SUMMARY
The presence of DNA in the cytosol of mammalian cells is an unusual event that is often associated with genotoxic stress or viral infection. The enzyme cGAS is a sensor of cytosolic DNA that induces interferon and inflammatory responses that can be protective or pathologic, depending on the context. Along with other cytosolic innate immune receptors, cGAS is thought to diffuse throughout the cytosol in search of its DNA ligand. Herein, we report that cGAS is not a cytosolic protein but rather localizes to the plasma membrane via the actions of an N-terminal phosphoinositide-binding domain. This domain interacts selectively with PI(4,5)P2, and cGAS mutants defective for lipid binding are mislocalized to the cytosolic and nuclear compartments. Mislocalized cGAS induces potent interferon responses to genotoxic stress, but weaker responses to viral infection. These data establish the subcellular positioning of a cytosolic innate immune receptor as a mechanism that governs self-nonself discrimination.
Graphical abstract
In Brief
The innate immune receptor cGAS interacts with PI(4,5)P2 in order to localize to the plasma membrane, which is critical to prevent aberrant interferon responses to self-DNA under conditions of genotoxic stress, as well as to properly sense viral infections.
INTRODUCTION
The ability to discriminate between self and nonself is critical for recognition and response to pathogens. In mammals, numerous proteins serve as sensors of foreign motifs, or pathogen-associated molecular patterns (PAMPs) (Takeuchi and Akira, 2010). Some PAMPs, such as bacterial lipopolysaccharide, are exclusively nonself, in which no cognate molecule exists in the host organism (Takeuchi and Akira, 2010). However, other PAMPs, such as viral nucleic acids, bear strong similarities to molecules found in the host cell. In the case of RNA, structural differences between host and viral RNA allow for discrimination between self and nonself (Goubau et al., 2014; Hornung et al., 2006; Kato et al., 2006). Yet with DNA, the distinction between host-derived and pathogen-derived molecules is less clear. Despite this, several DNA sensors are essential for clearance of infections, including Toll-like Receptor 9 (TLR9), the AIM2-like receptors (ALRs), and cyclic GMP-AMP Synthase (cGAS) (Bhat and Fitzgerald, 2014).
Of these receptors, cGAS has emerged as a pattern recognition receptor (PRR) that is implicated in the detection of self-and nonself-DNA. cGAS surveys the intracellular space for DNA and generates interferon (IFN) and inflammatory responses upon detection (Sun et al., 2013). cGAS recognizes double-stranded, B-form DNA independent of its sequence through contact with the sugar-phosphate backbone (Kranzusch et al., 2013). Upon DNA binding, cGAS dimerizes, assembles into large liquid droplets, and produces the secondary messenger 2′3′-cyclic GMP-AMP (cGAMP) (Ablasser et al., 2013; Du and Chen, 2018; Zhang et al., 2013). This molecule binds to the endoplasmic reticulum (ER) resident protein STING, leading to its activation and the subsequent expression of IFNs and other inflammatory mediators (Ishikawa et al., 2009; Sun et al., 2013). Because cGAS does not recognize specific DNA sequences, it is essential for the detection and control of many pathogenic infections (Chen et al., 2016). Notably, cGAS also regulates immune responses in the absence of infection through the detection of endogenous (self) DNA. For instance, cGAS promotes IFN responses to genotoxic stress induced by DNA damaging agents, micronuclei formation, and cellular senescence (Dou et al., 2017; Glück et al., 2017; Harding et al., 2017; Härtlova et al., 2017; Mackenzie et al., 2017; Pépin et al., 2017; Yang et al., 2017). cGAS is therefore not only a sensor of pathogens but also a sensor of cellular stress and genomic integrity.
While the ability of cGAS to detect pathogen DNA promotes beneficial responses during infection, its ability to detect self-DNA is associated with immunopathology. Indeed, the cGAS-STING signaling pathway is a driver of pathologies associated with autoinflammatory diseases (Gao et al., 2015; Gray et al., 2015). Genetic analysis of human patients suffering from various interferonopathies revealed loss of function mutations in cytosolic nucleases that hydrolyze DNA or RNA-DNA hybrids, both of which are cGAS ligands (Bartsch et al., 2017; Crow et al., 2015; Mankan et al., 2014). These observations support the view that the maintenance of low cytosolic DNA concentrations is critical to prevent inappropriate cGAS activation. Whether additional mechanisms exist to prevent inappropriate activation of cGAS remains unknown.
Though some have noted nuclear localization (Orzalli et al., 2015; Yang et al., 2017), the subcellular positioning of cGAS at steady state is loosely defined as within the cytosol, where it encounters its ligands through diffusion (Sun et al., 2013). Since cGAS lacks a transmembrane domain, the possibility of its specific positioning within the cytoplasm is unexplored. However, work over the last decade identified several innate immune regulators that were first considered cytosolic but are now recognized to associate with membranes through electrostatic interactions. These proteins include the mammalian proteins TIR domain containing adaptor protein (TIRAP) and TRIF-related adaptor molecule (TRAM), and the Drosophila protein dMyD88, which regulate TLR and Toll pathway signaling, respectively (Kagan and Medzhitov, 2006; Kagan et al., 2008; Marek and Kagan, 2012). Each of these proteins contain a phosphoinositide phosphate (PIP)-binding domain, enabling their positioning at the cell surface. Mutant alleles lacking these domains are mislocalized and are consequently defective for TLR or Toll signaling. To date, the use of PIP binding proteins to regulate receptor-proximal innate immune responses is a feature unique to the TLR family; whether this aspect of regulation extends to other pathways is unknown.
In this study, we report that cGAS is not a cytosolic protein but is rather a peripheral membrane protein that primarily resides on the plasma membrane of human and mouse macrophages. Membrane association is mediated by an N-terminal phosphoinositide-binding domain that interacts selectively with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), a plasma membrane resident lipid. A mutant allele of cGAS lacking this domain is mislocalized to the cytosol and nucleus and is hyper-responsive to genotoxic stress. However, mislocalized cGAS does not generate a similarly potent response to viral infection. These findings identify a strategy of self-nonself discrimination, whereby positioning of cGAS at the plasma membrane prevents the recognition of self-DNA. These findings also establish subcellular positioning through PIP binding as a common feature of the TLR and cGAS signaling pathways to optimize signaling potential.
RESULTS
cGAS Is Located at the Plasma Membrane of Human and Mouse Phagocytes
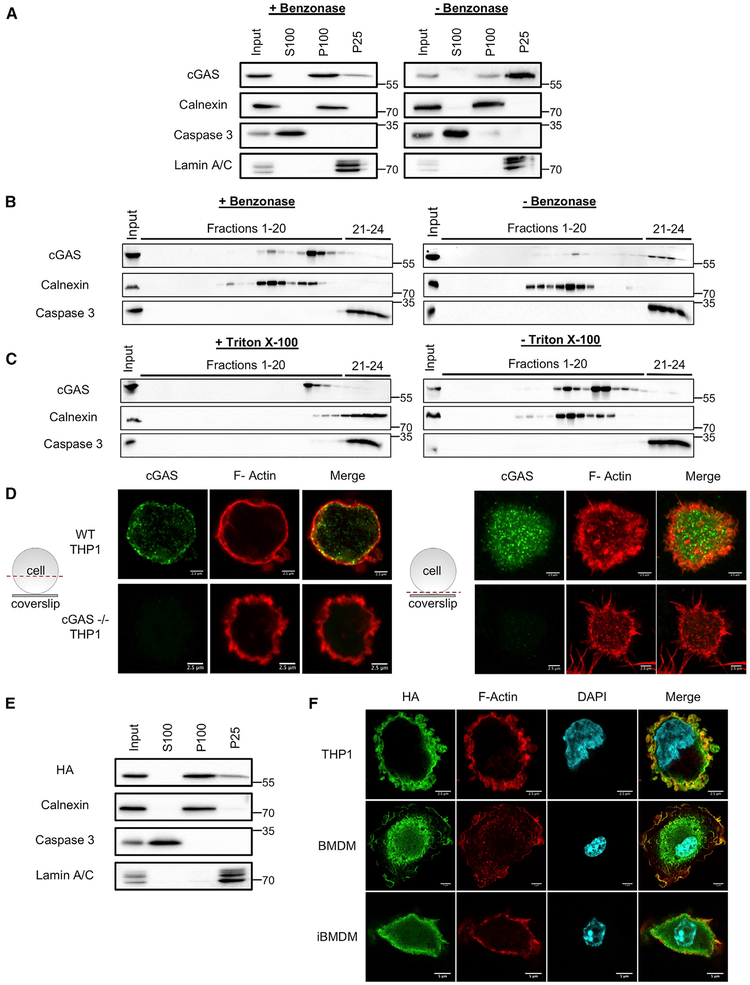
We sought to identify the site of cGAS residence in unstimulated macrophages, a cell type critical for immune responses to infection and tissue injury. We examined the distribution of cGAS in human THP1 monocytes through subcellular fractionation by differential ultracentrifugation. Fractionation fidelity was verified by detection of the cytosolic protein caspase-3 in the soluble fraction (S100), the ER-localized transmembrane protein calnexin in the insoluble, membrane fraction (P100), and Lamins A and C in the nuclear fraction (P25). We utilized a cGAS antibody, validated on cGAS knockout (KO) cells, to ensure accuracy (Figures S1A and S1B). The nuclease benzonase was included in all buffers to minimize post-lysis DNA binding by cGAS. This possibility was important to consider, as cGAS forms liquid droplets upon DNA binding that co-sediment with nuclei (Du and Chen, 2018). The bulk of endogenous cGAS was detected in the membranous P100 fraction along with a small amount present in the nuclear P25 fraction (Figure 1A). No cGAS was detected in the S100 cytosolic fractions (Figure 1A). These data suggest that cGAS is not a cytosolic protein, but rather associates with membranes and nuclei.
Figure 1. cGAS Associates with the Plasma Membrane.
(A) Subcellular fractionation of THP1 cells in the presence or absence of benzonase. Western blot analysis was used to probe the cytosolic fraction (S100), membrane fraction (P100), and nuclear (P25) fractions for the indicated proteins.
(B) Membrane floatation assays of THP1 post-nuclear lysates on a 10%–45% Optiprep gradient in the presence or absence of benzonase. Fractions 1–24 were taken from the top to the bottom of the gradient, and western blot analysis was used to probe these fractions for the indicated proteins.
(C) Membrane floatation assays of THP1 post-nuclear lysates on a 10%–45% Optiprep gradient in the presence of benzonase in the presence or absence of 1% Triton X-100.
(D) Confocal micrographs cGAS in WT and cGAS−/− THP1 cells treated with 1,000 U/mL recombinant IFN-β1 for 4 h. In diagrams to the left of micrographs, red dashed lines indicate the plane in view (not to scale).
(E) Subcellular fractionation of THP1-expressing cGAS-HA cells in the presence of benzonase.
(F) Confocal micrographs of various cell lines expressing cGAS-HA. Experiments shown are representative of n = 3 biological replicates.
See also Figure S1.
Whereas the presence of cGAS in the nucleus is consistent with established literature (Orzalli et al., 2015; Yang et al., 2017), its predominant distribution in the membrane fraction warranted further investigation. Membrane-bound organelles should not only sediment after ultracentrifugation, but also migrate from dense to light fractions of a bottom-loaded density gradient, known as a membrane flotation assay. We therefore performed membrane flotation assays on Optiprep gradients bottom-loaded with post-nuclear supernatants of THP1 cells. As expected, the membrane protein calnexin floated from the site of gradient loading (fractions 21–24) into several contiguous fractions throughout the gradient (Figure 1B). Similar observations were made with cGAS, with nearly the entire population of this protein migrating into lighter membrane fractions (Figure 1B). These complementary procedures indicate that cGAS is present on a membrane-bound organelle.
If we eliminated benzonase from our buffers, cGAS fractionation patterns were distinct from those described above. In sedimentation assays, the absence of benzonase resulted in a greater amount of cGAS fractionating with nuclei (Figure 1A). Based on the knowledge that cGAS forms liquid droplets that approach the size of nuclei upon DNA binding, it is reasonable to suggest that post-lysis DNA binding causes cGAS to appear as a predominantly nuclear protein in biochemical assays. Consistent with this idea, elimination of benzonase from our flotation-based assays resulted in virtually all cGAS remaining in the heaviest fractions 21–24, where the gradient was loaded (Figure 1B). These findings support the conclusion that in the absence of DNA exposure, cGAS is predominantly a membrane protein and highlight the importance of considering post-lysis DNA binding in any assay for cGAS function.
To complete our biochemical analysis, we determined whether the flotation of cGAS into light membrane fractions was sensitive to the detergent Triton X-100. Whereas treatment of cell lysates with Triton X-100 prevented calnexin flotation, cGAS retained the ability to float into low-density fractions (Figure 1C). cGAS is therefore present on a membrane insensitive to solvation by Triton X-100. Triton X-100-resistant membranes include lipid rafts, which are subdomains of the plasma membrane (Brown and Rose, 1992; Lingwood and Simons, 2010). These subdomains are rich in several lipids, including PI(4,5) P2, which will be discussed later.
Confocal microscopy was used to complement these analyses and identify the precise subcellular residency of cGAS. Staining of wild-type (WT) or cGAS KO THP1 monocytes was performed to identify conditions suitable for endogenous protein detection, using two different cGAS antibodies. Neither antibody generated a staining pattern that is present in resting WT cells but absent in KO cells (Figures S1C and S1D). However, a short treatment (4 h) of cells with IFN-β increased the abundance of cGAS and permitted detection of the endogenous protein. The staining observed was concentrated at the plasma membrane (Figure 1D). Cross-section confocal slices revealed a circumferential staining pattern that coincided with filamentous actin (F-actin), whereas confocal slices of the ventral side of the cell (the site of attachment to the coverslip) revealed cGAS concentration in multiple foci scattered throughout the plasma membrane (Figure 1D). Quantification revealed that over 90% of cells displayed cGAS staining at the site of coverslip contact (Figure S1E). cGAS KO cells completely lacked staining with this antibody (Figure 1D). We reasoned that if the cGAS antibody used (from Cell Signaling Technology [CST]) truly detected cGAS at the plasma membrane, then cells expressing a C-terminally hemagglutinin (HA)-tagged cGAS (cGAS-HA) allele should display extensive colocalization when stained with antibodies specific for cGAS and HA. This possibility was tested by generating THP1 cells that stably express cGAS-HA (Figures S1F and S1G). Consistent with our studies of endogenous cGAS, cGASHA fractionated primarily with membranes in the presence of benzonase and localized to the plasma membrane (Figures 1E and 1F).
Also consistent with our microscopic studies of endogenous cGAS, the CST and HA antibodies displayed extensive plasma membrane co-distribution (Figure S1G). In contrast, the cGAS antibody from Sigma was unable to detect endogenous cGAS or cGAS-HA (Figure S1G). The abundance of cGAS-HA in our stable lines was modestly greater than the endogenous protein (Figure S1F), but these cells exhibited no evidence of cGAS activation, as basal IFN-β expression was comparable to what was observed in WT THP1 cells, as discussed below. These data, combined with our biochemical analysis, support the conclusion that, in its inactive state, cGAS is a membrane protein that resides primarily at the surface of THP1 cells.
To determine whether our findings extended to other cell populations, we examined the staining of cGAS-HA in primary murine bone marrow-derived macrophages (BMDMs), where the transgene was expressed at levels that were lower than the endogenous protein. Consistent with this statement, total cGAS abundance in WT BMDMs expressing cGAS-HA was no greater than cGAS abundance in cells containing no transgene (Figure S1F). Staining for HA in these cells revealed a clear localization of cGAS to the plasma membrane, with a concentration in actin-rich membrane ruffles (Figure 1F). Additionally, immortal BMDMs (iBMDMs) stably expressing cGAS-HA displayed similar staining patterns, with prominent cGAS staining detected at the cell surface in actin-rich ruffles (Figure 1F).
Several studies have examined the localization of cGAS in non-phagocytes and have not observed plasma membrane localization (Liang et al., 2014; Orzalli et al., 2015; Sun et al., 2013; Zhang et al., 2014). To standardize our studies with those of others, we introduced the same cGAS-HA cDNA into THP1 cells, iBMDMs, HeLa cells, and L929 cells, the latter two of which were examined previously (Liang et al., 2014; Sun et al., 2013; Zhang et al., 2014). Whereas cGAS was almost singularly located at the plasma membrane of THP1 cells and iBMDMs, a heterogeneous distribution of cGAS was observed in HeLa and L929 cells (Figures S1H and S1I). While cGAS localization was predominantly nuclear in L929 cells and highly varied in HeLa cells, all cell types displayed some proportion of plasma membrane localized cGAS (Figures S1H and S1I). Therefore, cell-type-specific factors influence cGAS localization.
Others have observed cGAS to be concentrated in cytosolic spots following DNA transfection (Sun et al., 2013). Similarly, we found that within 30 min of DNA transfection, cGAS was no longer concentrated at the cell surface but was rather detected in various cytoplasmic puncta (Figure S1J). These puncta are thought to be sites of cGAS-DNA interaction, in which cGAS oligomerizes and synthesizes cGAMP (Du and Chen, 2018). Collectively, these findings indicate that, within resting phagocytic cells, cGAS primarily resides on the plasma membrane.
The N-Terminal Domain of cGAS Is Necessary and Sufficient for cGAS Localization to the Plasma Membrane
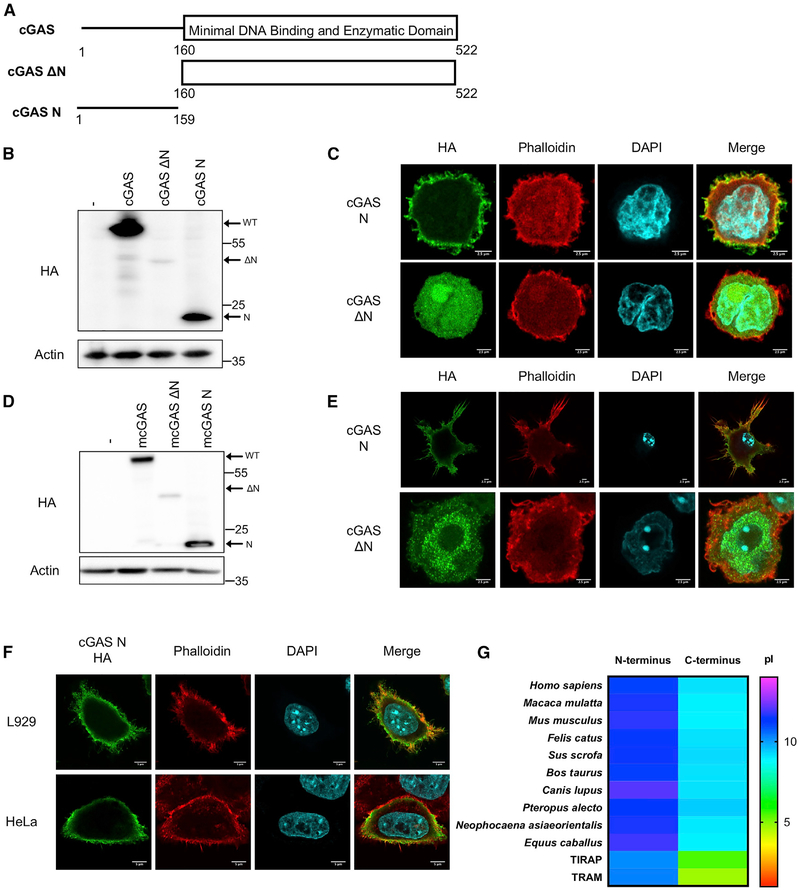
Human cGAS is a 522-amino-acid protein with an unstructured N terminus of 159 amino acids (Kranzusch et al., 2013). The structurally defined C terminus (residues 160–522) is sufficient to recognize DNA and produce cGAMP in vitro (Civril et al., 2013; Kranzusch et al., 2013) and induce IFN-β upon overexpression within cells (Sun et al., 2013). The N terminus of cGAS is poorly characterized but contributes to DNA binding and to efficient DNA-induced assembly of lipid droplets (Du and Chen, 2018; Sun et al., 2013). To identify regions of cGAS that mediate plasma membrane localization, deletion analysis was performed. We separated cGAS into its N-terminal domain (residues 1–159 human; residues 1–147 murine) and its C-terminal domain (residues 160–522 human; residues 148–507 murine) (Figure 2A). The human alleles were stably expressed as C-terminally HA-tagged proteins in THP1 cells. Western analysis verified production of each cGAS protein (Figure 2B). The allele lacking its N terminus (cGASΔN) was expressed at lower levels than the WT protein or N terminus alone (cGAS N) (Figure 2B).
Figure 2. The N Terminus of cGAS Is Necessary and Sufficient for Plasma Membrane Association.
(A) Schematic of human cGAS domain architecture and truncation mutants.
(B) Western blot analysis of mutant cGAS expression in THP1 cells.
(C) Confocal micrograph of THP1 cells expressing human cGAS N or cGASΔN.
(D) Western blot analysis of murine mutant cGAS expression in iBMDMs.
(E) Confocal micrographs of iBMDMs expressing murine cGAS N or cGASΔN.
(F) Confocal micrographs of L929 and HeLa cells expressing cGAS N.
(G) Heatmap showing the relative pI of the N- and C-terminal domains of cGAS from various species (n = 10) alongside innate immune adaptors TIR-domain-containing adaptor protein (TIRAP) and TRIF-related adaptor molecule (TRAM) with N-terminal PIP-binding domains. Experiments shown are representative of n = 3 biological replicates.
See also Figure S2.
cGAS N phenocopied the behavior of the WT protein, localizing to the cell surface (Figure 2C). In contrast, cGASΔN was not located at the plasma membrane but was rather found throughout the cytoplasm and nucleus (Figure 2C). Similar to human cGAS, murine cGASΔN was expressed at lower levels than WT cGAS and cGAS N (Figure 2D). Furthermore, murine cGAS N localized to the plasma membrane, while cGASΔN did not associate with the plasma membrane (Figure 2E). Quantification revealed that all cells expressing the cGAS N terminus displayed plasma membrane localization, while none of the cells expressing cGASΔN showed such localization (Figure S2A). Pseudo-3D renderings of WT cGAS and these mutants further illustrated the conclusions obtained from confocal slices, as WT and cGAS N proteins were concentrated at the plasma membrane and absent from the interior of the cell (Figure S2B). cGASΔN, in contrast, was concentrated in the cytosol and nucleus (Figure S2B).
As described above, cGAS localization varies by cell type. However, this heterogeneity of cGAS localization was not observed when we examined the distribution of the N-terminal domain. This domain was uniformly localized at the plasma membrane of all cell types examined (Figure 2F). These findings indicate the N terminus of cGAS is necessary and sufficient for positioning at the cell surface, acting as a bona fide localization domain.
Alignment of the domains within cGAS from ten vertebrate animals revealed strong conservation of the catalytic domain, and while all species examined contained an N-terminal domain, this region of the protein was less conserved. Despite this weak degree of conservation, the N-termini of all species examined were highly charged, as compared to the catalytic domain, with an isoelectric point (pI) that was at least 2 pH units higher than the catalytic domain (Figure 2G). Thus, while the specific amino acids that comprise the cGAS N terminus are not well conserved, the pI of this localization domain is conserved.
The cGAS N Terminus Binds PI(4,5)P2 to Mediate Plasma Membrane Localization
The pI of the cGAS N terminus was comparable to that of the localization domains from TIRAP and TRAM, with a pI of 11.07 in the human cGAS N terminus (Figure 2G). The high net positive charge of these domains mediates subcellular positioning through electrostatic interactions with negatively charged PIPs and is a common feature of PIP-binding localization domains (Balla, 2013). Considering the features of the cGAS N terminus, we hypothesized that cGAS localizes to the plasma membrane through interactions with a PIP.
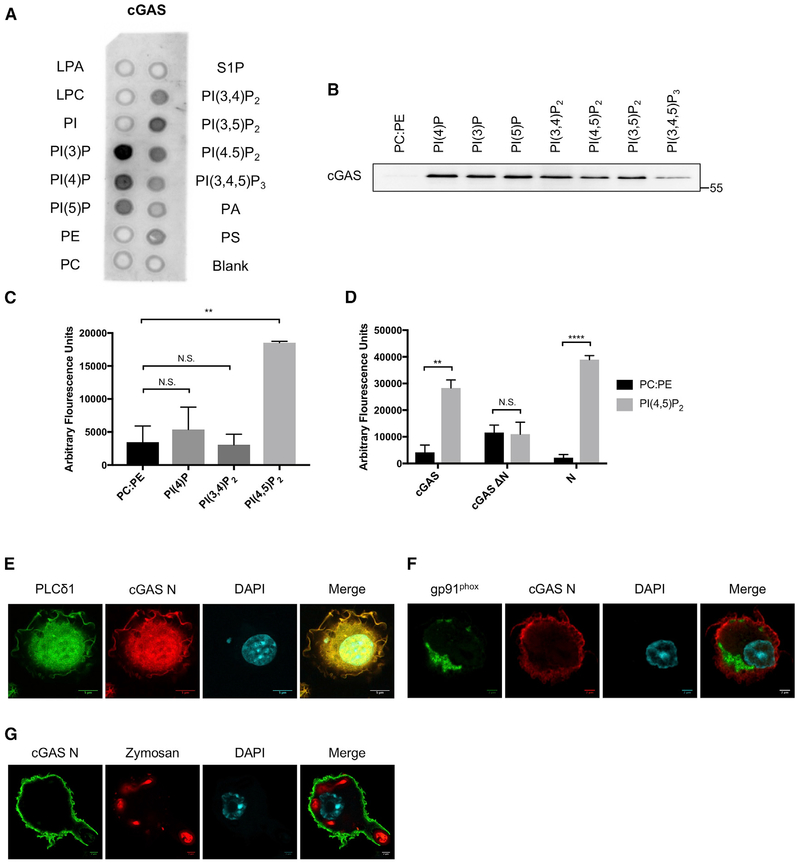
We examined the ability of recombinant cGAS to bind a panel of lipids immobilized on hydrophobic membranes (PIP strips). Far western analysis revealed interactions between cGAS and several PIPs, but not the unphosphorylated lipid phosphatidylinositol (Figure 3A). No interactions were observed between cGAS and other membrane lipids (Figure 3A). To corroborate these findings, we examined the ability of recombinant cGAS to bind PIPs within lipid bilayers, the physiological context for protein-PIP interactions. We constructed liposomes consisting of phosphatidylcholine (PC) and phosphatidylethanolamine (PE), abundant lipids present naturally in the plasma membrane, and select PIPs. Recombinant cGAS interacted with liposomes containing 18% of various PIPs, but not with liposomes lacking PIPs (Figure 3B). These experiments indicate that cGAS promiscuously interacts with PIPs but does not associate with other membrane lipids.
Figure 3. cGAS Binds PI(4,5)P2 via Its N Terminus.
(A) PIP strip analysis of recombinant cGAS. Recombinant cGAS was incubated with a membrane spotted with the indicated lipids and analyzed by far western.
(B) Cosedimentation assay for cGAS interactions with liposomes containing 18% of the specified PIPs on a 3:1 PC:PE backbone. Recombinant cGAS was incubated with liposomes, after which the liposomes were isolated by ultracentrifugation and probed by western blot for cGAS association.
(C) Fluorescent liposome pull-down assay. 6xHis-tagged cGAS was incubated with fluorescent liposomes containing the specified PIPs. cGAS was isolated by nickel affinity resin, and cGAS-lipid interaction was measured by the amount of fluorescence pulled down with the resin..
(D) Same as (C), but 6x-His-tagged cGAS, cGASΔN, and cGAS N were probed for binding to PI(4,5)P2.
(E) Confocal micrographs of electroporated primary BMDMs expressing cGAS N-HA with PLCδ1-PH-YFP.
(F) Confocal micrographs of electroporated primary BMDMs expressing gp91phox-PX-YFP.
(G) Confocal micrograph of an iBMDM expressing cGAS N phagocytosing a fluorescent zymosan particle.
Experiments shown are representative of or averages of n = 3 biological replicates. Data are represented as a mean ± SEM, and statistical analysis was performed using a Student’s t test with asterisk coding as follows: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Quantitative analysis was performed using fluorescently labeled liposomes to determine whether cGAS displays preference for individual PIPs. For these studies, we reduced the concentration of PIPs present in each liposome to a physiological 2%. Given the localization of cGAS, we focused on PIPs found at the plasma membrane at steady state: PI(4)P and PI(4,5)P2 (Balla, 2013), and another di-phosphorylated PIP found only on the plasma membrane in an inducible manner: PI(3,4)P2 (Hawkins and Stephens, 2016). This quantitative analysis demonstrated that cGAS preferentially binds PI(4,5)P2 (Figure 3C).
Using the deletion mutants described above, we determined which domain within cGAS interacts with PI(4,5)P2. We found that the N-terminal localization domain phenocopied WT cGAS, as both proteins interacted with liposomes containing PI(4,5)P2, but not with liposomes containing only PC:PE (Figure 3D). In contrast, cGASΔN interacted weakly with PC:PE liposomes and displayed no preference for PI(4,5)P2 (Figure 3D). The cGAS N terminus also displayed preferential co-localization with PI(4,5)P2 within cells. We observed extensive colocalization in primary BMDMs expressing the cGAS N terminus and the PI(4,5)P2-specific PH domain of PLCδ1 (Figure 3E) (Marek and Kagan, 2012). These experimental conditions resulted in partial mislocalization of both proteins as a result of competition for PI(4,5)P2, which is observed when two PI(4,5)P2-binding proteins are co-expressed (Jost et al., 1998). In contrast, cGAS did not colocalize with the PI(3)P-binding phox (PX) domain from gp91phox (Marek and Kagan, 2012) (Figure 3F). Furthermore, the cGAS N terminus displays a canonical behavior of PI(4,5)P2 distribution on forming phagosomes (Botelho, et al. 2000). cGAS was detected on phagocytic protrusions, but was absent from the base of the phagocytic cup (Figure 3G). These independent assays demonstrate that interactions with PI(4,5)P2 direct cGAS to the plasma membrane.
Plasma Membrane Localization of cGAS Is Important to Prevent Recognition of Self-DNA
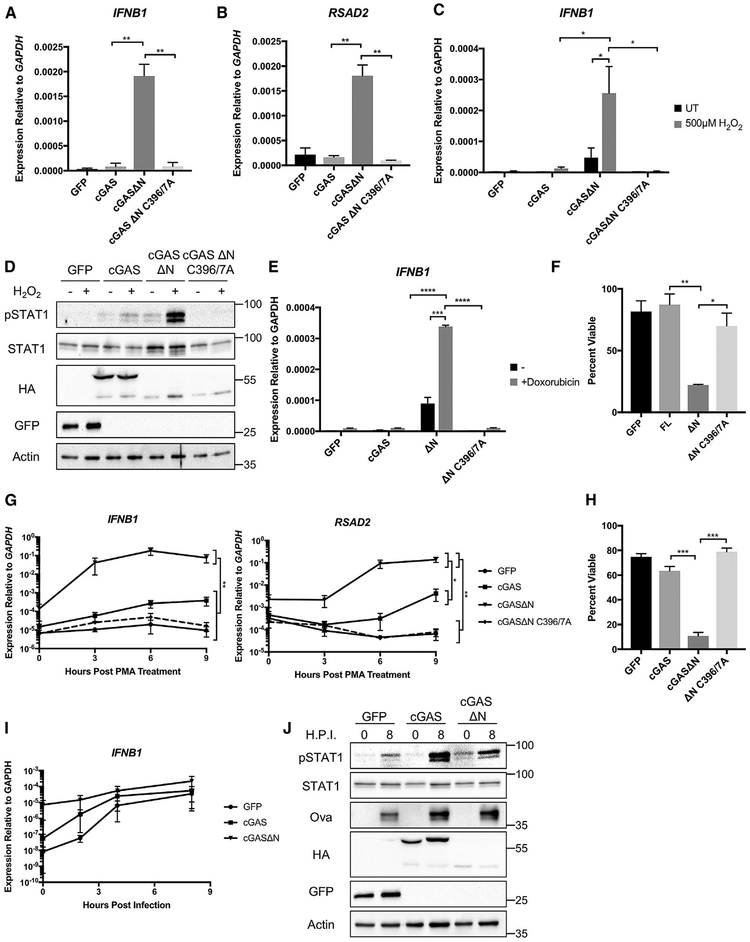
cGAS has dual functions in innate immunity; it must minimize detection of self-DNA while simultaneously being capable of pathogenic DNA detection. To determine whether either of these tasks are influenced by cGAS membrane localization, functional analysis was performed. For these studies, we aimed to utilize a cellular system that closely matched physiological contexts. Tonic signaling via the cGAS pathway sets the expression level of IFNs and IFN-stimulated genes (ISGs), and cGAS-deficient cells express low levels of these genes (Schoggins et al., 2014). Expression of WT cGAS in THP1 cells did not influence basal IFN and ISG levels (Figures 4A and 4B), allowing us to perform functional studies in this genetic background. In addition to WT cGAS, we engineered THP1 cell lines stably expressing the N-terminal localization domain (cGAS N) or the mislocalized cGASΔN. Additional THP1 lines were generated that expressed GFP, a mutant cGAS allele defective for DNA-binding (C396/397A) (Civril et al., 2013) or a cGASΔN allele defective for DNA binding (C396/397A). Western analysis verified the presence of each protein, with cGASΔN produced at lower levels than other proteins (Figure S3A). The expression of WT cGAS did not influence the basal expression of IFNβ1 or the ISG RSAD2 (Figures 4A and 4B). Thus, the transgenic cells generated by this approach behave similarly to WT cells in their resting state and are suitable for functional analysis.
Figure 4. Loss of the cGAS N Terminus Leads to Heightened Basal IFN Signaling and Increased Responses to Genotoxic Stress.
(A) qPCR analysis of IFNβ1 expression in THP1 cell lines at steady state.
(B) qPCR analysis of RSAD2 expression in THP1 cell lines at steady state.
(C) qPCR analysis of IFNβ1 expression following a 6-h treatment with 500 μM H2O2 in the indicated THP1 cell lines.
(D) Western blot analysis for the indicated proteins before and after 6-h treatment with 500 μM H2O2.
(E) qPCR analysis of IFNβ1 expression following a 14-h treatment with 500 nM Doxorubicin in the indicated THP1 cell lines.
(F) Viability of THP1 cell lines following a 14-h treatment with 500 nM Doxorubicin, shown as a percentage of untreated controls, as measured by intracellular ATP content.
(G) qPCR analysis of IFNβ1 and RSAD2 expression at the indicated time points following treatment with 50 ng/mL PMA in the indicated THP1 cell lines.
(H) Viability of THP1 cell lines following overnight treatment with 50 ng/mL PMA, as described in (F).
(I) qPCR analysis of IFNβ1 expression at the indicated time points following infection with MVA-Ova (MOI 3) in the indicated THP1 cell lines.
(J) Western blot analysis of the indicated proteins before and after infection with MVA-Ova (MOI 3) in the indicated cell lines.
H.P.I., hours post-infection. Experiments shown are representative of or averages of n = 3 biological replicates. Data are shown as a mean ± SEM. Statistical analysis comparing two data points was performed using a Student’s t test, and statistical analysis of time-course experiment trends in (G) and (I) were performed using two-way ANOVA with asterisk coding as follows: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
See also Figure S3.
Although the cGASΔN allele was expressed at lower levels than its WT counterpart (Figure S3A), cells producing this protein displayed high expression of IFNβ1 and RSAD2 (Figures 4A and 4B). Cells producing the DNA-binding-deficient cGASΔN (C396/397A) allele did not display a similar phenotype (Figures 4A and 4B), indicating that high basal IFN expression was due to detection of self-DNA. These data suggest that localization of cGAS is important to avoid detection of self-DNA.
If this prediction was correct, then cells expressing cGASΔN should be more sensitive to genotoxic stress. Indeed, the cGAS pathway is activated in several experimental settings of chronic DNA damage (Dou et al., 2017; Glück et al., 2017; Harding et al., 2017; Härtlova et al., 2017; Mackenzie et al., 2017; Pépin et al., 2017; Yang et al., 2017). This idea was tested by treating cells with the DNA damaging agent hydrogen peroxide (H2O2). Treatment with H2O2 for 6 h induced minimal IFNβ1 expression in most of the transgenic THP1 lines examined (Figure 4C). In contrast, cells expressing cGASΔN induced substantial increases in IFNβ1 expression in response to H2O2 (Figure 4C). This H2O2-induced IFN response was not observed in cells expressing the DNA-binding-deficient cGASΔN C396/397A (Figure 4C). Western analysis demonstrated that cells expressing cGASΔN responded to H2O2 by inducing the phosphorylation of STAT1 (Figure 4D), a hallmark of an IFN response. No other transgene induced potent STAT1 phosphorylation in response to H2O2, suggesting that loss of localization leads to heightened sensitivity to genotoxic stress.
We performed similar analyses with two additional DNA-damage inducing agents: doxorubicin and phorbol myristate acetate (PMA) (Emerit and Cerutti, 1981; Tewey et al., 1984). Treatment with doxorubicin induced IFNβ1 expression in THP1 cells expressing cGASΔN but not cGASΔN C396/7A or WT cGAS (Figure 4E). Notably, this upregulation was accompanied by the death of cGASΔN-expressing cells. At 14 h post-treatment with doxorubicin, only 22.2% of cGASΔN-expressing cells remained viable, as compared to untreated controls (Figure 4F). This phenotype was not observed in cells expressing WT cGAS or cGASΔN C396/7A (Figure 4F).
Treatments with PMA also demonstrated a stark contrast between cells expressing WT cGAS and cGASΔN. PMA stimulations for as little as 3 h induced a 2–3 log increase in IFNβ1 expression in cells expressing cGASΔN (Figure 4G). Expression of the ISG RSAD2 correlated with IFN-β expression, although with delayed kinetics (Figure 4G). This increase in IFN and ISG expression was not observed in cells expressing cGASΔN C396/397A (Figure 4G), an observation that further implicates DNA detection as the cause of this response. Cells expressing WT cGAS also induced an IFN and ISG response to PMA treatment, but this response was not evident for several hours and the extent of IFN and ISG induction was lower than observed in cells expressing cGASΔN (Figure 4G).
As observed with doxorubicin treatment, overnight stimulation demonstrated specific toxicity of PMA to cells expressing cGASΔN (Figures 4H and S3B). Whereas all other transgenic cell populations attached to the tissue culture plate upon PMA treatment, cells expressing cGASΔN remained unattached and only 10.82% of cells remained viable (Figures 4H and S3B). Cells expressing WT cGAS or cGASΔN C396/397A did not display these phenotypes (Figures 4H and S3B). These data support the idea that mislocalized cGAS cannot avoid detection of self-DNA and triggers unusually high IFN responses that correlate with loss of viability upon genotoxic stress.
Plasma Membrane Localization of cGAS Is Important to Efficiently Detect Viral Infection
The above findings suggest that cGAS localization to the plasma membrane is necessary to prevent self-DNA detection. However, these findings could also be explained if the N-terminal domain of cGAS operated as an intrinsic inhibitor of catalytic activity. If this alternative explanation were correct, then any DNA stimulus should induce a more robust response from cells expressing cGASΔN than those expressing WT cGAS. This possibility was examined by infecting cells with Modified Vaccinia Ankara (MVA), a DNA virus that stimulates cGAS-dependent IFN responses (Dai et al., 2014).
We infected THP1 cells with an MVA strain expressing an Ovalbumin (Ova) transgene, which we utilized as a reporter for the establishment of infection (Albrecht et al., 2008). Upon MVA infection, cells expressing GFP or WT cGAS upregulated IFNβ1 expression (Figure 4I). This increase in IFNβ1 expression correlated with increases in the intracellular abundance of phospho-STAT1 (Figure 4J). However, cGASΔN-expressing cells were less responsive to MVA infection. Whereas MVA induced a multi-log increase in IFNβ1 transcripts in cells expressing GFP or WT cGAS, minimal and delayed changes in IFNβ1 transcript abundance were observed in cells expressing cGASΔN (Figure 4I). Additionally, STAT1 phosphorylation was less robust in cGASΔN-expressing cells, as compared to cells expressing WT cGAS (Figure 4J). Expression of the virus-encoded Ova transgene was observed across all cells examined (Figure 4J).
The mislocalized cGASΔN allele is therefore potently responsive to cell intrinsic DNA ligands, as compared to the WT allele, yet a similar hyper-responsiveness is not observed with stimulation by foreign DNA through MVA infection. The differential sensitivity of mislocalized cGASΔN to genotoxic stresses and viral infections rules out the possibility that the N terminus serves as an intrinsic catalytic inhibitor. Rather, these findings demonstrate the importance of plasma membrane localization via PI(4,5)P2 for cGAS activities.
Molecular Basis for cGAS Interactions with PI(4,5)P2
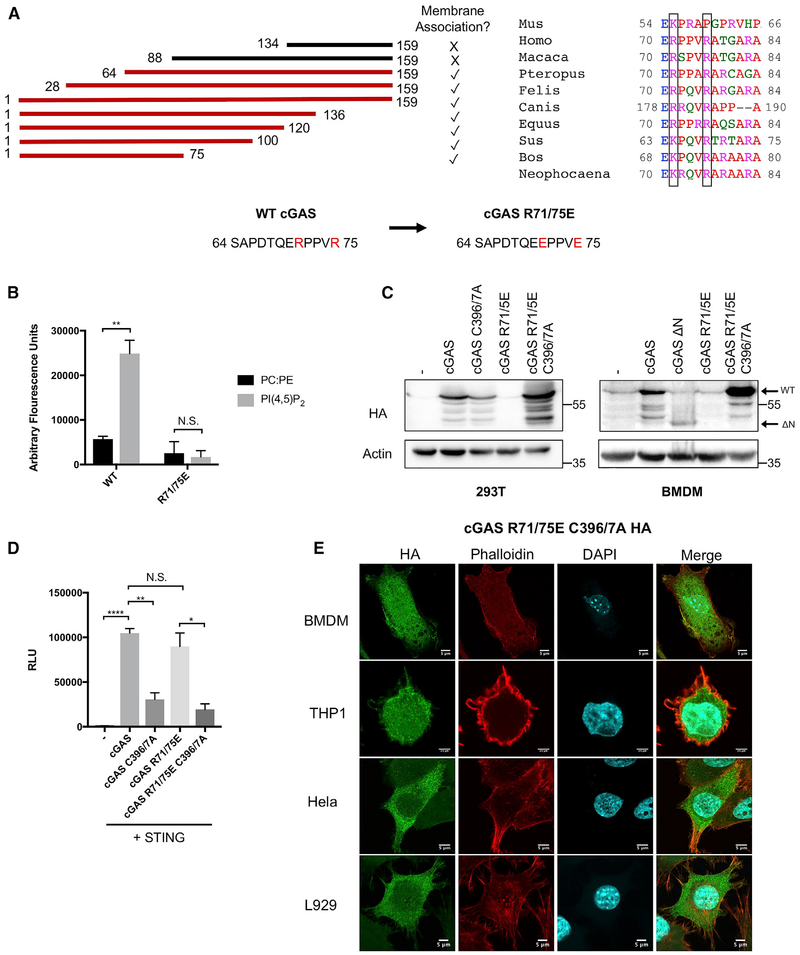
As PI(4,5)P2 is the first lipid identified to interact with cGAS, additional mechanistic insight into this interaction was of interest. As PI(4,5)P2 binding correlates with membrane localization, deletion analysis was performed within the cGAS N terminus to identify a minimal region necessary for plasma membrane association (Figure 5A). This analysis identified amino acids 64 to 75 as important for plasma membrane localization (Figure 5A). Within this region are two arginine residues that are conserved or charge-conserved between humans, mice, and several other mammalian species (Figure 5A). As phosphoinositide interactions are often electrostatic, these residues were of interest. We mutated both arginine residues to glutamic acid in the context of the full-length human cGAS protein (cGAS R71/75E) and examined its ability to localize in cells, induce IFN responses, and bind PI(4,5)P2.
Figure 5. The cGAS R71/75E Mutant Does Not Bind PI(4,5)P2 and Does Not Associate with the Plasma Membrane.
(A) Left: schematic of truncation mutants screened in the cGAS N terminus in HeLa cells and their ability to associate with the plasma membrane. Bottom center: residues identified as essential for cGAS membrane association with mutated residues indicated in red and the indicated point mutations. Right: alignment of mutated residues in several species (same as Figure 2G) with R71 and R75 boxed.
(B) Fluorescent liposome pull-down assay with the indicated mutants, as described in Figure 3C.
(C) Western blot analysis of expression of indicated cGAS mutants in transiently transfected 293T cells and BMDMs stably expressing mutants via lentiviral transduction.
(D) ISRE luciferase assay of 293Ts expressing the indicated constructs 24 h post-transfection. – indicates cells transfected with the ISRE reporter construct alone. All cGAS constructs were co-transfected with STING to enable pathway signaling.
(E) Confocal micrographs of indicated cell lines expressing cGAS R71/75E C396/7A.
Experiments shown are representative of or averages of n = 3 biological replicates. Data are shown as a mean ± SEM, and statistical analysis was performed using a Student’s t test with asterisk coding as follows: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
We found that recombinant cGAS R71/75E protein was completely defective for PI(4,5)P2 interactions in vitro, thus indicating an essential role of the mutated amino acids in protein-lipid interactions (Figure 5B). Notably, this protein was difficult to produce in 293T cells, as compared to its WT counterpart (Figure 5C). Despite the low abundance of cGAS R71/75E, expression of this gene drove IFN stimulatory response element (ISRE) reporter activation to levels comparable to WT cGAS (Figure 5D). The high signaling activity of low amounts of cGAS R71/75E is consistent with the idea that PI(4,5)P2 interactions prevent access of cGAS to cytosolic DNA. Direct evidence in support of this model was provided when we generated a point mutant deficient for plasma membrane association and DNA binding (cGAS R71/75E C396/7A). This mutant was unable to induce ISRE responses upon expression in 293T cells and was produced at high abundance (Figures 5C and 5D). Similar protein stability trends were observed in primary cells, as transduction of murine marrow with cGAS R71/75E did not yield any detectable protein after a week of macrophage differentiation, but parallel transductions demonstrated the robust expression of the DNA-binding-deficient cGAS R71/75E C396/7A allele (Figure 5C). This mutant did not associate with the plasma membrane upon stable expression in primary BMDMs, THP1 cells, and L929 cells and upon transient transfection of HeLa cells (Figure 5E). Thus, amino acids 71 and 75 are critical for cGAS interactions with PI(4,5)P2 and are consequently critical for plasma membrane localization within cells. Interfering with these interactions, either by deletion or amino acid substitution, renders cGAS responsive to cytosolic DNA, even when produced at very low levels.
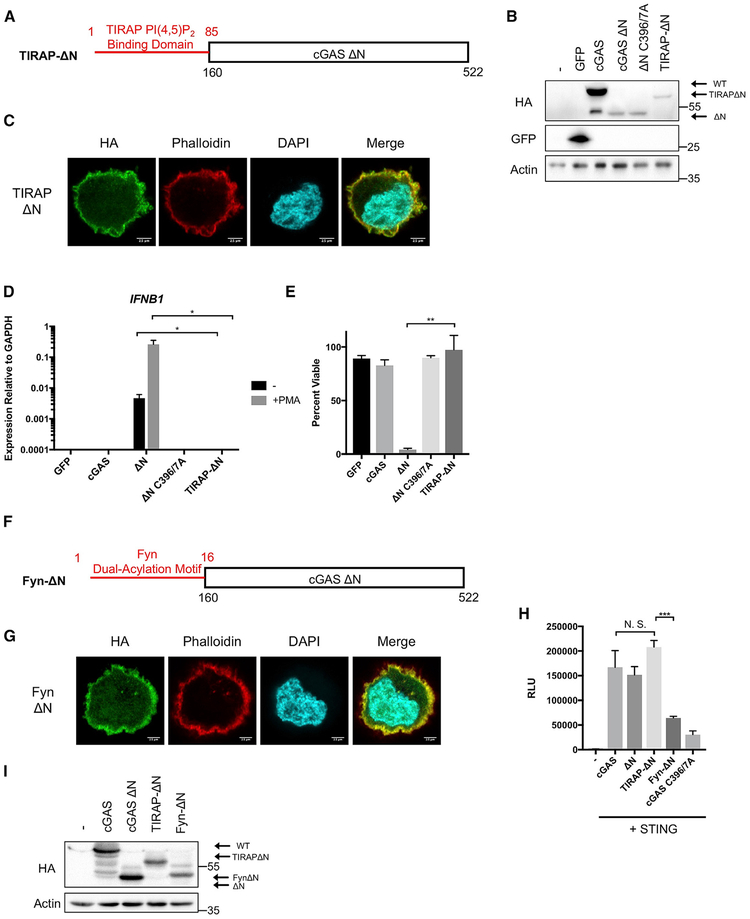
Thus far, all experiments indicate that cGAS interactions with PI(4,5)P2 are necessary for localization and function. To determine whether these interactions are sufficient for plasma membrane localization and prevention of self-DNA recognition, we replaced the N terminus of cGAS with a heterologous PIP-binding domain from the TLR adaptor TIRAP (TIRAP-ΔN; Figure 6A). Like the cGAS N terminus, this domain directs TIRAP to the plasma membrane via interactions with PI(4,5)P2. We generated stable THP1 lines expressing this cDNA as well as WT cGAS, cGASΔN, or cGASΔN C396/7A (Figure 6B). The TIRAP localization domain was sufficient to direct cGASΔN to the plasma membrane (Figure 6C). Functionally, the TIRAP localization domain reversed the basal IFN expression and lethal PMA-induced IFN responses associated with cGASΔN in THP1 cells. Resting cells expressing cGASΔN displayed high basal IFN-β and ISG expression, as compared to cells expressing TIRAP-ΔN or WT cGAS (Figure 6D). Moreover, whereas cGASΔN responded to PMA by producing copious amounts of IFN-β transcripts and subsequently dying, cells expressing TIRAP-ΔN or WT cGAS did not induce such responses (Figures 6D and 6E).
Figure 6. Artificial Localization of cGASΔN to the Plasma Membrane through the TIRAP PI(4,5)P2 Binding Domain Rescues Localization and Prevents Hyperresponsiveness to Genotoxic Stress.
(A) Schematic of the synthetic TIR domain containing adaptor protein (TIRAP) PI(4,5)P2-binding domain fused to cGASΔN.
(B) Western blot analysis of THP1 cells stably expressing the indicated mutants.
(C) Confocal micrograph of THP1 cells stably expressing TIRAP-ΔN-HA.
(D) qPCR analysis of IFNβ1 expression 3 h post-treatment with 50 ng/mL PMA in the indicated THP1 cell lines.
(E) Viability of THP1 cell lines following overnight treatment with 50 ng/mL PMA, shown as a percentage of untreated controls, as measured by intracellular ATP content.
(F) Schematic of the synthetic Fyn dual-acylation motif fused to cGASΔN.
(G) Confocal micrograph of THP1 cells stably expressing Fyn-ΔN-HA.
(H) ISRE luciferase assay of 293Ts expressing the indicated constructs 24 h post-transfection, as described in Figure 5D.
(I) Western blot analysis of 293T cells expressing indicated constructs.
Experiments shown are representative of or averages of n = 3 biological replicates. Data are shown as a mean ± SEM, and statistical analysis was performed using a Student’s t test with asterisk coding as follows: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
To determine whether any mechanism of plasma membrane localization was sufficient to ensure WT-like functions to cGASΔN, we generated a second cGASΔN fusion protein, Fyn-ΔN, which replaced the natural N terminus with the dual acylation SH4 domain from the kinase Fyn (Figure 6F). This lipidation motif differs from a PIP-binding domain in that the lipid anchor should prevent protein release into the cytosol. Like TIRAP-ΔN, Fyn-ΔN localized to the plasma membrane of THP1 cells (Figure 6G). Once this activity was confirmed, we transfected 293T cells with either of these mutants, along with plasmids encoding STING and the ISRE reporter (Figures 6H and 6I). While TIRAP-ΔN stimulated ISRE reporter activity to similar levels as WT cGAS, reporter activity in cells expressing Fyn-ΔN was nearly absent (Figure 6H). Despite these differences in activation, these constructs were expressed at comparable levels (Figure 6I). These results support the idea that cGAS must not only localize to the plasma membrane to prevent aberrant activation, but it must be released from the cell surface in order to signal. Combined with the loss-function analysis performed with cGAS R71/75E and cGAS R71/75E C396/7A, these data establish that PI(4,5)P2 binding is an activity within the N-terminal domain that is necessary and sufficient for cGAS self-nonself discrimination.
DISCUSSION
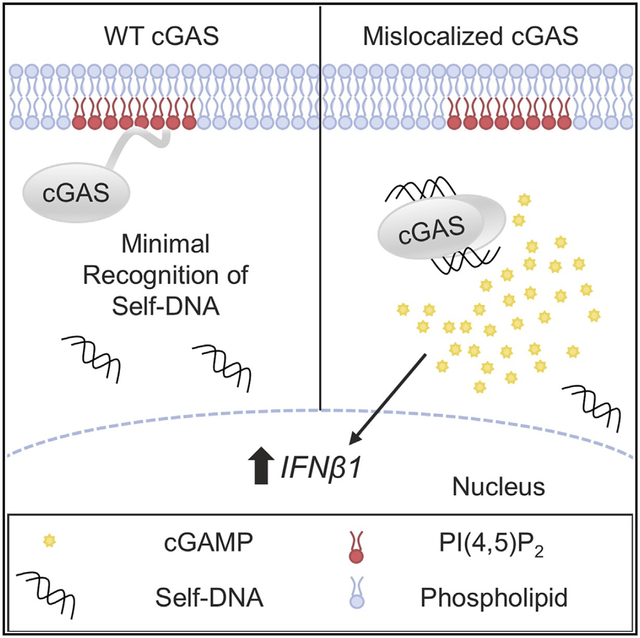
The importance of localization for proteins lacking transmembrane domains is often overlooked, and it is generally assumed that innate immune regulators lacking such domains must operate as cytosolic proteins. In this study, we tested the predictions of this model through a detailed analysis of the subcellular positioning and functions of cGAS. Several lines of evidence support the conclusion that cGAS is not a cytosolic protein but is rather a plasma membrane protein whose localization is mediated by PI(4,5)P2. First, subcellular fractionation of endogenous cGAS reveals that a majority of cGAS cofractionates with membranes through both sedimentation and floatation, while being present at low levels in the nucleus and absent from the cytosol. Second, confocal microscopy of endogenous and tagged cGAS show that it is concentrated at the plasma membrane of human and murine monocytes and macrophages. Third, using multiple independent assays for protein-lipid interactions, cGAS interacts with acidic PIPs and selectively binds PI(4,5)P2. Finally, subcellular localization and the PI(4,5)P2-binding activity of cGAS map to the same amino acids in the N terminus of this protein, providing a mechanism to explain how cGAS can be positioned at the plasma membrane of resting cells. cGAS can therefore be considered primarily a plasma membrane protein, whose localization is dictated by protein-lipid interactions.
Although we have found cGAS to be located at the plasma membrane, we also observed a pool of nuclear cGAS biochemically and microscopically. While the mechanisms of nuclear localization (or function) are unclear, some technical considerations must be made. First, our use of benzonase in fractionations was critical to prevent post-lysis DNA binding, which could drive cGAS oligomerization into nucleus-sized aggregates (Du and Chen, 2018). Second, microscopic detection of lipid-binding proteins is sensitive to methods of cell fixation. For example, paraformaldehydes immobilize lipid-binding proteins within cells, whereas methanol fixatives extract lipids and their associated proteins. These latter conditions may exaggerate the appearance of selective pools of intracellular cGAS. With these statements made, we do not consider cGAS to be absent from the nucleus, but rather consider the plasma membrane to be the most common site of its residence in phagocytes. While we found that the frequency of nuclear- or plasma membrane-localized cGAS varied among cell types, the cGAS N terminus localizes to the plasma membrane regardless of cell type. These findings suggest this association is a fundamental feature of the N terminus that is actively modulated by cells and is dependent on cGAS activity. We suggest that cGAS localization may emerge as a mechanism of regulation that varies in a context-dependent manner.
While our analysis identified the N terminus of cGAS as the domain responsible for lipid binding and localization, this domain also facilitates DNA binding and assembly of cGAS into liquid droplets (Du and Chen, 2018). DNA binding and liquid droplet formation are cornerstone features of active cGAS, whereas PI(4,5) P2-binding and plasma membrane localization are activities consistent with inactive cGAS. Based on this collection of data, we propose a unifying model whereby the cGAS N terminus serves distinct functions in its resting and active states. In resting cells, the N terminus positions cGAS at the cell surface, where it is least likely to detect self-DNA and prevent aberrant activation. Upon DNA detection, the N terminus may release from PI(4,5) P2 and facilitate liquid droplet formation and signaling in the cytosol. Consistent with this idea is our finding that DNA transfection results in the movement of cGAS from the cell surface to cytosolic foci and that restricting cGAS release from the plasma membrane prevents signal transduction.
Our phylogenetic analysis revealed that the high pI of the cGAS N terminus relative to its C terminus is conserved across several species. Electrostatic interactions with PIPs may therefore be conserved as well. In further support of this concept, we determined that the two residues essential for cGAS interaction with PI(4,5)P2 and membrane localization (R71/R75) were conserved or charge-conserved across these species. We therefore suggest plasma membrane localization of cGAS is a fundamental and evolutionarily conserved feature of this PRR.
To date, access to DNA ligands has been the focal point of discussions surrounding the ability of cGAS to maintain the balance of self-nonself discrimination (Ablasser et al., 2014; Stetson et al., 2008). The cytosol of mammalian cells should be DNA-free, with genomic DNA and mitochondria DNA confined to distinct subcellular sites. Any DNA released into the cytosol is hydrolyzed by well-defined nucleases, and genetic defects in these nucleases lead to cGAS-dependent pathologies (Chen et al., 2016). Thus, current models predict that the sole mechanism of preventing inappropriate cGAS activation is by restricting access of DNA to the cytosol. It is with this model in mind that our findings may be most notable, as we can now propose that DNA is not only hidden from cGAS, but cGAS via membrane localization is also hidden from mislocalized DNA that escapes nuclease-mediated hydrolysis.
By positioning cGAS at the cell periphery, the most distal site from the nucleus in the cell, this PRR may be localized to a site that is free of DNA entirely. Under such a model, we speculate that mislocalized cGAS is no longer geographically restricted from DNA that leaks from the nucleus. Thus, mislocalized cGAS is prone to induce IFN responses in the absence of infection or major genomic damage. Why mislocalized cGAS did not induce similarly potent IFN responses to viral DNA is also worth considering. Positioning at the plasma membrane places cGAS near portals of pathogen entry, which may enable it to sense foreign DNA as it enters the cell. Alternatively, the defect in virus detection by cGASΔN may not be linked directly with its inability to localize properly but instead may be related to its inefficient ability to assemble into liquid droplets upon DNA binding. Indeed, cGASΔN is defective for signaling in response to DNA transfection (Du and Chen, 2018). While future work is necessary to dissect the relative activities present in the cGAS N terminus, our functional studies with TIRAP-ΔN and Fyn-ΔN suggest that PI(4,5)P2 binding is an activity necessary for cGAS function.
Finally, it has not escaped our attention that the similarities in subcellular positioning of TLR adaptors and cGAS suggest an evolutionary theme may have been uncovered by our study. The use of PIP-binding domains to position a sensor of viral infection (cGAS) and sensors of activated TLRs (TIRAP, TRAM, and dMyD88) raises the possibility that other innate immune pathways may use similar strategies to ensure signaling fidelity. This study provides a mandate to explore this possibility, as well as the localization of other seemingly cytosolic PRRs that may operate by principles similar to those described for cGAS.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jonathan C. Kagan (Jonathan.Kagan@childrens.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture and Recombinant Cell Lines
Immortalized bone marrow derived macrophages (iBMDMs), HeLa cells, HEK293Ts, and L929 fibroblasts were cultured in DMEM supplemented with 10% FBS, Penicillin and Streptomycin (Pen+Strep), L-glutamine, and sodium pyruvate, referred to as complete DMEM, at 37°C in 5% CO2. For passage, iBMDMs were lifted using PBS supplemented with 2.5mM EDTA and plated at dilution 1:10. HEK293T, L929, and HeLa cells were grown under the same conditions as iBMDMs but were passaged by washing with PBS and lifting with 0.25% trypsin with a 1:10 dilution factor. THP1 cells were grown in suspension culture using RPMI-1640 media supplemented with 10% FBS, Pen+Strep, L-glutamine, and sodium pyruvate, referred to as complete RPMI-1640, at 37°C in 5% CO2. For passage, cells were washed in PBS and plated at a dilution of 1:4. For experiments examining the effects of PMA-induced differentiation of THP1 cell lines, cells were treated for the indicated times with PMA at a concentration of 50ng/mL. For intracellular DNA stimulation, iBMDMs were transfected with 1:1 Lipofectamine 2000: CT-DNA at the specified concentrations.
To generate lentiviral particles for the stable expression of transgenes, HEK293T cells were transfected with the packaging plasmids psPAX2 and pCMV-VSV-G along with the transgene in pLenti-CMV-GFP-Puro. All genes of interest were subcloned into the GFP site, and all mutant variants of cGAS were generated by overlap-extension PCR. For the production of TIRAP-GFP retroviral particles for stable expression, the packaging plasmids pCL-Eco and pCMV-VSV-G were used in addition to the target construct pMSCV2.2-TIRAP-GFP-IRES-hCD2tm. Plasmids were transfected into 10cm2 dishes of HEK293Ts at 50%–80% confluency using polyethylenimine (PEI) at a ratio of 3:1 PEI:DNA. Media was changed on transfected 293Ts 16 hours after transfection, and virus-containing supernatants were harvested 24 hours following the media change. Viral supernatants were passed through a 0.45 mm filter to remove any cellular debris. Filtered viral supernatants were placed directly onto target cells for 24 hours and then replaced with the appropriate complete media. Transgene expression was assessed by western analysis and confocal microscopy.
Primary Macrophage Culture and Manipulation
For the production of primary bone marrow derived macrophages, the leg bones of dead, female C57BL/6 mice were cleaned, cut with scissors, and flushed with sterile PBS through a syringe. The resultant isolated marrow was passed through a 70 μm cell strainer to remove any debris. Cells were plated into non-cell culture treated dishes in macrophage differentiation media, complete DMEM containing 30% L929 M-CSF conditioned media. Three days after isolation, cells were fed additional macrophage differentiation media. On day 7, cells were lifted using PBS containing 2.5mM EDTA and plated for further experiments. Primary mouse macrophages were transfected using the Amaxa Nucleofector II in coordination with the Mouse Macrophage Nucleofector Kit and were used according to the manufacturer’s instructions.
For the production of immortalized Cgas −/− cells, primary macrophages were differentiated from isolated bone marrow, as described above, and immortalized using J2 retrovirus from supernatants collected from CREJ2 cell lines. Cgas −/− C57BL/6 leg bones were kindly provided by Dr. D. Stetson. Following differentiation, Cgas −/− primary macrophages were incubated with macro-phage differentiation media containing 50% CREJ2 supernatant for seven days with a media replacement after the first three days of culture. Following viral transduction, cells were cultured until confluent in complete DMEM containing 30% L929 supernatant, split 1:2 into complete DMEM containing 25% L929 supernatant, and then slowly weaned from L929 supernatant with each passage into fresh complete DMEM with 5% less L929 supernatant than the previous. Once cells were able to grow in complete DMEM lacking any L929 supernatant, the immortalization process was complete.
For the production of CRISPR/Cas9 mediated generation of THP1 cGAS −/− cells, a cGAS-specific guide RNA was cloned into the previously described the pRRL-Cas9-Puro vector, kindly provided by Dr. D. Stetson, and transduced into THP1 cells, as described above. Cells were then single cell cloned, tested for cGAS expression by western blot, and tested for loss of intracellular DNA sensing by transfection of 1 μg/mL CT-DNA.
METHOD DETAILS
Viral Infection
The MVA-Ova strain was generously provided Dr. N. Chevrier. For infection, viral stocks were diluted to a MOI of 3 in serum-free medium and incubated with cells for 1 hour at 37°C in 5% CO2 with frequent agitation. After the period of infection, viral media was aspirated and replaced with complete DMEM and incubated at 37°C in 5% CO2. At the indicated time points, cells were lysed with 1× Laemmli Buffer for western analysis or lysed for RNA isolation for RT-qPCR analysis.
H2O2, Doxorubicin, and PMA Treatments
THP1 monocytic cells were incubated with either 500 μMH2O2 or 50ng/mL PMA or 500nM Doxorubicin in complete DMEM and incubated at 37°C in 5% CO2. At the indicated time points, cells were lysed with 1× Laemmli Buffer for western analysis or lysed for RNA isolation for RT-qPCR analysis. With PMA and Doxorubicin treatments, cells were incubated with 50ng/mL PMA for 24 hours and then analyzed using the cell viability assay described below.
Subcellular Fractionation
Benzonase (10U/mL) was used in all subcellular fractionation experiments to prevent post-lysis DNA-cGAS interactions unless otherwise stated. To separate cell membranes from the soluble cellular components, cells were washed once in hypotonic buffer (10mM Tris-HCl pH 7.4; 10mM KCl; 1.5mM MgCl) supplemented with a protease inhibitor cocktail (Roche), incubated on ice in hypotonic buffer, and lysed by dounce homogenization. Lysates were centrifuged at 4°C for 5 minutes at 2,500xg to remove nuclei and cellular debris. This nuclear pellet was then washed 3X in hypotonic buffer supplemented with 0.3% NP40 to remove any contaminating organelles and fully isolate nuclei, resulting in the nuclear fraction (P25). Supernatants were centrifuged at 100,000xg for 1 hour at 4°C. The resultant pellets (P100) were resuspended in lysis buffer volumes equal to those of the supernatants (S100), stored with the addition of 6× Laemmli Buffer, and analyzed by western blot.
For membrane floatation assays, post nuclear supernatants were collected and described above, mixed with Optiprep™-supplemented hypotonic buffer to yield a final concentration of 45% optiprep at laid at the bottom of an Optiprep™ step gradient ranging from 10% (top) to 45% (bottom), and spun at 52,000xg for 90 minutes. Gradient was then fractionated into 24 fractions and analyzed by western blot. For gradients run in the presence of Triton X-100, post nuclear supernatants were mixed with a 10% Triton X-100 solution to achieve a final concentration of 1%.
Confocal Microscopy
Cells were plated onto glass coverslips, treated as described, and fixed for 1.5 hours in a periodate-lysine-paraformaldehyde fixation buffer (20mM MES; 70mM NaCl; 5mM KCl; 70mM Lysine-HCl; 5mM MgCl2; 2mM EGTA; 10mM NaIO4; 2% Paraformaldehyde; 4.5% sucrose). Following fixation, cells were blocked and permeabilized in blocking buffer (0.1% saponin; 25mM Tris-HCl pH 7.5; 150mM NaCl; 4.5% sucrose; 2% goat serum; 50mM NH4Cl), incubated overnight at 4°C with the indicated primary antibody, and incubated for 1 hour at RT with secondary antibodies and stains. Cells were mounted onto glass slides with Prolong Gold Antifade (Thermo) and imaged using a 63× oil immersion objective on the LSM 880 with Airyscan (Zeiss). Images were processed using ZEN software (Zeiss) and ImageJ (NIH).
For imaging THP-1 cells, cells were attached to coverslips coated with Alcian Blue by incubation in serum free media for 20 minutes at 37°C in 5% CO2. For imaging of zymosan phagocytosis, cells were incubated for 10 minutes at 37°C in 5% CO2 in media containing zymosan at a MOI of 5. For quantification of cGAS localization, at least 200 cells per replicate were counted of three replicates in total. Plasma membrane localization was characterized as co-localization with Phalloidin, while nuclear localization was characterized as co-localization with DAPI.
Real-Time Quantitative PCR
RNA was isolated from cells using Qiashredder (QIAGEN) homogenizers and the PureLink Mini RNA Kit (Life Technologies) and subsequently DNase I (Thermo) treated to remove genomic DNA. Relative mRNA expression was analyzed using the TaqMan RNA-to-Ct 1-Step Kit (Applied Biosystems) with indicated probes on a CFX384 Real-Time Cycler (BioRad).
Cell Viability Assay
Cell viability was measured using CellTiter-Glo (Promega), a luminescent assay for ATP in living cells. Assays were performed as described by the manufacturer and untreated or untransduced cells were used as a positive control for cell viability, considered as the 100% viable benchmark compared to treated or transduced cells. Luminescent outputs were read on a Tecan plate reader.
Phylogenetic Analysis
Ten vertebrate animals’ cGAS sequences were analyzed by Clustal ω software (https://www.ebi.ac.uk/Tools/msa/clustalo/). The N- and C-termini were defined according to their alignments with human cGAS, and the PI of each domain was calculated using the ExPASy Compute pI/MW tool (https://web.expasy.org/compute_pi/). Percent identity was calculated as the ratio of conserved residues to the total number of residues in the protein, using human cGAS as the reference.
Recombinant Protein Purification
Recombinant cGAS protein and corresponding variants were purified as previous described (Zhou et al., 2018). Briefly, human cGAS and cGAS R71/75E variants were cloned into a custom pET vector for expression as an N-terminal 6 × His-SUMO2-fusion protein using PCR amplification and standard cloning techniques. The pET-cGAS plasmids were transformed into E. coli BL21 DE3 (Aglient) bacteria harboring a pRARE2 tRNA plasmid. E. coli were grown in M9ZB media at 37°C and protein expression was induced by IPTG for ~ 16 h at 16°C. Bacterial pellets expressing cGAS were re-suspended in lysis buffer and then lysed by sonication. After centrifugation, the supernatants were collected and recombinant protein was isolated using Ni-NTA (QIAGEN) chromatography. The His-SUMO2 tag was then removed by digestion with human SENP2 protease except when used as His-tagged proteins for the PI-binding pulldown assay. Recombinant cGAS proteins were further purified by Heparin HP ion-exchange (GE Healthcare) and 16/600 S75 size-exclusion chromatography (GE Healthcare). Final cGAS proteins were concentrated in storage buffer (20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM TCEP), flash-frozen in liquid nitrogen, and stored at −80°C.
Recombinant human cGASΔN and cGAS N were expressed using the pET50b vector containing an N-terminal 6xHis-NusA tag (gift from Dr. Sun Hur) in BL21 (DE3) E. coli at 16°C for 20 hours after induction with 5mM IPTG. Cells were lysed using an Emulsiflex-C5 (Avestin), and protein was batch purified using Ni-NTA affinity chromatography. Proteins were dialyzed overnight into a storage buffer (20mM HEPES pH 7.5; 150mM KCl; 10% Glycerol; 1mM TCEP) (Kranzusch et al., 2013) and snap frozen in liquid nitrogen for use in biochemical assays. Preparation efficacy and purity was assessed by SDS-PAGE and SimplyBlue SafeStain (Thermo).
PIP Strip Lipid Binding Assay
Lipid binding assays were performed as described (Kagan and Medzhitov, 2006). Briefly, PIP strips (Echelon) were blocked in (10mM Tris pH 8; 150mM NaCl; 0.1% Tween-20; 0.1% ovalbumin) for 1 hour, then incubated for 2 hours with purified hcGAS (500ng/mL) in the presence of an anti-hcGAS antibody (Sigma), washed 3 times with blocking buffer, and then probed with an HRP-conjugated secondary antibody in blocking buffer for 30 minutes. Lipid bound protein was detected using enhanced chemiluminescence.
Liposome Preparation and Assays
Liposome preparation and subsequent binding assays were performed described (Kagan and Medzhitov, 2006). In brief, PC:PE were mixed at a 3:1 ratio in a solution of 2:1 chloroform:methanol in borosilicate glass tubes and gently dried into a lipid film using inert nitrogen gas. For liposomes containing PIPs, the 3:1 PC:PE ratio was maintained with the addition of the specified percentage of PIP. Dried lipids were resuspended in 300mM sucrose and vortexed aggressively to yield liposomes at a final concentration of 1mg/mL.
For sedimentation assays, 2 μg of liposomes were incubated with 5 mg of recombinant cGAS in a cytosol buffer (25mM HEPES pH7.2; 25mM KCl; 2.5mM Magnesium Acetate; 150mM Potassium Glutamate) for 15 minutes at RT. Following incubation, samples were centrifuged at 100,000xg, and the pellets were analyzed by western blot for cGAS binding.
For fluorescent liposome pulldown assays, liposomes were prepared as described above, except fluorescently tagged PC (NBDPC) was used in place of PC. 25 μg of liposomes were mixed with 25 μg of recombinant protein in cytosol buffer in the presence of Ni-NTA resin and incubated with gentle mixing for 30 minutes at RT. Ni-NTA resin was pelleted and washed two times before elution of liposomes with 10% SDS. Resultant fluorescence recovered was quantified by spectrofluorimetry using a Tecan plate reader. Interactions between the beads and liposomes alone were subtracted from all samples to account for nonspecific binding.
Luciferase Reporter Assays
293T cells were transiently transfected with the indicated plasmids using PEI. After 24h, cells were lysed and incubated with Bright-Glo reagent (Promega) according to manufactuer’s instructions, and luminescence was read on a Tecan plate reader.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed using Graphpad Prism7 software. All experiments were performed in triplicate as three independent biological replicates. For comparison in which multiple variables were tested with multiple time points, two-way ANOVA analysis was performed. For comparison of two data points, the student’s two-tailed t test was used. Asterisk coding is as follows: * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001. Data with error bars depict the average with the SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-HA (3F10) | Sigma | Cat# 11867431001; RRID: AB_390916 |
| Rabbit monoclonal anti-cGAS (human specific) (D1D3G) | Cell Signaling Technology | Cat#15102S; RRID: AB_2732795 |
| Rabbit monoclonal anti-cGAS (mouse specific) (D3080) | Cell Signaling Technology | Cat# 31659S |
| Rabbit polyclonal anti-beta actin (4967) | Cell Signaling Technology | Cat# 4967S; RRID: AB_330288 |
| Rabbit polyclonal anti-MD21D1 (cGAS) | Sigma | Cat# HPA031700; RRID: AB_10601693 |
| Rabbit polyclonal anti-calnexin (H-70) | Santa Cruz | Cat# sc-11397; RRID: AB_2243890 |
| Rabbit polyclonal anti-caspase-3 (H-277) | Santa Cruz | Cat# sc-7148; RRID: AB_637828 |
| Rabbit monoclonal anti-GFP (JL-8) (microscopy) | Clontech | Cat# 632381; RRID: 2313808 |
| Rabbit polyclonal anti-GFP (western blot) | Clontech | Cat# 632592; RRID: AB_2336883 |
| Mouse monoclonal anti-HSV1 ICP4 antibody | Virusys | Cat# P1101 |
| Mouse monoclonal anti-Ovalbumin (6C8) | Abcam | Cat# ab17293; RRID: AB_443790 |
| Mouse monoclonal anti-GST (6G9C6) | Sigma | Cat# SAB5300159 |
| Mouse Anti-Stat1 (pY701) | BD Biosciences | Cat# 612233; RRID: AB_399556 |
| Rabbit monoclonal anti-Stat1 (total) (9172) | Cell Signaling Technology | Cat# 9172S; RRID: AB_2198300 |
| Bacterial and Virus Strains | ||
| Modified Vaccinia Virus Ankara Ovalbumin (MVA-Ova) | Albrecht et al., 2008 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Alexaflour 568 Phalloidin | Invitrogen | Cat# A12380 |
| OptiPrep Density Gradient Medium | Sigma | Cat# D1556 |
| Triton X-100 | Sigma | Cat# X-100 |
| IGEPAL CA-630 (NP-40) | Sigma | Cat# I8896 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Invitrogen | Cat# D1306 |
| PMA (Phorbol 12-myristate 13-acetate) | Sigma | Cat# P8139 |
| Alcian Blue | ThermoFisher | Cat# ICN203450 |
| 16:0-14:0 PC (1-palmitoyl-2-myristoyl-sn-glycero-3-phosphocholine) (PC) | Avanti Polar Lipids | Cat# 850454 |
| 16:0-12:0 NBD PC (1-palmitoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphocholine) (NBD-PC) | Avanti Polar Lipids | Cat# 810131 |
| Heart PE (L-α-phosphatidylethanolamine) (PE) | Avanti Polar Lipids | Cat# 840025 |
| Phosphatidylinositol 3-phosphate diC16 (PI(3)P diC16) | Echelon Biosciences | Cat# P-3016 |
| Phosphatidylinositol 4-phosphate diC16 (PI(4)P diC16) | Echelon Biosciences | Cat# P-4016 |
| Phosphatidylinositol 5-phosphate diC16 (PI(5)P diC16) | Echelon Biosciences | Cat# P-5016 |
| Phosphatidylinositol 3,4-bisphosphate diC16 (PI(3,4)P2 diC16) | Echelon Biosciences | Cat# P-3416 |
| Phosphatidylinositol 4,5-bisphosphate diC16 (PI(4,5)P2 diC16) | Echelon Biosciences | Cat# P-4516 |
| Phosphatidylinositol 3,5-bisphosphate diC16 (PI(3,5)P2 diC16) | Echelon Biosciences | Cat# P-3516 |
| Phosphatidylinositol 3,4,5-trisphosphate diC16 (PI(3,4,5)P3 diC16) (PIP3) | Echelon Biosciences | Cat# P-3916 |
| Polyethylenimine (PEI) | Polysciences | Cat# 23966-2 |
| Lipofectamine 2000 | ThermoFisher | Cat# 11668019 |
| cOmplete Protease Inhibitor Cocktail | Sigma | Cat# 11697498001 |
| Zymosan A S. cerevisiae BioParticles, Texas Red conjugate | ThermoFisher | Cat# Z2843 |
| ProLong Gold Antifade Mountant | ThermoFisher | Cat# P10144 |
| Calf Thymus DNA (CT-DNA) | Sigma | Cat# D1501 |
| PI(4,5)P2 Grip (PLC-δ1-PH) | Echelon Biosciences | Cat# G-4501 |
| Ni-NTA Agarose | QIAGEN | Cat# 30230 |
| Tris-HCl (Trizma hydrochloride) | Sigma | Cat# T3253-5KG |
| Tris-Base (Trizma base) | Sigma | Cat# T1503-5KG |
| HEPES | VWR | Cat# 45000-694 |
| KCl | Amresco | Cat# 0395-1kg |
| NaCl | Sigma | Cat# S9625-5KG |
| Saponin | Sigma | Cat# S4521-25G |
| Tween-20 | Fisher | Cat# BP337-500 |
| Ovalbumin | Sigma | Cat# A5253-500G |
| Chloroform | Sigma | Cat# C7559-5VL |
| Methanol | Fisher | Cat# A412-4 |
| Magnesium acetate tetrahydrate | Sigma | Cat# M5661-250G |
| L-Glutamic acid potassium salt monohydrate | Sigma | Cat# G1501-500G |
| TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) | Sigma | Cat# CA4706-2G |
| MES (2-(N-Morpholino)ethanesulfonic acid) hemisodium salt | Sigma | Cat# M0164 |
| Lysine-HCl | Sigma | Cat# L5626-500G |
| NaIO4 (Sodium (meta)periodate) | Sigma | Cat# 769517-100G |
| EGTA (Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid) | Sigma | Cat# E3889-100G |
| EDTA (Ethylenediaminetetraacetic acid) | Sigma | Cat# 798681-100G |
| Paraformaldehyde | Electron Microscopy Services | Cat# 15710 |
| Simply Blue Safe Stain | Thermo | Cat# LC6060 |
| Recombinant SENP2 protease | Enzo Life Sciences | Cat# BML-UW9765-0100 |
| Sucrose | Sigma | Cat# S0389-1KG |
| DNase I, Rnase-free | ThermoFisher | Cat# EN-0521 |
| Doxorubicin | Cell Signaling Technology | Cat# 5927S |
| IPTG (Isopropyl-β-D-thiogalactoside) | Sigma | Cat# 10724815001 |
| Benzonase Nuclease, Purity > 99% | Millipore | Cat# 70664 |
| Critical Commercial Assays | ||
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat# G7570 |
| Bright-Glo Luciferase Assay System | Promega | Cat# E2610 |
| Taqman RNA-to-Ct 1-step Kit | ThermoFisher | Cat# 4392938 |
| PureLink Mini RNA Kit | ThermoFisher | Cat# 12183018A |
| QIAshredder | QIAGEN | Cat# 79656 |
| PIP Strips | Echelon Biosciences | Cat# P-6001 |
| Amaxa Mouse Macrophage Nucleofector Kit | Lonza | Cat# VVPA-1009 |
| Experimental Models: Cell Lines | ||
| THP1 | Jonathan Kagan Laboratory | N/A |
| HeLa | Jonathan Kagan Laboratory | N/A |
| L929 | Jonathan Kagan Laboratory | N/A |
| HEK293T | Jonathan Kagan Laboratory | N/A |
| CREJ2 Cells (Producing J2 Retrovirus) | Jonathan Kagan Laboratory | N/A |
| Immortalized bone marrow-derived macrophages (iBMDMs) | Jonathan Kagan Laboratory | N/A |
| Cgas−/− Immortalized bone marrow-derived macrophages (iBMDMs) | This Paper | N/A |
| Cgas−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing cGAS-HA | This Paper | N/A |
| Cgas−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing cGASΔN-HA | This Paper | N/A |
| Cgas−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing cGAS-N-HA | This Paper | N/A |
| THP1 expressing cGAS-HA | This Paper | N/A |
| THP1 expressing cGAS-HA C396/7A | This Paper | N/A |
| THP1 expressing cGASΔN-HA | This Paper | N/A |
| THP1 expressing cGASΔN-HA C396/7A | This Paper | N/A |
| THP1 expressing cGAS-N-HA | This Paper | N/A |
| THP1 expressing cGAS-HA R71/75E | This Paper | N/A |
| THP1 expressing cGAS-HA R71/75E C396/7A | This Paper | N/A |
| THP1 expressing TIRAP-cGASΔN-HA | This Paper | N/A |
| THP1 cGAS−/− expressing Cas9 | This Paper | N/A |
| Experimental Models: Organisms/Strains | ||
| E. coli BL21 (DE3) | Stratagene | Cat# 230130 |
| Mouse: C57BL/6J | The Jackson Laboratory | Cat# 000664 |
| Mouse: Cgas−/− C57BL/6J | Daniel Stetson Laboratory (Gray et al., 2015) | N/A |
| Oligonucleotides | ||
| Ifnb1 Taqman Probe (Mouse) | ThermoFisher | Mm00439552_s1 |
| Rsad2 Taqman Probe (Mouse) | ThermoFisher | Mm00491265_m1 |
| Gapdh Taqman Probe (Mouse) | ThermoFisher | Mm99999915_g1 |
| Ifnb1 Taqman Probe (Human) | ThermoFisher | Hs01077958_s1 |
| Rsad2 Taqman Probe (Human) | ThermoFisher | Hs00369813_m1 |
| Gapdh Taqman Probe (Human) | ThermoFisher | Hs02758991_g1 |
| gRNA targeting cGAS (antisense) 5′ CCCCGATGGATCC CACCGAGTCT 3′ | Jønsson et al., 2017 | N/A |
| Recombinant DNA | ||
| pLenti-CMV-GFP-Puro | Campeau et al., 2009 | Addgene # 17448 |
| psPAX2 | Didier Torno Laboratory | Addgene# 12260 |
| pCMV-VSV-G | Stewart et al., 2003 | Addgene # 8454 |
| pCL-Eco | Naviaux et al., 1996 | Addgene# 12371 |
| pMSCV2.2-TIRAP-GFP-IRES-hCD2tm | Kagan and Medzhitov, 2006 | N/A |
| pRRL-Cas9-Puro | Eckard et al., 2014 | N/A |
| pRRL-gRNA(anti-cGAS)-Cas9-Puro | This paper | N/A |
| pET-50b(+)-cGAS | This Paper | N/A |
| pET-50b(+)-cGASΔN | This Paper | N/A |
| pET-50b(+)-cGAS-N | This Paper | N/A |
| pET-50b(+)-cGAS R71/75E | This Paper | N/A |
| pLenti-CMV-cGAS-HA | This Paper | N/A |
| pLenti-CMV-cGASΔN-HA | This Paper | N/A |
| pLenti-CMV-cGAS-N-HA | This Paper | N/A |
| pLenti-CMV-cGAS-HA (murine) | This Paper | N/A |
| pLenti-CMV-cGASΔN-HA (murine) | This Paper | N/A |
| pLenti-CMV-cGAS-N-HA (murine) | This Paper | N/A |
| pLenti-CMV-cGAS C396/7A-HA | This Paper | N/A |
| pLenti-CMV-cGASΔN C396/7A-HA | This Paper | N/A |
| pLenti-CMV-cGAS R71/75E-HA | This Paper | N/A |
| pLenti-CMV-cGAS R71/75E C396/7A-HA | This Paper | N/A |
| pLenti-CMV-TIRAP-cGASΔN-HA | This Paper | N/A |
| pLenti-CMV-Fyn-cGASΔN-HA | This Paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Zen Software | Ziess | https://www.zeiss.com/microscopy/int/downloads.html?vaURL=www.zeiss.com/microscopy/int/downloads/zen.html |
| Fiji / ImageJ version 2.0.0 | NIH | https://imagej.net/Fiji |
| Clustal ω | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Compute Molecular Weight/pI | ExPASy | https://web.expasy.org/compute_pi/ |
| Other | ||
| Heparin HP ion-exchange column | GE Healthcare | Cat# 17040601 |
| 16/600 S75 size-exclusion chromatography column | GE Healthcare | Cat# 28989333 |
Highlights.
Biochemical and microscopic evidence indicates cGAS is a plasma membrane protein
The cGAS N terminus interacts with PI(4,5)P2 to mediate membrane localization
Mislocalized cGAS mutants drive lethal interferon responses to genotoxic stress
Mislocalized cGAS mutants are poorly responsive to DNA virus infection
ACKNOWLEDGMENTS
We would like to thank members of the Kagan lab for helpful discussions. J.C.K. is supported by NIH grants AI093589, AI116550, and P30 DK34854 and an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. K.C.B. is supported by NIH grant F31AI131469. J.M.C.-S. is supported by Universidad Complutense-Santander Universidades predoctoral contract grant CT45/15-CT46/15. P.J.K. is supported by the Claudia Adams Barr Program for Innovative Cancer Research and Richard and Susan Smith Family Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.cell.2019.01.049.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, and Hornung V (2013). cGAS produces a 2′−5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, and Hornung V (2014). TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol 192, 5993–5997. [DOI] [PubMed] [Google Scholar]
- Albrecht M, Suezer Y, Staib C, Sutter G, Vieths S, and Reese G (2008). Vaccination with a Modified Vaccinia Virus Ankara-based vaccine protects mice from allergic sensitization. J. Gene Med 10, 1324–1333. [DOI] [PubMed] [Google Scholar]
- Balla T (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev 93, 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch K, Knittler K, Borowski C, Rudnik S, Damme M, Aden K, Spehl-mann ME, Frey N, Saftig P, Chalaris A, and Rabe B (2017). Absence of RNase H2 triggers generation of immunogenic micronuclei removed by auto-phagy. Hum. Mol. Genet 26, 3960–3972. [DOI] [PubMed] [Google Scholar]
- Bhat N, and Fitzgerald KA (2014). Recognition of cytosolic DNA by cGAS and other STING-dependent sensors. Eur. J. Immunol 44, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Teruel M, Dierckman R, Anderson R, Wells A, York JD, Meyer T, and Grinstein S (2000). Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol 151, 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, and Rose JK (1992). Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, and Kaufman PD (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, and Chen ZJ (2016). Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol 17, 1142–1149. [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, and Hopfner KP (2013). Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GMA, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. (2015). Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A 167A, 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li XD, Chen Z, Merghoub T, et al. (2014). Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog 10, e1003989–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, et al. (2017). Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, and Chen ZJ (2018). DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, and Stetson DB (2014). The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat. Immunol 15, 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerit I, and Cerutti PA (1981). Tumour promoter phorbol-12-myristate-13-acetate induces chromosomal damage via indirect action. Nature 293, 144–146. [DOI] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, and Chen ZJ (2015). Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 112, E5699–E5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glück S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, Bridgeman A, Rehwinkel J, Zender L, and Ablasser A (2017). Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol 19, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, et al. (2014). Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, and Stetson DB (2015). Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi–Goutières Syndrome. J. Immunol 195, 1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, and Greenberg RA (2017). Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kröger A, Nilsson JA, et al. (2017). DNA Damage Primes the Type I Interferon System via the Cytosolic DNA Sensor STING to Promote Anti-Microbial. Innate Immun 42, 1–13. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, and Stephens LR (2016). Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways. Biochem. Soc. Trans 44, 307–314. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. (2006). 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, and Barber GN (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønsson KL, Laustsen A, Krapp C, Skipper KA, Thavachelvam K, Hotter D, Egedal JH, Kjolby M, Mohammadi P, Prabakaran T, et al. (2017). IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun 8, 14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Simpson F, Kavran JM, Lemmon MA, and Schmid SL (1998). Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr. Biol 8, 1399–1402. [DOI] [PubMed] [Google Scholar]
- Kagan JC, and Medzhitov R (2006). Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell 125, 943–955. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, and Medzhitov R (2008). TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol 9, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee ASY, Berger JM, and Doudna JA (2013). Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep 3, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, et al. (2014). Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15, 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, and Simons K (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. [DOI] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Höning K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, and Hornung V (2014). Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J 33, 2937–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek LR, and Kagan JC (2012). Phosphoinositide binding by the Toll adaptor dMyD88 controls antibacterial responses in Drosophila. Immunity 36, 612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, and Verma IM (1996). The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses.J. Virol 70, 5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, and Knipe DM (2015). cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. USA 112, E1773–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pépin G, Nejad C, Thomas BJ, Ferrand J, McArthur K, Bardin PG, Williams BRG, and Gantier MP (2017). Activation of cGAS-dependent anti-viral responses by DNA intercalating agents. Nucleic Acids Res 45, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. (2014). Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, and Medzhitov R (2008). Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, and Akira S (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. [DOI] [PubMed] [Google Scholar]
- Tewey KM, Rowe TC, Yang L, Halligan BD, and Liu LF (1984). Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 226, 466–468. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Ren J, Chen Q, and Chen ZJ (2017). cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 114, E4612–E4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, and Chen ZJ (2013). Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, and Nagarajan UM (2014). The DNA Sensor, Cyclic GMP–AMP Synthase, Is Essential for Induction of IFN-β during Chlamydia trachomatis Infection. J. Immunol 193, 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, Gray NS, Mekalanos JJ, and Kranzusch PJ (2018). Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 174, 300–311.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.