Abstract
NMR spectroscopy is particularly adept at site-specifically monitoring dynamic processes in proteins, such as protein folding, domain movements, ligand binding, and side-chain rotations. By coupling the favorable spectroscopic properties of highly dynamic side-chain methyl groups with transverse-relaxation-optimized spectroscopy (TROSY), it is now possible to routinely study such dynamic processes in high-molecular-weight proteins and complexes approaching 1 MDa. In this Perspective, we describe many elegant methyl-based NMR experiments that probe slow (second) to fast (picosecond) dynamics in large systems. To demonstrate the power of these methods, we also provide interesting examples of studies that utilized each methyl-based NMR technique to uncover functionally important dynamics. In many cases, the NMR experiments are paired with site-directed mutagenesis and/or other biochemical assays to put the dynamics and function into context. Our vision of the future of structural biology involves pairing methyl-based NMR spectroscopy with biochemical studies to advance our knowledge of the motions large proteins and macromolecular complexes use to choreograph complex functions. Such studies will be essential in elucidating the critical structural dynamics that underlie function and characterizing alterations in these processes that can lead to human disease.
Graphical Abstract
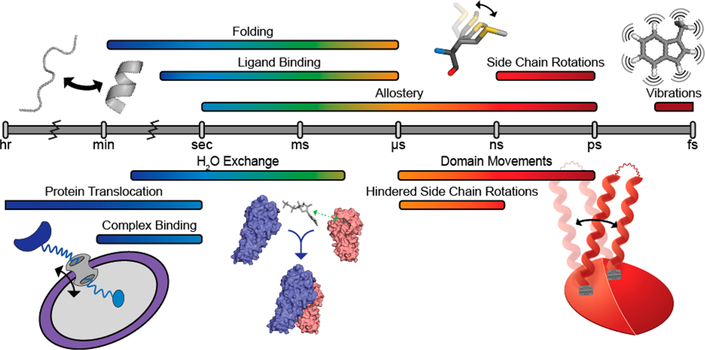
Molecular motions over a variety of time scales are required for biological macromolecules to maintain a diverse and efficient response to stimuli.1–4 Processes such as ligand recognition and binding, domain movements and folding, allosteric regulation, and catalysis are all intimately linked to structural dynamics in proteins.1–4 Moreover, the molecular motions of side-chain conformers in biomolecular systems are directly related to the conformational entropy of the system, which has also been shown to be important in ligand binding and allostery.5,6 Thus, the molecular motions of proteins over a variety of length and time scales, as illustrated in Figure 1, are well-established as key components in the overall behavior of proteins. However, these dynamics are often difficult to characterize in large proteins and large protein complexes and therefore generally remain uncharacterized.
Figure 1.
Timeline for dynamic processes in proteins, ranging from the hour to the femtosecond time scale.1 Visuals of biologically relevant macromolecular motions are shown above (protein folding, side-chain rotations, vibrations) and below (protein translocation, complex and ligand binding, and domain movements) the time scale bar.
NMR spectroscopy is among the most widely used techniques for studying dynamics in biomolecules.2,7 NMR spectroscopy can site-specifically probe macromolecular motions on the so-called slow (hour-to-microsecond) to fast (nanosecond-to-picosecond) time scales, as depicted in Figure 1.2,7 In the past, solution-state NMR spectroscopic studies were limited by the size of the molecule because relaxation and the resulting signal broadening become more efficient with the slower molecular tumbling associated with larger size. In fact, traditional amide-based (1H−15N) NMR methods are generally limited to proteins with masses of ~30 kDa or less. However, advances in transverse-relaxation-optimized spectroscopy (TROSY),8,9 labeling techniques including deuteration and side-chain methyl group isotopic labeling,10,11 and ultra-high field magnets equipped with cryogenically cooled probes have pushed that size limit closer to 1 MDa.9,12−14 The focus of this Perspective is to provide an overview of several NMR techniques that can be used to probe the dynamics of side-chain methyl groups on the second-to-picosecond time scale. It should be noted that many of these experiments were originally developed for the protein backbone amide spin system but were then modified to take advantage of the properties of the methyl group described below. We will also discuss the application of each method in elucidating critical dynamic processes in large proteins, where traditional amide-detected NMR experiments are not suitable. When paired with biochemical studies, these techniques provide a powerful framework for describing the role of structural dynamics in protein function.
Methyl Groups as Spectroscopic Probes.
The side-chain methyl groups of proteins have many features that are beneficial for NMR spectroscopy. First, they are located at the ends of their respective amino acids. This allows the methyl groups to have a local “tumbling” time that is much shorter than that for the overall macromolecular tumbling of the biomolecule, providing a more resolved signal.9,12,15 Second, methyl groups have three protons from which magnetization can originate, whereas the backbone amide nitrogen has only one attached proton.12 This property effectively triples the initial magnetization, leading to greater signal. Third, methyl groups are located throughout the tertiary structure of proteins in the hydrophobic core, catalytic active sites, protein−protein interfaces, and even flexible loops.16 From this nearly uniform distribution of methyl-containing amino acids and the fact that methyl groups are highly sensitive to their chemical environment, detection of very subtle changes in protein structure (e.g., domain movements, side-chain rotations, orientation and distance relative to an aromatic group, etc.) is feasible. When these features are paired with the spectroscopic advantages of the methyl-TROSY technique,9,15 a scheme that causes destructive interference between the intramethyl proton dipolar relaxation to provide greater sensitivity and resolution, our ability to study large protein systems is even further enhanced.
Labeling schemes and the conditions for cell growth and heterologous expression for the production of highly deuterated, methyl-labeled protein samples to be used in the NMR experiments described below have been previously reviewed.17–19 While not all of the methods depicted below require highly deuterated, site-specifically 13CH3-labeled protein samples, such samples are absolutely required for studies on high-molecular-weight proteins and complexes.12 Thus, we will focus on techniques that utilize these types of labeled samples, with the exception of 2H relaxation studies. However, it is straightforward to prepare side-chain methyl group-labeled samples containing the other isotopomers (i.e.,13CHD2 or 13CH2D) through the incorporation of these specific amino acid precursors during protein expression or through fractional deuteration.17 Isotopomers containing 13C, 1H, and 2H probes have some advantages, such as complete labeling of both methyl groups and the ability to measure experiments that cover a wide range of dynamics time scales.17 However, there are also drawbacks, such as the absence of a methyl-TROSY effect and one-third the sensitivity in the case of 13CHD2-labeled methyl groups, which make this probe ill-suited for large macromolecular systems. Finally, detailed explanations of experimental theory and design, setup, and data analysis for each method can be found in the referenced literature. Our goal here is to provide an idea of the power of methyl-based NMR techniques to probe structural dynamics over a range of biologically relevant time scales.
SECOND-TO-MICROSECOND TIME SCALE DYNAMICS
Chemical exchange is the kinetic process in which a nucleus transitions from one state to another (e.g., A ⇆ B), where each state exists in a different chemical environment and therefore has a different chemical shift.20,21 Motions associated with conformational changes, protein−ligand binding, local unfolding, and allostery are some examples where chemical exchange to a functionally relevant “excited” state could be observed via NMR spectroscopy.7,20 Moreover, NMR spectroscopy is uniquely capable of simultaneously determining the populations of both chemical states (pA and pB, where A and B are the major and minor states, respectively), the rate of exchange between states (kex = kA→B + kB→A), and the magnitude of chemical shift change between states (Δω = ωB − ωA).20,21 These observables offer critical information on the thermodynamics, kinetics, and structure of the exchanging system and can be combined with biochemical data to provide a more comprehensive view of the underlying structure−dynamics− function relationship.
To detect and quantify chemical exchange via NMR spectroscopy, four conditions must be satisfied: (1) the rate of exchange between the chemical states must be much higher than the rate of relaxation while chemical exchange is developing (kex ≫ R1 or R2);22 (2) the population of the minor state(s) must be significant enough to be monitored by the experiment that is being employed; (3) the chemical shift of the minor state(s) must be different than that of the major state; and (4) the chemical exchange must occur on the time scale of the method used to manipulate its effect in the NMR experiment.20,21 If one or more of these requirements are not met, the resulting data will appear as if no chemical exchange is occurring in the molecule.
ZZ-Exchange Spectroscopy Probes Molecular Motions on the Second-to-Millisecond Time Scale.
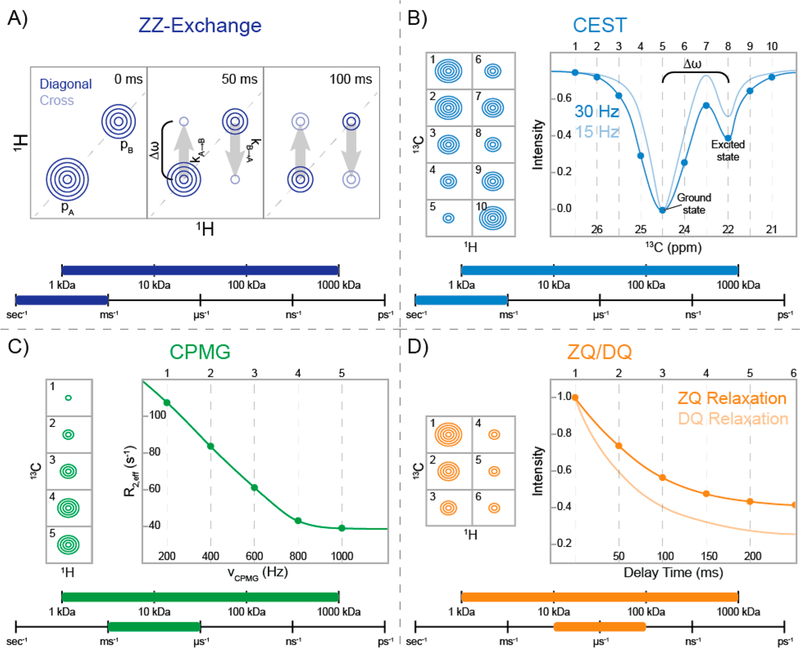
When chemical exchange is slow on the NMR chemical shift time scale (i.e., kex ≪ Δω) and pB is significant enough to detect, ZZ-exchange spectroscopy is the appropriate technique for quantifying chemical exchange.21,23 2D ZZ-exchange spectra display off-diagonal exchange peaks correlating the on-diagonal signals of the observable ground and “excited” states. Varying the time that mixing between the two states via chemical exchange occurs influences the intensity of the cross-peaks; thus, the exchange cross-peaks experience an initial buildup over time before losing intensity as a result of relaxation. From this time dependence, pB and kex can be calculated.21,23−25 Figure 2A shows a representation of ZZ-exchange spectra at several different time points highlighting the buildup of the exchange cross-peaks over time.
Figure 2.
Schematic representations of NMR experiments used to probe slow (second-to-microsecond) time scale chemical exchange dynamics in side-chain methyl groups. For each experiment, a range of applicable molecular weights and time scales are also provided. (A) Example of ZZ-exchange spectra21,23 for a methyl group with two states having populations pA and pB. Diagonal peaks arise from magnetization of each state, whereas the cross-peaks are due to magnetization that exchanged from pA to pB or vice versa during the indicated delay. Second-to-millisecond exchange can be observed as the buildup of exchange cross-peaks at different delay times. (B) Schematic of the ground-state peak intensities at different B1 positions for a chemical exchange saturation transfer (CEST) experiment (left).22 Spectra 1–10 provide the B1-offset-dependent intensities of the CEST profile for the fitted curves (right). The B1 field is on-resonance with the ground and “excited” states in spectra 5 and 8, respectively. CEST profiles at 30 and 15 Hz (dark and light blue, respectively) illustrate the improved resolution from a weaker B1. (C) Representative ground-state peak intensities from a Carr–Purcell–Meiboom–Gill (CPMG) experiment (left)21,29 are shown for a side-chain methyl group experiencing millisecond-to-microsecond time scale dynamics at five different refocusing pulse frequencies (νCPMG). The effective relaxation rate (R2,eff) is shown as a function of νCPMG (right). The relaxation dispersion can be fit to obtain the exchange parameters. (D) Representative example of zero-quantum (ZQ) and double-quantum (DQ) relaxation30,31 for a side-chain methyl group. The decreasing peak intensities at different delay times (left) result in the relaxation decay curves (right). ZQ and DQ relaxation curves are represented as dark- and light-orange lines, respectively.
In a ZZ-exchange experiment, the effect of chemical exchange is allowed to manifest itself while magnetization is stored along the z axis, where relaxation occurs at the spin− lattice or longitudinal relaxation rate (R1).21,23 For methyl groups in large, highly deuterated biomolecules, longitudinal relaxation can take many seconds to cause complete loss of signal. As stated above, the exchange rate between states must therefore be much higher than R1, which means that ZZ-exchange is able to probe chemical exchange on the second-to-hundreds of milliseconds time scale. Additionally, the mixing of the ground and “excited” states is most often performed while magnetization is associated with a heteronucleus (in this case, the methyl carbon). This is done to minimize complications in the spectra, which will occur because of efficient 1H−1H dipolar interactions (i.e., the nuclear Overhauser effect (NOE)).24,26 Finally, because signals arising from pB are observed in the same correlation spectra as signals from pA, spectral crowding leads to extensive overlap between states. This makes the crowded isoleucine, leucine, and valine regions complicated to analyze unless the signals from pA and pB are well-resolved in the spectra. However, methionine residues typically have superior resolution and are therefore most often used in ZZ-exchange experiments on larger proteins.
Methyl-based ZZ-exchange experiments have been used on several large macromolecular systems to characterize “excited” states.14,27,28 For example, Sprangers and Kay utilized this technique in their study of ClpP, an ~300 kDa tetradecameric protease responsible for protein degradation in bacteria.27 The protease active sites are located in an enclosed chamber into which chaperones translocate unfolded substrates for degradation. They found that the interface between heptameric rings, a region hypothesized to be critical for releasing hydrolyzed amino acids, exchanges between two chemically distinct states. The chemical exchange was eliminated by introducing a disulfide bond at the interface via site-directed mutagenesis, which also decreased product release as measured by a separate biochemical assay. Moreover, a temperature series (0.5–40 °C) revealed an increase in kex, while pB remained relatively unchanged. From these experiments, the entropy and enthalpy for the opening of the pore responsible for product release were calculated.27 Notably, the NMR spectra remained high-quality even at 0.5 °C despite the slow global tumbling time of ~400 ns, highlighting the strength of methyl-based NMR techniques in studying large macromolecular complexes.27
Kay and co-workers have also extensively utilized ZZ-exchange spectroscopy on the N-terminal methionine residues of the gating region of the archaeal 20S proteasome to characterize dynamics on the second time scale.14 By comparing the proteolysis rate in the wild-type 20S proteasome with those in a series of mutants with single amino acid substitutions and deletion mutations, they showed that the dynamics associated with the gate regulates the uptake of unfolded polypeptides into the proteolytic active site. These mutations, which eliminated the structural dynamics (i.e., locked the gate in the open state) or removed a portion of the gate, resulted in enhanced protease activity, with the mutants being up to ~500-fold more active than the wild-type protein. Importantly, the activity of the largest deletion mutant (Δ3–11) was comparable to that of a point mutation that locked the gate in the open state, illustrating how the second time scale dynamics is absolutely required for protease function.14
As a final example, Sprangers and co-workers utilized ZZ-exchange spectroscopy to study domain motions in the dimeric yeast DcpS enzyme, which is responsible for removing the 7-methylguanosine monophosphate cap from the 5′-end of mRNA.32 These motions are induced upon asymmetric binding of the substrate to the enzyme and are used by the enzyme to sequester the substrate in the active site for catalysis. By comparing the exchange rates for domain movement from the closed to the open state and vice versa to the rate of product formation, they were able to suggest that domain motions are important for substrate binding and product release. Interestingly, they were also able to show that these motions actually impaired catalytic efficiency (i.e., the motions slow the formation of product). This restriction could be alleviated through a mutation that uncoupled the motions from substrate binding to the second site and decreased the rate for the domain motions, leading to hyperactivity of the decapping enzyme.32
Chemical Exchange Saturation Transfer Spectroscopy (CEST) Can Detect Sparsely Populated States Possessing Slow-Time-Scale Dynamics.
Like ZZ-exchange, CEST is also sensitive to slow (second-to-hundreds of milliseconds) time scale chemical exchange while the magnetization is along the z axis.22 Whereas ZZ-exchange is dependent on an observable population of pB in the correlation spectra, CEST can report on sparsely populated, invisible “excited” states (pB ≳ 0.5%). To achieve this, a weak radiofrequency field (B1; 5–50 Hz) is swept through the frequency range of the resonances of interest. When B1 is on-resonance for pA (ωA), the incoherence leads to a dramatic loss in signal, as illustrated in Figure 2B (point 5). If a nucleus is experiencing chemical exchange, then when B1 is on-resonance with the chemical shift of the “excited” state (ωB), a loss in the intensity is also observed for the major state because the saturation has been transferred via chemical exchange (point 8 in Figure 2B). The CEST profile is then a plot of the intensity of the major state versus the frequency of B1. Subsequently, the CEST data can be fit to the Bloch–McConnell equations,33 which describe the evolution of the magnetization in the presence of chemical exchange, to calculate pB, kex, and Δω.22
Similar to ZZ-exchange experiments, CEST experiments were initially designed to use chemical-exchange-mediated transfer of saturation for a heteronucleus.22,34 Again, since exchange is monitored while the signal is stored along the z axis, complications can arise because of 1H−1H NOEs. However, a number of new experiments to circumvent this problem have recently been described.35−37 An excellent example of the application of methyl-1H CEST comes from Kay and co-workers, who used the technique to observe the effect of Hsp70 on the invisible “excited” states of side-chain methyl groups in 13CH3-labeled drkN SH3 domain, which slowly interconverts between observable folded and unfolded states (i.e., peaks from both states are observed in 2D correlation spectra).35 Before this study, the model for binding of the Hsp70 chaperone protein to native and/or unfolded proteins was not well understood. Methyl-1H CEST on the SH3 domain in the absence of Hsp70 showed the expected interconversion of the folded state with the unfolded state when CEST was observed on the folded state (and of the unfolded state with the folded state when CEST was observed on the unfolded state). However, an additional “excited” state at a unique chemical shift was also observed to be interconverting with the unfolded state but not with the native state. The addition of Hsp70 generated a large flux to this alternate minor conformer of the unfolded state, which provided evidence that Hsp70 preferred to bind the unfolded protein over the native state. Moreover, calculation of the rates of exchange for each state offered a clear kinetic description of the possible binding events.35 This elegant study established a conformational selection mechanism for binding of Hsp70 to a client protein and emphasized the importance of slow-time-scale structural dynamics in Hsp70 substrate detection.
Dark-state exchange saturation transfer spectroscopy (DEST) is another method to study slowly exchanging systems while magnetization is stored along the z axis.38,39 As in CEST, a saturating radiofrequency field (B1; 100−250 Hz) is applied at various offsets, which causes a loss of signal when B1 is on-resonance with the chemical shift of the ground state. However, unlike CEST, DEST, which employs stronger B1 fields, does not require a significantly different chemical shift for the “excited” (i.e., dark) state. Instead, a substantial difference in the transverse relaxation rates of the ground (R2,A) and “excited” (R2,B) states is necessary.38,40 As an excellent example of the technique, Clore and co-workers used side-chain methyl group 13C-DEST to study the initial steps of fibril formation for the peptide amyloid β (Aβ), which is the fibril plaque buildup in Alzheimer’s disease. Aβ exchanges between a monomeric peptide (<5 kDa) and a protofibrillar aggregate (2–80 MDa), leading to a situation where R2,A ≪ R2,B because of the immense reduction of global tumbling of Aβ in the “excited” protofibrillar state. By employing 50 and 240 μM samples of side-chain methyl group 13C-labeled Aβ, where the monomeric species is ~95% and ~40%, respectively, they were able to identify the key methyl-bearing amino acids participating in protofibril formation. Significantly, the DEST data were then used to calculate the exchange rates for the monomer to the “tethered” (i.e., partially bound) and “contact” (i.e., fully bound) states of Aβ protofibrils. This work offers critical structural and kinetic insight into the mechanism of Aβ protofibril formation, which leads to the protein aggregates diagnostic of Alzheimer’s disease.
Carr–Purcell–Meiboom–Gill (CPMG) Relaxation Is Sensitive to the Millisecond Time Scale.
The CPMG experiment is another exquisite example of an NMR experiment that can detect invisible “excited” states.21,41–43 Unlike ZZ-exchange and CEST experiments, the CPMG method is limited by the spin−spin or transverse relaxation rate (R2), since chemical exchange influences the signal while it is in the xy plane: slow chemical exchange on the hundreds of milliseconds-to-millisecond time scale causes an additional loss of signal as R2,eff = R2 + Rex.21,41 In a CPMG experiment, a series of spin-echo sequences are applied, and the rate of pulsing (νCPMG) is arrayed such that the time between pulses decreases.29 As νCPMG increases, the loss of signal from chemical exchange decreases, as demonstrated in Figure 2C, and R2,eff approaches R2 as νCPMG approaches infinity.21 The CPMG profile can then be fit to the Bloch−McConnell equations, allowing pB, kex, and Δω to be extracted.33,44,45 Figure 2C illustrates the CPMG profile of a methyl group experiencing chemical exchange.
Notably, the relevant time scales that can be probed by CPMG experiments are limited by a few factors. If the exchange is too slow (kex < ms−1), there is simply not enough interconversion between states to cause a significant change in signal.22 Increasing the total time that the CPMG spin echoes are applied to allow slower-exchange regimes to process would lead to complete relaxation since transverse relaxation is a much more efficient process than longitudinal relaxation.46 The upper limit (kex > μs−1) is related to how much power the hardware can handle from increasing the pulsing frequency.47 Furthermore, accurate determination of the exchange parameters (i.e., pB, kex, and Δω) requires collecting CPMG data at a minimum of two static magnetic field strengths, as Δω varies with the field.21,41 Finally, it is also important to note that CPMG studies must be carried out on isolated spin systems—that is, 13CH3 CPMG experiments cannot be acquired on a uniformly 13C-labeled sample, for example—since the scalar couplings evolve during the spin-echo period. For the applications described herein, this is not an issue because isolated 13CH3-labeled methyl groups are readily obtained using the methods referenced above.
The CPMG method is the most routinely applied methyl-based NMR method for studying slow dynamics in proteins.48–51 In one example of the methyl-based CPMG method, Ahn, Pardi, and co-workers described the dependence of the structural dynamics in ERK2, a kinase responsible for cell growth and differentiation, on post-translational modification.50 An increase in millisecond time scale dynamics upon phosphorylation of ERK2 was previously described by 1H/2H exchange mass spectrometry, but a description of the thermodynamics, kinetics, and structure of the exchanging state was missing.52 Methyl-based CPMG studies confirmed the appearance of millisecond-to-microsecond time scale dynamics upon dual phosphorylation of ERK2. Importantly, the CPMG data revealed that phosphorylation shifted 80% of the population to a state that is critical for activity. Moreover, a hinge mutant caused similar chemical exchange in the unphosphorylated state, revealing the importance of this domain for regulating internal motions in wild-type ERK2.50
Methyl-based CPMG studies on the nucleosome core particle (NCP), recently described by Kay and co-workers, have uncovered plasticity within the ~210 kDa DNA−protein complex.51 The NCP sterically hinders enzymes from interacting with bound chromosomal DNA, thereby regulating critical processes such as replication and transcription. They analyzed millisecond-to-microsecond time scale dynamics in a series of mutations that have increased transcription rates in cells.53 A clear increase in side-chain dynamics was present in the mutants compared with the wild-type protein. A comparison of the calculated Δω values from the CPMG data for each of the mutants showed that the chemical exchange involved populating the same “excited” state, which appears to be important for transcription in vivo.51
CPMG methodologies have been recently described by Kay and co-workers that exploit a unique 1H triple-quantum (TQ) coherence that can be probed in 13CH3 methyl groups attached to large macromolecules (described below).54 In these 1H TQ coherences, the chemical shift evolves as the sum of the methyl proton chemical shifts (i.e., 3 times the normally observed 1H chemical shift); therefore, Δω is up to 3 times larger than in the single-quantum (SQ) experiments described above. The greater Δω produces larger differences in the CPMG dispersion profiles, making subtle changes in R2,eff more pronounced. Moreover, this effect extends the range of kex where CPMG is sensitive into a faster regime (ms−1 ≤ kex ≤ μs−1) without needing to increase νCPMG. Despite the advantages of TQ CPMG, it is mostly limited to low-molecular-weight proteins (<30 kDa) or highly dynamic spin systems as the transverse relaxation is also more efficient for TQ coherences. In addition to the enhanced dispersions observed for the folding transitions of the 8.2 kDa FF domain, a phosphopeptide-binding four-helix bundle from human HYPA/FBP11 that has been used as a model system for studying protein folding via CPMG experiments,55−57 Kay and co-workers were also able to detect relaxation dispersions at 50 °C using the TQ CPMG experiment (where the SQ experiments showed only weak dispersions) for well-resolved and highly dynamic methionine residues in the flexible N-terminus of a half 20S proteasome, a 360 kDa protein complex.54
Zero-Quantum (ZQ) and Double-Quantum (DQ) Relaxation Can Provide Enhanced Resolution and Probe the Microsecond Time Scale.
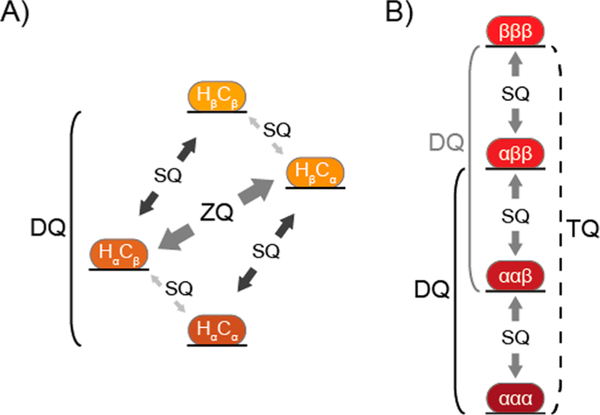
The normal route for studying conformational exchange with rates higher than the CPMG window is through the use of spin-lock-based rotating-frame relaxation (R1ρ) measurements.2,21 However, these experiments are problematic for 13CH3-labeled methyl groups, as applying the spin lock can lead to mixing of the slow and fast relaxation components, resulting in a loss of the methyl-TROSY effect.15,58 R1ρ experiments can be applied to 13CHD2-labeled proteins,59 but with the associated loss of sensitivity and methyl-TROSY effect noted earlier for this isotopomer. As an alternative approach, it has been known for ~25 years that the difference in relaxation rates for DQ and ZQ coherences (ΔRMQ = [RDQ − RZQ]/2) and the relaxation rate for multiple-quantum (MQ) coherence (RMQ = [RDQ + RZQ]/2) are sensitive to chemical exchange ( and .)60−63 A simplified energy level diagram for a 1H−13C spin system that highlights these transitions is shown in Figure 3A. Building on work by Kay and co-workers,61,64 Gill and Palmer30 described a pair of experiments to measure the DQ and ZQ relaxation rates in highly deuterated 13CH3-labeled protein systems of any molecular weight. A schematic representation of RZQ and RDQ data for a methyl group is shown in Figure 2D.
Figure 3.
Simplified energy level diagrams for intramethyl 1H−13C and 1H transitions within a side-chain methyl group. In each, α and β represent the low- and high-energy states being manipulated in NMR experiments. (A) Four energy levels are shown for the simplified 1H–13C spin system. The single-quantum (SQ), double-quantum (DQ), and zero-quantum (ZQ) transitions are indicated. The ZQ/DQ relaxation experiments shown in Figure 2D monitor the relaxation of the ZQ and DQ coherences depicted here. (B) Four energetic states are shown for intramethyl proton transitions,65 where the SQ, DQ, and triple-quantum (TQ) transitions are indicated. SQ transitions between adjacent energy levels (gray arrows) are allowed. The 1H DQ (solid lines) and TQ (dashed line) coherences are traditionally “forbidden” but can be accessed in side-chain methyl groups in macromolecular systems since their relaxation is nonzero.
Shimada and co-workers have employed these ZQ/DQ relaxation experiments to study the dynamics in Gα-GDP, a heterotrimeric G-protein involved in eukaryotic cell signaling.31 Initial characterization of chemical exchange was challenging with CPMG methods because kex > 103 −104 s−1, which is outside the sensitivity window for that experiment. Therefore, they extended the ZQ/DQ methods of Gill and Palmer,30 utilizing the static external magnetic field (B0) dependence of the RMQ relaxation rates, to probe the faster-time-scale motions sampled by Gα-GDP. TQ intramethyl 1H−1H dipolar cross-correlated relaxation rates (described below) were also measured to extract information on the picosecond-to-nanosecond time scale and supplement their analysis of chemical exchange.65 From these data, kex and Δω were calculated for the dynamics of the β1-strand, which is associated with GDP binding. They extended these studies by applying this technique to an oncogenic mutant of Gα that displays a lower affinity and higher rate of dissociation for GDP.66 Interestingly, the mutant exhibited a greater B0 dependence of RMQ for methyl groups in the β1-strand, indicating an increase in the population of the “excited” state. This impressive work combined CPMG, RMQ, and cross-correlated relaxation experiments to elucidate millisecond-to-picosecond dynamics in a Gα-GTPase. Moreover, these results highlight how changing the dynamics in the β1-strand region of Gα-GDP via mutagenesis can be associated with a gain-of-function activity linked with various cancer types.31
Shimada and co-workers also described a novel method to extract conformational exchange information from ΔRMQ based on heteronuclear double resonance (HDR) pulses.67 In this technique, spin-lock pulses are applied simultaneously to the 1H- and 13C-coupled nuclei, during which time ZQ and DQ relaxation rates are measured. If the spin-lock pulses are applied faster than the exchange process, then the exchange contribution to RMQ is reduced (similar to the CPMG experiment described above). Initial simulations suggested that the methyl-TROSY effect is maintained during the HDR pulses. After validating this method with experimental data on maltose binding protein (MBP) and the folding intermediate found in the FF domain, they applied the technique to the prokaryotic inwardly rectifying K+ channel KirBac1.1 in detergent micelles. Interestingly, they found significant ΔRMQ exchange contributions for Ileδ1 residues in the trans-membrane helix and cytoplasmic regions, which are thought to be important in allosteric regulation of K+ gating.67
NANO- TO PICOSECOND TIME SCALE DYNAMICS
Nanosecond-to-picosecond motions of a bond vector in relation to the B0 field produce locally fluctuating random magnetic fields.2,68,69 When these local fields are at certain frequencies, NMR transitions occur, giving rise to relaxation of the NMR signal. For spin-1/2 nuclei (e.g., 1H, 13 C, and 15N), dipole–dipole interactions and chemical shift anisotropy are the dominant mechanisms that give rise to the random fields.2,68,69 For the spin-1 nucleus 2H, the quadrupolar interaction is the dominant relaxation process. The model-free formalism is the most common framework for describing the motions that underlie the relaxation process.68,70–72 In model-free analysis, the dynamics is separated into contributions from the local fluctuations of the bond vector, which occur on the picosecond time scale, and global reorientation of the molecule, which occurs on the nanosecond time scale.71,73 The local and global motions contribute to the R1 and R2 relaxation rates.
Two parameters that describe the local fast-time-scale molecular motions are generally obtained from the analysis of side-chain methyl group relaxation rates.73 The first and most widely used is the amplitude of the motion of the methyl group about its threefold symmetry axis, which is known as the order parameter ().74 ranges between 0 and 1 for a completely flexible or rigid methyl group bond vector, respectively.71,73,74 Studies of methyl group on a wide variety of proteins and their complexes revealed that this order parameter can be binned into three distributions: J, α, and ω. Motions of side-chain methyl groups within these bins have been ascribed to jumps between rotameric states (J; ~ 0.35), large-amplitude motions within rotamer states (α; ~ 0.6), and relatively small amplitude motions within a rotameric well (ω; ~ 0.85).72,75 Furthermore, the determination of side-chain methyl group order parameters is an extremely valuable measurement used to calculate the conformational entropy of a system.5,6,76,77 By measurement of under different conditions, for example in the absence/presence of a ligand or upon mutation, the contribution of conformational entropy in a reaction can be determined.5,6 The second, less used parameter is the picosecond time scale lifetime (τe) for the local reorientation of the side-chain methyl group.68,73,74
“Forbidden” Multiple-Quantum Transitions Can Be Used as a Tool To Study Picosecond-to-Nanosecond Time Scale Dynamics.
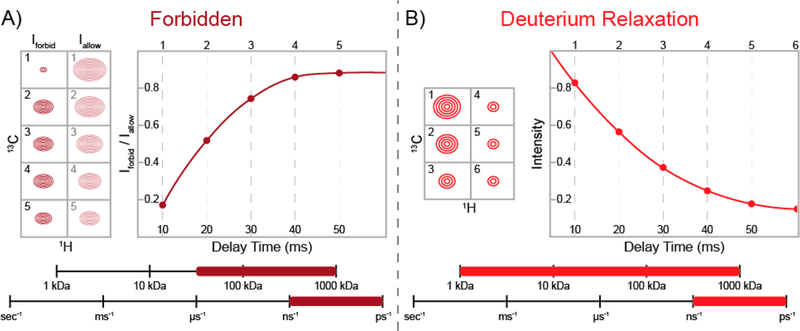
Relaxation-violated coherence transfer takes advantage of a peculiar phenomenon of methyl groups in macromolecular systems and can be used to study the fast (nanosecond-to-picosecond) time scale side-chain dynamics.65,78,79 In a methyl group attached to a large molecule and rapidly rotating around its symmetry axis, relaxation-violated methyl proton DQ and TQ coherences can be prepared because the relaxation of these states is not zero. The intramethyl 1H–1H dipolar cross-correlated relaxation rate (η) for these “forbidden” 1H DQ or TQ coherences, illustrated in the simplified methyl 1H energy level diagram shown in Figure 3B, directly reports on the amplitude of fast-time-scale motions in a methyl group () and the global correlation time .65,79 The TQ experiment is 50% more sensitive than the DQ method;65 however, the TQ experiment requires slower macromolecular tumbling (i.e., a larger τc). Varying the time where the “forbidden” coherence can develop produces a buildup curve, an example of which is shown in Figure 4A, that can be used to calculate η and, if τc is known, . Interestingly, the “forbidden” experiment is a rare case in solution-state NMR spectroscopy where a larger biomolecule actually improves the signal, as increasing the correlation time yields larger η values.
Figure 4.
Schematic representations of NMR experiments used to probe fast (nanosecond-to-picosecond) time scale dynamics in side-chain methyl groups. The ranges of protein molecular weights and time scales where each experiment is sensitive are shown at the bottom of each panel. (A) Schematic of the relaxation-violated methyl 1H “forbidden” experiment.65,79 The raw peak intensities for the “forbidden” (DQ or TQ) coherences (Iforbid) and the allowed (SQ) coherences (Iallow) are shown at five different delay times (left). The ratio of “forbidden” and allowed coherences (Iforbid/Iallow) produces a buildup curve (right), which can be fit to obtain methyl group order parameters reporting on the amplitude of fast-time-scale dynamics. (B) Representative example of deuterium relaxation for a partially deuterated side-chain methyl group (i.e., 13CHD2 or 13CH2D).74 The decreasing peak intensities at different delay times (left) produce a decay curve (right), which reports on fast-time-scale dynamics.
Recently, our research group has employed methyl 1H TQ “forbidden” experiments on Pyrococcus furiosus Rad50NBD, an ~42 kDa ABC ATPase essential for DNA double-strand break repair, to characterize a dynamic allosteric network within the enzyme.80 We examined a set of mutations in the hinge region that altered the rigid ground state of Rad50NBD by interrupting an interaction, the so-called “basic switch”,81 between the N-and C-terminal lobes. The 2D methyl correlation spectra of wild-type and mutant proteins showed a linear movement of chemical shifts away from the wild-type positions upon mutation, implying an allosteric network within Rad50NBD. By comparing η values from methyl 1H TQ “forbidden” experiments, we found that the mutations altered the fast-time-scale dynamics landscape of side-chain methyl groups within Rad50NBD such that the mutated proteins were generally more dynamic than the wild-type protein. We also showed that this allosteric network was key to regulating Rad50NBD ATP-induced dimerization and hydrolysis, with ATP hydrolysis catalytic efficiencies increased by up to ~8-fold in the mutants. We hypothesized that the hinge mutations preemptively perturb the basic switch, which locks Rad50NBD into a rigid state, and populate a more dynamic state. We further proposed that this dynamic state of Rad50NBD is on-pathway for ATP-induced dimerization, a function critical for DNA double-strand break repair.80
Giedroc and co-workers also employed relaxation-violated spectroscopy to study allostery within the transcriptional regulator CzrA.82 In a pathogenic organism, such as Staphylococcus aureus, CzrA binds Zn2+ ions and releases promoter DNA, allowing the transcription of genes to mitigate a potentially toxic concentration of Zn2+ ions within the cell. CzrA displays strong negative cooperativity between binding of Zn2+ ions and promoter DNA; however, the mechanism of the negative cooperativity was not understood because the structures of apo and Zn2+-bound CzrA are nearly superimposable. By calculating via methyl 1H DQ “forbidden”experiments for the apo, Zn2+-bound, and DNA-bound states, they described a decrease in side-chain flexibility upon Zn2+ binding—that is, the DNA-bound state was the more flexible conformer. On the basis of this inverse relationship, they proposed that the Zn2+-bound state is not able to sample the flexible conformers necessary for gene regulation. To test this model, the fast-time-scale dynamics of a series of L/V-to-A mutations, for many of which Zn2+ binding does not hinder DNA binding, were measured. While the slow-time-scale dynamics, monitored by CPMG, was quenched in both the wild-type and mutant CzrA upon Zn2+ binding, the fast-time-scale dynamics of the mutants did not change, and the protein stayed in the more dynamic state. Impressively, the degree of allosteric regulation from the Zn2+-bound state could be accurately estimated solely from the values. This study provides further evidence of the relationship between side-chain dynamics and allostery, which is emerging as a prevalent trend in biochemistry.
A recent study by Gardner, Rosenbaum, and co-workers employed TQ “forbidden” experiments to probe fast-time-scale dynamics of isoleucine side chains in A2AR, a G-protein-coupled receptor (GPCR) that is involved in extracellular signal transduction pathways.83 Because GPCRs react to various ligands differently, the cellular response can be extremely diverse. To describe the effect of an agonist versus an inverse agonist on the dynamics of A2AR, they recorded methyl 1H TQ “forbidden” data in the presence of different ligands. By comparing the η values for each methyl group against those for the apo conformation, there was a clear increase in rigidity in the presence of an inverse agonist. Conversely, the side-chain dynamics increased in the agonist-bound state, as revealed by the lower η values. GPCRs are challenging systems, as they are exceedingly hard to express and purify in the high yields that NMR studies require.84 However, this impressive example illustrates a significant step toward understanding the dynamic landscape of GPCRs. Future methyl-based NMR studies could further characterize the differences in side-chain methyl group dynamics in disease mutation, ligand binding, and pharmaceutical studies.83
Deuterium Quadrupolar Relaxation in 13CH2D and 13CHD2 Methyl Groups Probes Fast Dynamics.
Methods that measure deuterium relaxation were among the first experiments to probe nanosecond-to-picosecond dynamics in methyl groups.74 The quadrupolar interaction, which is much more efficient than the dipolar and chemical shift anisotropy mechanisms that lead to 1H and 13C relaxation and does not suffer from exchange contributions, dominates in deuterium relaxation.74 Thus, quantification of fast-time-scale dynamics via 2H relaxation studies is more straightforward and robust than 1H- and 13C-based measurements. Moreover, five different deuterium relaxation rates can be measured for a 13CH2D-labeled methyl group, allowing for a more thorough analysis of the data: each of these relaxation rates reports on the fast-time-scale dynamics via its relationships to the amplitude of motion of the methyl group () and the correlation time, providing a more thorough determination of the side-chain dynamics in complex systems.85 It should be noted that to determine side-chain methyl group values from 2H relaxation studies, an estimate of τc, usually obtained from backbone amide 15N relaxation studies, is necessary. Figure 4B illustrates the single-exponential behavior indicative of any deuterium relaxation NMR data for a fractionally deuterated methyl group.
Many studies have employed deuterium relaxation to measure fast dynamics in proteins. Pioneering work by the groups of Wand5,75 and Kay76,86 have demonstrated the power of these methods to quantify the energetics of various protein systems. Recently, Akke and co-workers employed side-chain methyl group deuterium relaxation experiments in FKBP12, a 12 kDa isomerase that is the target of many immunosuppressive drugs.87 FKBP12 forms a complex with FK506 to inhibit calcineurin, causing a loss of T-lymphocyte signal transduction and interleukin-2 transcription. This study validated previously reported backbone amide order parameters and extended the dynamic view further by determining methyl values for both the apo FKBP12 and FK506-bound conformations. Subsequently, molecular dynamics simulations were performed for each state to supplement the NMR data and estimate the total conformational entropy of ligand binding. These studies revealed a shift in the distribution of accessible conformers upon ligand binding, leading to a significant decrease in the total entropy upon ligand binding. Interestingly, a significant portion of this change in total entropy comes from the difference between the conformational entropies of the apo and ligand-bound states.87
USING METHYL GROUPS TO PROBE OTHER MOTIONS OF PROTEINS
Real-Time NMR Spectroscopy Can Monitor Non-equilibrium Processes.
Since the height of the NMR signal is related to the concentration of the species in solution giving rise to that signal, NMR data can be used to determine the relative concentrations of different species in solution. Real-time NMR experiments leverage this fact by collecting many spectra to quantify the change in species over time (i.e., unfolded → folded, apo → ligand-bound, or substrate → product).88,89 The reaction can be initiated by pressure, pH, or temperature jump or through rapid mixing with the substrate.90–93 Since the time resolution is limited by the length of the NMR experiment, methods have been developed that allow 2D and 3D NMR data to be collected in a fraction of the time of more conventional multidimensional spectra (~5 s vs ~5 min for 2D data sets).94,95 The time dependence of the disappearance of peaks from the reactant and the appearance of new peaks from the product can then be fitted to extract the rate constant for the reaction.
Boisbouvier and co-workers have recently used real-time NMR spectroscopy on 13CH3-labeled alanine samples of TET2, a 500 kDa homododecameric complex that hydrolyzes small peptides into amino acids.96 Under acidic conditions (pH ~4), the quaternary structure of the complex disassembles. After a jump to pH ~8, which promotes self-assembly, 2D correlation spectra were recorded with a time resolution of 15 s. As expected, destabilized monomeric TET2 peaks disappeared over time as peaks arising from the complex appeared. However, an intermediate set of peaks appeared then disappeared prior to full complex assembly. They confirmed that these intermediate peaks were indicative of an ensemble of oligomeric intermediates through negative-stain electron microscopy (EM) and native mass spectrometry. Finally, analysis of the calculated rate constants allowed them to determine that the initial transition from the monomer to the oligomeric intermediate is faster than the transition from the intermediate to the full dodecamer. Thus, their data are highly suggestive of many parallel paths toward full self-assembly.
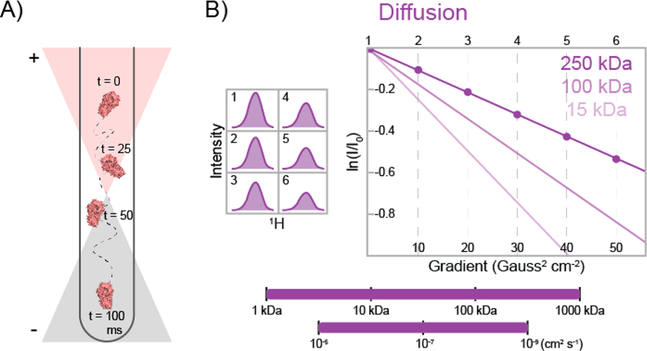
Protein Diffusion Can Be Measured with Pulsed Field Gradient (PFG) Experiments.
PFG diffusion methods can be useful for characterizing protein−protein interactions, large conformational changes, or binding to a large ligand that significantly changes the size of the complex (e.g., a large DNA strand). In PFG diffusion experiments, a spatially dependent magnetic field (i.e., the pulsed field gradient) is applied along an axis, usually the z axis.97,98 This additional field effectively encodes the positions of the biomolecules on the basis of their initial positions in the sample (Figure 5A), thereby defining the position of the macromolecule in solution. During a delay time, in which magnetization is stored along the z axis, the molecule diffuses to a different position in the NMR tube. This new position is subsequently decoded with another gradient pulse. Because the diffusing molecule is in a different position for each gradient pulse, the effect of the spatially dependent magnetic field on the magnetization does not cancel out, leading to a loss of signal. Smaller molecules diffuse farther, resulting in greater signal loss compared with larger molecules. Increasing the delay time enhances the signal decay due to diffusion because the molecule has had more time to move. The loss of signal versus delay time can subsequently be fit to calculate the average diffusion rate of the molecule in solution. Figure 5B illustrates the decay curves for proteins of different sizes in a PFG diffusion experiment. Recently, a diffusion method based on TQ coherences (see above) has been developed for improved sensitivity in exceedingly large, slowly diffusing systems.99
Figure 5.
Schematic representation of a pulsed field gradient diffusion experiment.97 (A) Cartoon depicting a 13CH3-labeled protein in an NMR tube at four different delay times. The position of the protein is encoded and then decoded by pulsed field gradients, whose effect on the sample is dependent on the position of the protein in the tube. The orange and gray triangles represent the applied gradient field, which adds to or subtracts from the main static magnetic field. (B) Representative methyl group 1D peak intensities are shown at six different gradient strengths (left). The natural logarithm of the relative peak intensity vs the square of the gradient strength is then plotted (right). Dark-, medium-, and light-purple curves illustrate the expected diffusion curves for proteins with molecular masses of 250, 100, and 15 kDa, respectively. Methyl-based diffusion experiments are sensitive to diffusion rates of 10−5−10−9 cm2 s−1 and can be employed for nearly any protein size (bottom).
PFG diffusion experiments are most routinely used on large biomolecular systems to ensure that the macromolecular complex of interest has actually been formed. A simple example would be a multi-subunit protein complex. In this case, it is important that the individual proteins form a stable complex to ensure that all of the downstream studies are valid. Diffusion rates of individual proteins can be compared with that of the complex, for which a lower diffusion rate would be expected.
Another use of PFG diffusion methods is to determine the viscosity of an NMR sample. For example, Kay and co-workers paired PFG diffusion experiments with ZZ-exchange spectroscopy (see above) on the single ring of the 20S proteasome.28 Here, the viscosity of the sample was increased via stepwise additions of glycerol or sucrose, and ZZ-exchange was measured at each step to determine the rate of chemical exchange as a function of viscosity. Since an exact measurement of the viscosity at each step was critical in their calculations, PFG diffusion studies were also employed. The goal of this study was to map the “roughness” of the energy landscape100 in the dynamic gating process, which allows the substrate to enter the active site. Monitoring the dynamic process of gate opening and closing at different viscosities allowed the role of collisions by water molecules and the length scale of this chemical exchange process to be determined, providing an in-depth description of the motions involved in proteasome gating.
CONCLUDING REMARKS
In this Perspective, we have highlighted many elegant methyl-based NMR experiments to probe protein dynamics in large protein systems. Slow-time-scale dynamics studies, which are examined with ZZ-exchange, CEST, CPMG, and ZQ/DQ experiments, are useful for observing protein complex binding, folding, ligand binding, and allostery. Fast-time-scale dynamics studies, which are probed by intramethyl 1H relaxation-violated “forbidden” coherence and deuterium relaxation experiments, can probe allostery, domain movements, and side-chain rotations. The exciting, functionally relevant examples of studies we have used to illustrate each method can act as guides for future investigations into the structural dynamics of large protein complexes.
The advancement of biochemistry will continue to depend on melding structural biology and biochemical/biophysical functional studies to ascertain a complete understanding of the structures and dynamics that underlie the activities of macromolecular systems. As the methods described herein are applied to increasingly more complicated systems, there will be a need to combine methyl-based NMR spectroscopy with other structural techniques, such as cryo-EM, small-angle X-ray scattering, and molecular dynamics simulations.96,101,102 Thus, a vision for the future of biochemical structure/dynamics/function studies is one of hybrid approaches that make use of complementary techniques to drive our understanding of macromolecular function and increase our understanding of the precise mechanisms proteins use to direct a decisive response to stimuli. This will move us ever closer to understanding the biochemical underpinnings for a variety of human diseases.
ACKNOWLEDGMENTS
The authors thank Marella D. Canny (TTU) for critical reading of this manuscript.
Funding
Research in the Latham laboratory is supported by grants from the Welch Foundation (D-1876), the Cancer Prevention Research Institute of Texas (RP180553), and the National Institute of General Medical Sciences, NIH (1R35GM128906).
ABBREVIATIONS
- NMR
nuclear magnetic resonance
- TROSY
transverse-relaxation-optimized spectroscopy
- kDa
kilodalton
- MDa
megadalton
- pA(B)
population of the ground (excited) state
- kex
rate of exchange between states
- ωA(B)
chemical shift of the ground (excited) state
- Δω
difference in chemical shift between states
- R1
spin−lattice or longitudinal relaxation rate
- R2
spin–spin or transverse relaxation rate
- Rex
exchange contribution to R2
- 2D
two-dimensional
- NOE
nuclear Overhauser effect
- CEST
chemical exchange saturation transfer
- CPMG
Carr–Purcell–Meiboom–Gill
- TQ
triple quantum
- SQ
single quantum
- DQ
double quantum
- ZQ
zero quantum
- B0
static magnetic field
order parameter
- τe
picosecond time scale local correlation time
- τc
nanosecond time scale global correlation time
- η
intramethyl proton− proton dipolar cross-correlated relaxation rate
- PFG
pulsed field gradient
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Skjærven L, Reuter N, and Martinez A (2011) Dynamics, flexibility and ligand-induced conformational changes in biological macromolecules: A computational approach. Future Med. Chem 3, 2079–2100. [DOI] [PubMed] [Google Scholar]
- (2).Palmer AG III. (2004) NMR Characterization of the Dynamics of Biomacromolecules. Chem. Rev 104, 3623–3640. [DOI] [PubMed] [Google Scholar]
- (3).Karplus M, and Kuriyan J (2005) Molecular dynamics and protein function. Proc. Natl. Acad. Sci. U. S. A 102, 6679–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Torchia DA (2011) Dynamics of biomolecules from picoseconds to seconds at atomic resolution. J. Magn. Reson 212, 1–10. [DOI] [PubMed] [Google Scholar]
- (5).Frederick KK, Marlow MS, Valentine KG, and Wand AJ (2007) Conformational entropy in molecular recognition by proteins. Nature 448, 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tzeng S-R, and Kalodimos CG (2012) Protein activity regulation by conformational entropy. Nature 488, 236–240. [DOI] [PubMed] [Google Scholar]
- (7).Mittermaier AK, and Kay LE (2009) Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci 34, 601–611. [DOI] [PubMed] [Google Scholar]
- (8).Pervushin K, Riek R, Wider G, and Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U. S. A 94, 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tugarinov V, Hwang PM, Ollerenshaw JE, and Kay LE (2003) Cross-Correlated Relaxation Enhanced 1 H- 13 C NMR Spectroscopy of Methyl Groups in Very High Molecular Weight Proteins and Protein Complexes. J. Am. Chem. Soc 125, 10420–10428. [DOI] [PubMed] [Google Scholar]
- (10).Kay LE, and Gardner KH (1997) Solution NMR spectroscopy beyond 25 kDa. Curr. Opin. Struct. Biol 7, 722–731. [DOI] [PubMed] [Google Scholar]
- (11).Religa TL, and Kay LE (2010) Optimal methyl labeling for studies of supra-molecular systems. J. Biomol. NMR 47, 163–169. [DOI] [PubMed] [Google Scholar]
- (12).Sprangers R, Velyvis A, and Kay LE (2007) Solution NMR of supramolecular complexes: providing new insights into function. Nat. Methods 4, 697–703. [DOI] [PubMed] [Google Scholar]
- (13).Sprangers R, and Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445, 618–622. [DOI] [PubMed] [Google Scholar]
- (14).Religa TL, Sprangers R, and Kay LE (2010) Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 328, 98–102. [DOI] [PubMed] [Google Scholar]
- (15).Ollerenshaw JE, Tugarinov V, and Kay LE (2003) Methyl TROSY: explanation and experimental verification. Magn. Reson. Chem 41, 843–852. [Google Scholar]
- (16).Janin J, Miller S, and Chothia C (1988) Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol 204, 155–164. [DOI] [PubMed] [Google Scholar]
- (17).Tugarinov V, and Kay LE (2005) Methyl Groups as Probes of Structure and Dynamics in NMR Studies of High-Molecular-Weight Proteins. ChemBioChem 6, 1567–1577. [DOI] [PubMed] [Google Scholar]
- (18).Tugarinov V, Kanelis V, and Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc 1, 749–754. [DOI] [PubMed] [Google Scholar]
- (19).Kerfah R, Plevin MJ, Sounier R, Gans P, and Boisbouvier J (2015) Methyl-specific isotopic labeling: A molecular tool box for solution NMR studies of large proteins. Curr. Opin. Struct. Biol 32, 113–122. [DOI] [PubMed] [Google Scholar]
- (20).Palmer AG III. (2014) Chemical exchange in biomacromolecules: Past, present, and future. J. Magn. Reson 241, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Palmer AG III, Kroenke CD, and Loria JP (2001) Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol 339, 204–238. [DOI] [PubMed] [Google Scholar]
- (22).Vallurupalli P, Bouvignies G, and Kay LE (2012) Studying “Invisible” Excited Protein States in Slow Exchange with a Major State Conformation. J. Am. Chem. Soc 134, 8148–8161. [DOI] [PubMed] [Google Scholar]
- (23).Jeener J, Meier BH, Bachmann P, and Ernst RR (1979) Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys 71, 4546–4553. [Google Scholar]
- (24).Farrow NA, Zhang O, Forman-Kay JD, and Kay LE (1994) A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734. [DOI] [PubMed] [Google Scholar]
- (25).Kloiber K, Spitzer R, Grutsch S, Kreutz C, and Tollinger M (2011) Longitudinal exchange: An alternative strategy towards quantification of dynamics parameters in ZZ exchange spectroscopy. J. Biomol. NMR 51, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Montelione GT, and Wagner G (1989) 2D Chemical Exchange NMR Spectroscopy by Proton-Detected Heteronuclear Correlation. J. Am. Chem. Soc 111, 3096–3098. [Google Scholar]
- (27).Sprangers R, Gribun A, Hwang PM, Houry WA, and Kay LE (2005) Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proc. Natl. Acad. Sci. U. S. A 102, 16678–16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Latham MP, Sekhar A, and Kay LE (2014) Understanding the mechanism of proteasome 20S core particle gating. Proc. Natl. Acad. Sci. U. S. A 111, 5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, and Kay LE (2001) Slow Dynamics in Folded and Unfolded States of an SH3 Domain. J. Am. Chem. Soc 123, 11341–11352. [DOI] [PubMed] [Google Scholar]
- (30).Gill ML, and Palmer AG III. (2011) Multiplet-filtered and gradient-selected zero-quantum TROSY experiments for 13C1H3 methyl groups in proteins. J. Biomol. NMR 51, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Toyama Y, Kano H, Mase Y, Yokogawa M, Osawa M, and Shimada I (2017) Dynamic regulation of GDP binding to G proteins revealed by magnetic field-dependent NMR relaxation analyses. Nat. Commun 8, 14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Neu A, Neu U, Fuchs AL, Schlager B, and Sprangers R (2015) An excess of catalytically required motions inhibits the scavenger decapping enzyme. Nat. Chem. Biol 11, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).McConnell HM (1958) Reaction Rates by Nuclear Magnetic Resonance. J. Chem. Phys 28, 430–431. [Google Scholar]
- (34).Bouvignies G, and Kay LE (2012) A 2D 13C-CEST experiment for studying slowly exchanging protein systems using methyl probes: an application to protein folding. J. Biomol. NMR 53, 303–310. [DOI] [PubMed] [Google Scholar]
- (35).Sekhar A, Velyvis A, Zoltsman G, Rosenzweig R, Bouvignies G, and Kay LE (2018) Conserved conformational selection mechanism of Hsp70 chaperone–substrate interactions. eLife 7, e32764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Yuwen T, Huang R, and Kay LE (2017) Probing slow timescale dynamics in proteins using methyl 1H CEST. J. Biomol. NMR 68, 215–224. [DOI] [PubMed] [Google Scholar]
- (37).Bouvignies G, and Kay LE (2012) Measurement of Proton Chemical Shifts in Invisible States of Slowly Exchanging Protein Systems by Chemical Exchange Saturation Transfer. J. Phys. Chem. B 116, 14311–14317. [DOI] [PubMed] [Google Scholar]
- (38).Fawzi NL, Ying J, Ghirlando R, Torchia DA, and Clore GM (2011) Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature 480, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Fawzi NL, Libich DS, Ying J, Tugarinov V, and Clore GM (2014) Characterizing methyl-bearing side chain contacts and dynamics mediating amyloid β protofibril interactions using 13C-(methyl)-DEST and lifetime line broadening. Angew. Chem., Int. Ed 53, 10345–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Fawzi NL, Ying J, Torchia DA, and Clore GM (2012) Probing exchange kinetics and atomic resolution dynamics in high-molecular-weight complexes using dark-state exchange saturation transfer NMR spectroscopy. Nat. Protoc 7, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sekhar A, and Kay LE (2013) NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. U. S. A 110, 12867–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Carr HY, and Purcell EM (1954) Effects of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments. Phys. Rev 94, 630–638. [Google Scholar]
- (43).Meiboom S, and Gill D (1958) Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev. Sci. Instrum 29, 688–691. [Google Scholar]
- (44).Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, and Kay LE (2004) Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430, 586–590. [DOI] [PubMed] [Google Scholar]
- (45).Vallurupalli P, Hansen DF, Stollar E, Meirovitch E, and Kay LE (2007) Measurement of bond vector orientations in invisible excited states of proteins. Proc. Natl. Acad. Sci. U. S. A 104, 18473–18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Stone MJ (2001) NMR Relaxation Studies of the Role of Conformational Entropy in Protein Stability and Ligand Binding. Acc. Chem. Res 34, 379–388. [DOI] [PubMed] [Google Scholar]
- (47).Reddy JG, Pratihar S, Ban D, Frischkorn S, Becker S, Griesinger C, and Lee D (2018) Simultaneous determination of fast and slow dynamics in molecules using extreme CPMG relaxation dispersion experiments. J. Biomol. NMR 70, 1–9. [DOI] [PubMed] [Google Scholar]
- (48).Kleckner IR, Gollnick P, and Foster MP (2012) Mechanisms of Allosteric Gene Regulation by NMR Quantification of Microsecond–Millisecond Protein Dynamics. J. Mol. Biol 415, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lisi GP, East KW, Batista VS, and Loria JP (2017) Altering the allosteric pathway in IGPS suppresses millisecond motions and catalytic activity. Proc. Natl. Acad. Sci. U. S. A 114, E3414–E3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Xiao Y, Lee T, Latham MP, Warner LR, Tanimoto A, Pardi A, and Ahn NG (2014) Phosphorylation releases constraints to domain motion in ERK2. Proc. Natl. Acad. Sci. U. S. A 111, 2506–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kitevski-LeBlanc JL, Yuwen T, Dyer PN, Rudolph J, Luger K, and Kay LE (2018) Investigating the Dynamics of Destabilized Nucleosomes Using Methyl-TROSY NMR. J. Am. Chem. Soc 140, 4774–4777. [DOI] [PubMed] [Google Scholar]
- (52).Hoofnagle AN, Resing KA, Goldsmith EJ, and Ahn NG (2001) Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc. Natl. Acad. Sci. U. S. A 98, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hsieh F-K, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, and Studitsky VM (2013) Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A 110, 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yuwen T, Vallurupalli P, and Kay LE (2016) Enhancing the Sensitivity of CPMG Relaxation Dispersion to Conformational Exchange Processes by Multiple-Quantum Spectroscopy. Angew. Chem., Int. Ed 55, 11490–11494. [DOI] [PubMed] [Google Scholar]
- (55).Korzhnev DM, Religa TL, Lundström P, Fersht AR, and Kay LE (2007) The folding pathway of an FF domain: characterization of an on-pathway intermediate state under folding conditions by (15)N, (13)C(alpha) and (13)C-methyl relaxation dispersion and (1)H/(2)H-exchange NMR spectroscopy. J. Mol. Biol 372, 497–512. [DOI] [PubMed] [Google Scholar]
- (56).Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, and Kay LE (2010) A transient and low-populated protein-folding intermediate at atomic resolution. Science 329, 1312–1316. [DOI] [PubMed] [Google Scholar]
- (57).Latham MP, and Kay LE (2014) A Similar In Vitro and In Cell Lysate Folding Intermediate for the FF Domain. J. Mol. Biol 426, 3214–3220. [DOI] [PubMed] [Google Scholar]
- (58).Ishima R, Louis JM, and Torchia DA (1999) Transverse 13 C Relaxation of CHD 2 Methyl Isotopmers To Detect Slow Conformational Changes of Protein Side Chains. J. Am. Chem. Soc 121, 11589–11590. [Google Scholar]
- (59).Weininger U, Blissing AT, Hennig J, Ahlner A, Liu Z, Vogel HJ, Akke M, and Lundström P (2013) Protein conformational exchange measured by 1H R1ρ relaxation dispersion of methyl groups. J. Biomol. NMR 57, 47–55. [DOI] [PubMed] [Google Scholar]
- (60).Orekhov VY, Korzhnev DM, and Kay LE (2004) Double- and Zero-Quantum NMR Relaxation Dispersion Experiments Sampling Millisecond Time Scale Dynamics in Proteins. J. Am. Chem. Soc 126, 1886–1891. [DOI] [PubMed] [Google Scholar]
- (61).Tugarinov V, Sprangers R, and Kay LE (2004) Line Narrowing in Methyl-TROSY Using Zero-Quantum 1 H- 13 C NMR Spectroscopy. J. Am. Chem. Soc 126, 4921–4925. [DOI] [PubMed] [Google Scholar]
- (62).Konrat R, and Sterk H (1993) Cross-correlation effects in the transverse relaxation of multiple-quantum transitions of heteronuclear spin systems. Chem. Phys. Lett 203, 75–80. [Google Scholar]
- (63).Norwood TJ, Tillett ML, and Lian LY (1999) Influence of cross-correlation between the chemical shift anisotropies of pairs of nuclei on multiple-quantum relaxation rates in macromolecules. Chem. Phys. Lett 300, 429–434. [Google Scholar]
- (64).Tugarinov V, and Kay LE (2004) 1 H, 13 C- 1 H, 1 H Dipolar Cross-correlated Spin Relaxation in Methyl Groups. J. Biomol. NMR 29, 369–376. [DOI] [PubMed] [Google Scholar]
- (65).Sun H, Kay LE, and Tugarinov V (2011) An Optimized Relaxation-Based Coherence Transfer NMR Experiment for the Measurement of Side-Chain Order in Methyl-Protonated, Highly Deuterated Proteins. J. Phys. Chem. B 115, 14878–14884. [DOI] [PubMed] [Google Scholar]
- (66).Leyme A, Marivin A, Casler J, Nguyen LT, and Garcia-Marcos M (2014) Different biochemical properties explain why two equivalent Gα subunit mutants cause unrelated diseases. J. Biol. Chem 289, 21818–21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Toyama Y, Osawa M, Yokogawa M, and Shimada I (2016) NMR Method for Characterizing Microsecond-to-Millisecond Chemical Exchanges Utilizing Differential Multiple-Quantum Relaxation in High Molecular Weight Proteins. J. Am. Chem. Soc 138, 2302–2311. [DOI] [PubMed] [Google Scholar]
- (68).Jarymowycz VA, and Stone MJ (2006) Fast Time Scale Dynamics of Protein Backbones: NMR Relaxation Methods, Applications, and Functional Consequences. Chem. Rev 106, 1624–1671. [DOI] [PubMed] [Google Scholar]
- (69).Kempf JG, and Loria JP (2003) Protein dynamics from solution NMR: theory and applications. Cell Biochem. Biophys 37, 187–211. [DOI] [PubMed] [Google Scholar]
- (70).Lipari G, and Szabo A (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc 104, 4546–4559. [Google Scholar]
- (71).Lipari G, and Szabo A (1982) Model-Free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 2. Analysis of Experimental Results. J. Am. Chem. Soc 104, 4559–4570. [Google Scholar]
- (72).Igumenova TI, Frederick KK, and Wand AJ (2006) Characterization of the Fast Dynamics of Protein Amino Acid Side Chains Using NMR Relaxation in Solution. Chem. Rev 106, 1672–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, and Gronenborn AM (1990) Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc 112, 4989–4991. [Google Scholar]
- (74).Muhandiram DR, Yamazaki T, Sykes BD, and Kay LE (1995) Measurement of 2H T1 and T1.rho. Relaxation Times in Uniformly 13C-Labeled and Fractionally 2H-Labeled Proteins in Solution. J. Am. Chem. Soc 117, 11536–11544. [Google Scholar]
- (75).Lee AL, and Wand AJ (2001) Microscopic origins of entropy, heat capacity and the glass transition in proteins. Nature 411, 501–504. [DOI] [PubMed] [Google Scholar]
- (76).Yang D, Mok Y-K, Forman-Kay JD, Farrow NA, and Kay LE (1997) Contributions to protein entropy and heat capacity from bond vector motions measured by NMR spin relaxation. J. Mol. Biol 272, 790–804. [DOI] [PubMed] [Google Scholar]
- (77).Akke M, Brueschweiler R, and Palmer AG III. (1993) NMR order parameters and free energy: an analytical approach and its application to cooperative Ca2+ binding by calbindin D9k. J. Am. Chem. Soc 115, 9832–9833. [Google Scholar]
- (78).Kay LE, and Prestegard JH (1987) Methyl group dynamics from relaxation of double quantum filtered NMR signals. Application to deoxycholate. J. Am. Chem. Soc 109, 3829–3835. [Google Scholar]
- (79).Tugarinov V, Sprangers R, and Kay LE (2007) Probing Side-Chain Dynamics in the Proteasome by Relaxation Violated Coherence Transfer NMR Spectroscopy. J. Am. Chem. Soc 129, 1743–1750. [DOI] [PubMed] [Google Scholar]
- (80).Boswell ZK, Rahman S, Canny MD, and Latham MP (2018) A dynamic allosteric pathway underlies Rad50 ABC ATPase function in DNA repair. Sci. Rep 8, 1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, and Tainer JA (2011) ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol 18, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Capdevila DA, Braymer JJ, Edmonds KA, Wu H, and Giedroc DP (2017) Entropy redistribution controls allostery in a metalloregulatory protein. Proc. Natl. Acad. Sci. U. S. A 114, 4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Clark LD, Dikiy I, Chapman K, Rödström KEJ, Aramini J, LeVine MV, Khelashvili G, Rasmussen SGF, Gardner KH, and Rosenbaum DM (2017) Ligand modulation of sidechain dynamics in a wild-type human GPCR. eLife 6, e28505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Clark L, Dikiy I, Rosenbaum DM, and Gardner KH (2018) On the use of Pichia pastoris for isotopic labeling of human GPCRs for NMR studies. J. Biomol. NMR 71, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Millet O, Muhandiram DR, Skrynnikov NR, and Kay LE (2002) Deuterium Spin Probes of Side-Chain Dynamics in Proteins. 1. Measurement of Five Relaxation Rates per Deuteron in 13 C-Labeled and Fractionally 2 H-Enriched Proteins in Solution. J. Am. Chem. Soc 124, 6439–6448. [DOI] [PubMed] [Google Scholar]
- (86).Yang D, and Kay LE (1996) Contributions to Conformational Entropy Arising from Bond Vector Fluctuations Measured from NMR-Derived Order Parameters: Application to Protein Folding. J. Mol. Biol 263, 369–382. [DOI] [PubMed] [Google Scholar]
- (87).Solomentsev G, Diehl C, and Akke M (2018) Conformational Entropy of FK506 Binding to FKBP12 Determined by Nuclear Magnetic Resonance Relaxation and Molecular Dynamics Simulations. Biochemistry 57, 1451–1461. [DOI] [PubMed] [Google Scholar]
- (88).Smith MJ, Marshall CB, Theillet F-X, Binolfi A, Selenko P, and Ikura M (2015) Real-time NMR monitoring of biological activities in complex physiological environments. Curr. Opin. Struct. Biol 32, 39–47. [DOI] [PubMed] [Google Scholar]
- (89).Kumar A, and Balbach J (2015) Real-time protein NMR spectroscopy and investigation of assisted protein folding. Biochim. Biophys. Acta, Gen. Subj 1850, 1965–1972. [DOI] [PubMed] [Google Scholar]
- (90).Buck J, Fürtig B, Noeske J, Wöhnert J, and Schwalbe H (2007) Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. U. S. A 104, 15699–15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Roche J, Dellarole M, Caro JA, Norberto DR, Garcia AE, Garcia-Moreno B, Roumestand C, and Royer CA (2013) Effect of Internal Cavities on Folding Rates and Routes Revealed by Real-Time Pressure-Jump NMR Spectroscopy. J. Am. Chem. Soc 135, 14610–14618. [DOI] [PubMed] [Google Scholar]
- (92).Marshall CB, Ho J, Buerger C, Plevin MJ, Li G-YY, Li Z, Ikura M, and Stambolic V (2009) Characterization of the intrinsic and TSC2-GAP - Regulated GTPase activity of rheb by real-time nmr. Sci. Signaling 2, ra3. [DOI] [PubMed] [Google Scholar]
- (93).Charlier C, Alderson TR, Courtney JM, Ying J, Anfinrud P, and Bax A (2018) Study of protein folding under native conditions by rapidly switching the hydrostatic pressure inside an NMR sample cell. Proc. Natl. Acad. Sci. U. S. A 115, E4169–E4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Schanda P, and Brutscher B (2005) Very Fast Two-Dimensional NMR Spectroscopy for Real-Time Investigation of Dynamic Events in Proteins on the Time Scale of Seconds. J. Am. Chem. Soc 127, 8014–8015. [DOI] [PubMed] [Google Scholar]
- (95).Rennella E, Cutuil T, Schanda P, Ayala I, Forge V, and Brutscher B (2012) Real-Time NMR Characterization of Structure and Dynamics in a Transiently Populated Protein Folding Intermediate. J. Am. Chem. Soc 134, 8066–8069. [DOI] [PubMed] [Google Scholar]
- (96).Macek P, Kerfah R, Erba EB, Crublet E, Moriscot C, Schoehn G, Amero C, and Boisbouvier J (2017) Unraveling self-assembly pathways of the 468-kDa proteolytic machine TET2. Sci. Adv 3, e1601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Stejskal EO, and Tanner JE (1965) Spin diffusion measurements: Spin echoes in the presence of a time-dependent field gradient. J. Chem. Phys 42, 288–292. [Google Scholar]
- (98).Johnson CS Jr. (1999) Diffusion ordered nuclear magnetic resonance spectroscopy: principles and applications. Prog. Nucl. Magn. Reson. Spectrosc 34, 203–256. [Google Scholar]
- (99).Huang R, Brady JP, Sekhar A, Yuwen T, and Kay LE (2017) An enhanced sensitivity methyl 1H triple-quantum pulse scheme for measuring diffusion constants of macromolecules. J. Biomol. NMR 68, 249–255. [DOI] [PubMed] [Google Scholar]
- (100).Kramers HA (1940) Brownian motion in a field of force and the diffusion model of chemical reactions. Physica 7, 284–304. [Google Scholar]
- (101).Kitevski-LeBlanc JL, Fradet-Turcotte A, Kukic P, Wilson MD, Portella G, Yuwen T, Panier S, Duan S, Canny MD, van Ingen H, Arrowsmith CH, Rubinstein JL, Vendruscolo M, Durocher D, and Kay LE (2017) The RNF168 paralog RNF169 defines a new class of ubiquitylated-histone reader involved in the response to DNA damage. eLife 6, e23872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Huang R, Ripstein ZA, Augustyniak R, Lazniewski M, Ginalski K, Kay LE, and Rubinstein JL (2016) Unfolding the mechanism of the AAA+ unfoldase VAT by a combined cryo-EM, solution NMR study. Proc. Natl. Acad. Sci. U. S. A 113, E4190–E4199. [DOI] [PMC free article] [PubMed] [Google Scholar]