Abstract
To understand the mechanism of transcriptional down-regulation of BRCA1 by promoter methylation, we screened 51 breast cancer cell lines and identified HCC38 as another BRCA1 promoter-methylated cell line in addition to UACC3199. There was low expression of BRCA1 mRNA and BRCA1 protein in both cell lines as measured by quantitative RT-PCR and western blot analysis. After transient treatment with 5-aza-2′-deoxycytidine (5-aza-CdR) and trichostatin A (TSA), re-expression of BRCA1 mRNA and BRCA1 protein was detected in UACC3199 cells, but not in HCC38 cells. Another demethylating agent, zebularine, did not induce BRCA1 re-expression in either cell line. To test the hypothesis that methylation of CpG sites may affect accessibility of the BRCA1 promoter to transcription factors and consequently cause down-regulation of BRCA1, we analyzed the binding of four transcription factors (CTCF, Sp1, E2F1 and E2F6) to the BRCA1 promoter using chromatin immunoprecipitation assay (ChIP) and quantitative PCR. CTCF and E2F1 were enriched at the unmethylated BRCA1 promoter in MCF-7 cells. In contrast, these two transcription factors were not enriched at the methylated BRCA1 promoter in UACC3199 and HCC38 cells. Following demethylating drug treatment, E2F1 was enriched at the BRCA1 promoter in the demethylated UACC3199 cells. This indicates that reduced accessibility of transcription factors to the methylated promoter is one of the mechanisms for down-regulation of BRCA1 in heavily methylated cancer cells.
Keywords: Methylation, BRCA1, Promoter, ChIP assay, Transcription factor
Introduction
Approximately 5% of all breast cancers are caused by inherited mutations in breast cancer susceptibility genes such as BRCA1 and BRCA2 [1, 2]. Despite the fact that somatic mutations of the BRCA1 and BRCA2 genes are rare, several lines of evidence implicate reduced BRCA1 gene expression in sporadic tumors [3, 4]. Epigenetic regulation, especially promoter methylation, is an important mechanism for down-regulating tumor suppressor genes in human cancer cells [5–7]. Methylation of the BRCA1 promoter has been reported in sporadic breast cancer with proportions ranging from 11 to 31% [4, 8]. In addition, BRCA1 promoter methylation has been linked to reduced mRNA and protein expression in primary breast tumors and cancer cell lines [3, 9–13]. Previously, we observed that inactivation of BRCA1 by promoter methylation is associated with reduced BRCA1 gene copy number, reduced transcripts and chromosome 17 aneusomy, as has been observed in tumors from BRCA1 mutation carriers. We concluded that BRCA1 promoter methylation contributes to a subset of sporadic breast cancers, with the resulting molecular and clinicopathologic phenotype similar to that of hereditary BRCA1-associated breast cancers [14].
The utilization of drugs that target specific enzymes involved in the epigenetic regulation of gene expression is emerging as an effective and valuable approach to treatment as well as prevention of cancer [15]. 5-aza-CR and 5-aza-CdR are two widely used DNA methyltransferase inhibitors and have been approved by the FDA for the treatment of patients with leukemia [15]. Despite their potency in inhibiting DNA methylation, both drugs are unstable in aqueous solution and show high cytotoxicity [16–18]. Zebularine is an analogue of 5-aza-CR/5-aza-CdR, but it is more stable and less toxic than 5-aza-CR and 5-aza-CdR [19–21]. Exploring the effectiveness of zebularine and other compounds as DNA methylation inhibitors could allow for the development of novel cancer therapies. Tumor suppressors, such as p16, p21 and MLH1, have been extensively examined for re-expression following demethylating drug treatment [7]. However, restoring BRCA1 expression in cancer cells by demethylating agents is still under investigation.
We speculate from the results of bisulfite sequencing of the BRCA1 promoter that CpGs at positions –355 and –21 from the transcription start site are critical in the re-expression of BRCA1 [14]. It has been reported that Specific protein 1(Sp1) and CCCTC binding factor (CTCF) bind to the BRCA1 promoter and play a crucial role in maintaining a methylation-free state of the functional BRCA1 promoter region [22]. The Sp1 binding site includes –355 CpG. In addition, E2F transcription factors bind to the BRCA1 promoter in the region including –21 CpGs and affect BRCA1 promoter-driven reporter activity and BRCA1 expression [23–25]. However, it is largely unknown how methylation affects the accessibility of the BRCA1 promoter to transcription factors. In this study, we screened 51 cell lines and identified HCC38 as another BRCA1 promoter methylated cell line. Subsequently, we evaluated the effect of 5-aza-CdR and zebularine on BRCA1 expression in both cell lines. Finally, we analyzed the binding of four transcription factors (CTCF, Sp1, E2F1 and E2F6) to the BRCA1 promoter and demonstrated that reduced accessibility of the methylated promoter to transcription factors is one of the mechanisms for BRCA1 down-regulation in heavily methylated cancer cells.
Materials and methods
Cell lines
DNA from 51 previously characterized breast cancer cell lines were obtained from Dennis Slamon (Supplemental Table 1). MCF-7 and HCC38 cell lines were obtained from American Type Culture Collection (Rockville, MD). UACC3199 was obtained from the University of Arizona Cancer Center (Tucson, AZ). MCF-7 was cultured in DMEM (Fisher Scientific, Hanover Park, IL). HCC38 and UACC3199 were grown in RPMI 1640 (Invitrogen, Grand Island, NY). The media were supplemented with 10% FBS and 1% penicillin/streptomycin.
Demethylation of HCC38 and UACC3199 cells
Cells were seeded at a density of 1 × 106 per 100-mm plate and exposed to 5-aza-CdR or zebularine (Sigma, St. Louis, MO) at different dose levels for 96 h (with a change of media at 48 h). For UACC3199, 0.2 μM trichostatin A (TSA; Sigma) was added in the last 12 h. Mock-treated cells were used as a control.
Cell growth assay
UACC3199 or HCC38 cells were plated at 5 × 105 per plate and treated with demethylating drugs. Cell growth was measured by counting live cells after staining with trypan blue. Growth inhibition was calculated as: 100 – the number of cells in treated plate/number of cells in untreated plate.
DNA isolation and bisulfite modification
Genomic DNA was extracted from cultured cells with the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN), followed by phenol–chloroform (Invitrogen) extraction. The sodium bisulfite reaction was carried out as described [26].
BRCA1 promoter methylation analysis
Methylation analysis was done using a methylation-specific PCR (MSP) as described previously [27]. The final PCR products of methylated and unmethylated DNA are 84 and 104 bp, respectively.
Sodium bisulfite genomic sequencing of the BRCA1 promoter
The BRCA1 promoter was sequenced for cytosine methylation using a previously described method [14]. The percentage of methylated CpGs was calculated from ten clones. DNA from untreated and demethylated cells was analyzed.
RNA isolation and quantitative RT-PCR
Total cellular RNA was isolated from cultured cells using the Trizol reagent (Invitrogen) and cleaned using the RNAeasy mini kit (Qiagen, Montgomery, MD). Reverse transcriptase reactions were done using the SuperScript III first strand synthesis system (Invitrogen) with 1 μg of RNA and 1 μl of oligo (dT) 12–18 as the reverse transcription primer. Real-time PCR was performed in a Light-Cycler apparatus (Roche Diagnostics, Indianapolis, IN) using the Light-Cycler-FastStart DNA Master plus SYBR Green I Kit (Roche Diagnostics). cDNA primers for BRCA1 were used as described [3]. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a normalization control [14]. At the end of the PCR cycles, melting curve analyses and electrophoresis of the products on 2% agarose gels were performed to validate the generation of the specific PCR product expected. The fold change in BRCA1 cDNA (target gene) relative to the GAPDH control was determined by 2−ΔΔCt method [28].
Protein extraction and western blot
Cells were lysed in RIPA buffer (50 mM Tris pH 7.4; 150 mM NaCl; 1% NP-40; 1 mM EDTA; 10 mM NaF; 1 mM sodium orthovanadate) supplemented with 1 mM PMSF and a protease inhibitor cocktail (Roche, Nutley, NY). About 50 μg of the total protein of each cell lysate was run on a 7.5% acrylamide gel and the proteins were transferred to an Immobilon-P PVDF membrane (Millipore, Bedford, MA). The ECL kit (Amersham Bioscience, Buckinghamshire, UK) was used for detection.
Chromatin immunoprecipitation (ChIP) assays
To examine in vivo transcription factor binding to specific DNA sequences in the BRCA1 promoter, chromatin immunoprecipitation (ChIP) assays were performed following the manufacturer’s protocol (Millipore, Temecula, CA) using 3 × 106 of cells. Each antibody was added to the sonicated and precleared supernatant fraction and incubated overnight at 4°C with rotation. As a control, the addition of antibody was omitted. Conventional or quantitative PCR analysis of ChIP assays was performed using 1/50 of immunoprecipitate or 1% of input DNA. The annealing temperature was 57°C for CTCF/Sp1and 60°C for E2F, respectively. Accumulation of fluorescent products was monitored by real-time PCR and threshold cycles (CTs) were calculated by the Roche software. The specificity of amplification was monitored by the melting-temperature profiles of final products. Relative promoter occupancy was calculated with a previously described equation [29] and fold of enrichment was measured as antibody added over control. Products from both conventional PCR and real-time PCR were run on 2% agarose gel. The following primers were used to amplify the immuno-precipitated DNA fragment. For CTCF and Sp1, forward 5′-GGA TGG GAA TTG TAG TCT CCC T-3′ and reverse 5′-GGA AGC TGG TAA GGA AGC AG-3′ were used. For amplification of E2F site, forward 5′-CGA GAG ACG CTT GGC TCT TTC TGT-3′ and reverse 5′-GCC CAG TTA TCT GAG AAA CCC CAC-3′ primers were used.
Antibodies
Anti-CTCF antibody (sc-15914x), anti-E2F1 (sc-193x), anti-E2F6 (sc-8366x), anti-actin (sc-1616), anti-Dnmt1 and anti-Dnmt3a were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-BRCA1 monoclonal antibody AB-4 was from EMD (San Diego, CA). Anti-Dnmt3b antibody was purchased from Novus Biologicals (Littleton, CO). Anti-Sp1 rabbit antibody was from Millipore (Temecula, CA).
Statistical analyses
After logarithmic transformation, BRCA1 mRNA level was analyzed using two-factor analysis of variance (ANOVA). The first factor was the study group (e.g., UACC3199 vs. MCF-7, or different concentration of 5-aza-CdR) and the second factor was the indicator of two independent experiments. Then, the regression coefficients were transformed back to obtain the fold change or geometric mean difference between study groups. Similarly, enrichment of transcription factors was analyzed using three-factor ANOVA (cell line, antibody versus control, replicate experiment) after logarithmic transformation.
Results
BRCA1 promoter methylation in cultured cancer cells
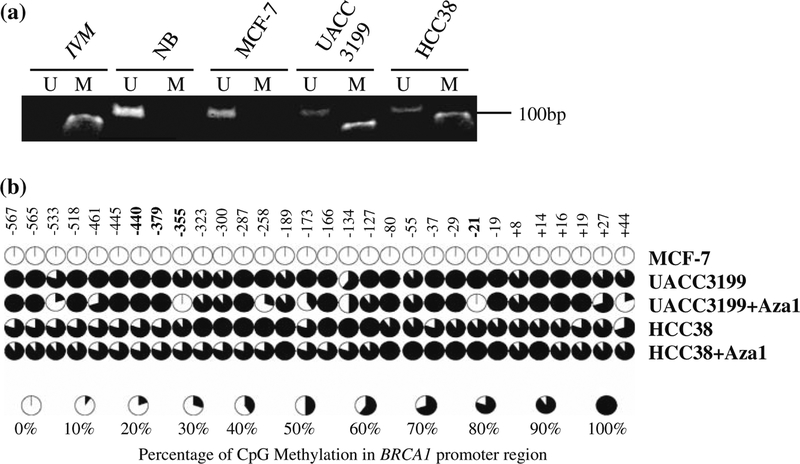
In screening DNA from 51 cultured cancer cell lines by methylation-specific PCR, we identified HCC38 as another BRCA1 promoter methylated cell line in addition to UACC3199. Interestingly, both cell lines have a basal-like pattern of gene expression (data not shown). As shown in Fig. 1a, methylated product was only observed in HCC38 and UACC3199 DNA; in contrast, only unmethylated product was observed in DNA of normal breast tissue or MCF-7. We further confirmed MSP results by bisulfite sequencing of 658 bp proximal BRCA1 promoter, which contains 30 CpG islands. The 30 CpGs were completely unmethylated in MCF-7 cells. In contrast, all of the 30 CpGs were methylated in HCC38 and UACC3199 cells (Fig. 1b).
Fig. 1.
Methylation analyses of BRCA1 in breast cancer cell lines. a Methylation-specific PCR analysis of BRCA1 promoter with DNA from MCF-7, UACC3199, and HCC38 cell lines. In vitro methylated DNA (IVM) was used as a positive control and DNA from normal breast tissue (NB) was used as a negative control. M lanes contain methylated products; U lanes contain unmethylated products. b Bisulfite genomic DNA sequencing of the BRCA1 promoter in MCF-7, UACC3199, and HCC38 cells with or without demethylation. The treatment of 5-aza-CdR and TSA led to demethylation of certain CpG islands of BRCA1 promoter in UACC3199 cells but not in HCC38 cells
Effects of 5-aza CdR and zebularine on cell growth and DNMT protein levels
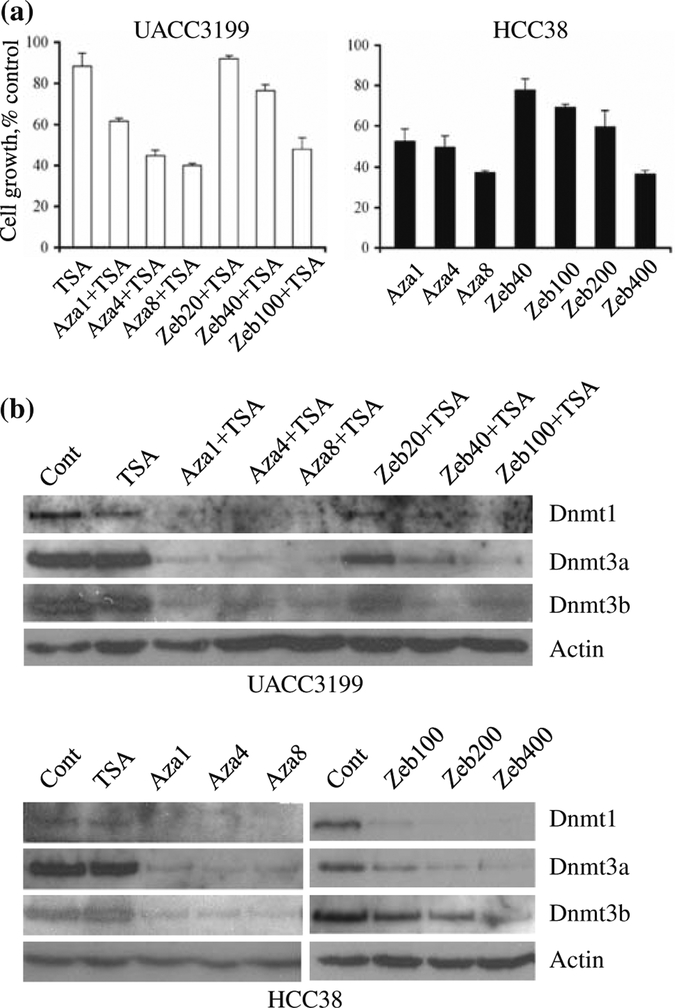
To optimize demethylation condition, we analyzed cell growth and DNMT levels after drug treatment. Cell growth was measured by trypan blue exclusion assay. UACC3199 cells were treated with 1–8 μM 5-aza-CdR or 20–100 μM zebularine for 96 h combined with 0.2 μM TSA. We observed a 38–60% growth reduction in UACC3199 treated with 5-aza-CdR/TSA and 8–52% growth reduction with zebularine/TSA treatment compared with untreated cells (Fig. 2a). The combination of 5-aza-CdR or zebularine with TSA led to a dramatic decrease in the growth of HCC38 cells (data not shown). Hence, HCC38 cells were treated with 1–8 μM 5-aza-CdR or 40–400 μM zebularine for 96 h. HCC38 showed 48–63% growth reduction with 5-aza-CdR and 22–63% growth reduction with zebularine (Fig. 2a).
Fig. 2.
Response of UACC3199 and HCC38 cells to 5-aza-CdR and zebularine. a Effect of 5-aza-CdR and zebularine on growth of UACC3199 and HCC38 cells. UACC3199 and HCC38 cells were treated with the indicated dose of 5-aza-CdR or zebularine for 96 h and cell growth was monitored by trypan blue exclusion assay. Results are represented as the average of two independent experiments with triplicates. Bars represent SE. b Effect of 5-aza-CdR or zebularine on the protein levels of DNMT1, DNMT3a, and DNMT3b in UACC3199 and HCC38 cells. UACC3199 or HCC38 cells were treated with the indicated dose of 5-aza-CdR or zebularine for 96 h, and the soluble protein levels of DNMTs were analyzed by western blot
The effect of 5-aza-CdR, zebularine and HDAC inhibitor TSA on DNMT protein levels was analyzed by western blot. HDAC inhibitor TSA alone had no effect on soluble DNMT protein levels in both UACC3199 and HCC38 cells. Soluble DNMT1, DNMT3a and DNMT3b were depleted by treatment with 1 μM or more of 5-aza-CdR for 96 h in both UACC3199 and HCC38 cells (Fig. 2b). UACC3199 was more sensitive to zebularine treatment compared to HCC38. The DNMTs were reduced by 40 μM zebularine and depleted by 100 μM zebularine in UACC3199 cells. In HCC38 cells, DNMT1 and DNMT3a were depleted by 200 μM zebularine; DNMT3b was reduced by 200 μM zebularine and depleted by 400 μM zebularine.
Demethylation and BRCA1 gene expression in cancer cells
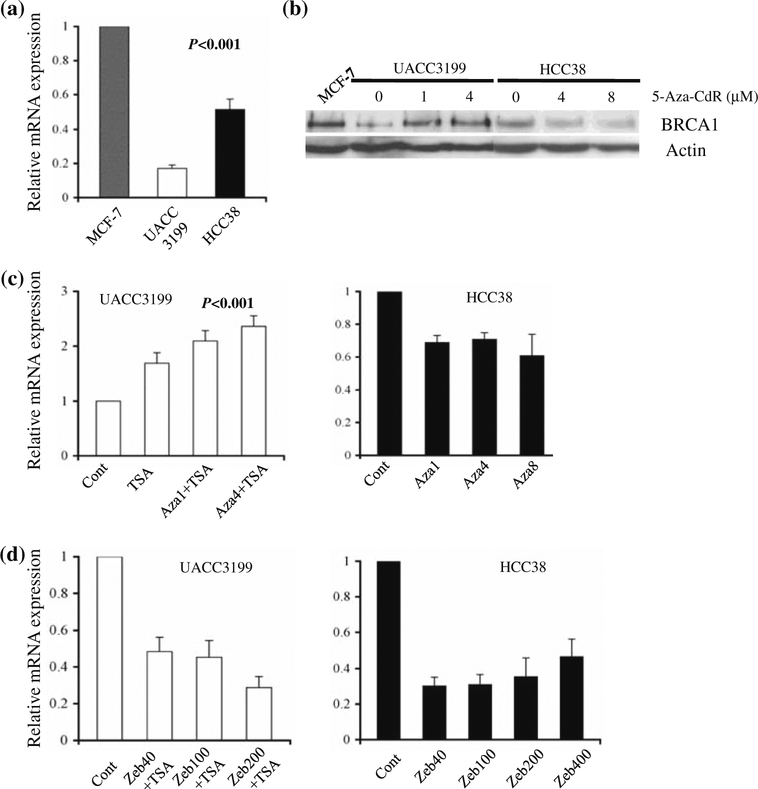
BRCA1 gene expression was analyzed in untreated MCF-7, HCC38 and UACC3199 cells by quantitative RT-PCR and western blot. As shown in Fig. 3a, the amount of BRCA1 mRNA in UACC3199 and HCC38 was 20 and 50%, respectively, of that in MCF-7. Little protein expression was detected in UACC3199 and HCC38 cells (Fig. 3b). After treatment with 5-aza-CdR and TSA, we observed a two to three fold increase in BRCA1 mRNA expression in UACC3199 cells (Fig. 3c). Concordantly, a combination of 1 μM 5-aza-CdR and 0.2 μM TSA was enough to induce BRCA1 protein re-expression in demethylated UACC3199 cells (Fig. 3b). Surprisingly, 5-aza-CdR treatment was unable to induce BRCA1 re-expression in HCC38 cells (Fig. 3c). As measured by real-time RT-PCR, BRCA1 mRNA expression decreased 30–40% in HCC38 cells treated with 1–8 μM 5-aza-CdR compared with untreated cells, despite DNMTs being reduced or depleted by the drug treatment. DNA was extracted from HCC38 cells after 5-aza-CdR treatment and BRCA1 promoter was analyzed by bisulfite sequencing. There was no change in BRCA1 promoter methylation after drug treatment in HCC38 cells in contrast to that observed in UACC3199 (Fig. 1b).
Fig. 3.
Down-regulation of BRCA1 gene expression by promoter methylation in UACC3199 and HCC38 cells. a Quantitative RT-PCR analysis of BRCA1 mRNA expression in untreated MCF-7, UACC3199 and HCC38 cells. Real-time PCR was performed in triplicate in two independent experiments. Bar shows the mean ± SE. b Western blot analysis of BRCA1 protein expression in untreated and demethylated UACC3199, and HCC38 cells. MCF-7 was used as a positive control and Actin served as a loading control. c BRCA1 mRNA expression in UACC3199 and HCC38 cells after treatment with indicated dose of 5-aza-CdR. d BRCA1 mRNA expression in UACC3199 and HCC38 cells after treatment with indicated dose of zebularine
Since zebularine has been shown to be effective in reactivating tumor suppressors, such as p16 [21], we treated both UACC3199 and HCC38 cells with a series of doses of zebularine with and without TSA and measured BRCA1 mRNA expression by q-RT-PCR. Zebularine failed to induce BRCA1 expression in both cell lines; instead, we observed lower BRCA1 expression after zebularine treatment (Fig. 3d).
CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors
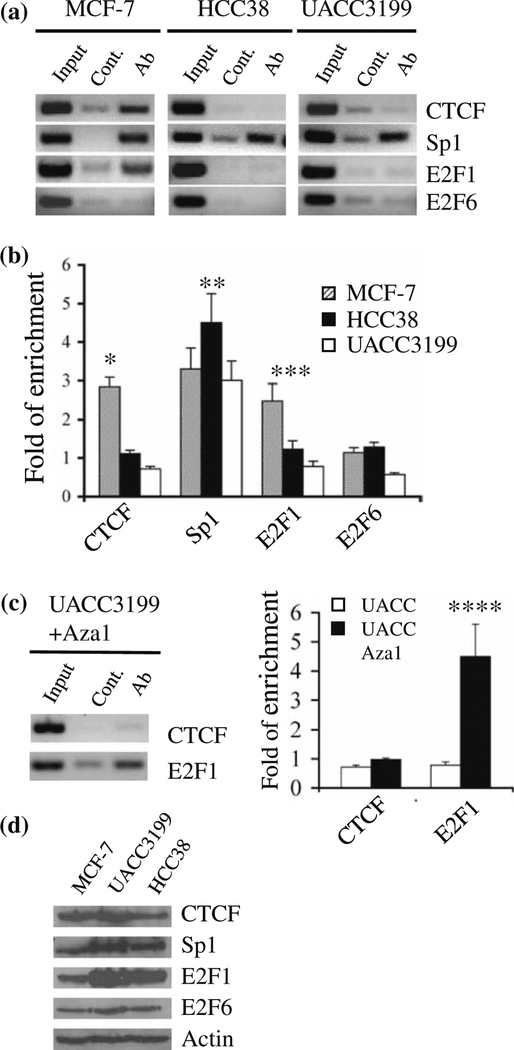
Previous work has showed that transcription factors such as Sp1 and E2F1 bind to the proximal BRCA1 promoter region that spans the CpG dinucleotides located at positions –355 and –21, respectively. Those CpG sites were completely unmethylated in MCF-7 cells. In contrast, both CpGs were highly methylated in UACC3199 and HCC38 cells. This indicates that methylation of those CpG islands may affect accessibility of BRCA1 promoter to transcription factors. To understand how methylation affects the accessibility of the BRCA1 promoter to transcription factors, we analyzed the binding of four transcription factors (CTCF, Sp1, E2F1 and E2F6) to the BRCA1 promoter using chromatin immunoprecipitation assay (ChIP) and quantitative real-time PCR. As shown in Fig. 4a, b, CTCF and E2F1 were enriched at the unmethylated BRCA1 promoter in MCF-7 cells. In contrast, these two transcription factors were not enriched at the methylated BRCA1 promoter in UACC3199 and HCC38 cells even though there was more E2F1 expression in those cell lines (Fig. 4d). Interestingly, E2F1 was enriched at the demethylated BRCA1 promoter in UACC3199 cells treated with 1 μM 5-aza-CdR/0.2 μM TSA, while CTCF enrichment was not detected at the BRCA1 promoter with the same treatment (Fig. 4c). Enrichment of Sp1 at the BRCA1 promoter was observed in all three cell lines, indicating that the binding of Sp1 to the BRCA1 promoter is methylation insensitive. No E2F6 enrichment at the BRCA1 promoter was detected in any cell line that had been tested. To prove that the enrichment was not due to protein expression level differences in the cell lines, western blot was performed using MCF-7, UACC3199 and HCC38 cell extracts probed with antibodies against the four transcription factors. As shown in Fig. 4d, protein expression levels of CTCF, Sp1 and E2F6 were roughly equal in the three cell lines, while E2F1 was more abundant in UACC3199 and HCC38 cells.
Fig. 4.
CpG island methylation affects accessibility of the BRCA1 promoter to transcription factors. a Representative gel images of BRCA1 amplification products obtained by conventional PCR with DNA from ChIP assays. ChIP assays were carried out with MCF-7, HCC38, and UACC3199 using CTCF, Sp1, E2F1, and E2F6 antibodies. b Quantitative analysis of BRCA1 promoter occupancy in untreated MCF-7, HCC38, and UACC3199 cells by real-time PCR with two independent ChIP assays. The fold change was calculated as antibody versus control. Enrichment of transcription factors was analyzed using a three factor ANOVA (cell line, antibody vs. control, replicate experiment) after logarithmic transformation. P values were calculated based on the differences among the three cell lines for each transcription factor. * P < 0.001; ** P = 0.21; *** P < 0.001. c Conventional PCR and quantitative analysis of BRCA1 promoter occupancy in demethylated UACC3199 cells with two independent ChIP assays. P value was calculated based on the differences between untreated and demethylated UACC3199. ****P < 0.001. d Protein expression of CTCF, Sp1, E2F1, and E2F6 in MCF-7, UACC3199, and HCC38 cells by western blot analysis. Actin was used as a loading control
Discussion
There are multiple factors involved in the transcriptional regulation of BRCA1 [4]. However, it is not known how methylation affects accessibility of the proximal BRCA1 promoter to transcription factors and contributes to the down-regulation of BRCA1 gene expression in breast cancer cells. In this study, we have identified HCC38 as another BRCA1 methylated cell line. We evaluated the effects of demethylating agents on induction of BRCA1 re-expression in two BRCA1 promoter-methylated cell lines. Using the ChIP assay, we demonstrated that reduced accessibility of the methylated proximal BRCA1 promoter to transcription factors is one of the mechanisms accounting for down-regulation of BRCA1 in heavily methylated UACC3199 and HCC38 cancer cells.
CTCF is required for the enhancer blocking activity of vertebrate insulators and CTCF binding is largely sensitive to CpG methylation [30–33]. CTCF-dependent chromatin insulation is linked to epigenetic remodeling [34]. It has been suggested that Sp1 and CTCF may play roles as insulators in maintaining a methylation-free state of the proximal BRCA1 promoter region in unmethylated cells, such as MCF-7 or normal breast tissue [22]. There is no CpG within the CTCF binding site, but there are two CpGs (–440 and –379) flanking the CTCF binding site. We observed that CTCF only binds to unmethylated BRCA1 promoter but not the methylated promoter. This indicates that CTCF binding is affected by the methylation status of the flanking region. In contrast, Sp1 binding to the BRCA1 promoter was methylation insensitive. Taken together, this suggests that only CTCF is involved in protecting the adjacent sequences against de novo CpG methylation at the proximal BRCA1 promoter.
Disruption of CTCF binding by methylation in UACC-3199 and HCC38 cells may facilitate aberrant methylation of the proximal BRCA1 promoter and, consequently, affect other transcription activator binding. Indeed, we observed E2F1 binding to the unmethylated BRCA1 promoter in MCF-7 cells but not in the methylated UACC3199 and HCC38 cells. It has been reported that E2F1 is a positive transcriptional regulator of BRCA1 [23–25]. The methylation within the E2F binding site may inhibit binding of E2F1 transcription factors to the BRCA1 promoter and, consequently, cause down-regulation of BRCA1 expression in those cells. Our results demonstrate that is the case since we observed concordant BRCA1 up-regulation and E2F1 enrichment at the BRCA1 promoter after demethylation.
E2F6 has been identified as a transcriptional repressor of BRCA1, and depletion of E2F6 resulted in the recruitment of E2F1 to the BRCA1 promoters in 293 cells [24]. Therefore, we thought E2F6 might play a role in regulating BRCA1 in breast cancer cells. Enrichment of E2F6 at the BRCA1 promoter was not detected in MCF-7, UACC3199 and HCC38 cells in the present study, suggesting that E2F6 is not involved in regulation of BRCA1 expression in those cell lines.
5-aza-CdR is being used for treatment of leukemia [35–37]. We successfully obtained BRCA1 protein re-expression using a combination of 5-aza-CdR and the HDAC inhibitor TSA in breast cancer cell line UACC3199. This suggests that down-regulation of BRCA1 by methylation is reversible, and demethylation may be applied clinically in the treatment or prevention of breast cancers. However, 5-aza-CdR failed to demethylate BRCA1 promoter and induce BRCA1 re-expression in HCC38 cells, despite DNMTs being reduced or depleted by the drug treatment. While the exact mechanism is still elusive, the different responses of cell lines to demethylating drugs may be reflective of different responses patients have to demethylating drugs, which should be addressed in future studies.
Zebularine has been considered as a potential demethylating drug because of its high stability and less toxicity compared with 5-aza-CdR [38, 39]. In a previous study, five of seven cancer cell lines treated with zebularine showed p16 induction [21]. Even though we observed lower toxicity of zebularine compared with 5-aza-CdR, as measured by growth inhibition assay, we could not detect up-regulation of BRCA1 with up to 400 μM and a 9-day treatment with HCC38 cells. Other non-nucleoside demethylation drugs, such as EGCG and procaine, were also not effective on HCC38 cells (data not shown). The mechanism for non-responsiveness in HCC38 deserves further study.
Supplementary Material
Acknowledgments
Grant support: National Institute of Environmental Health Sciences grant P50 ES012382, NIH-National Cancer Institute Cancer Center Support grant 3P30 CA 23074, the Breast Cancer Research Foundation, the National Women’s Cancer Research Alliance, and the Falk Medical Research Trust (O.I. Olopade). We thank Olopade lab members for helpful discussion. We also thank Michelle Porcellino and Lisa Sveen for critical reading of the manuscript.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-009-0422-1) contains supplementary material, which is available to authorized users.
Contributor Information
Jinhua Xu, Department of Medicine, Center for Clinical Cancer Genetics, University of Chicago, Chicago, IL 60637-1463, USA.
Dezheng Huo, Department of Health Studies, University of Chicago, Chicago, IL 60637-1463, USA.
Yinghua Chen, Department of Medicine, Center for Clinical Cancer Genetics, University of Chicago, Chicago, IL 60637-1463, USA.
Chika Nwachukwu, Department of Medicine, Center for Clinical Cancer Genetics, University of Chicago, Chicago, IL 60637-1463, USA.
Cindy Collins, Department of Medicine, Center for Clinical Cancer Genetics, University of Chicago, Chicago, IL 60637-1463, USA.
Janelle Rowell, Department of Medicine, Center for Clinical Cancer Genetics, University of Chicago, Chicago, IL 60637-1463, USA.
Dennis J. Slamon, Department of Medicine, University of California at Los Angeles, Los Angeles, CA 90095, USA
Olufunmilayo I. Olopade, Department of Human Genetics, University of Chicago, Chicago, IL 60637-1463, USA Department of Medicine, Center for Clinical Cancer Genetics, University of Chicago, Chicago, IL 60637-1463, USA.
References
- 1.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y et al. (1994) BRCA1 mutations in primary breast and ovarian carcinomas. Science 266(5182):120–122. doi: 10.1126/science.7939630 [DOI] [PubMed] [Google Scholar]
- 2.Newman B, Mu H, Butler LM, Millikan RC, Moorman PG, King MC (1998) Frequency of breast cancer attributable to BRCA1 in a population-based series of American women. JAMA 279(12): 915–921. doi: 10.1001/jama.279.12.915 [DOI] [PubMed] [Google Scholar]
- 3.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA et al. (2000) Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92(7):564–569. doi: 10.1093/jnci/92.7.564 [DOI] [PubMed] [Google Scholar]
- 4.Mueller CR, Roskelley CD (2003) Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Res 5(1):45–52. doi: 10.1186/bcr557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3(6):415–428 [DOI] [PubMed] [Google Scholar]
- 6.Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349(21):2042–2054. doi: 10.1056/NEJMra023075 [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128(4):683–692. doi: 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catteau A, Morris JR (2002) BRCA1 methylation: a significant role in tumour development? Semin Cancer Biol 12(5):359–371. doi: 10.1016/S1044-579X(02)00056-1 [DOI] [PubMed] [Google Scholar]
- 9.Rice JC, Massey-Brown KS, Futscher BW (1998) Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene 17(14):1807–1812. doi: 10.1038/sj.onc.1202086 [DOI] [PubMed] [Google Scholar]
- 10.Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D et al. (1999) Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet 21(2):236–240. doi: 10.1038/6029 [DOI] [PubMed] [Google Scholar]
- 11.Rice JC, Ozcelik H, Maxeiner P, Andrulis I, Futscher BW (2000) Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis 21(9):1761–1765. doi: 10.1093/carcin/21.9.1761 [DOI] [PubMed] [Google Scholar]
- 12.Okochi E, Miyamoto K, Wakazono K, Shima H, Sugimura T, Ushijima T (2002) Reduced Brca1 protein expression in 2-amino-1-methyl-6-phenylimidazo[4, 5-b]pyridine-induced rat mammary carcinomas. Mol Carcinog 34(4):211–218. doi: 10.1002/mc.10065 [DOI] [PubMed] [Google Scholar]
- 13.Matros E, Wang ZC, Lodeiro G, Miron A, Iglehart JD, Richardson AL (2005) BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res Treat 91(2):179–186. doi: 10.1007/s10549-004-7603-8 [DOI] [PubMed] [Google Scholar]
- 14.Wei M, Grushko TA, Dignam J, Hagos F, Nanda R, Sveen L, Xu J, Fackenthal J, Tretiakova M, Das S et al. (2005) BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res 65(23):10692–10699. doi: 10.1158/0008-5472.CAN-05-1277 [DOI] [PubMed] [Google Scholar]
- 15.Yoo CB, Jones PA (2006) Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5(1):37–50. doi: 10.1038/nrd1930 [DOI] [PubMed] [Google Scholar]
- 16.Lin KT, Momparler RL, Rivard GE (1981) High-performance liquid chromatographic analysis of chemical stability of 5-aza-2′-deoxycytidine. J Pharm Sci 70(11):1228–1232. doi: 10.1002/jps.2600701112 [DOI] [PubMed] [Google Scholar]
- 17.Juttermann R, Li E, Jaenisch R (1994) Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA 91(25):11797–11801. doi: 10.1073/pnas.91.25.11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F (2006) Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res 66(5):2794–2800. doi: 10.1158/0008-5472.CAN-05-2821 [DOI] [PubMed] [Google Scholar]
- 19.Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU (2003) Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst 95(5):399–409 [DOI] [PubMed] [Google Scholar]
- 20.Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, Marquez VE, Jones PA (2004) Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol 24(3):1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, Marquez VE, Greer S, Orntoft TF, Thykjaer T et al. (2004) Preferential response of cancer cells to zebularine. Cancer Cell 6(2):151–158. doi: 10.1016/j.ccr.2004.06.023 [DOI] [PubMed] [Google Scholar]
- 22.Butcher DT, Mancini-DiNardo DN, Archer TK, Rodenhiser DI (2004) DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int J Cancer 111(5):669–678. doi: 10.1002/ijc.20324 [DOI] [PubMed] [Google Scholar]
- 23.Wang A, Schneider-Broussard R, Kumar AP, MacLeod MC, Johnson DG (2000) Regulation of BRCA1 expression by the Rb-E2F pathway. J Biol Chem 275(6):4532–4536. doi: 10.1074/jbc.275.6.4532 [DOI] [PubMed] [Google Scholar]
- 24.Oberley MJ, Inman DR, Farnham PJ (2003) E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem 278(43):42466–42476. doi: 10.1074/jbc.M307733200 [DOI] [PubMed] [Google Scholar]
- 25.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM (2005) Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res 65(24):11597–11604. doi: 10.1158/0008-5472.CAN-05-2119 [DOI] [PubMed] [Google Scholar]
- 26.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93(18): 9821–9826. doi: 10.1073/pnas.93.18.9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei M, Xu J, Dignam J, Nanda R, Sveen L, Fackenthal J, Grushko TA, Olopade OI (2008) Estrogen receptor alpha, BRCA1, and FANCF promoter methylation occur in distinct subsets of sporadic breast cancers. Breast Cancer Res Treat 111(1):113–120. doi: 10.1007/s10549-007-9766-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 29.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15(16): 2069–2082. doi: 10.1101/gad.906601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98(3):387–396. doi: 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- 31.Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ (2001) CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet 28(4):335–343. doi: 10.1038/ng570 [DOI] [PubMed] [Google Scholar]
- 32.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405 (6785):486–489. doi: 10.1038/35013106 [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa H, Chadwick RB, Peltomaki P, Plass C, Nakamura Y, de La Chapelle A (2001) Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci USA 98(2):591–596. doi: 10.1073/pnas.011528698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishihara K, Oshimura M, Nakao M (2006) CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23(5):733–742. doi: 10.1016/j.molcel.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 35.Wijermans P, Lubbert M, Verhoef G, Bosly A, Ravoet C, Andre M, Ferrant A (2000) Low-dose 5-aza-2’-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: a multicenter phase II study in elderly patients. J Clin Oncol 18(5):956–962 [DOI] [PubMed] [Google Scholar]
- 36.Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J et al. (2004) Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103(5):1635–1640. doi: 10.1182/blood-2003-03-0687 [DOI] [PubMed] [Google Scholar]
- 37.Issa JP, Gharibyan V, Cortes J, Jelinek J, Morris G, Verstovsek S, Talpaz M, Garcia-Manero G, Kantarjian HM (2005) Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol 23(17):3948–3956. doi: 10.1200/JCO.2005.11.981 [DOI] [PubMed] [Google Scholar]
- 38.Yoo CB, Cheng JC, Jones PA (2004) Zebularine: a new drug for epigenetic therapy. Biochem Soc Trans 32(Pt 6):910–912. doi: 10.1042/BST0320910 [DOI] [PubMed] [Google Scholar]
- 39.Chuang JC, Yoo CB, Kwan JM, Li TW, Liang G, Yang AS, Jones PA (2005) Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol Cancer Ther 4(10):1515–1520. doi: 10.1158/1535-7163.MCT-05-0172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.