SUMMARY
Neuronal and synaptic loss is characteristic in many neurodegenerative diseases, such as frontotemporal dementia and Alzheimer’s disease. Recently, we showed that inducing gamma oscillations with visual stimulation (Gamma ENtrainment Using Sensory stimuli, GENUS) reduced amyloid-plaques and phosphorylated-tau in multiple mouse models. Whether GENUS can impact neurodegeneration or cognitive performance remains unknown. Here, we demonstrate that GENUS can entrain gamma oscillations in the visual cortex, hippocampus, and prefrontal cortex in Tau P301S and CK-p25 mouse models of neurodegeneration. Tau P301S and CK-p25 mice subjected to chronic daily GENUS from the early stages of neurodegeneration showed a preservation of neuronal and synaptic density across multiple brain areas and modified cognitive performance. Our transcriptomic and phosphoproteomic data suggest that chronic GENUS shifts neurons towards a less degenerative state, improving synaptic function, enhancing neuroprotective factors and reducing DNA damage in neurons while also reducing inflammatory response in microglia.
Graphical Abstract
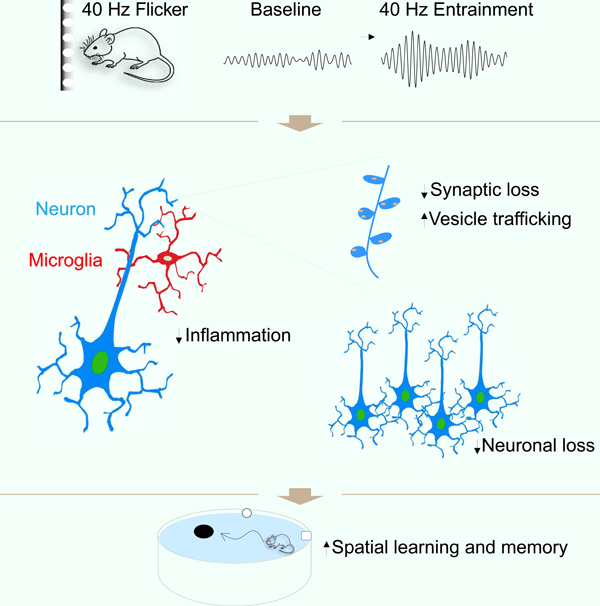
eTOC Blurb
Chronic application of patterned visual stimulation in neurodegeneration mouse models to entrain gamma oscillations results in preservation of neuronal and synaptic density across multiple brain regions.
INTRODUCTION
Neurodegenerative diseases, such as Alzheimer’s disease (AD), are characterized by deterioration of the brain and cognitive dysfunction (Canter et al., 2016; Palop and Mucke, 2016). Multiple factors contribute to the pathogenesis of these diseases, including amyloid-β deposition, tau accumulation, microglia- and astrocyte-mediated inflammation, loss of neurons and synapses, and altered network oscillations (for reviews see, Canter et al., 2016; Heneka et al., 2015; Palop and Mucke, 2016). Human AD patients show reduced power of oscillations in the gamma frequency band (~30–120 Hz) (Guillon et al., 2017; Koenig et al., 2005; Ribary et al., 1991; Stam et al., 2008; Stam et al., 2002), a phenotype that is replicated in multiple AD and AD-risk mouse models (Gillespie et al., 2016; Iaccarino et al., 2016; Verret et al., 2012). Moreover, changing neural activity has been shown to impact AD pathology, such as amyloid-β and tau accumulation, in multiple mouse models (Bero et al., 2011; Cirrito et al., 2005; Cirrito et al., 2008; Tampellini et al., 2010; Wu et al., 2016; Yamada et al., 2014; Yamamoto et al., 2015). Recent studies thus investigate whether manipulating neural oscillations can be effective in ameliorating AD pathology (Iaccarino et al., 2016; Kastanenka et al., 2017; Martinez-Losa et al., 2018; Verret et al., 2012).
Increasing gamma oscillations through genetic modification of Nav1.1 expression in parvalbumin-positive (PV+) cells or interneuron progenitors reduced epileptiform activity and cognitive decline in hAPP-J20 mice (Martinez-Losa et al., 2018; Verret et al., 2012). Optogenetic activation of PV+ interneurons at 40 Hz, which induces robust gamma oscillations (Cardin et al., 2009; Sohal et al., 2009), has also been shown to reduce amyloid load in 5XFAD mice (Iaccarino et al., 2016). In the latter study, we also utilized the well-studied phenomenon that neural oscillations can be induced in cortical regions by patterned sensory stimuli (Herrmann, 2001; Pastor et al., 2003; Rager and Singer, 1998). We applied a light flickering at 40 Hz and showed that this non-invasive approach of Gamma ENtrainment Using Sensory stimuli (GENUS) effectively decreased amyloid levels in the visual cortex of young 5XFAD (Iaccarino et al., 2016). Promisingly, extending 1h of GENUS to 1h/d for 7-days reduced not only amyloid levels (Aβ1–40 and Aβ1–42), but also plaque pathology in the visual cortex in 6-months old 5XFAD (Iaccarino et al., 2016). GENUS also modified microglia morphology, consistent with increased phagocytic activity (Iaccarino et al., 2016; Wang et al., 2015). The potential for a stronger response with prolonged GENUS raises the possibility that this approach may have an impact on neurodegeneration.
In this study, we determined whether longer-term application of GENUS could elicit a neuroprotective effect and possibly modify behavioral performance, using two neurodegenerative disease mouse models, CK-p25 and Tau P301S. We found that 40 Hz visual stimulation (GENUS) significantly increased gamma power in visual cortex and higher order brain areas, including hippocampus and prefrontal cortex, and induced functional binding at low gamma frequencies across these structures. In both neurodegeneration mouse models we tested, we found that chronic GENUS reduced the loss of neuronal and synaptic density in these regions and improved performance in the water maze task. Our cell type-specific transcriptomic profiling and phosphoproteomic analysis revealed an association of GENUS with reduced microglia-mediated inflammation and DNA damage-associated cytotoxicity in neurons, and enhanced synaptic function in neurons.
RESULTS
GENUS recruits higher order brain areas
We first determined if 40 Hz visual stimulation modulates neuronal activity in brain areas beyond primary visual cortex by performing c-Fos immunostaining as a marker of neuronal activation in C57BL/6J mice (Figure 1A). We used a custom-made LED device to deliver visual stimuli at specified frequencies with a 50% duty cycle (Iaccarino et al., 2016; Singer et al., 2018). 40 Hz visual stimulation increased the number of c-Fos+ cells in visual cortex (V1) (Figure 1A, 1B and Figure S1A, S1B), as well as in somatosensory cortex (SS1), hippocampal area CA1 and dentate gyrus (DG), and cingulate cortex (CC) of the prefrontal cortex (PFC) (Figure 1B and Figure S1A, S1B).
Figure 1. 40 Hz visual stimulation entrains gamma oscillations beyond visual cortex.
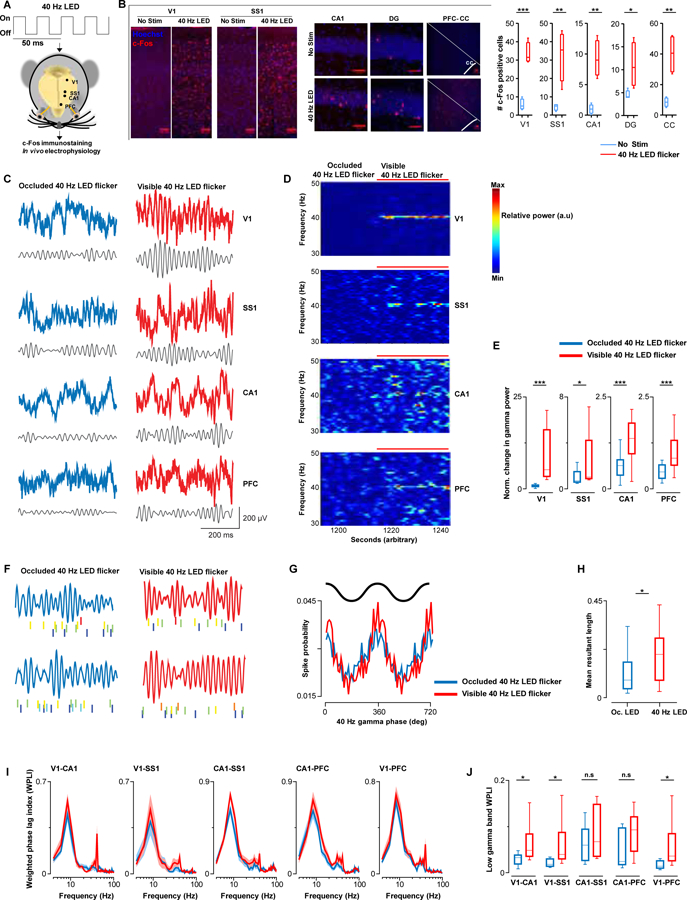
(A) Experimental protocol, and target regions for analyses is indicated.
(B) Representative c-Fos (red) immunostaining images, and Hoechst labeling of cell nuclei (blue). Scale bar 50 μm. 40 Hz visual stimulation increased c-Fos expression (N = 4 mice/group. T-test; V1, t = 8.312, p = 0.0002; SS1, t = 4.071, p = 0.006; CA1, t = 5.33, p = 0.0018; DG, t = 2.56, p = 0.042; and CC, t = 4.771, p = 0.003).
(C) Representative raw and filtered (30–50 Hz) LFP traces recorded concurrently in V1, SS1, CA1, and PFC with the LED delivering 40 Hz visual stimulation occluded (blue) and visible (red).
(D) Representative spectra of LFPs recorded simultaneously from V1, SS1, CA1, and PFC.
(E) Normalized group gamma power (see also, Figure S1C) (N = 7 mice. Wilcoxon-Ranksum test; V1, Z = 5.9, p < 0.0001; SS1, Z = 2.4, p = 0.018; CA1, Z = 3.4, p < 0.0001; and PFC, Z = 3.3, p < 0.0001).
(F) Raster plots of single CA1 units (labeled in different colors) with concurrently recorded LFP (band-pass filtered for 30–50 Hz) from two representative mice.
(G) Spike probability of all isolated CA1 units across 40 Hz phase.
(H) Phase locking strength of neuronal spikes to local LFP analyzed by mean resultant length (N = 24 cells from 4 mice. Wilcoxon-Ranksum, Z = 2.5, p = 0.011). Mean firing rate of single CA1 units did not differ between occluded (2.0 ± 0.12 Hz) and visible 40 Hz stimulation (2.1 ± 0.13 Hz) (Z = 0.55, p = 0.58).
(I) LFP coherence between pairs of recording sites, as indicated, quantified using WPLI (N = 7 mice; 40 Hz visual stimulation occluded (blue) and visible (red)).
(J) Group changes in low gamma band (30–50 Hz) WPLI, related to Figure 1I (Wilcoxon-Ranksum; V1-CA1, p = 0.03; V1-SS1, p = 0.021; and V1-PFC, p = 0.014; n.s. = not significant).
To understand how GENUS might alter ongoing neural activity across these areas, we next implanted C57BL/6J mice with a multi-electrode probe to record local field potentials (LFP) concurrently from V1, SS1, CA1, and PFC. Acute 40 Hz visual stimulation increased the power of 40 Hz gamma oscillations (peak frequency at 40 Hz) in V1 (Figure 1C–1E and Figure S1C, S1D). Significant increases in gamma power were also observed in CA1, SS1, and PFC (Figure 1C–1E and Figure S1C, S1D). To assess entrainment of neuronal firing outside of V1, we recorded single unit activity using tetrodes targeted to hippocampal area CA1 (Figure 1F). During baseline conditions when the light was occluded, CA1 pyramidal cells showed phase-locking to low gamma with preferential discharge at the peak (Figure 1F, 1G) (Middleton and McHugh, 2016). With the 40 Hz visual stimulation visible, we observed no change in the preferred phase of individual CA1 neurons but robust phase-locking, quantified as a significant increase in mean resultant length across the population (Figure 1H), suggesting that 40 Hz stimulation drives populations of hippocampal neurons in a more temporally organized manner. We hypothesized that GENUS might act to coordinate neuronal activity between V1, SS1, CA1, and PFC. To test this, we calculated coherence across these brain regions using the weighted phase lag index (WPLI) method which minimizes potential contamination through volume conduction (Vinck et al., 2011). Acute 40 Hz stimulation increased 30–50 Hz gamma coherence between V1-CA1, V1-SS1, and V1-PFC (Figure 1I, 1J).
As we planned to examine the effects of a chronic GENUS paradigm (with the longest at 6 weeks’ duration), we next carried out long-term multi-site LFP recordings as mice were exposed to 40 Hz stimulation for 6-weeks to determine if gamma entrainment persists after repeated exposure to the stimuli. As we observed with acute 40 Hz stimulation, LFPs recorded on day 43 from V1, SS1, CA1, and PFC showed significantly enhanced 40 Hz gamma power during visible 40 Hz stimulation (Figure S1E), and significantly increased 30–50 Hz gamma WPLI between V1-CA1, V1-SS1, V1-PFC, and CA1-PFC, compared to light-occluded periods (Figure S1F). As a whole, these data show that 40 Hz stimulation enhances local as well as coordinated inter-areal oscillatory activities across V1, CA1, SS1 and PFC. To determine if this increase in low gamma power and coherence is specific to 40 Hz stimulation, we exposed C57BL/6J mice to 80 Hz stimulation (50% duty cycle) (Figure S1G). We observed no significant changes in neither 40 Hz nor 80 Hz spectral power in V1 during 80 Hz stimulation compared to light occluded periods (Figure S1H, S1I, S1J).
GENUS reduces neurodegeneration
To test if GENUS can provide neuroprotection in models of neurodegeneration, we utilized Tau P301S (PS19) mice, which show synaptic and neuronal loss, and cognitive deficits by 8-months old (Yoshiyama et al., 2007). Although we aimed to test the potential for neuroprotection early in pathological progression, we first confirmed that 40 Hz stimulation could entrain neural oscillations even in conditions of neurodegeneration. We found significantly enhanced LFP gamma power in 8-months old P301S in response to 40 Hz stimulation in V1, CA1, and PFC (Figure 2A and Figure S2A–S2C). To explore the potential for chronic GENUS to impact neurodegeneration, we subjected a cohort of P301S to either no stimulation (No Stim) or GENUS 1h/d for 22-days, starting at 7.5-months old (the time point when neuronal loss begins) and assessed neurodegeneration at 8-months old (Figure 2B). Immunohistochemical (IHC) analysis with the neuronal marker, NeuN (Supplemental video1), confirmed that No Stim P301S have reduced NeuN+ cells in V1, and CA1 (Figure 2B, 2C). In contrast, the number of NeuN+ cells in GENUS P301S did not differ from wildtype (WT) littermates in V1, and CA1 (Figure 2B, 2C and Figure S2D). We obtained similar results using fluorescence activated cell sorting (FACS) to isolate and quantify NeuN+ nuclei. No Stim P301S showed a significant reduction in the percentage of NeuN+ nuclei in V1, whereas GENUS P301S did not differ from WT controls (Figure S2E). As our IHC analysis revealed that GENUS reduced the loss of NeuN+ cells outside of V1, we examined ventricle size, which expands in P301S mice with neurodegeneration (Yoshiyama et al., 2007; Shi et al., 2017). We found a moderate increase in ventricle size in No Stim P301S at 8-months old, but no difference in GENUS P301S from WT littermates following GENUS (Figure S2F). To determine if GENUS interferes with mutant Tau transgene expression, we examined Tau protein levels, and confirmed that Tau expression is similarly elevated in No Stim and GENUS P301S, compared to WT controls (Figure S2G, S2H). To test if the neuroprotective effect is specific to visual stimulation at 40 Hz, we next subjected P301S to 80 Hz stimulation. We found that 22-days of 80 Hz stimulation did not alter neuronal loss in V1 in P301S compared to No Stim P301S (Figure S2I).
Figure 2. Chronic 40 Hz stimulation reduces neurodegeneration in Tau P301S and CK-p25 mice.
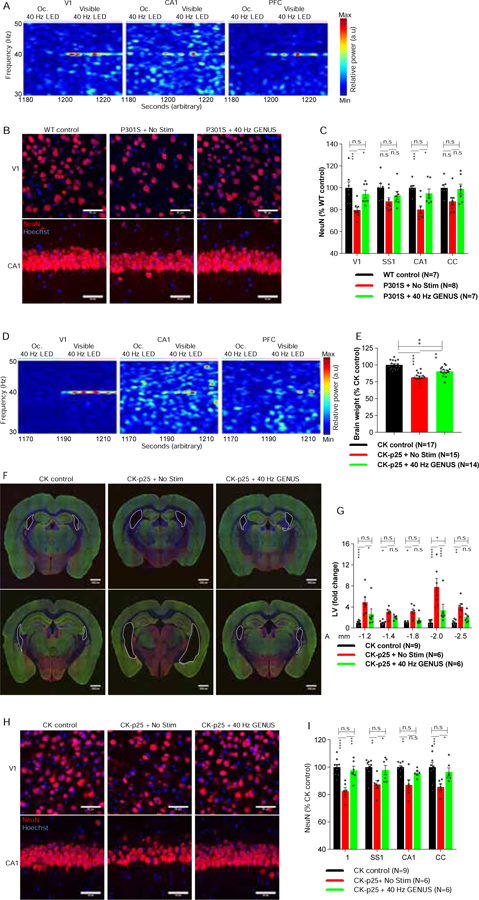
(A) Representative LFP spectra from V1, CA1, and PFC from 8-month-old P301S (see Figure S2A–S2C).
(B) Representative images for NeuN (red) from V1 and CA1 in No Stim or GENUS P301S, with Hoechst labeling of cell nuclei (blue) (Scale bar 50 μm).
(C) Group data quantifying NeuN+ cells (N = 7–8 mice/group. Two-way ANOVA F (2, 76) = 21.8, p < 0.0001. Post-hoc test; WT control Vs No Stim P301S: V1, p = 0.0005; SS1, p = 0.051; CA1, p = 0.0006; CC, p = 0.0500; WT control Vs GENUS P301S: V1, p = 0.844; SS1, p = 0.573; CA1, p > 0.999; CC, p > 0.999; No Stim Vs GENUS P301S: V1, p = 0.016; SS1, p = 0.859; CA1, p = 0.0145; CC, p = 0.090).
(D) Representative LFP spectra from V1, CA1, and PFC from CK-p25 mice after 6 weeks of p25 induction (see Figure S2J–S2L).
(E) Bar graph of brain weight (N = 14–17 mice/groups. ANOVA F (2, 43) = 26.6, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: p < 0.0001; CK control Vs GENUS CK-p25: p = 0.001; No Stim Vs GENUS CK-p25: p = 0.005).
(F) Representative IHC images with vGlut1 (green), GAD65 (red), and Hoechst (blue) of the lateral ventricles (outlined) at anterior-posterior (AP) −1.2 (top row) and −2.0 mm (bottom row) from bregma.
(G) Fold change of size of lateral ventricles (N = 6–9 mice/group. Two-way RM-ANOVA, between groups F (2, 18) = 31.51, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: AP from bregma (mm): −1.2, p < 0.0001; −1.4, p = 0.042; −1.8, p = 0.033; −2.0, p < 0.0001, −2.5, p = 0.002; CK control Vs GENUS CK-p25: −1.2, p = 0.123; −1.4, p = 0.421; −1.8, p = 0.801; −2.0, p = 0.0137; −2.5, p = 0.393; No Stim Vs GENUS CK-p25: −1.2, p = 0.035; − 1.4, p = 0.523; −1.8, p = 0.194; −2.0, p < 0.0001; −2.5, p = 0.085).
(H) Representative images for NeuN (red) in V1 and CA1, and Hoechst labeling of cell nuclei (blue) (Scale bar 50 μm).
(I) Group data quantifying NeuN+ cells (N = 6–9 mice/group. Two-way ANOVA F (2, 72) = 31.38, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: V1, p < 0.0001; SS1, p = 0.001; CA1, p = 0.002; CC, p = 0.0007; CK control Vs GENUS CK-p25: V1, p > 0.99; SS1, p > 0.99; CA1, p = 0.765; CC, p > 0.999; No Stim CK-p25 Vs GENUS CK-p25: V1, p = 0.0001; SS1, p = 0.035; CA1, p = 0.106; CC, p = 0.026; n.s.= not significant).
To further test the potential for GENUS in impacting neurodegeneration, we utilized the CK-p25 mouse model of severe neurodegeneration (Cruz et al., 2003). CK-p25 mice exhibit extensive neuronal and synaptic loss with cognitive impairment evident 6 weeks after p25 induction (Cruz et al., 2003; Fischer et al., 2005). First, we confirmed that 40- Hz stimulation was able to significantly enhance gamma oscillations in V1, CA1, and PFC of CK-p25 mice even at the 6-week endpoint (Figures 2D and S2J–S2L). Then, we examined whether GENUS applied from the early stages of neurodegeneration could influence disease progression and measured brain atrophy, cortical shrinkage, aberrant ventricle expansion, and neuronal loss as previously reported in this model (Cruz et al., 2003). We simultaneously induced p25 expression in CK-p25 mice while also exposing them to 1h/d No Stim or GENUS for 6-weeks. No Stim CK-p25 showed reduced brain weight and cortical thickness (Cruz et al., 2003), whereas CK-p25 with GENUS during p25 induction showed no difference compared to CK controls (CaMKIIα promoter, tTA) (Figures 2E, S2M, and S2N). Cortical shrinkage is also tightly correlated with ventricular expansion in CK-p25 (Cruz et al., 2003), which was profoundly reduced in GENUS CK-p25 (Figure 2F, 2G). No Stim CK-p25 have reduced neuronal density (Cruz et al., 2003), with significantly fewer NeuN+ cells in V1, SS1, CA1, and CC, whereas GENUS CK-p25 showed no significant difference from CK controls across these brain regions (Figure 2H, 2I and Figure S2O). In agreement with these IHC results, FACS analysis revealed a significantly lower percentage of NeuN+ nuclei in V1 of No Stim CK-p25, but no difference in GENUS CK-p25, compared with CK controls (Figure S2P, S2Q). We examined p25 levels, confirming that GENUS did not alter transgene expression in CK-p25 (Figure S2R–S2U). Together, these results demonstrate that GENUS is effective at reducing neuronal loss in multiple brain regions in both CK-p25 and P301S models.
GENUS modifies behavioral performance
Next, we evaluated whether GENUS had any impact on behavioral indices. Mice were tested in an open field (OF) and assessed for changes in anxiety and activity levels, followed by a one-trial hippocampus-independent novel object recognition (NOR) test of short-term memory. Separate cohorts of mice were also tested for hippocampus-dependent spatial learning and memory using the Morris water maze (MWM), a task where learning is gradually acquired across repeated trials. CK-p25 received either No Stim or GENUS during 6-weeks of p25 expression, as before, and underwent OF and NOR tests on the final two days (Figure 3A). GENUS did not affect the time spent in the center of the OF arena, total distance traveled, body weight, nor plasma corticosterone levels in CK-p25 compared to No Stim CK-p25 (Figure 3B–3E and Figure S3A), suggesting no changes in anxiety or stress. In NOR, GENUS CK-p25 showed a non-significant trend towards increased preference for the novel object compared to No Stim CK-p25 (Figure S3B). In the cohort for MWM, spatial learning and memory were assessed during the final week of GENUS (or No Stim) (Figure 3F). GENUS significantly improved learning, with reduced latencies to find the platform over training days (Figure 3G), with no impact on swimming velocity (Figure 3H). In a probe test performed 24h after the last training day, GENUS CK-p25 showed significantly improved spatial memory, with more visits to the platform location and more time in the target quadrant compared with No Stim CK-p25 (Figure 3I, 3J).
Figure 3. Chronic GENUS modifies behavior in mouse models of neurodegeneration.
(A) Behavior schedule of CK-p25 with No Stim or GENUS during 6-weeks of p25 induction, with OF (day 40) test.
(B) Representative occupancy heat maps from OF session.
(C) Time spent in the center of OF (N= 15 mice/group. t = 0.467, p = 0.644).
(D) Distance travelled in the OF test (t = 0.1179, p = 0.907).
(E) Plasma corticosterone levels (t = 0.9314, p = 0.360).
(F) MWM schedule of p25-induced CK-p25 with or without GENUS for 6-weeks.
(G) Latency to find the platform in the training (Two-way RM-ANOVA, effect of days, F (5, 168) = 14.05, p < 0.001; effect between groups, F (1, 168) = 20.47, p < 0.001. D1, t = 1.768, p = 0.259; D2, t = 2.859, p = 0.039; D3, t = 0.682, p = 0.500; D4, t = 1.867, p = 0.259; D5, t = 3.089, p = 0.0269; D6, t = 1.687, p = 0.259).
(H) Swimming velocity during MWM (Two-way RM-ANOVA, effect between groups, F (1, 168) = 0.05377, p = 0.816).
(I) Number of platform crossings in the probe test (Mann-Whitney U = 53, p = 0.0100).
(J) Time spent in the target quadrant (t = 2.754, p = 0.010).
(K) Behavior schedule of P301S with No Stim or GENUS for 22-days, with OF (day 20) test.
(L) Representative occupancy heat maps from OF session.
(M) Time spent in the OF center (t = 0.8801, p = 0.388).
(N) Distance travelled during OF (t = 0.3122, p = 0.757).
(O) Plasma corticosterone levels following No Stim or GENUS (t = 0.9232, p = 0.364).
(P) MWM schedule of P301S with or without GENUS for 22-days.
(Q) Latency to the platform during training (Two-way RM-ANOVA, effect of days, F (4, 150) = 9.702, p < 0.0001; effect between groups, F (1, 150) = 7.096, p = 0.008. D1, t = 0.160, p = 0.873; D2, t = 0.488, p = 0.862; D3, t = 0.7342, p = 0.849; D4, t = 2.266, p = 0.03; D5, t = 2.465, p = 0.019).
(R) Swimming velocity during training (Two-way RM-ANOVA, effect between groups F (1, 150) = 1.4, p = 0.238).
(S) Number of platform crossings in the probe test (Mann-Whitney U = 83.5, p = 0.091).
(T) Time in target quadrant in the probe test (t = 2.603, p = 0.014).
We next subjected 8-months old P301S to GENUS for 22-days, and assessed behavioral performance (Figure 3K). GENUS did not alter OF center time or total distance travelled, body weight, nor plasma corticosterone levels compared to No Stim P301S (Figure 3L–3O and Figure S3C), suggesting no changes in anxiety. In NOR, both No Stim and GENUS P301S showed preference for the novel object over the familiar object, with no significant difference between the two groups (Figure S3D). We performed MWM during the final week of GENUS (or No Stim) (Figure 3P). GENUS P301S showed reduced escape latency over training days compared to No Stim P301S (Figure 3Q), with no differences in swimming velocity (Figure 3R). In the probe test, GENUS P301S exhibited slightly improved spatial memory, with a trend towards a higher number of platform crossings and significantly more time spent in the target quadrant, compared with No Stim P301S (Figure 3S, 3T). We additionally tested MWM in 9-months old 5XFAD (Oakley et al., 2006) with No Stim or GENUS for 22-days. We found no difference in the latency to find the platform nor swimming speed over training days (Figure S3E, S3F). During the probe test GENUS 5XFAD showed a partial improvement in spatial memory after GENUS, with no difference in target quadrant preference during the probe test, but significantly more platform crossings compared to No Stim 5XFAD (Figure S3G, S3H).
To test the effect of GENUS on modifying behavior in wildtype mice, we subjected young 3.5-months old C57BL/6J mice to GENUS for 7-days. We found no change in body weight or plasma corticosterone levels (Figure S3I, S3J). GENUS for 7-days also did not affect time in center or distance travelled in the open field test compared to No Stim C57BL/6J (Figure S3K, S3L), although it did increase the time spent in the open arms in the elevated plus maze (EPM) (Figure S3M, S3N). We carried out MWM after 7-days of GENUS, and found no effect on the latency to find the platform over training days, nor target platform location visits or time in target quadrant in the probe test, suggesting no change in spatial MWM learning and memory in the C57BL/6J (Figure S3O–S3Q). Thus, 7-days of GENUS did not lead to strong behavioral modifications in young C57BL/6J. We extended the GENUS regime to 5-weeks to evaluate its effects on behavior in aged 17-months old C57BL/6J mice. We observed an anxiolytic effect, as GENUS aged C57BL/6J spent more time in the center of the arena in OF, and more time in the open arms in EPM, compared to No Stim aged C57BL/6J (Figure S3R – S3U). We performed MWM in aged C57BL/6J during the final week of GENUS (or No Stim). GENUS reduced the latency to find the platform over training days, but did not affect either behavioral measures in the probe test compared to No Stim aged C57BL/6J (Figure S3V–S3X), suggesting an improvement in spatial learning but not memory. Overall, these data suggest that GENUS can modify behavioral performance in mouse models of neurodegeneration as well as aging.
GENUS reduces inflammatory response
Using 40 Hz optogenetic or visual stimulation in 5XFAD, Iaccarino et al. (2016) found that increasing gamma oscillations induces gene expression changes and morphological transformation of microglia, suggesting that GENUS may induce microglia to play a beneficial role in disease pathology. We thus examined the effect of GENUS on microglia-mediated inflammatory response, which has been previously reported in CK-p25 and P301S (Gjoneska et al., 2015; Mathys et al., 2017; Yoshiyama et al., 2007, Dejanovic, 2018). To gain insight into the transcriptomic changes underlying the microglial response to GENUS, we performed microglia-specific RNA-seq from V1 following No Stim or GENUS for 6-weeks (during p25 induction) in CK-p25, and for 22-days in P301S. We found significantly elevated numbers of CD11b and CD45 double positive cells in No Stim CK-p25, which was reduced with daily GENUS during p25 induction (Figure S4A, S4B). RNA-seq revealed large differences in microglial transcription between CK controls and No Stim CK-p25 with 2333 upregulated genes and 2019 downregulated (Figure 4A). Gene ontology (GO) analysis revealed that many upregulated genes were involved in immune response (Figure 4A, 4B) (Mathys et al., 2017), while many downregulated genes related to processes involved homeostatic microglia function (Figure 4A, 4B). Compared to No Stim CK-p25, we found that GENUS led to the upregulation of 358 genes, many associated with protein synthesis and membrane trafficking (Figure 4B) and the downregulation of 518 genes, including many related to immune response and MHC-1 mediated antigen processing presentation (Figure 4B).
Figure 4. Chronic GENUS reduces inflammatory response in microglia.
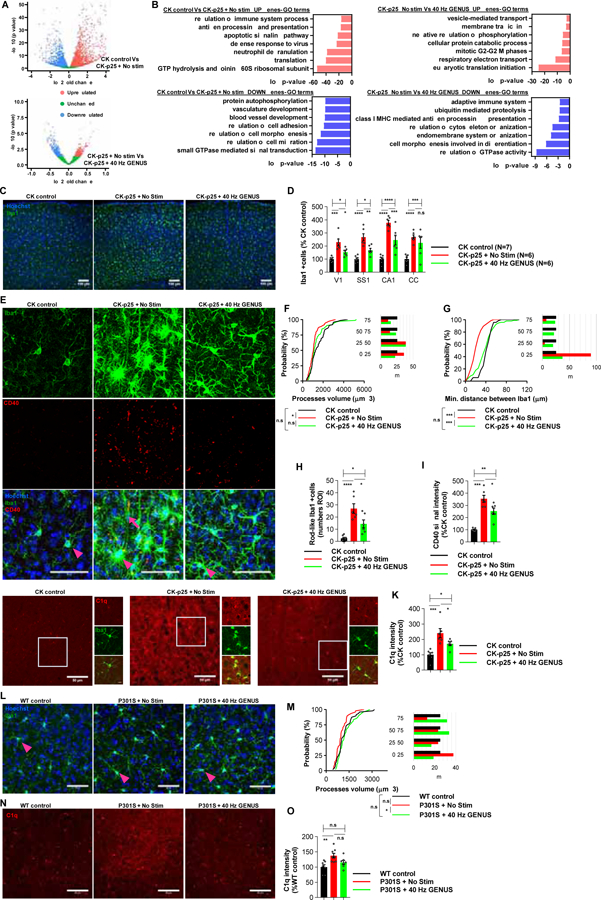
(A) RNA-sequencing of FACS-isolated CD11b and CD45 double-positive microglia RNAs from CK control, No Stim CK-p25, and GENUS CK-p25. Volcano plots of DEGs (N = 4 mice/group).
(B) Top 7 selected GO terms for biological processes associated with the DEGs.
(C) Representative images of microglia stained with Iba1 (green) and Hoechst labeling of cell nuclei (blue). Scale bar, 100 μm.
(D) Bar graph of number of Iba1+ cells (N = 6–7 mice/group. Two-way ANOVA F (2, 64) = 80.35, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: V1, p = 0.0001; SS1, p < 0.0001; CA1, p < 0.0001; CC, p < 0.0001; CK control Vs GENUS CK-p25: V1, p = 0.050; SS1, p = 0.049; CA1, p < 0.0001; CC, p = 0.0002; No Stim Vs GENUS CK-p25: V1, p = 0.046; SS1, p = 0.005; CA1, p = 0.0002. CC, p = 0.474).
(E) Representative images of microglia (Iba1; green), CD40 (red), and Hoechst labeling of cell nuclei (blue). Arrowheads and arrow indicate bushy arborization and rod-shaped ramified processes, respectively (N = 6–7 mice/group. Scale bar 50 μm).
(F) Distribution plot of volume of processes of microglia (excluding rod like microglia). Right: bar chart of microglia processes volume binned into four categories based on the distribution of CK controls: minimum-25% percentile, 25%-median, 75% percentile and upper 75% percentile-maximum (N = 73 microglia/group from 6–7 mice/group. Kruskal-Wallis test, H = 9.224, p = 0.009. Dunn’s multiple comparisons; CK control Vs No Stim CK-p25: p = 0.008; CK control Vs GENUS CK-p25: p = 0.908; No Stim Vs GENUS CK-p25: p = 0.150).
(G) Distribution plot of minimum distance between microglia. Right: bar graph of microglia distance, binned as in Figure 4F (N = 68, 131 and 95 microglia from 9 CK control, 6 No Stim and 6 GENUS CK-p25 mice, respectively. Kruskal-Wallis test, H = 100.1, p < 0.0001. Dunn’s multiple comparisons; CK control Vs No Stim CK-p25: p < 0.0001; CK control Vs GENUS CK-p25: p = 0.1731; No Stim Vs GENUS CK-p25: p < 0.0001).
(H) Bar graph of number of rod like microglia (N = 6–9 mice/group. ANOVA F (2, 18) = 24.05, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: p < 0.0001; CK control Vs GENUS CK-p25: p = 0.0100; No Stim Vs GENUS CK-p25: p = 0.0113).
(I) Bar graph of CD40 signal intensity (ANOVA F (2, 16) = 36.84, p < 0.0001. N = 6–7 mice/group. Post-hoc test; CK control Vs No Stim CK-p25: p < 0.0001; CK control Vs GENUS CK-p25: p = 0.0003; No Stim Vs GENUS CK-p25: p = 0.01).
(J) Representative C1q (red) and Iba1 (green) images (N = 6–7 mice/groups. Scale bar 50 μm (10 μm inset)).
(K) Bar graph of C1q signal intensity (ANOVA F (2, 16) = 13.39, p = 0.0004).
(L) Representative images of microglia (Iba1; green) and Hoechst labeling of cell nuclei (blue). Arrowhead indicates the complexity of microglia processes (scale bar, 50 μm).
(M) Distribution plot of the volume of processes of microglia, and (right) based on WT controls binned as in Figure 4F (N = 7–8 mice/group; 58 microglia/group. Kruskal Wallis test H = 7.895, p = 0.0193. Dunn’s multiple comparisons; WT control Vs No Stim P301S: p = 0.210; WT control Vs GENUS P301: p > 0.99; No Stim Vs GENUS P301S: p = 0.0170).
(N) Representative C1q (red) images (N = 7–8 mice/group. Scale bar 50 μm).
(O) Bar graph of C1q signal intensity (ANOVA F (2, 19) = 6.887, p = 0.005). Post-hoc test in D, H, I, K, & O ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, n.s. = not significant.
In No Stim P301S, we found 331 upregulated and 292 downregulated genes relative to WT littermates (Figure S4E–S4F). GO analysis associated upregulated genes with protein synthesis and inflammatory/immune response, whereas downregulated genes were related to cell migration and cytoskeleton organization (Figure S4F). After GENUS, microglia from P301S displayed 238 upregulated and 244 downregulated genes compared to No Stim P301S. Upregulated genes were associated with cellular catabolic proteolysis, and membrane trafficking, while downregulated genes were involved in gene expression and interferon response (Figure S4F). Changes in microglial transcription in No Stim CK-p25 and P301S were largely distinct, and while differentially expressed genes (DEGs) between the two neurodegeneration models following GENUS also did not overlap significantly (Figure S4G, S4H), we observed a convergence onto related GO terms in the two models, including membrane trafficking and immune response. Our data suggest that GENUS modified common biological functions of microglia albeit via unique sets of genes in each of these models.
We next performed IHC to validate our gene expression analysis. Immunostaining results with the microglia marker, Iba1, were in accord with previous report (Mathys et al., 2016) and FACS analysis (Figure S4B), revealing significant microgliosis in V1, SS1, CA1, and CC of No Stim CK-p25 compared to CK controls, which was reduced in GENUS CK-p25 (Figure 4C, 4D). Furthermore, microglia in No Stim CK-p25 displayed a complex ‘bushy’ arborization pattern (Figure 4E; lower panels - arrowheads) that is associated with axonal and terminal synaptic degeneration (Jensen et al., 1994; Jørgensen et al., 1993), with significantly reduced volume of processes (Figure 4E, 4F). Microglia from No Stim CK-p25 were also in closer physical proximity to each other compared to CK controls, as analyzed by measuring the minimum distance between microglia soma (Figure 4G). This indication of dysregulated territories contrasts with homeostatic resting states where each microglia has its own region of occupation, with little overlap between neighboring territories (Nimmerjahn et al., 2005). In contrast, GENUS CK-p25 showed no significant difference in the total volume of microglial processes (Figure 4F) and the minimum distance between microglia in GENUS CK-p25 was comparable to that in CK controls (Figure 4G), suggesting the preservation of microglial territories with GENUS. Additionally, a large fraction of microglia in No Stim CK-p25 displayed an elongated rod-like body without polarized processes (Figure 4E; lower center panel - arrow), a phenotype reported in rats and in human subjects following brain injury (Bachstetter et al., 2017; Taylor et al., 2014). GENUS reduced the number of rod-microglia in CK-p25 (Figure 4H).
Interferon response gene CD40 is a key regulator of the neuro-immune response (Benveniste et al., 2004) and CD40-CD40L signaling has been linked to microglial activation and amyloidosis (Tan et al., 2002). IHC revealed elevated CD40 expression in microglia from No Stim CK-p25 mice (Mathys et al., 2017), and this was reduced by GENUS (Figure 4E, 4I). Immunostaining patterns for C1q, which has been implicated in synaptic loss, was consistent with other neurodegeneration mouse models (Hong et al., 2016; Dejanovic et al., 2018), showing elevated signal intensity in No Stim CK-p25, some of which co-localized with Iba1 as well as appearing as diffuse signal (Figure 4J). This elevation in C1q signal was reduced following GENUS (Figure 4K), supporting a role for GENUS in mitigating the immune response in CK-p25.
Neuroinflammatory indicators were also impacted by GENUS in the P301S model. The volume of microglial processes in GENUS P301S was increased compared to No Stim P301S, which were similar to WT controls (Figure 4L, 4M). As in the CK-p25 model, we observed elevated C1q signal in No Stim P301S compared to WT controls (Dejanovic et al., 2018). This increase was abolished following GENUS (Figure 4N, 4O). We also investigated whether GENUS affects neuro-inflammatory markers in aged C57BL/6J mice, and found no significant change in C1q and CD40 signal intensity and microglia number or processes volume in V1 (Figure S4I–S4M). These data demonstrate that microglia response to GENUS may vary depending on disease state or wildtype background.
GENUS modifies synaptic function
To gain insight into potential molecular mechanisms of GENUS, we examined neuronal gene expression by performing RNA-seq on isolated NeuN+ nuclei from V1 of CK-p25 and P301S following No Stim or GENUS for 6-weeks (during p25 induction) in CK-p25, and for 22-days in P301S (Figure S2P). RNA-seq revealed that more genes were downregulated in CK-p25 (618 genes) and P301S (351 genes) than were upregulated (CK-p25: 565 genes; P301S: 229 genes), compared to their respective CK and WT controls (Figure 5A, 5C). GENUS resulted in a similar number of upregulated versus downregulated genes in CK-p25 (409 up; 422 down) and P301S (220 up; 221 down), compared to No Stim CK-p25 and P301S, respectively (Figure 5A, 5C). Downregulated genes in No Stim CK-p25 compared to CK control included those involved in synaptic transmission and intracellular transport, whereas GENUS CK-p25 showed an upregulation of genes associated with these same biological processes relative to No Stim CK-p25 (Figure 5B and Figure S5A). Downregulated genes in No Stim P301S compared to WT controls were also mainly involved in synaptic transmission and intracellular transport, and these processes were among the top biological functions associated with upregulated genes in GENUS P301S compared to No Stim P301S (Figure 5D). These data suggest that even across two mechanistically distinct mouse models of neurodegeneration (CK-p25 and P301S), the altered patterns of gene expression converge on many similar cellular and biological functions - including downregulation of synaptic function, intracellular transport, and apoptotic regulation – that ultimately promote neuronal demise. In contrast, GENUS upregulated many genes that are involved in synaptic transmission, and intracellular and vesicle-mediated transport. There was significant overlap of DEGs between P301S and CK-p25, with 25 genes commonly upregulated and 28 downregulated (Figure S5A, S5B), indicating some common transcriptomic changes in neurons after GENUS. However, there was no enrichment for any specific biological terms associated with these overlap genes.
Figure 5. Chronic GENUS modifies synaptic function and reduces DNA damage in neurons.
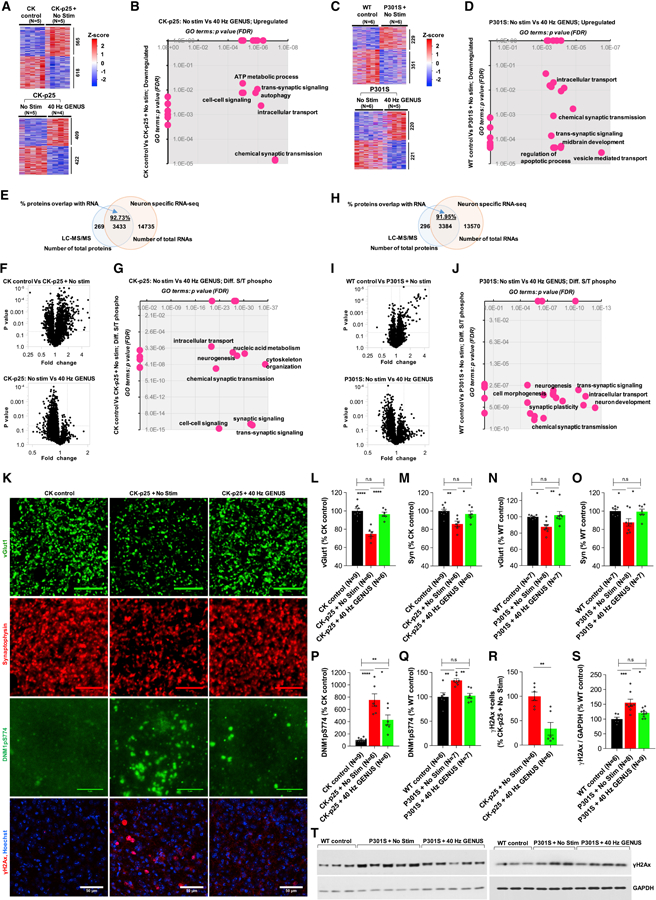
(A) RNA-sequencing of FACS-isolated NeuN+ RNAs from CK control, No Stim CK-p25, and GENUS CK-p25. Heat-maps of the DEGs. Number of DEGs is indicated to the right (N = 4–5 mice/group).
(B) GO terms associated with the DEGs.
(C) P301S with No Stim or GENUS followed by NeuN+ RNA-seq. Heat-maps of the DEGs (N = 5–6 mice/group).
(D) GO terms associated with the DEGs.
(E) S/T phosphorylated proteins analysis in CK-p25 using LC-MS/MS. Venn diagram of overlap of total RNAs from neuron specific RNA-seq (Figure 5A) and total proteins from LC-MS/MS (N = 3–4 mice/group).
(F) Volcano plot of differentially S/T phosphorylated proteins.
(G) GO terms associated with the differentially S/T phosphorylated proteins.
(H) S/T phosphorylated proteins analysis in P301S using LC-MS/MS. Overlap of total RNAs from neuron specific RNA-seq (Figure 5C) and total proteins from LC-MS/MS (N = 3–4 mice/group).
(I) Volcano plot of differentially S/T phosphorylated proteins.
(J) GO terms associated with the differentially S/T phosphorylated proteins.
(K) Representative images of vGlut1 (green), synaptophysin (Syn; red), DNM1pS774 (green), γH2Ax (red), and Hoechst labeling of cell nuclei (blue) (N = 6–9 mice/group. Scale bar represents 10 μm, 10 μm, 5 μm and 50 μm for vGlut1, Syn, DNM1pS774, and γH2Ax, respectively).
(L) Bar graph of vGlut1 puncta (ANOVA between groups effect F (2, 18) = 17.9, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: p < 0.0001; CK control Vs GENUS CK-p25: p = 0.577; No Stim Vs GENUS CK-p25: p < 0.0001).
(M) Bar graph of synaptophysin puncta (ANOVA F (2, 18) = 7.693, p = 0.003. Post-hoc test; CK control Vs No Stim CK-p25: p = 0.003; CK control Vs GENUS CK-p25: p > 0.99; No Stim Vs GENUS CK-p25: p = 0.042).
(N) Bar graph of vGlut1 puncta (ANOVA F (2, 19) = 6.681, p = 0.006. Post-hoc test; WT control Vs No Stim P301S: p = 0.02; WT control Vs GENUS P301S: p = 0.879; No Stim Vs GENUS P301S: p = 0.008).
(O) Bar graph of synaptophysin puncta (Two-way ANOVA F (2, 19) = 5.099, p = 0.016. Post-hoc test; WT control Vs No Stim P301S: p = 0.034; WT control Vs GENUS P301S: p > 0.99; No Stim Vs GENUS P301S: p = 0.046).
(P) Bar graph of DNM1pS774 expression (ANOVA F (2, 18) = 27.89, p < 0.0001. Post-hoc test; CK control Vs No Stim CK-p25: p < 0.0001; CK control Vs GENUS CK-p25: p = 0.0046; No Stim Vs GENUS CK-p25: p = 0.0102).
(Q) Bar graph of DNM1pS774 expression (ANOVA F (2, 17) = 10.084, p = 0.0009. Post-hoc test; WT control Vs No Stim P301S: p = 0.0022; WT control Vs GENUS P301S: p = 0.9575; No Stim Vs GENUS P301S: p = 0.002).
(R) Bar graph of γH2Ax+ cells. γH2Ax+ cells was not detectable in CK controls (N = 6 mice/group. t = 4.224, p = 0.001).
(S) Bar graph of expression of γH2Ax (ANOVA F (2, 20) = 10.02, p = 0.0010. Post-hoc test; WT control Vs No Stim P301S: p = 0.0010; WT control Vs GENUS P301S: p = 0.341; No Stim Vs GENUS P301S: p = 0.019).
(T) Immunoblots of γH2Ax and GAPDH.
We next carried out phosphoproteomic (LC-MS/MS) analysis of V1 from CK-p25 and P301S to identify proteins affected following GENUS. The vast majority of identified proteins (CK-p25: 92.75%; P301S: 91.95%) mapped to genes in the neuron-specific RNA-seq data (Figure 5E, 5H), implicating their role in neuronal function. Comparison of No Stim CK-p25 and P301S with their respective CK and WT controls, revealed an overall increase in S/T-phosphorylated proteins in both models (Figure 5F, 5I), indicating aberrant modification of functional proteins in P301S and CK-p25. Consistent with the neuron-specific RNA-seq data, similar GO terms were identified, including synaptic function and intracellular vesicle transport (Figure 5G, 5J). GENUS resulted in an overall reduction of S/T phosphorylation of proteins in both CK-p25 and P301S, compared to their respective No Stim controls (Figure 5F, 5I). S/T-modified proteins affected by GENUS (CK-p25: 202 proteins; P301S: 75 proteins) included those involved in synaptic function (Extended File 1), regulation of intracellular vesicle transport, and nucleic acid metabolism (Figure 5G, 5J). We additionally examined the effect of GENUS on the phosphoproteomic profile in aged C57BL/6J mice and found that GENUS modified S/T phosphorylation status of 172 proteins. Enrichment analysis again revealed that these modified proteins were involved in synaptic function and neuronal structural organization (Figure S5C, S5D). Overall, we find that the effects of GENUS converge on multiple genes and proteins that regulate synaptic functions and vesicle transport, independent of neurodegenerative background.
Hyper-phosphorylation of tau is evident in the P301S mouse model (Yoshiyama et al., 2007). Unbiased S/T phosphoproteomics revealed a significant effect of GENUS on overall phosphorylation levels of tau in P301S (Figure S5E), with significant S/T modification of tau protein at S491, S494;T497, S502, S502;S506, S704, S705 and T58 residues compared to No Stim P301S (Figure S5E and Extended File 1).
A comparison of the differentially S/T-phosphorylated proteins across GENUS CK-p25, P301S, and aged C57BL/6J (Figure S5F) revealed many related to synaptic transmission and synaptic plasticity (Figure S5G). We thus carried out IHC for synaptic marker synaptophysin (Syn) and vesicular glutamate transporter 1 (vGlut1), and for vesicle trafficking protein dynamin 1 (DNM1), which are all involved in vesicle and neurotransmitter transport, synaptic transmission, and learning and memory (Armbruster et al., 2013; Balschun et al., 2009). We found that vGlut1 and Syn puncta were reduced in No Stim CK-p25 compared to CK controls (Figure 5K, 5L, 5M), and that GENUS ameliorated these differences (Figure 5K, 5L, 5M). Similarly, the expression of vGlut1 and Syn puncta were reduced in P301S compared to WT controls, and GENUS ameliorated these differences (Figure 5N, 5O). Outside of V1, GENUS also reduced the loss of vGlut1 puncta in CA1 and CC of both CK-p25 and P301S, maintaining them at levels comparable to their respective CK and WT controls (Figure S5H, S5I). These results, along with the neuron-specific transcriptomic analysis, suggest that GENUS regulates synaptic proteins, leading us to speculate that GENUS might also modulate synaptic connectivity and spine morphology. To examine this possibility, we used a viral-mediated approach in Fos-CreERT2 mice to label active cortical neurons with EYFP (see STAR Methods), and then assessed spine morphology in mice with No Stim or GENUS (Figure S5J, S5K). While there was no change in the total number of spines with GENUS, we observed reduced thin and stubby spines and increased mushroom spine density (Figure S5L–S5M), a morphology associated with higher expression of AMPA receptors which mediate fast glutamatergic synaptic transmission (Matsuzaki et al., 2001).
On comparing commonly S/T-modified proteins across GENUS P301S, CK-p25, and aged C57BL/6J mice, we observed that DNM1 was differentially phosphorylated in similar sites (Extended File 1). We examined phosphorylation at Ser774 in dynamin1 (pS774DNM1), one of many residues that was found to be hyper-phosphorylated in No Stim CK-p25 and P301S, and which was reduced with GENUS (Figure S5N, S5O). Reduced phosphorylation at Ser774 is required for endocytosis of synaptic vesicles and synaptic transmission (Clayton et al., 2010). Accordingly, IHC/immunoblotting showed increased phosphorylation levels of pS774DNM1 in No Stim CK-p25 and P301S, which was reduced with GENUS (Figure 5K, 5P, 5Q and Figure S5Q, S5R). Similarly, GENUS also reduced pS774DNM1 levels in C57BL/6J (Figure S5S). These data suggest that GENUS regulates the phosphorylation state of neuronal and synaptic proteins to impact their function regardless of neurodegeneration or wildtype context.
The observed changes in nucleic acid metabolism in No Stim CK-p25 and P301S are consistent with previous literature demonstrating increased DNA damage in neurons in these mouse models (Dobbin et al., 2013; Kim et al., 2008). We performed IHC for yH2Ax, a marker of DNA double stranded breaks, as well as the expression of proteins involved in ameliorating DNA damage-associated neurotoxicity, such as histone 3.3 (H3F3) (Frey et al., 2014; Maze et al., 2015) and mesencephalic astrocyte-derived neurotrophic factor (MANF/ARMET) (Airavaara et al., 2009; Hellman et al., 2011). We observed significantly more yH2Ax-positive neurons in V1 of No Stim CK-p25 compared to CK controls (Figure 5K, 5R) (Kim et al., 2008), and reduced DNA damage in GENUS CK-p25 (Figure 5R). GENUS also increased the expression of H3F3 and MANF in CK-p25 compared to No Stim CK-p25 (Figure S5T). Similarly, we observed significantly higher protein levels of yH2Ax in No Stim P301S compared to WT controls, with a reduction following GENUS compared to No Stim P301S (Figure 5S, 5T). These results suggest that GENUS preserves synaptic density, reduces DNA damage, and increases neuroprotective factors, thus possibly shifting neurons towards a less degenerative state in CK-p25 and P301S mice.
DISCUSSION
In the current study, we demonstrate that 40 Hz visual stimulation is effective at entraining gamma oscillations across broad brain regions in mouse models of neurodegeneration. With long-term daily exposure, GENUS reduced the loss of neurons and synapses in V1, as well as hippocampus and PFC, and improved behavioral performance in P301S and CK-p25 mice. Moreover, GENUS resulted in an upregulation of cytoprotective proteins and reduction in DNA damage, suggesting a shift in neurons towards a less degenerative state. Together with our prior study (Iaccarino et al., 2016), we show that GENUS is capable of ameliorating several key neuropathologies, such as amyloid plaques, tau phosphorylation, neuronal and synaptic loss in multiple mouse models of AD (5XFAD, APP/PS1, P301S and CK-p25 mice), and point to an overall neuroprotective effect. The GENUS stimulation regimen in our study was initiated in the early stages of respective pathologies in these mice. Based on our in vivo electrophysiological data, which showed 40 Hz entrainment even in the later stages of neurodegeneration in P301S and CK-p25, it is possible that GENUS may also offer neuroprotection in more advanced disease stages.
We found that GENUS impacted microglia in CK-p25, P301S, and 5XFAD (Iaccarino et al., 2016) to varying degrees, but not in aged C57BL/6J mice. The observations of different microglia morphologies likely reflect different stages of neurodegeneration in the different mouse models. For instance, CK-p25 and P301S are known to develop neuronal loss, while 5XFAD at 6-months and aged C57BL/6J old do not (Calhoun et al., 1998; Cruz et al., 2003; Oakley et al., 2006; Yoshiyama et al., 2007). GENUS reduced neuroinflammatory markers in P301S and CK-p25, whereas neuroinflammatory markers in aged wildtype mice were not altered. In CK-p25, GENUS restored microglia territorial domains to homeostatic conditions by reducing microglia numbers as well as the distance between microglia. The precise manner and consequences of microglial change by GENUS remains to be determined. Nonetheless, our data support the growing evidence that reducing neuroinflammation may offer beneficial effects in neurodegeneration (Heneka et al., 2015; Shi et al., 2017; Spangenberg et al., 2016).
A unifying link between GENUS and microglia response across CK-p25, P301S, and wildtype mice remains unknown, although we observed several commonalities across these models apparently independent of a neuropathology context. While some of the effects of GENUS to modify neuronal, synaptic, and intracellular transport proteins may simply reflect an increased preservation of neuronal and synaptic function in CK-p25 and P301S mice, consistent findings in wildtype mice suggest that this is not the sole reason. The observed changes in spine morphology in wildtype mice, with an increase of mushroom spines still evident 24h after the final GENUS session, is intriguing as it suggests the potential for long-lasting impact on synaptic transmission. Indeed, reduced phosphorylation of DNM1, which is observed during bouts of elevated neuronal activity (Clayton et al., 2010), was evident with GENUS in CK-p25, P301S, as well as wildtype mice. Although the above findings were focused in V1, we observed increased functional coupling during GENUS using measures of weighted phase lag index, which is less susceptible to volume conductance from uncorrelated noise sources (Vinck et al., 2011), across broader brain regions such as CA1, SS1, and PFC. A key question that our data pose is how does long-term visual stimulation impact neural activity across these broader brain regions?
We examined the effects of GENUS with a range of behavioral tests for changes in anxiety (OF), activity level (OF and MWM), short-term recognition memory (NOR), and spatial learning and memory (MWM). While GENUS did not affect short-term recognition memory nor general activity levels in any of the three models, GENUS improved spatial water maze learning and memory in the CK-p25 severe neurodegeneration model. P301S mice showed improved learning curves and performance on one of the two memory probe measures. The behavioral effects of GENUS in aged wildtype mice and 5XFAD were more equivocal. Aged wildtype showed enhanced performance during training but no effect in the memory probe trial. 5XFAD showed no difference in learning curves and during the memory probe trial, we saw increased number of platform crossings, but no effect on time spent in the target quadrant. However, a recent study applying GENUS with auditory stimuli in younger 5XFAD mice observed improvement in MWM and NOR (Martorell et al., 2019), underscoring the overall potential for GENUS to modify cognitive performance. The mechanisms underlying the beneficial effects of GENUS on behavior are likely numerous and multifaceted. Preservation of neurons and synapses, reduced neuroinflammation, enhanced synaptic transmission and synaptic plasticity gene expression, and/or enhanced coherent gamma oscillations, may contribute to the improved cognitive processes. Overall, our findings support the notion that manipulating neural network oscillations may represent a promising strategy to alleviate pathological changes and behavioral performance deficits associated with neurological disorders (Cho et al., 2015; Iaccarino et al., 2016; Kastanenka et al., 2017; Martinez-Losa et al., 2018; Verret et al., 2012).
STAR METHODS
DATA AND RESOURCE SHARING
The accession number for the RNA sequencing data reported in this paper is GSE115244. Further information and requests for resources and reagents should be directed to the Lead Contact, Li-Huei Tsai (lhtsai@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal models
All the experiments were approved by the Committee for Animal Care of the Division of Comparative Medicine at the Massachusetts Institute of Technology (MIT), and carried out at MIT. C57BL6, Tg(Camk2a-tTA), Tg(APPSwFlLon, PSEN1*M146L*L286V), Tg(Prnp-MAPT*P301S)PS19, and Fos-tm1.1(cre/ERT2) were obtained from the Jackson laboratory. Tg(tetO-CDK5R1/GFP) was generated in our lab. All the transgenic mice were bred and maintained in our animal facility. Mice were bred and maintained by a female experimenter. We used male Tau P301S mice that were 7.5- to 8-months old at the start of the experiments, an age when synaptic and neuronal loss first become apparent (Yoshiyama et al., 2007). P301S mice were exposed to No Stim or GENUS for 22 days. CK-p25 mice are generated by breeding CaMKIIα promoter-tTA mice (CK controls) with tetO-CDK5R1/GFP mice, and raised on doxycycline-containing food to repress p25 expression. We used adult (6 to 9-month-old) CK-p25 mice of both sexes for experiments: male mice were used for RNA-sequencing, behavior & western blotting; female mice were used for immunohistochemistry. Normal rodent diet is given to induce p25-GFP transgene expression over 6 weeks (Cruz et al., 2003; Fischer et al., 2005). Thus, CK-p25 mice were exposed to No Stim or GENUS for 6 weeks during p25 induction. We used male 5XFAD mice at 9-months old. We used 5.5 months old male Fos-CreERT2 mice. All young (3.5-months old) C57BL/6J mice were male. We used male aged C57BL/6J (17 months old), with the exception of MWM test. All mice were group housed (2– 5 mice per cage) except those that were implanted with electrophysiology probes. All experiments were done using age-matched littermates.
METHOD DETAILS
Blinding and randomization
Figure 1 and Figure S1: An experimenter not blind to the treatment condition did c-Fos immunohistochemical staining and quantification. All electrophysiological results described were performed in two cohorts, using mice to the total number as defined in figure legends. Two researchers analyzed the data and were not blind to the experimental condition.
Figure 2 and Figure S2: Neuronal counts described were performed twice using mice to the total number as defined in figure legends by experimenters blind to the treatment condition. Electrophysiological results described were performed in two cohorts, using mice to the total number as defined in figure legends. Two researchers analyzed the data and were not blind to the experimental condition.
Figure 3 and Figure S3: All the behavioral experiments were conducted by experimenters blind to the treatment condition, and analyzed after un-blinding the treatment conditions.
Figure 4, Figure S4: RNA-sequencing was performed by experimenters not blind to the treatment condition. Experimenters blind to the treatment conditions performed all the immunohistochemical staining and quantification. Researchers who analyzed the aged C57BL/6J data (Figure S4) were not blind to the experimental condition.
Figure 5 and Figure S5: RNA-sequencing and phosphoproteomics were performed by experimenters not blind to the treatment condition. Two independent experiments, with one experiment conducted experimenter blind to the treatment condition, was combined for vGlut1 and synaptophysin analysis. ȖH2Ax immunohistochemical staining and western blotting were not done blinded. Experimenter who was blind to the treatment condition imaged, classified, and quantified the total number and types of spines.
Box plot in Figure 1 represent median, and upper (75%) and lower (25%) quartile range. Unless noted, all bar and line plots in Figure 2 to Figure 5 with error bars are reported as mean ± SEM. Unless otherwise noted all samples are reported as number of mice (N).
No statistical methods were used to pre-determine sample sizes, instead we opted to use group sizes similar to previously published studies from our lab (Iaccarino et al., 2016; Kim et al., 2008; Middleton and McHugh, 2016; Nott et al., 2016).
Light flicker stimulation
Light flicker stimulation was delivered as previously described (Iaccarino et al., 2016; Singer et al., 2018). Mice were transported from the holding room to the flicker room, located on adjacent floors of the same building. Mice were habituated under dim light for 1 h before the start of the experiment, and then introduced to the test cage (similar to the home cage, except without bedding and three of its sides covered with black sheeting). All GENUS protocols were administered on a daily basis for 1h/d for the number of days as specified. Mice were allowed to freely move inside the cage but did not have an access to food or water during the 1 h light flicker. An array of light emitting diodes (LEDs) was present on the open side of the cage and was driven to flicker at a frequency of 40 Hz (or 80 Hz where specified) with a square wave current pattern using an Arduino system. The luminescence intensity of light that covered inside the total area of GENUS stimulation cage varied from ~ 200 – 1000 lux as measured from the back and front of the cage (mice were free to move in the cage). After 1h of light flicker exposure, mice were returned to their home cage and allowed to rest for a further 30 min before being transported back to the holding room. No-stimulation mice underwent the same transport and were exposed to similar cages with similar food and water restriction in the same room, but experienced only normal room light (of similar lux as 40 Hz or 80 Hz stimulation) for the 1h duration.
Experimenters who stimulated the mice were male.
Tissue preparation
Immunohistochemistry:
Mice were transcardially perfused with 40 ml of ice cold phosphate buffered saline (PBS) followed by 40 ml of 4% paraformaldehyde (PFA; Electron Microscopy Sciences, Cat#15714-S) in PBS. Brains were removed and post-fixed in 4% PFA overnight at 4°C and transferred to PBS prior to sectioning.
Western blotting:
Visual cortex was dissected out and snap frozen in liquid nitrogen and stored in a −80°C freezer until processing. Samples were h omogenized using a glass homogenizer with RIPA (50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) buffer which contains protease and phosphatase inhibitor. The concentration of proteins in samples were quantified using a Bio-Rad protein assay. Equal concentrations of proteins were prepared and added with SDS-sample buffer.
Immunohistochemistry
Brains were mounted on a vibratome stage (Leica VT1000S) using superglue and sliced into 40 μm sections. Slices were subsequently washed with PBS and blocked using 5% normal donkey serum prepared in PBS containing 0.3% Triton-X100 (PBST) for 2 h at room temperature. Blocking buffer was aspirated out and the slices were incubated with the appropriate primary antibody (prepared in fresh blocking buffer) overnight at 4°C on a shaker. Slices then were washed three times (10 minutes each) with the blocking buffer and then incubated with the Alexa Fluor 488, 555, 594 or 647 conjugated secondary antibodies for 2 h at room temperature. Following three washes (15 minutes each) with blocking buffer and one final wash with PBS (10 minutes), slices were mounted with fluromount-G (Electron microscopic Sciences).
The following combination of secondary antibodies were used: (1) Alexa Fluor 488, 594 and 647, (2) Alexa Fluor 555 and 647, (3) Alexa Fluor 594 and 647, or (4) Alexa Fluor 488 and 647.
Imaging and quantification
Images were acquired using either LSM 710 or LSM 880 confocal microscopes (Zeiss) with 5×, 10×, 20×, 40× or 63× objectives at identical settings for all conditions. Images were quantified using either ImageJ 1.42q or Imarisx64 8.1.2 (Bitplane, Zurich, Switzerland). For each experimental condition, two coronal sections per mouse from the indicated number of animals were used. The averaged values from the two to four images per mouse was used for quantification.
NeuN, and p25:GFP positive cell counting:
All images were acquired in Z-stacks- 10 per image (step of 4 µm) and were quantified. An average of every two (excluding stack 1 and 10) and sum of all the counts was computed using ImageJ.
c-Fos positive cell counting:
LSM 710, with a 20× objective, plus cropping and tiling tools were used to cover the entire cortical column, was used to acquire the images from V1 and SS1. LSM 710 with a 40× or 20× objective was used to acquire the images from CA1 or CC-PFC, respectively. The multi-point tool in ImageJ was used to count cells manually.
γH2Ax positive cell counting:
The multi-point tool in ImageJ was used to count cells manually.
vGlut1, synaptophysin puncta:
LSM 880, with a 63× objective and further zoom of 3 times was used to acquire the images. The deep layers (primarily 4 and 5) from visual and somatosensory cortices, CA1 stratum radiatum and layer 5 of the ventral cingulate cortex were all targeted. A single plane image was acquired. The particle count plugin in ImageJ was used to quantify the number of vGlut1 puncta.
C1q intensities:
Using a LSM 710 with a 40× objective z-stacks of the entire slice thickness 40 μm (40 images from each field) were acquired. All images were compressed/collapsed and the signal intensity was measured in ImageJ.
CD40:
An LSM 880 with a 63× objective was used to obtain z-stacks of the entire slice thickness 40 μm (40 images from each field). All images were compressed/collapsed and the signal intensity was measured in ImageJ.
Lateral ventricles:
LSM 710 microscope with a 5× objective was used to image complete coronal slices (using image tiling) at −1.2, −1.4, −1.8, −2.0, and −2.5 AP relative to bregma. For each coordinate we took images from two adjacent slices per mouse. Outlines covering the entire area of the lateral ventricles were drawn using the freehand selection tool from ImageJ and the area of the LV was measured. Representative images shown in Figure 2 was tiled from multiple 5x images per section.
Microglia:
Iba1 immunoreactive cells were considered microglia. Using a LSM 710 with a 40× objective z-stacks of the entire slice thickness 40 μm (40 images from each field) were acquired. Imaris was used for 3D rendering of images to quantify the total volume of soma and processes microglia. Iba1 aggregation analysis was performed in Image J 3D rendering plugin. Minimum distance between Iba1 was calculated for every microglia from the images. All images were compressed/collapsed and Image J was used to quantify the total number of Iba1 positive cells. Rod-like microglia was identified and the total numbers was counted using Image J.
Cortical thickness:
LSM 710 microscope with a 5× objective was used to image complete cortical columns. The distance between outer cortical boundary and cortical side of corpus callosum was measured using ImageJ.
Brain weight measurement:
Mice were transcardially perfused with PBS followed by 4% PFA and the brain was post-fixed overnight in 4% PFA. Brains were washed in PBS and any excess PBS was removed before being weighed in a wet lab high precision scale (Mettler Toledo, accurate to 1mg). Separate cohorts of mice were sacrificed, brain was flash frozen in liquid nitrogen and brain weight was measured. Brain weight was normalized to CK control brains in each of these two independent measures, and combined.
Spine analysis:
Fos-CreERT2 mice were anaesthetized with avertin for AAV5-DIO-ChR2-EYFP (obtained from UNC viral core) virus infusion. A small hole was made in the skull above the visual cortex (relative to Bregma: AP −3.1 and ML ± 2.20), and a Hamilton syringe with 33-gauge needle was lowered to −0.5 DV where 300 Șl of virus was infused with a flow rate of 50 Șl per minute into each hemisphere of V1. The needle was withdrawn, following a 10-min post-injection period, and mice were allowed to recover for 4 days. In Fos-CreERT2 mice, the tamoxifen-dependent expression of Cre-recombinase (CreERT2) is driven from the loci of the immediate early gene Fos, allowing genetic access to neurons that are active during a time window of less than 12h. Three weeks post-injection, mice were injected with tamoxifen (intraperitoneal) and 1h later were exposed to No Stim or GENUS. 24h after the final session of 7d of daily GENUS, mice were perfused and their brains removed. Dendritic spines labelled with EYFP signals were amplified using anti-EGFP. Images were acquired using a LSM 880 with a 63× objective plus a further 2x zoom. Z-stacks of the entire slice thickness of 40 μm (40 images from each plane/field) were taken, and analyzed manually using 3D video in ImageJ.
Western blotting
Six to ten μg of protein was loaded onto 8% or 10% polyacrylamide gels and electrophoresed. Protein was transferred from acrylamide gels to nitrocellulose membranes (Bio-Rad) at 100 V for 120 min. Membranes were blocked using milk (5% w/v) diluted in PBS containing 0.1% Tween-20 (PBSTw), then incubated in primary antibodies overnight at 4°C. The following day, they were washed three times with PBSTw and incubated with horseradish peroxidase-linked secondary antibodies (GE Healthcare) at room temperature for 60 min. After three further washes with PBSTw, membranes were treated with chemiluminscence substrates and the films were developed. Signal intensities were quantified using ImageJ 1.46q and normalized to values of loading control such as β-actin, GAPDH or total proteins of the corresponding phosphorylated proteins analyses.
Plasma Corticosterone
Blood samples were collected in EDTA coated tubes (BD medical, Cat# 367856), centrifuged at 12,000g for 10 min at 4°C, and plasma was separated. Corticosterone levels were analyzed using corticosterone ELISA kit (Enzolifesciences, Cat# ADI-900–097) according to the manufacturers’ instruction.
Electrode probe implantation and in vivo electrophysiology
Multi-electrode probes:
Multi-site LFP probes were custom built using a 3D printed base with perfluoroalkoxy coated tungsten wire electrodes (50 μm bare diameter, 101.6 coated diameter; A-M Systems) and Neuralynx electrode interface board (EIB-36). Polyimide tubes were used to protect electrodes and reduce electrical noise. Electrodes were arranged to target layer 3 or 4 of visual cortex (co-ordinates relative to bregma; AP, −3.1; ML, + 2.5), SS1 (AP, −2.0; ML, + 2.3), CA1 region of the hippocampus (AP, −1.8; ML, +1.5) and cingulate area of the prefrontal cortex (AP, +1.0; ML, +0.2). Reference electrode was placed in the cerebellum.
Multi-tetrode probes:
Custom probes contain four nichrome tetrodes (14 mm; California Fine Wire Company), gold-plated (Neuralynx) to an impedance of 200 to 250 kOhm, arranged in one row (running along the CA3 to CA1 axis of the dorsal hippocampus) was implanted to target dorsal CA1 pyramidal layer (AP, −1.8). Reference electrode was targeted to the white matter tract above the hippocampus.
Surgery:
Mice were anaesthetized with avertin, restrained in a stereotactic apparatus and craniotomies were made exposing the visual, hippocampus, somatosensory, and prefrontal cortices, as specified. Probes were implanted and slowly lowered to the target depth. Mice were allowed to recover for a period of 4 days.
In vivo electrophysiology:
Following a 2–3 day habitation period to the chamber, recordings commenced with the animal allowed to move freely in a small open field. Recording sessions were composed of a 10–15 min period in which the LED was flashing at 40 Hz but was completely occluded by a black acrylic polypropylene sheet, immediately followed by a further 10 minutes with the sheet removed and animals exposed to the flickering LED. Data were acquired using Neuralynx SX system (Neuralynx, Bozeman, MT, USA) and signals were sampled at 32,556 Hz. Position of animals was tracked using red light-emitting diodes affixed to the probes. At the conclusion of the experiment, mice underwent terminal anesthesia and electrode positions were marked by electrolytic lesioning of brain tissue with 50 µA current for 10 s through each electrode individually, to confirm their anatomical location.
Spikes:
Single units was manually isolated by drawing cluster boundaries around the 3D projection of the recorded spikes, presented in SpikeSort3D software (Neuralynx). Cells were considered pyramidal neurons if mean spike width exceeded 200 μs and had a complex spike index (CSI) ≥5.
Data analyses:
LFPs were first filtered to the Nyquist frequency of the target sampling rate then down sampled by a factor of 20 (to 1628 Hz). Power spectral analyses were performed using the pwelch function in Matlab using a 500 ms time window with a 50% overlap, only segments of LFP data where animal velocity remained > 4 cm/s were included in the analyses. The weighted phase lag index (WPLI), a measure of coherence more suited to small rodent brains and which reduces potential contamination of coherence by volume conducted signals, was used as previously described. WPLI was calculated for pairs of electrodes in anatomically distinct regions, only for periods in which animal speed was > 4 cm/s. All analyses were performed using Matlab.
Pyramidal Cell 40 Hz Modulation:
The relationship between spike firing times and 40 Hz LFP phase was calculated as previously described (Middleton and McHugh, 2016), using the Circular Statistics Toolbox. Briefly, spikes were sorted and LFP traces were filtered using the continuous wavelet transform with a cmor 1.5–1 wavelet centered on 40 Hz, returning the instantaneous signal phase and amplitudes. Spike times were linearly interpolated to determine phase, with peaks and troughs of gamma defined as 0 and 180 degrees respectively. The resulting phase values were binned to generate firing probabilities, for each 20-degree interval. Cells were considered to be phase-locked only if they had a distribution significantly different from uniform (p < 0.05 circular Rayleigh test), with the strength of phase locking calculated as the mean resultant length.
RNA sequencing
Isolation of microglia:
Mice were transcardially perfused with ice cold phosphate buffered saline. The visual cortex was rapidly dissected and placed in ice cold Hanks’ balanced salt solution (HBSS) (Gibco by Life Technologies, catalogue number 14175–095). The tissue was enzymatically digested using the Neural Tissue Dissociation Kit (P) (Miltenyi Biotec, catalogue number 130–092-628) according to the manufacturer’s protocol, with minor modifications. Specifically, the tissue was enzymatically digested at 37°C for 15 min instead of 35 min and the resulting cell suspension was passed through a 40 µm cell strainer (Falcon Cell Strainers, Sterile, Corning, product 352340) instead of a MACS SmartStrainer, 70 µm. The resulting cell suspension was then stained using allophycocyanin (APC)-conjugated CD11b mouse clone M1/70.15.11.5 (Miltenyi Biotec, 130–098-088) and phycoerythrin (PE)-conjugated CD45 antibody (BD Pharmingen, 553081) according to the manufacturer’s (Miltenyi Biotec) recommendations. FACS was then used to purify CD11b and CD45 positive microglial cells. Standard, strict side scatter width versus area and forward scatter width versus area criteria were used to discriminate doublets and gate only singlets. Viable cells were identified by staining with propidium iodide (PI) and gating only PI-negative cells. CD11b and CD45 double positive ells were sorted into 1.5ml centrifuge tubes which contains 500 μl of RNA lysis buffer (QIAGEN, catalogue number 74134) with 1% β-mercaptoethanol (Sigma-Aldrich, catalogue number M6250). RNA was extracted using RNeasy Plus Mini Kit (QIAGEN, catalogue number 74134) according to the manufacturer’s protocol. RNA was eluted and then stored at −80°C until whole transcriptome amplification, library preparation and sequencing.
Isolation of neurons:
Visual cortex was homogenized in 0.5mL ice cold PBS with protease inhibitors and the suspension was centrifuged at 1600 g for 10 minutes. Pellet was resuspended in 5 mL NF-1 hypotonic buffer (0.5% Triton X-100, 0.1M Sucrose, 5mM MgCl2, 1mM EDTA, 10mM Tris-HCl, pH 8.0), incubated for 5 min, and then Dounce-homogenized (pestle A) with 30 strokes. 5mL NF-1 buffer was added to the suspension, washed pestle with 10 ml NF-1 buffer, for a combined total of 20 mL. Next, collected all in a 50 conical tube and filtered homogenate with 40 μm mesh filter, pelleted nuclei at 3,000 rpm (1,600 x g) for 15 min and resuspended in 30 mL NF-1 buffer and mixed well. Resuspended homogenate was rocked at 4 °C for 1 hour. Next, pellet was washed once with 2 0 mL NF-1 buffer, centrifuged at 3,000 rpm for 15 min and resuspended in 2–5 mL PBS+1%BSA+protease inhibitors without disturbing the pellet, on ice for 20 min. Alexa flour 488 or Alexa flour 647 conjugated NeuN antibodies (1:500) were added to the tubes, mixed, and incubated for 20 minutes at 4 °C in a rotating shaker. Unbound antibodies were washed out with suspending and centrifuging with PBS+1%BSA+protease inhibitors. Nuclei were spun down at 3,000 rpm for 15 min and resuspended in 0.5 mL (PBS+protease inhibitors), then filtered with 40 μm mesh filter for FACS sorting. Two drops of nucblue live ready probes reagent (Thermo Fischer Scientific; Cat. No. R37605) was added for nuclei gating. NeuN positive cells were sorted into 1.5 ml centrifuge tubes which contained 500 μl of RNA lysis buffer (QIAGEN, catalogue number 74134) with 1% β-mercaptoethanol (Sigma-Aldrich, catalogue number M6250). RNA was extracted using RNeasy Plus Mini Kit (QIAGEN, catalogue number 74134) according to the manufacturer’s protocol. RNA was eluted and then stored at −80°C u ntil whole transcriptome amplification, library preparation, and sequencing.
RNA library preparation:
Extracted total RNA was subjected to QC using an Advanced Analytical-fragment Analyzer before library preparation. SMARTer Stranded Total RNA-Seq Kit - Pico Input was used for the P301S neuronal, CK-p25 neuronal, and P301S microglial RNA-seq. SMART-Seq v4 Ultra Low Input RNA Kit was used for CK-p25 microglia specific RNA-seq. Libraries were prepared according to the manufacturer’s instructions, and sequenced on the Illumina Nextseq 500 platform at the MIT BioMicro Center.
RNA-seq analysis:
The raw fastq data of 40-bp paired-end sequencing reads were aligned to the mouse mm9 reference genome using STAR2.4. The total number of reads and the percentage of reads aligned are as follows: CK-p25 neurons- total reads 18094674.857 ± 827050.464; percentage aligned 85.323 ±0.414. P301S neurons- total reads 22792713.18 ± 6308817.636; percentage aligned 84.45 ± 0.548. CK-p25 microglia- total reads 28694964.5 ± 435841.6674; percentage aligned 89.43 ± 0.25. P301S microglia- total reads 18990369.2 ± 1667316.196; percentage aligned 27.243 ± 4.04. The mapped reads were processed by Cufflinks2.2 using mm9 reference gene annotation to estimate transcript abundances with library-type as fr-second strand (for stranded neuron data). Gene differential expression test between groups was performed using Cuffdiff module with p-value < 0.05 (for neuron data) (Trapnell et al., 2012). Geometric method was chosen as the library normalization method for Cuffdiff. For microglia data, featureCounts tool was used to quantify gene exonic counts from the non-stranded RNA-Seq data, and DESeq2 was employed to calculate statistical significance. Color-coded scatter plots were used to visualize group FPKM values for differentially expressed genes and other genes.
Z-scores of replicate expression FPKM values for differentially expressed genes were visualized in heatmaps for different sample groups. Gene ontology for microglia-specific DEGs was performed using Metascape tool, while neuron-specific DEGs was performed using TOPPGENE.
Phospho-proteomics (LC-MS/MS)
LC-MS/MS experiments were carried out in a proteomics core facility at Koch institute for integrative cancer research at MIT. 24h after the final No stim or GENUS, visual cortex was dissected out and snap frozen in liquid nitrogen and stored in a −80°C freezer until further use. Samples were subsequently homogenized using a plastic hand-held motor driven homogenizer with freshly prepared 8 M urea solution. The concentration of proteins in samples was quantified using a Bio-Rad protein assay. Samples containing 1 mg of protein per 1 ml were prepared, aliquoted and stored at −80°C freezer until further use. Proteins were reduced with 10 mM dithiothreitol (DTT) for 1 h at 56°C, alkylated wit h 50 mM iodoacetamide for 1 h at room temperature (RT) and diluted to less than 1M urea with 100 mM ammonium acetate at pH 8.9. Proteins were digested using sequencing grade trypsin (Promega; 1 μg trypsin per 50 μg protein) overnight at RT. Enzyme activity was quenched by acidification of the samples with acetic acid. The peptide mixture was desalted and concentrated on a C18 Sep-Pak Plus cartridge (Waters) and eluted with 50% acetonitrile, 0.1% formic acid and 0.1% acetic acid. Solvent was evaporated in a SpeedVac vacuum centrifuge. 400 µg aliquots of each sample were aliquoted and frozen in liquid nitrogen for 5 min, lyophilized and stored at −80°C.
TMT labeling:
TMT labeling and phosphopeptide enrichment: Lyophilized peptides were labeled with TMT-10-plex Mass Tag Labeling Kits (Thermo). For each TMT multiplex, a pooled sample was included consisting of a combination of equal amounts of peptides from WT, No Stim and GENUS mice, allowing for relative quantification to a normalization channel. For TMT labeling, nine samples from 9 mice peptide aliquots and one normalization channel (400 ȝg peptide for each channel) were resuspended in 100 ȝL of 70% (vol/vol) ethanol, 30% (vol/vol) 0.5 M triethyl-ammonium bicarbonate at pH 8.5, and incubated with TMT reagent resuspended in 40 ȝL anhydrous acetonitrile at RT for 1 h. The samples were concentrated, combined, and concentrated to dryness using a SpeedVac vacuum centrifuge.
Peptide Fractionation:
The TMT-labeled peptide pellet was fractioned via high-pH reverse phase HPLC. Peptides were resuspended in 100uL buffer A (10mM TEAB, pH8) and separated on a 4.6mm x 250 mm 300Extend-C18, 5um column (Agilent) using an 90 minute gradient with buffer B (90% MeCN, 10mM TEAB, pH8) at a flow rate of 1ml/min. The gradient was as follows: 1–5% B. (0–10min), 5–35% B (10–70min), 35–70% B (70–80min), 70% B (80–90min). Fractions were collected over 75 minutes at 1 minute intervals from 10 min to 85 min. The fractions were concatenated into 15 fractions non-contiguously (1+16+31+46+61, 2+17+32+47+62, etc). The fractions then underwent speed-vac (Thermo Scientific Savant) to near dryness.
Phosphopeptide enrichment:
Phosphopeptides were enriched from each of the 15 fractions using the High-Select Fe-NTA phosphopeptide enrichment kit (Thermo) per manufacturer’s instructions.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS):
Peptides were separated by reverse phase HPLC (Thermo Easy nLC1000) using a precolumn (made in house, 6 cm of 10 μm C18) and a self-pack 5 μm tip analytical column (12 cm of 5 μm C18, New Objective) over a 140 minutes’ gradient before nanoelectrospray using a QExactive Plus mass spectrometer (Thermo). Solvent A was 0.1% formic acid and solvent B was 80% MeCN/0.1% formic acid. The gradient conditions were 0–10% B (0–5 min), 10–30% B (5–105 min), 30–40% B (105–119 min), 40–60% B (119–124 min), 60–100% B (124–126 min), 100% B (126–136 min), 100–0% B (136–138 min), 0% B (138–140 min), and the mass spectrometer was operated in a data-dependent mode. The parameters for the full scan MS were: resolution of 70,000 across 350–2000 m/z, AGC 3e6 and maximum IT 50 ms. The full MS scan was followed by MS/MS for the top 10 precursor ions in each cycle with a NCE of 34 and dynamic exclusion of 30 s. Raw mass spectral data files (.raw) were searched using Proteome Discoverer (Thermo) and Mascot version 2.4.1 (Matrix Science). Mascot search parameters were: 10 ppm mass tolerance for precursor ions; 15 mmu for fragment ion mass tolerance; 2 missed cleavages of trypsin; fixed modification were carbamidomethylation of cysteine and TMT 10plex modification of lysines and peptide N-termini; variable modifications were methionine oxidation, tyrosine phosphorylation, and serine/threonine phosphorylation. TMT quantification was obtained using Proteome Discoverer and isotopically corrected per manufacturer’s instructions and were normalized to the mean of each TMT channel. Only peptides with a Mascot score greater than or equal to 25 and an isolation interference less than or equal to 30 were included in the data analysis.
Raw mass spectral data files were loaded into Proteome Discoverer version 1.4.1.14 (DBversion: 79) (Thermo) and searched against the mouse SwissProt database using Mascot version 2.4 (Matrix Science). TMT reporter quantification was extracted and isotope corrected in Proteome Discoverer. Tandem mass spectra were matched with an initial mass tolerance of 10 ppm on precursor masses and 15 mmu for fragment ions. Cysteine carbamidomethylation, TMT-labeled lysine and protein N-terminal were searched as fixed modifications. Minimal peptide length was seven amino acids. The data sets were filtered by ion score >20 for all peptides to ensure high confidence in peptide identification and phosphorylation localization and to achieve an (FDR) below 1% for peptides. To identify differentially expressed and phosphorylated peptides with significantly regulated ratios, we chose an arbitrary cutoff of ± 20 % difference with an adjusted p value of < 0.05.
Behavior
Elevated plus maze (EPM):
Mice were introduced into the center area of the EPM (ANY-Maze dimensions: arm length = 35cm, width = 5cm) and were tracked using Noldus program for 10 minutes. Time spent in each arm and the center of EPM was calculated offline.
Open Field (OF) and novel object recognition (NOR) in CK-p25 and P301S mice:
For OF, mice were introduced into an open field box (dimensions: length = 460mm, width = 460mm and height = 400mm; TSE-Systems) and were tracked using Noldus (Ethovision) for 7 min, with time spent in the center and peripheral area of the arena measured. NOR occurred on the following day, when mice were re-introduced into the same open field box which now additionally contained two identical novel objects, and were allowed to explore the objects for 10 min. Mice were then placed back in their home cages for 10 min after the last exploration. They were then returned to the same arena, with one of the two objects replaced with a new object. Mouse behavior was monitored for 7 min. Time spent exploring both the familiar and novel objects was recorded using Noldus and computed offline. Percentage of novelty preference index was calculated as follows: time exploring novel object (N) divided by total time exploring novel and familiar (F) objects and presented in %- {[N/N+F]*100}.
Morris water maze (MWM):
Apparatus consisted of a circular pool (122 cm in diameter), filled with tap water (22°C-24°C) and a non-toxic white pa int added to make the solution opaque. An escape platform (10 cm in diameter) with a blunt protruding edge for better grip was submerged 1 inch under the water level. The pool was divided into four equal quadrants labeled N (north), E (east), S (south) and W (west). Mice were introduced in different maze quadrants, in a randomized order across trials, approximately 12.5 cm from the edge. Trials lasted 60 s, and the time required to find the hidden platform (latency) was recorded. The probe test was conducted 24h after the last training trial, with the submerged platform removed. All the training and probe test trials were recorded using Noldus.
CK-p25, P301S, and 5XFAD mice were subjected to GENUS in the morning (until 12pm at the latest) and then tested in the MWM in the afternoon (3–7pm), which limited behavioral testing time to only 4 hours per day. A minimum of 2 and a maximum of 4 trials per day were used during training to allow us to run all groups in the MWM while also maintaining a strict dark/light cycle. Thus, all transgenic mice used in MWM performed the same number of training trials (20 total), although over a different number of training days (CK-p25: 6 days; P301S: 5 days; 5XFAD: 7 days). Young C57BL/6J were trained over 16 trials (4 trials per day for 4 days), while aged C57BL/6J mice required 24 trials (2–4 trials per day). Due to variable training trial schedules, all comparisons are made between No Stim and GENUS groups, and not between different mouse strains.
Statistical analyses
Statistical analysis was conducted in SPSS, Matlab, R-package or Prism. Statistical significance was calculated as noted in the appropriate figure legends, using independent samples t test, Mann-Whitney U test, Kolmogorov Smirnov test, Wilcoxon Rank-sum test, one-way ANOVA or two-way repeated-measures ANOVA with a Bonferroni post hoc analysis and Krushkal Wallis test with Dunn’s post hoc test. Statistical significance was set at 0.05.
Supplementary Material
EXTENDED FILE 1
Related to Figure 5. Microsoft excel sheet with differentially serine/threonine phosphorylated proteins. Sheet 1, 2 and 3 has data from CK-p25, P301S and aged wild type mice cohorts, respectively.
SUPPLEMENTAL VIDEO 1
Related to Figure 2. Video showing the NeuN (red) and Hoechst labeling of cell nuclei (blue) in 1 μm steps across the entire 40 μm slice from a representative mouse.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| vGlut1 (1:1000) | Synaptic Systems | Cat# 135 303 |
| NeuN (1:2000) | Synaptic Systems | Cat# 266 004 |
| IBA1 (1:500) | Wako Chemicals | Cat# 019–19741 |
| C1q (1:500) | Abcam | Cat# ab182451 |
| CD40 (1:200) | Thermo Fisher Scientific | Cat# MA5–17852 |
| γH2Ax (1:500) | Millipore | Cat# 05–636 |
| GAD65 (1:500) | Abcam | Cat# ab26113, GAD-6 |
| GAPDH (1:2000) | Santa Cruz Biotechnology | Cat# sc-32233, 6C5 |
| c-Fos (1:500) | Santa Cruz Biotechnology | Cat# sc-52 |
| ARMET (1:1000) | Abcam | Cat# ab67271 |
| Total Tau (1: 1000) | Santa Cruz Biotechnology | Cat# sc-5587, H-150 |
| p35/p25 (1:1000) | In house made | N/A |
| EGFP (1:1000) | Thermo Fisher Scientific | Cat# A-11122 |
| β-Actin (1:3000) | Abcam | Cat# ab9485 |
| DNM-1 (1:1000) | Thermo Fisher Scientific | Cat# MA5–15285, 3G4B6 |
| Ser774-DNM-1 (1:1000) | Thermo Fisher Scientific | Cat# PA5–38112 |
| DNM-3 (1:1000) | Thermo Fisher Scientific | Cat# PA1–662 |
| Donkey anti-Rabbit, Alexa Fluor 488 | Invitrogen | Cat# A21206 |
| Donkey anti-Mouse, Alexa Fluor 488 | Invitrogen | Cat# A21202 |
| Donkey anti-Rabbit, Alexa Fluor 555 | Invitrogen | Cat# A-31572 |
| Donkey anti-Rabbit, Alexa Fluor 594 | Invitrogen | Cat# A11037 |
| Donkey anti-Goat, Alexa Fluor 594 | Invitrogen | Cat# A11058 |
| Donkey anti-Mouse, Alexa Fluor 594 | Invitrogen | Cat# A11032 |
| Donkey anti-Guinea pig, Alexa Fluor 647 | Invitrogen | Cat# A21450 |
| Donkey anti-Goat, Alexa Fluor 647 | Invitrogen | Cat# A21447 |
| Donkey anti-Mouse, Alexa Fluor 647 | Invitrogen | Cat# A31571 |
| Donkey anti-Rabbit, Alexa Fluor 647 | Invitrogen | Cat# A31573 |
| Experimental Models: Organisms/Strains | ||
| Mouse: wild type C57BL/6J | Jackson Laboratory | JAX: 000664 |
| B6;CBA-Tg(Camk2a-tTA)1Mmay/J | Jackson Laboratory | Stock No: 003010 |
| C57BL/6-Tg(tetO-CDK5R1/GFP)337Lht/J | Jackson Laboratory | Stock No: 005706 |
| 5XFAD: B6SJL-Tg(APPSwFlLon,PSEN1*M146L*L28 6V)6799Vas/Mmjax | Jackson Laboratory | Stock No: 34840-JAX |
| Tau P301S-tg: B6;C3-Tg(Prnp-MAPT*P301S)PS19Vle/J | Jackson Laboratory | Stock No: 008169 |
| B6.129(Cg)-Fostm1.1(cre/ERT2)Luo/j | Jackson Laboratory | Stock No: 021882 |
| Software and Algorithms | ||
| MATLAB | Mathworks | mathworks.com |
| IMARIS | Bitplane | bitplane.com |
| R studio | www.rstudio.com | |
| ImageJ | NIH | imagej.nih.gov/ij |
| EthoVision XT | Noldus | noldus.com |
| TSEsystem | TSE Systems | tse-systems.com |
| GraphPad Prism | GraphPad | graphpad.com |
| Proteome Discoverer version 1.4.1.14 | Thermofisher | tools.thermofisher.com |
| Ingenuity Pathway Analysis | Qiagen | qiagenbioinformatics.com |
| SPSS (version 24) | IBM Analytics | ibm.com |
| ZEN | Zeiss | zeiss.com |
HIGHLIGHTS.
40 Hz visual stimulation entrains gamma oscillations in V1, CA1, and PFC
GENUS reduces neuronal and synaptic loss in mouse models of neurodegeneration
GENUS modifies synaptic signaling and synaptic plasticity related proteins
GENUS improves spatial learning and memory in Tau P301S and CK-p25 mice
ACKNOWLEDGEMENTS
We thank N Dedic, AS Singer, S Barker, RW Komorowski, K Abdelaal, M Murdock, F Abdurrob, E Lockshin, RT Stott and the members of L-H.T lab for helpful discussion and suggestions. We thank E McNamara for mice colony maintenance. A Del Rosario (Koch institute for integrative cancer research) for help with Mass spectrometry. A Watson, T Ko, M Saturno-Condon (Koch institute) and P Autissier (Whitehead institute) for help with FACS. C.A. was supported by JPB foundation. This work was supported by NIH RF1 AG047661 and RF1 AG054321 to L-H.T, and grants to L-H.T from Halis Family Foundation, The JPB Foundation, and Robert A and Renee E Belfer Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
L.-H.T. and E.S.B are scientific co-founders and serve on the scientific advisory board of Cognito Therapeutics.
REFERENCES
- Airavaara M, Shen H, Kuo C, Peränen J, Saarma M, Hoffer B, and Wang Y (2009). Mesencephalic astrocyteǦderived neurotrophic factor reduces ischemic brain injury and promotes behavioral recovery in rats. J. Comp. Neurol 515, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster M, Messa M, Ferguson SM, De Camilli P, and Ryan TA (2013). Dynamin phosphorylation controls optimization of endocytosis for brief action potential bursts. Elife 2, e00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachstetter AD, Ighodaro ET, Hassoun Y, Aldeiri D, Neltner JH, Patel E, Abner EL, and Nelson PT (2017). Rod-shaped microglia morphology is associated with aging in 2 human autopsy series. Neurobiol. Aging 52, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balschun D, Moechars D, Callaerts-Vegh Z, Vermaercke B, Van Acker N, Andries L, and D’hooge R (2009). Vesicular glutamate transporter VGLUT1 has a role in hippocampal long-term potentiation and spatial reversal learning. Cerebral Cortex 20, 684–693. [DOI] [PubMed] [Google Scholar]
- Benveniste EN, Nguyen VT, Wesemann DR. (2004) Molecular regulation of CD40 gene expression in macrophages and microglia. Brain Behav Immun 18(1):7–12. [DOI] [PubMed] [Google Scholar]
- Bero AW, Yan P, Roh JH, Cirrito JR, Stewart FR, Raichle ME, Lee J, and Holtzman DM (2011). Neuronal activity regulates the regional vulnerability to amyloid-[beta] deposition. Nat. Neurosci 14, 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun ME, Kurth D, Phinney AL, Long JM, Hengemihle J, Mouton PR, Ingram DK, Jucker M. (1998) Hippocampal neuron and synaptophysin-positive bouton number in aging C57BL/6 mice. Neurobiol Aging 19(6):599–606. [DOI] [PubMed] [Google Scholar]
- Canter RG, Penney J, and Tsai L (2016). The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature 539, 187–196. [DOI] [PubMed] [Google Scholar]
- Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai L, and Moore CI (2009). Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 459, 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KK, Hoch R, Lee AT, Patel T, Rubenstein JL, and Sohal VS (2015). Gamma rhythms link prefrontal interneuron dysfunction with cognitive inflexibility in Dlx5/6 /− mice. Neuron 85, 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito JR, Yamada KA, Finn MB, Sloviter RS, Bales KR, May PC, Schoepp DD, Paul SM, Mennerick S, Holtzman DM. (2005). Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron 48(6):913–22. [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Kang JE, Lee J, Stewart FR, Verges DK, Silverio LM, Bu G, Mennerick S, Holtzman DM. (2008). Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron 58(1):42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JC, Tseng H, Goldman JA, Shih H, and Tsai L (2003). Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron 40, 471–483. [DOI] [PubMed] [Google Scholar]
- Clayton EL, Sue N, Smillie KJ, O’Leary T, Bache N, Cheung G, Cole AR, Wyllie DJ, Sutherland C, Robinson PJ, Cousin MA. (201) Dynamin I phosphorylation by GSK3 controls activity-dependent bulk endocytosis of synaptic vesicles. Nat Neurosci 13(7):845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejanovic B, Huntley MA, De Mazière A, Meilandt WJ, Wu T, Srinivasan K, Jiang Z, Gandham V, Friedman BA, Ngu H, Foreman O, et al. , (2018). Changes in the Synaptic Proteome in Tauopathy and Rescue of Tau-Induced Synapse Loss by C1q Antibodies. Neuron 100(6):1322–1336.e7. [DOI] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao P, Qiu Y, and Zhao Y (2013). SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci 16, 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Pang PT, Lu B, and Tsai L (2005). Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron 48, 825–838. [DOI] [PubMed] [Google Scholar]
- Frey A, Listovsky T, Guilbaud G, Sarkies P, Sale JE. (2014). Histone H3.3 is required to maintain replication fork progression after UV damage. Curr Biol 24(18):2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie AK, Jones EA, Lin Y, Karlsson MP, Kay K, Yoon SY, Tong LM, Nova P, Carr JS, and Frank LM (2016). Apolipoprotein E4 causes age-dependent disruption of slow gamma oscillations during hippocampal sharp-wave ripples. Neuron 90, 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M. (2015). Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518(7539):365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon J, Attal Y, Colliot O, La Corte V, Dubois B, Schwartz D, Chavez M, and Fallani FDV (2017). Loss of brain inter-frequency hubs in Alzheimer’s disease. Scientific reports 7, 10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman M, Arumae U, Yu LY, Lindholm P, Peranen J, Saarma M, and Permi P (2011). Mesencephalic astrocyte-derived neurotrophic factor (MANF) has a unique mechanism to rescue apoptotic neurons. J. Biol. Chem 286, 2675–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T, Vitorica J, and Ransohoff RM (2015). Neuroinflammation in Alzheimer’s disease. The Lancet Neurology 14, 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CS (2001). Human EEG responses to 1–100 Hz flicker: resonance phenomena in visual cortex and their potential correlation to cognitive phenomena. Experimental brain research 137, 346–353. [DOI] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, et al. (2016). Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, Mathys H, Seo J, Kritskiy O, Abdurrob F, et al. (2016). Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M, Gonzalez B, Castellano B, and Zimmer J (1994). Microglial and astroglial reactions to anterograde axonal degeneration: a histochemical and immunocytochemical study of the adult rat fascia dentata after entorhinal perforant path lesions. Experimental brain research 98, 245–260. [DOI] [PubMed] [Google Scholar]
- Jørgensen MB, Finsen BR, Jensen MB, Castellano B, Diemer NH, and Zimmer J (1993). Microglial and astroglial reactions to ischemic and kainic acid-induced lesions of the adult rat hippocampus. Exp. Neurol 120, 70–88. [DOI] [PubMed] [Google Scholar]
- Kastanenka KV, Hou SS, Shakerdge N, Logan R, Feng D, Wegmann S, Chopra V, Hawkes JM, Chen X, and Bacskai BJ (2017). Optogenetic Restoration of Disrupted Slow Oscillations Halts Amyloid Deposition and Restores Calcium Homeostasis in an Animal Model of Alzheimer’s Disease. PloS one 12, e0170275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, Guan JS, Lee BH, Moy LY, Giusti P, Broodie N, Mazitschek R, Delalle I, Haggarty SJ, Neve RL, Lu Y, Tsai LH.. (2008). Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron 60, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig T, Prichep L, Dierks T, Hubl D, Wahlund LO, John ER, and Jelic V (2005). Decreased EEG synchronization in Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 26, 165–171. [DOI] [PubMed] [Google Scholar]
- Martinez-Losa M, Tracy TE, Ma K, Verret L, Clemente-Perez A, Khan AS, Cobos I, Ho K, Gan L, and Mucke L (2018). Nav1. 1-Overexpressing Interneuron Transplants Restore Brain Rhythms and Cognition in a Mouse Model of Alzheimer’s Disease. Neuron 98, 75–89. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell AJ, Paulson AL, Suk HJ, Abdurrob F, Drummond G, Guan W, Young JZ, Kim DN, Kritskiy O, Barker SJ, et al. , (2019). Multi-sensory Gamma Stimulation Ameliorates Alzheimer’s-Associated Pathology and Improves Cognition. Advanced online publication: Cell 10.1016/j.cell.2019.02.014 [DOI] [PMC free article] [PubMed]
- Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, and Tsai L (2017). Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell reports 21, 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. (2001). Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci 4(11):1086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Wenderski W, Noh K, Bagot RC, Tzavaras N, Purushothaman I, Elsässer SJ, Guo Y, Ionete C, and Hurd YL (2015). Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton SJ, and McHugh TJ (2016). Silencing CA3 disrupts temporal coding in the CA1 ensemble. Nat. Neurosci 19, 945. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, and Helmchen F (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. [DOI] [PubMed] [Google Scholar]
- Nott A, Cheng J, Gao F, Lin Y, Gjoneska E, Ko T, Minhas P, Zamudio AV, Meng J, and Zhang F (2016). Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat. Neurosci 19, 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, et al. , (2006). Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci 26, 10129–10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, and Mucke L (2016). Network abnormalities and interneuron dysfunction in Alzheimer disease. Nature Reviews Neuroscience 17, 777–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor MA, Artieda J, Arbizu J, Valencia M, and Masdeu JC (2003). Human cerebral activation during steady-state visual-evoked responses. J. Neurosci 23, 11621–11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager G, and Singer W (1998). The response of cat visual cortex to flicker stimuli of variable frequency. Eur. J. Neurosci 10, 1856–1877. [DOI] [PubMed] [Google Scholar]
- Ribary U, Ioannides AA, Singh KD, Hasson R, Bolton JP, Lado F, Mogilner A, and Llinas R (1991). Magnetic field tomography of coherent thalamocortical 40-Hz oscillations in humans. Proc. Natl. Acad. Sci. U. S. A 88, 11037–11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, Gallardo G, et al. , (2017). ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549(7673):523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Chowdhury S, Ma R, Le KX, Hong S, Caldarone BJ, Stevens B, Lemere CA. (2017) Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci Transl Med 9(392). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AC, Martorell AJ, Douglas JM, Abdurrob F, Attokaren MK, Tipton J, Mathys H, Adaikkan C, and Tsai L (2018). Noninvasive 40-Hz light flicker to recruit microglia and reduce amyloid beta load. Nature protocols 1. [DOI] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, and Deisseroth K (2009). Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg EE, Lee RJ, Najafi AR, Rice RA, Elmore MR, Blurton-Jones M, West BL, and Green KN (2016). Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain 139, 1265–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam C, De Haan W, Daffertshofer A, Jones B, Manshanden I, van Cappellen van Walsum, Anne-Marie, Montez T, Verbunt J, De Munck J, and Van Dijk B (2008). Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer’s disease. Brain 132, 213–224. [DOI] [PubMed] [Google Scholar]
- Stam CJ, van Walsum, Anne Marie van Cappellen, Pijnenburg YA, Berendse HW, de Munck JC, Scheltens P, and van Dijk BW (2002). Generalized synchronization of MEG recordings in Alzheimer’s disease: evidence for involvement of the gamma band. Journal of Clinical Neurophysiology 19, 562–574. [DOI] [PubMed] [Google Scholar]
- Tampellini D, Capetillo-Zarate E, Dumont M, Huang Z, Yu F, Lin MT, Gouras GK. (2010). Effects of synaptic modulation on beta-amyloid, synaptophysin, and memory performance in Alzheimer’s disease transgenic mice. J Neurosci 30(43):14299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Town T, Crawford F, Mori T, DelleDonne A, Crescentini R, Obregon D, Flavell RA, and Mullan MJ (2002). Role of CD40 ligand in amyloidosis in transgenic Alzheimer’s mice. Nat. Neurosci 5, 1288. [DOI] [PubMed] [Google Scholar]
- Taylor SE, Morganti-Kossmann C, Lifshitz J, and Ziebell JM (2014). Rod microglia: a morphological definition. PloS one 9, e97096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret L, Mann EO, Hang GB, Barth AM, Cobos I, Ho K, Devidze N, Masliah E, Kreitzer AC, and Mody I (2012). Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in Alzheimer model. Cell 149, 708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinck M, Oostenveld R, Van Wingerden M, Battaglia F, and Pennartz CM (2011). An improved index of phase-synchronization for electrophysiological data in the presence of volume-conduction, noise and sample-size bias. Neuroimage 55, 1548–1565. [DOI] [PubMed] [Google Scholar]
- Wang WY, Tan MS, Yu JT, and Tan L (2015). Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann. Transl. Med 3, 136–5839.2015.03.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, Sanders DW, Cook C, Fu H, and Boonen RA (2016). Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci 19, 1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Holth JK, Liao F, Stewart FR, Mahan TE, Jiang H, Cirrito JR, Patel TK, Hochgrafe K, Mandelkow EM, and Holtzman DM (2014). Neuronal activity regulates extracellular tau in vivo. J. Exp. Med 211, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Tanei ZI, Hashimoto T, Wakabayashi T, Okuno H, Naka Y, Yizhar O,Fenno LE, Fukayama M, Bito H, et al. , (2015). Chronic optogenetic activation augments aβ pathology in a mouse model of Alzheimer disease. Cell Rep 11(6):859–865. [DOI] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang S, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, and Lee VM (2007). Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EXTENDED FILE 1
Related to Figure 5. Microsoft excel sheet with differentially serine/threonine phosphorylated proteins. Sheet 1, 2 and 3 has data from CK-p25, P301S and aged wild type mice cohorts, respectively.
SUPPLEMENTAL VIDEO 1
Related to Figure 2. Video showing the NeuN (red) and Hoechst labeling of cell nuclei (blue) in 1 μm steps across the entire 40 μm slice from a representative mouse.