 1p19q codeletion. These findings are hypothesis-generating, and further studies are needed to confirm these results.
Supplementary Material
Acknowledgements
This study was supported by Shenghua Yuying Project of Central South University to L.Y., National Science Foundation of China to XJL (81472594 and 81770781), and National Science and Technology Major Project to B.X. (2016YFC0904400).
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11060-018-2915-4) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest The authors declare no conflicts of interest.
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-oncology 14 Suppl(5):v1–v49. 10.1093/neuonc/nos218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G et al. (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N, Brat DJ, Verhaak RG et al. (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372 (26):2481–2498. 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw EG, Wang M, Coons SW et al. (2012) Randomized trial of radiation therapy plus procarbazine, lomustine, and vincristine chemotherapy for supratentorial adult low-grade glioma: initial results of RTOG 9802. J Clin Oncol 30(25):3065–3070. 10.1200/JC0.2011.35.8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckner JC, Shaw EG, Pugh SL et al. (2016) Radiation plus pro-carbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med 374(14):1344–1355. 10.1056/NEJMoa1500925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckel-Passow JE, Lachance DH, Molinaro AM et al. (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372(26):2499–2508. 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairncross G, Wang M, Shaw E et al. (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31(3):337–343. 10.1200/JC0.2012.43.2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van den Bent MJ, Brandes AA, Taphoorn MJ et al. (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 31(3):344–350. 10.1200/JCO.2012.43.2229 [DOI] [PubMed] [Google Scholar]
- 9.Boccard SG, Marand SV, Geraci S, Pycroft L, Berger FR, Pelletier LA (2015) Inhibition of DNA-repair genes Ercc1 and Mgmt enhances temozolomide efficacy in gliomas treatment: a pre-clinical study. Oncotarget 6(30):29456–29468. 10.18632/oncotarget.4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood RD, Mitchell M, Lindahl T (2005) Human DNA repair genes. Mutat Res 577(1–2):275–283. 10.1016/j.mrfmmm.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 11.Kamoun A, Idbaih A, Dehais C et al. (2016) Integrated multiomics analysis of oligodendroglial tumours identifies three sub-groups of 1p/19q co-deleted gliomas. Nat Commun 7:11263 10.1038/ncomms11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Jin F, Fan W et al. (2017) Gene expression meta-analysis in diffuse low-grade glioma and the corresponding histological subtypes. Sci Rep 7(1):11741 10.1038/s41598-017-12087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, Guo P, Kruh GD, Vicini P, Wang X, Gallo JM (2007) Predicting human tumor drug concentrations from a preclinical pharmacokinetic model of temozolomide brain disposition. Clin Cancer Res 13(14):4271–4279. 10.1158/1078-0432.CCR-07-0658 [DOI] [PubMed] [Google Scholar]
- 14.Jenkins RB, Blair H, Ballman KV et al. (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66(20):9852–9861. 10.1158/0008-5472.CAN-06-1796 [DOI] [PubMed] [Google Scholar]
- 15.Jackson SP, Helleday T (2016) DNA repair. drugging DNA repair. Science 352(6290):1178–1179. 10.1126/science.aab0958 [DOI] [PubMed] [Google Scholar]
- 16.Hegi ME, Diserens AC, Gorlia T et al. (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352(10):997–1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 17.Viel T, Monfared P, Schelhaas S et al. (2013) Optimizing glioblastoma temozolomide chemotherapy employing lentiviral-based anti-MGMT shRNA technology. Mol Ther 21(3):570–579. 10.1038/mt.2012.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Short SC, Giampieri S, Worku M et al. (2011) Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133 + tumor-derived cells. Neuro-oncology 13(5):487–499. 10.1093/neuonc/nor010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitange GJ, Mladek AC, Schroeder MA et al. (2016) Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep 14(11):2587–2598. 10.1016/jxelrep.2016.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFaline-Figueroa JL, Braun CJ, Stanciu M et al. (2015) Minor changes in expression of the mismatch repair protein MSH2 exert a major impact on glioblastoma response to temozolomide. Cancer Res 75(15):3127–3138. 10.1158/0008-5472.CAN-14-3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang L, White DW, Gross S et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462(7274):739–744. 10.1038/nature08617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulkowski PL, Corso CD, Robinson ND et al. (2017) 2-Hydroxy-glutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 9(375):eaal2463. 10.1126/scitranslmed.aal2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulsen ML, Bisgaard ML (2008) MUTYH associated polyposis (MAP). Curr Genomics 9(6):420–435. 10.2174/138920208785699562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Annovazzi L, Mellai M, Schiffer D (2017) Chemotherapeutic drugs: DNA damage and repair in glioblastoma. Cancers 9(6):57 10.3390/cancers9060057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang BC, Ross DA, Reed E (1995) Genomic copy number changes of DNA repair genes ERCC1 and ERCC2 in human gliomas. J Neuro-Oncol 26(1):17–23 [DOI] [PubMed] [Google Scholar]
- 26.Chen HY, Shao CJ, Chen FR, Kwan AL, Chen ZP (2010) Role of ERCC1 promoter hypermethylation in drug resistance to cisplatin in human gliomas. Int J Cancer 126(8):1944–1954. 10.1002/ijc.24772 [DOI] [PubMed] [Google Scholar]
- 27.Vago R, Leva V, Biamonti G, Montecucco A (2009) DNA ligase I and Nbs1 proteins associate in a complex and colocalize at replication factories. Cell Cycle 8(16):2600–2607. 10.4161/cc.8.16.9352 [DOI] [PubMed] [Google Scholar]
- 28.Sun D, Urrabaz R, Buzello C, Nguyen M (2002) Effects of cisplatin on expression of DNA ligases in MiaPaCa human pancreatic cancer cells. Biochem Biophys Res Commun 298(4):537–544 [DOI] [PubMed] [Google Scholar]
- 29.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T (1992) Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell 69(3):495–503 [DOI] [PubMed] [Google Scholar]
- 30.Prindle MJ, Loeb LA (2012) DNA polymerase delta in DNA replication and genome maintenance. Environ Mol Mutagen 53(9):666–682. 10.1002/em.21745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hocke S, Guo Y, Job A et al. (2016) A synthetic lethal screen identifies ATR-inhibition as a novel therapeutic approach for POLD1-deficient cancers. Oncotarget 7(6):7080–7095. 10.18632/oncotarget.6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazin AV, Mazina OM, Bugreev DV, Rossi MJ (2010) Rad54, the motor of homologous recombination. DNA Repair 9(3):286–302. 10.1016/j.dnarep.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahabir AG, Schaap M, Theunissen P et al. (2008) DNA-repair-deficient Rad54/Rad54B mice are more sensitive to clastogens than wild-type mice. Toxicol Lett 183(1–3):112–117. 10.1016/j.toxlet.2008.10.005 [DOI] [PubMed] [Google Scholar]
- 34.Ghamrasni SE, Cardoso R, Li L et al. (2016) Rad54 and Mus81 cooperation promotes DNA damage repair and restrains chromosome missegregation. Oncogene 35(37):4836–4845. 10.1038/onc.2016.16 [DOI] [PubMed] [Google Scholar]
- 35.Brandes AA, Nicolardi L, Tosoni A et al. (2006) Survival following adjuvant PCV or temozolomide for anaplastic astrocytoma. Neuro-Oncol 8(3):253–260. 10.1215/15228517-2006-005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


