Summary
In the yeast Saccharomyces cerevisiae the TOR complex 1 (TORC1) controls many growth-related cellular processes and is essential for cell growth and proliferation. Macrolide antibiotic rapamycin, in complex with a cytosol protein named FKBP12, specifically inhibits TORC1, causing growth arrest. The FKBP12-rapamycin complex interferes with TORC1 function by binding to the FRB domain of the TOR proteins. In an attempt to understand the role of the FRB domain in TOR function, we identified a single point mutation (Tor2W2041R) in the FRB domain of Tor2 that renders yeast cells rapamycin resistant and temperature sensitive. At the permissive temperature, the Tor2 mutant protein is partially defective for binding with Kog1 and TORC1 impaired for membrane association. At the restrictive temperature, Kog1 but not the Tor2 mutant protein, is rapidly degraded. Overexpression of ubiquitin stabilizes Kog1 and suppresses the growth defect associated with the tor2 mutant at the nonpremissive temperature. We find that ubiquitin binds non-covalently to Kog1, prevents Kog1 from degradation and stabilizes TORC1. Our data reveal a unique role for ubiquitin in regulation of TORC1 and suggest that Kog1 requires association with the Tor proteins for stabilization.
Keywords: Rapamycin, TOR, yeast, Kog1, Ubiquitin, TORC1
Introduction
The target of rapamycin (TOR) signaling controls cell growth spatially and temporally in response to nutrition, environmental stress, growth factor, and energy. Deregulation of Tor signaling contributes to tumorigenesis and other human diseases such as heart disease, diabetes and muscular atrophy (Zoncu et al., 2011). The structure and function of the Tor proteins are largely conserved from yeast to human (Jacinto & Hall, 2003, Wullschleger et al., 2006). In budding yeast Saccharomyces cerevisiae there are two Tor homologs, Tor1 and Tor2, which share ~67% sequence identity. The Tor proteins are large multi-domain polypeptides consisting of 20 copies of HEAT repeats at their N-terminus, a phosphatidylinositol kinase-like domain at its C-terminus, an FKBP12-rapamycin binding (FRB) domain immediately N-terminal to the kinase domain, a FAT domain before FRB, and a FATC motif at the C-terminus. The Tor proteins exist in two distinct multi-component complexes, Tor complex 1 (TORC1) and Tor complex 2 (TORC2) (Loewith et al., 2002). TORC1 consists of either Tor1 or Tor2 in complex with Kog1, Lst8, and Tco89, while TORC2 comprises Tor2, Avo1, Avo2, Avo3, Lst8, and Bit61. Hence, the function of TORC1 is shared by both Tor1 and Tor2 but that of TORC2 is Tor2 exclusive (Zheng et al., 1995). TORC1 regulates cell growth by regulating many growth-related processes, including translation, autophagy, and nutrient transporter turnover, and is rapamycin sensitive, whereas, TORC2 regulates the cell cytoskeleton and is rapamycin insensitive (Loewith et al., 2002, Wullschleger et al., 2006).
Rapamycin forms a complex with the cytoplasmic protein FKBP12, which in turn, blocks Tor function through binding to the FRB domain (Heitman et al., 1991, Koltin et al., 1991). Point mutations within the FRB domain have been found that abrogate the binding. However, these mutations do not appear to affect TOR function other than conferring yeast cell rapamycin-resistance, and thus are not useful for studying the role of this unique domain in TOR function (Freeman & Livi, 1996, Stan et al., 1994).
Ubiquitin is a highly conserved small protein which differs only in three residues of the 76-amino acid peptide between yeast and human. The conventional role for ubiquitin is in protein degradation through ubiquitin-proteasome pathway, in which attachment of ubiquitin to a target protein (ubiquitination) marks the protein for degradation by the proteasome machinery (Hershko, 1983). Ubiquitination is carried out in three sequential steps; activation, conjugation and ligation, performed, respectively, by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). This sequential cascade leads to ligation of ubiquitin to lysine residues on the protein substrate (Tanaka et al., 1998). In addition to its role in protein degradation, ubiquitin can also be attached to proteins and modifies protein conformation and activity in a way similar to phosphorylation. This non-proteolytic function of ubiquitin is involved in many cellular processes, such as internalization of membrane receptors, histone activity, DNA repair, virus budding, and nuclear export of p53. Unlike in proteasome pathway, the non-proteolytic regulatory functions of Ub are mainly involved with monoubiqitination rather than polyubiquitination(Hicke & Dunn, 2003, Sun & Chen, 2004, Mukhopadhyay & Riezman, 2007).
Here we report the isolation of a single point mutation in the FRB domain of Tor2 that renders yeast cells rapamycin resistant and temperature sensitive. By using this compromised tor2 allele, we subsequently identified ubiquitin as a high copy suppressor of the mutant. Our data suggest ubiquitin regulates Kog1 degradation and TORC1 stability through a unique non-covalent binding with Kog1.
Results
Isolation of the tor-2041 mutant that is both temperature sensitivity and rapamycin resistant
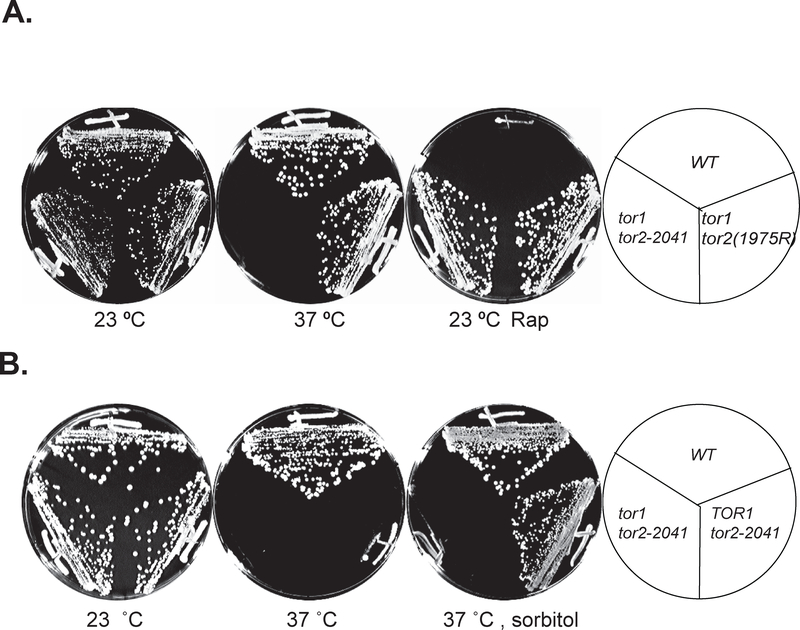
The rapamycin-FKBP12 complex inactivates the Tor proteins through binding to the rapamycin-FKBP12 binding (FRB) domain. To better understand the role of the domain in Tor function, we sought to determine how mutations within the domain affect Tor function. Accordingly, we created a library of tor2 mutants with mutations within the FRB domain in a tor1 deletion strain. This mutant library was then screened for mutants that were temperature sensitive and/or rapamycin resistant. Of the ~5,000 colonies screened, a surprisingly high portion (~70%) was found to be rapamycin resistant, among which a majority exhibited complete drug resistance yet without any obvious growth defects (data not shown). Nine colonies were identified that were also temperature sensitive, hence referred to as temperature sensitive and rapamycin (tr) mutants. One of the mutants, tor2-tr9, displayed a tight temperature sensitivity and complete rapamycin resistance. We thus selected this clone for further analysis. Sequencing the mutant tor2 gene in tor2-tr9 revealed that there were two point mutations within the FRB domain of the gene, including a serine-to-cysteine substitution at position 1784 (S1784C) and a tryptophan-to-arginine substitution at position 2041 (W2041R). To determine which mutation contributed to the temperature sensitivity and drug resistant phenotype, we generated single point mutations in TOR2 and tested the resulting mutants for temperature sensitivity and rapamycin resistance. We found that tor2S1784C behaved as the WT gene (data not shown), while tor2W2041R rendered yeast cells temperature sensitivity and drug resistance traits as did the original mutant (Fig.1A). Therefore, the point mutation W2041R within the FRB domain of TOR2 confers yeast cells both temperature sensitivity and drug resistance. Consistent with this finding, a previous study also reports that tor2W2041R is resistant to rapamycin (Huang et al., 2013). The mutant tor2 containing the single point mutation is hence referred as tor2–2041.
Figure 1. The tor2–2041 mutant is temperature sensitive and rapamycin resistant.
A. Temperature sensitive and rapamycin resistant traits of the tor2–2041 mutant. Wild type TOR2 control (Y818), tor1 tor2–2041 (Y812), and tor1 TOR2S1975R (Y031) mutant cells were placed on either YPD or YPD plate containing 100 nM of rapamycin. The plates were imaged after incubated for three days at either 23 or 37°C. B. The tor2–2041 mutant is defective for both the TOR-shared and TOR2-unique function. Wild type TOR2 (Y818), TOR1 tor2–2041 (Y817), and tor1 tor2–2041 (Y812) cells were placed on either YPD or YPD plate containing 1 M sorbitol. The plates were imaged after incubated for three days at either 23 or 37°C.
The tor2–2041 mutant is defective for both TOR-shared and TOR2-unique functions
As mentioned in the introduction, Tor2 has two distinct functions: the shared function with Tor1 and the Tor2-unique functions. Defects in the latter function cause a compromised cell wall, which can be rescued by including sorbitol, a cell wall stabilizer, in the growth media. To test if the TOR2-unique function is compromised in the tor2–2041 mutant, we created a tor2–2041 mutant in wild type TOR1 background and tested the mutant at 37 °C on a YEPD plate supplemented with 1 M sorbitol. As shown in Fig. 1B, the mutant cells grew in the presence of 1 M sorbitol but failed in the absence. This result indicates that the W2041R point mutation within the FRB domain affects TOR2 unique function, which cannot be recovered by TOR1. However, sorbitol did not restore the growth of tor2–2041 in the absence of TOR1 (Fig. 1B), indicating that the shared function is also compromised in the mutant.
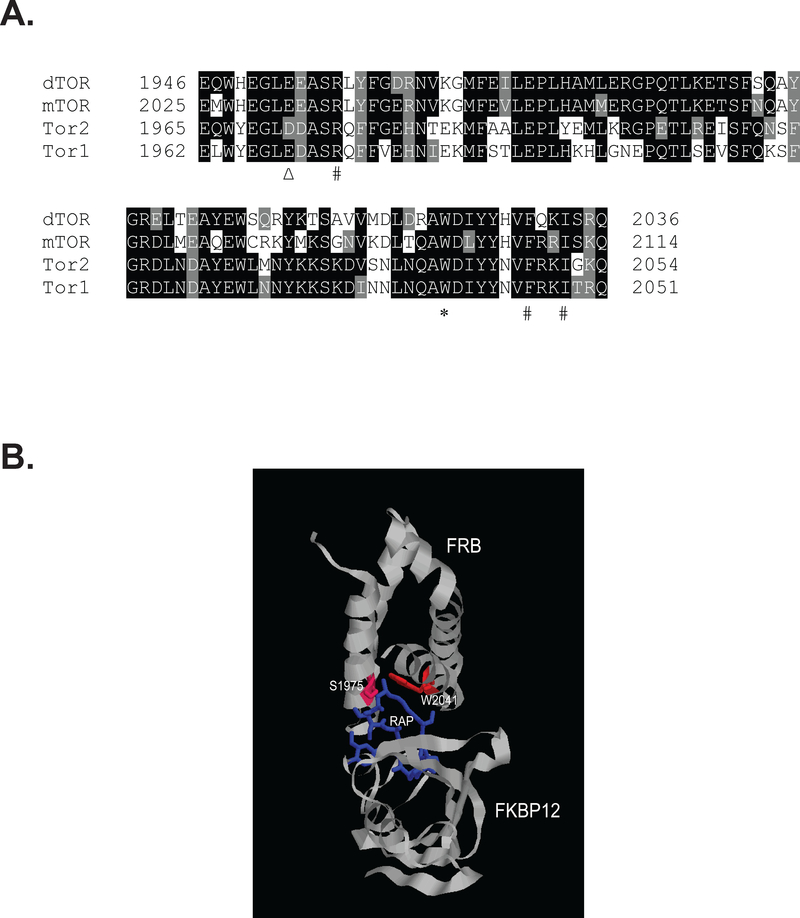
The tryptophan at 2041 is an invariant residue among all the Tor proteins in different organisms (Fig. 2A). Structural analysis revealed that the tryptophan residue is positioned at one of the two alpha helixes that serve as the binding packet of rapamycin, on the opposite of S1975, another residue whose replacement with bulk amino acids causes rapamycin resistance (Figs. 1A and 2B) (Choi et al., 1996). This position explains why changes in W2041, like those in S1975, are able to affect rapamycin binding. However, while changes in S1975 appear to affect only the drug binding, the W2041R mutation also impairs Tor2 function.
Figure 2. The W2041 residue is located within the rapamycin binding pocket of the FRB domain.
A. Sequence alignment of the FRB domains in Tor proteins. Δ indicates the conserved S1975 in Tor2 that confers yeast rapamycin resistant, but not temperature sensitive. ∗ designates the conserved W2041 in Tor2 that, when mutated to arginine, renders yeast rapamycin resistant and temperature sensitive. Other aromatic residues that contribute to rapamycin binding are underscored by #. dTOR: drosophila Tor. B. 3D structure of the FRB domain in complex with FKBP12 and rapamycin (PDB ID: 1FAP) with superimposed positions of W2041 and S1975.
The tor2–2041 mutant is defective for the membrane association of TORC1
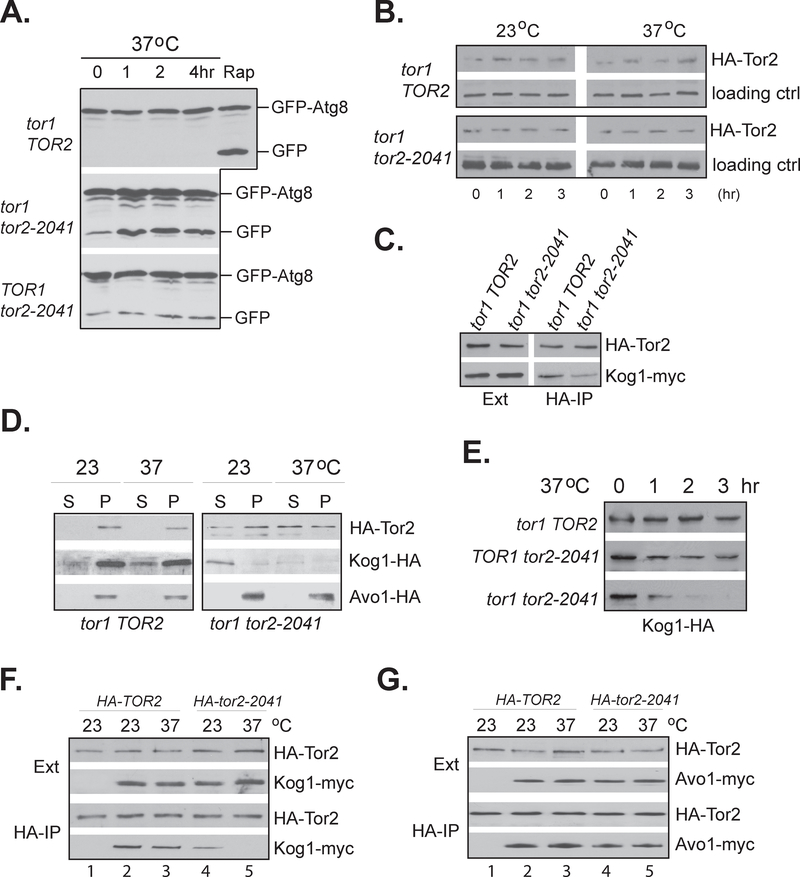
To determine how the mutation within the FRB domain of Tor2 affects its shared function, we examined the TORC1-dependent autophagy activity in the tor2–2041 mutant. In a tor1 deletion background, we found that the tor2–2041 mutant exhibited an elevated autophagy activity as indicated by enhanced cleavage of GFP-Atg8 at permissive temperature under normal growth condition (Fig 3A, mid panel). Upon shifting to the non-permissive temperature to inactivate the mutant Tor2, the autophagic cleavage of GFP-Atg8 was further enhanced. In contrast, the increased temperature had no effect on autophagy activity in wild type cells (Fig. 3A, upper panel). In a TOR1 background, we also found that the tor2–2041 mutant displayed an elevated autophagy activity. However, the presence of Tor1 blocked the temperature induced increase in the activity (Fig. 3A, lower panel). These observations suggest that the tor2–2041 mutant is defective for TORC1 function, which can be partially compensated by TOR1. Analysis of the level and stability of the mutant Tor2 protein did not reveal any obvious defects either at the permissive condition or at the restrictive temperature (Fig. 3B). However, the mutant Tor2 protein appeared to be partially defective for its association with Kog1 at the permissive temperature (Fig. 3C). This finding is consistent with the notion that the FRB domain of the Tor proteins is involved in binding with Kog1 (Adami et al., 2007). Because the function of both TORC1 and TORC2 requires their association with membrane, we further examined the membrane distribution of the complexes in the tor2-2041 mutant. As shown in Fig. 3D, in wild type cells grown at 23°C and 37°C, Tor2 was presented in the membrane fraction of the cell lysate (left panel). In contrast, in the mutant cells grown at 23°C, Tor2 was distributed in both the cytosol and membrane fractions and became more soluble upon shifting the cells to 37°C for two hours (right panel). This observation suggests that the mutant Tor2 protein is defective for its membrane association. Because Tor2 exists in both TORC1 and TORC2, we determined which complex was affected by the mutant Tor2 through examining the membrane association of Kog1 and Avo1, the unique components, respectively, to TORC1 and TORC2. In wild type cells grown at 23°C or 37°C, we found that Kog1 was largely in the membrane fraction (Fig. 3D, left panel). However, in the mutant cells grown at 23°C, Kog1 was mostly soluble, suggesting a defect in its membrane association. Upon shifting to 37°C for two hours, Kog1 became barely detectable (Fig. 3D, right panel). In contrast, Avo1, the TORC2 unique component, was found only in the membrane fractions of the cell extracts from both wild type and the tor2 mutant cells. Shifting the mutant cells to the non-permissive temperature had little effect on its membrane association and stability (Fig. 3D). These findings indicate that the tor2 mutant affects the membrane association of Kog1 but not Avo1.
Figure 3. The tor2–2041 mutant is defective for TORC1 function.
A. Exponentially growing wild type (Y818) and tor2 mutant (Y812 and Y817) cells expressing GFP-ATG8 were incubated at 23 and 37°C for indicated times or treated with 100 nM rapamycin for 1 hr at 23°C (Rap). The cell extracts were analyzed by western blotting with anti-GFP antibody for the autophagic cleavage of GFP-Atg8 fusion protein. B. Exponentially growing yeast wild type TOR2 (Y110) and mutant tor2–2041 (Y1035) cells expressing HA tagged wild type or mutant Tor2 protein were incubated at 23 and 37°C for indicated times. The expression levels of the HA tagged Tor2 protein and the loading control (Tpd3) was examined by western blotting. C. Cell extracts from exponentially growing wild type (Y110) and mutant tor2–2041 (Y1035) expressing KOG1-myc cells were precipitated with anti-HA antibody. The levels of HA-Tor2 and Kog1-myc in the extract and precipitates were determined by western blotting. D. Wild type TOR2 (Y110) and mutant tor2–2041 (Y1035) expressing control vector, KOG1-HA or AVO1-HA were shifted from 23 to 37°C for 2 hr. Cell extracts from cells before and after the shift were fractionated into pellet (P) and soluble fractions (S) by centrifugation at 100, 000 ×g for 1 h. The levels of HA-Tor2, Kog1-HA, or Avo1-HA in the soluble and pellet fraction were examined by western blotting. E. Wild type TOR2 (Y818) TOR1 tor2–2041 (Y817), and tor1 tor2–2041 (Y812) cells were shifted from 23 to 37°C. The levels of Kog1-HA in the cells collected at different time points after the shift were examined by western blotting. F. Wild type cells (Y662) were transformed with control vector (lane 1) or KOG1-myc (lanes 2–4) together with either HA-TOR2 (lanes 1–3) or HA-tor2–2041(lanes 4–5). Transformed cells were grown at 23°C and shifted to 37°C for 2 hr. Cell extracts were precipitated with anti-HA antibody and the levels of HA-Tor2 and Kog1-myc in the extracts (Ext) and precipitates (HAIP) were examined by western blotting. G. Wild type cells (Y662) were transformed with control vector (lane 1) or AVO1-myc (lanes 2–4) together with either HA-TOR2 (lanes 1–3) or HA-tor2–2041(lanes 4–5). Transformed cells were grown at 23°C and shifted to 37°C for 2 hr. Cell extracts were precipitated with anti-HA antibody and the levels of HA-Tor2 and Avo1-myc in the extracts (Ext) and precipitates (HA-IP) were examined by western blotting.
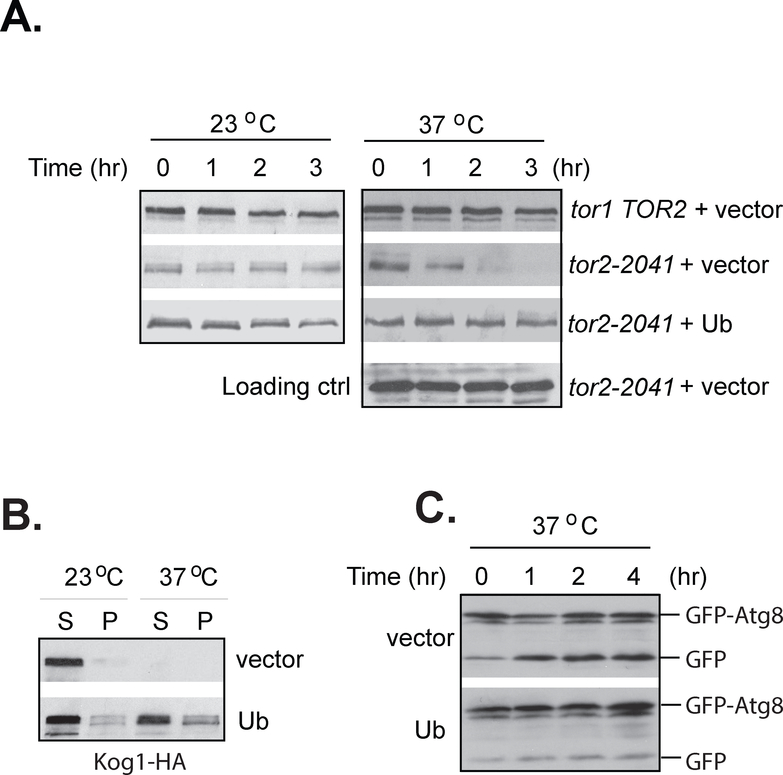
When the tor1 tor2–2041 cells were shifted to the nonpermissive temperature, Kog1 level was rapidly reduced (Fig. 3E, lower panel). The reduction was not observed in wild type cells after being shifted to the elevated temperature (Fig. 3E, top panel). This observation shows that Kog1 is unstable in the mutant cells at the elevated temperature. Interestingly, when TOR1 tor2–2041 cells were shifted to the elevated temperature, Kog1 level became stable after an initial reduction (Fig. 3E, middle panel), suggesting that the presence of wild type Tor1 stabilized Kog1 after Tor2 inactivation. Utilizing this feature, we further assayed the association of the Tor2 mutant protein with Kog1 or Avo1 at elevated temperature in wild type cells expressing HA tagged tor2–2041 mutant gene (HA-tor2–2041) together with Myc tagged Kog1 (Kog1-myc) or Avo1 (Avo1-myc). As expected, the level of the expressed Kog1-myc was largely unchanged upon shifting from 23 to 37°C in cells expressing HA-tor2–2041 (Fig. 3F, upper panels). However, the amount of Kog1-myc copurified with the mutant Tor2 protein was greatly reduced at the elevated temperature (Fig. 3F, lower panels). In contrast, the amount of Avo1 coprecipitated with the mutant Tor2 protein was not affected by the temperature shift in cells expressing HA-tor2–2041. This finding further confirms that the Tor2 mutant protein is defective for binding with Kog1 and that binding with the Tor proteins protects Kog1 from degradation.
Overexpression of ubiquitin suppresses the growth defect of tor2–2041 mutant cells
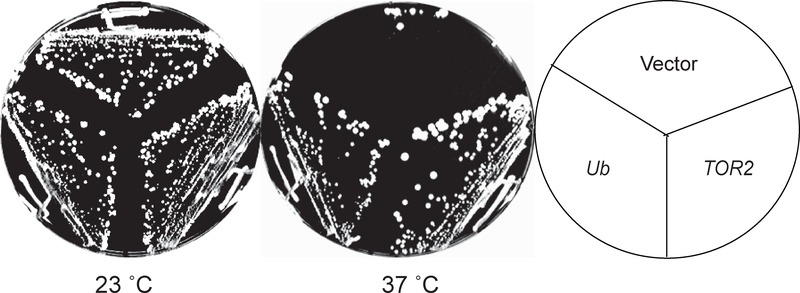
To further determine how the tor2 mutant protein affects TORC1 function, we screened a high-copy yeast genomic library for suppressors of the growth defect of the tor1 tor2–2041 mutant at the non-permissive temperature. This screen identified 80 clones that were able to rescue the growth defect of the mutant cells to various degrees under the restrictive temperature. Among these positive clones, 12 contained the TOR2 gene and ten other bore overlapping inserts. The only complete ORF in these overlapped regions is RPL40A. The RPL40A gene encodes a fusion protein, which is cleaved upon translation into ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit (Finley et al., 1987). To distinguish which part of the fusion protein is responsible for the suppression, we transformed the tor1 tor2–2041 mutant with a plasmid expressing a synthetic yeast ubiquitin gene (YEP96) and found that it restored the growth of the mutant cells at the non-permissive temperature (Fig. 4). This result shows that ubiquitin is a dosage suppressor of the tor1 tor2–2041 mutant. To identify suppressors for the defect for the TOR2 unique function, a similar screen was done in the wild type TOR1 background (TOR1 tor2–2041), which yielded several suppressor genes, including MSS4, RHO2, ROM1 and TUS1. The products of these genes are components of the cell wall integrity pathway that controls cell wall synthesis and remodeling. A previous study has shown that the pathway is a downstream event of the TOR2 unique function and responsible for the cell wall defects associated with tor2 unique mutants (Helliwell et al., 1994). This finding hence reinforces the notion that the mutant is defective in the Tor2 unique function, in addition to its defect in TORC1 function.
Figure 4. Overexpression of ubiquitin suppresses the growth defect of the tor2–2041 mutant.
The tor2–2041 cells (Y864) were transformed with control vector, or vectors expressing ubiquitin or wild type TOR2. The transformants were plated on SC-URA plates and imaged after incubation at 23 or 37°C for three days.
Overexpression of ubiquitin prevents Kog1 degradation in tor1 tor2–2041 cells
To understand how ubiquitin overexpression is able to restore the function of the mutant Tor2 protein, we examined the effect of the overexpression on the stability and membrane association of Kog1 in the mutant tor1 tor2–2041 cells grown under the restrictive temperature. We found that overexpression of ubiquitin prevented the degradation of Kog1 (Fig. 5A) and partially restored the membrane association of Kog1 in the mutant cells at the permissive temperature (Fig. 5B). Consistent with its effect on stabilization of Kog1, overexpression of ubiquitin also attenuated TORC1 inactivation induced by elevated temperature in the tor2–2041 cells, as indicated by the reduced cleavage of GFP-Atg8 in cells overexpressing ubiquitin (Fig. 5C).
Figure 5. Overexpression of ubiquitin prevents Kog1 degradation in the tor2–2041 mutant cells.
The tor1 tor2–2041 (Y864) cells expressing KOG1-HA were transformed with empty vector or vector expressing ubiquitin. The transformed cells were grown to exponential phase at 23°C and shifted to 37°C or remained at 23°C. A. The levels of Kog1-HA in the cell extracts from the cells collected at different time points after the shift were determined by western blotting. Wild type cells expressing KOG1-HA (Y985) were used as control. The levels of Tpd3 were used as the loading control. B. Cell extracts were partitioned into membrane (P) and soluble (S) by centrifugation at 100, 000 × g. The distribution of Kog1-HA in each fraction was examined by western blotting. C. The tor1 tor2–2041 (Y864) cells were transformed with the GFP-ATG8 gene together with a control vector or vector expressing ubiquitin. The transformed cells were grown to exponential phase at 23°C and shifted to 37°C for indicated times. The cell extracts were analyzed by western blotting with anti-GFP antibody for the autophagic cleavage of GFP-Atg8 fusion protein.
Polyubiquitination is not involved in the suppressive effect of ubiquitin
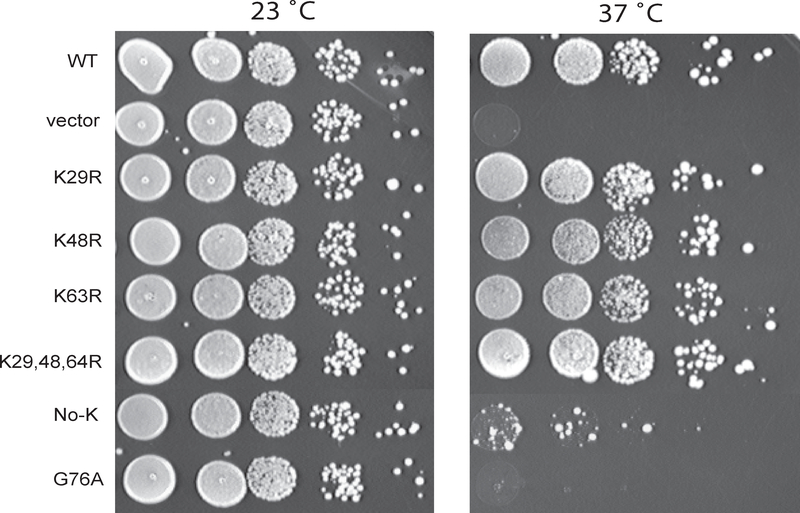
To determine how ubiquitin overexpression stabilized Kog1 in the tor2 mutant cells, we examined whether poly- or mono-ubiquitination was involved in the ubiquitin mediated suppression. Ubiquitination is achieved through a covalent isopeptide bond formed between the glycine 76 (G76) at the C-terminus of ubiquitin and a lysine residue in the targeted proteins (Pickart, 2001). Polyubiquitination requires lysine 48 (K48) on the proceeding ubiquitin for attachment of the ensuing ubiquitin. To test if polyubiquitination is involved in ubiquitin-mediated suppression of the growth defect of the tor1 tor2–2041 mutant, we examined if K48 of ubiquitin was required for the suppressor activity. We found a mutant ubiquitin with lysine 48 replaced with arginine (K48R) was as effective as its wild type version in suppressing the growth defect of the tor2 mutant (Fig. 6). Replacement of other potential ubiquitination sites, including K29 and K63, or triple substitution of K29, 48 and 63, did not affect the suppression ability of ubiquitin. In contrast, an ubiquitin mutant with all seven lysine residues replaced by arginine was found to be incapable of the suppression. Similarly, a mutant with G76 substituted with alanine was inactive (Fig. 6). These results suggest that polyubiquitination is not involved in regulation of Tor2 function
Figure 6. Ubiquitination is not involved in the effect of ubiquitin in suppression of the growth defect of the tor2–2041 mutant.
Exponentially growing tor1 tor2–2041 (Y864) cells expressing wild type or mutant ubiquitin were spotted on SC-TRP plates after a series of 10-fold dilutions. Plates were imaged after incubation at 23 or 37°C for 4 days.
We next tested the possibility that Tor or other components of the Tor complexes were modified by monoubiquitination. Accordingly, we immunopurified Tor1, Tor2, Kog1 Avo1 and Lst8 from yeast cells grown under various conditions and examined the ubiquitination by western blotting with anti-ubiquitin antibody. However, we were unable to detect ubiquitination on any of these proteins (data not shown), suggesting that a covalently attached ubiquitin does not play a direct role in regulation of Tor function.
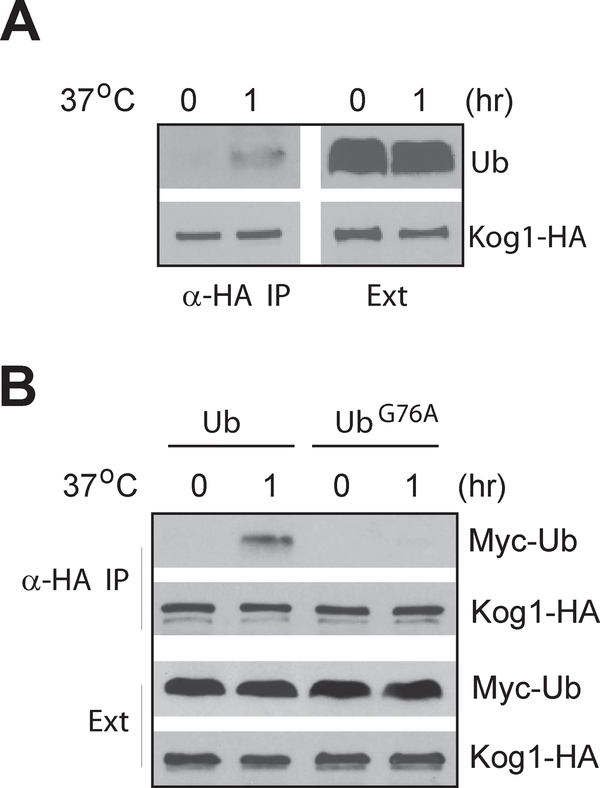
Ubiquitin has been found to be able to associate with Kog1 through the WD40 repeats at its C-terminal region (Pashkova et al., 2010), we thus investigated whether this non-covalent binding with ubiquitin was involved in regulation of Kog1 stability. In cells grown under normal growth condition, no obvious binding between Kog1 and ubiquitin was detected by co-immunoprecipitation. However, upon shifting yeast cells to 37°C degree for one hour, ubiquitin was found to associate with Kog1 (Fig. 7A). A similar result was obtained using myc tagged ubiquitin (Fig. 7B). In contrast, we found that the ubiquitinG76A was unable to associate with Kog1, suggesting that the mutation abrogated the interaction (Fig. 7B). This inability of ubiquitinG76A to bind with Kog1 correlates with its negligible effect in suppression of the growth defect of the tor2–2041 mutant (Fig. 6), indicating that the ability to bind with Kog1 is essential for the suppressor activity of ubiquitin.
Figure 7. Ubiquitin binds to Kog1 in response to temperature upshift.
A. Wild type yeast cells expressing KOG1-HA (Y985) and ubiquitin were subjected to heat stress for 1 hr. Cell extracts were precipitated with anti-HA antibody and the levels of Kog1-HA and ubiquitin in the cell extracts and precipitates were analyzed by western blotting. B. Wild type yeast cells expressing KOG1-HA (Y985), together with myc-tagged ubiquitin or ubiquitin G76A mutant were subjected to heat stress for 1 hr. Cell extracts were precipitated with anti-HA antibody and the levels of Kog1-HA and myc tagged ubiquitin in the extracts and precipitates were examined by western blotting.
Discussions
In the present study we identified a unique tor2 mutant, tor2–2041, that renders yeast cells both temperature sensitivity and rapamycin resistance. The rapamycin resistant trait of tor2–2041 is in agreement with the fact that the mutation is within the rapamycin binding domain. Chen et al. defined the region between amino acids 2025 to 2114 as the minimal domain on mTOR for binding with the rapamycin-FKBP12 complex, corresponding to amino acids 1965 to 2054 for yeast Tor2 (Chen et al., 1995). The crystal structure of the ternary complex of human FRB, rapamycin, and human FKBP12 show that rapamycin binds to FRB through a hydrophobic pocket that is located near the crossing of the α1 and α4 helices (Fig. 2B) (Choi et al., 1996). Six aromatic residues contribute to this rapamycin-binding hydrophobic pocket. The four aromatic residues Phe2039, Trp2101, Tyr2105 and Phe2108 appear to be important for binding of rapamycin. All these four aromatic residues are conserved from yeast to human (Fig. 2B). The side chain of Trp2101, the equivalent of yeast Trp2041, lines at the α4 side of the rapamycin binding pocket. Our finding that substitution of arginine for tryptophan at 2041 of Tor2 renders yeast rapamycin resistant substantiates the observation on crystal structure that Trp2101 of mTOR contributes to rapamycin binding. In accordance with the crystal structure, the conventional rapamycin-resistant allele at S1975 of Tor2, corresponding to S2035 of mTOR, also contacts with rapamycin in the ternary rapamycin-FKBP12-FRB complex. Substitution of S1975 with any residues that have a bulk side chain renders yeast rapamycin resistant {Stan, 1994 #13}. However, it does not appear to be the case for W2041. In addition to the tryptophan-to-arginine substitution, two other substitutions at the same position, W2041L and W2014C, were also found to block the interaction of Tor2 with the FKBP12-rapamycin complex and confer yeast cells rapamycin resistance, although the latter two did not appear to affect Tor2 function (Lorenz & Heitman, 1995). These resistance-conferring substituting residues (arginine, cysteine or leucine) have a smaller side chain than tryptophan does. Therefore, the drug resistance caused by changes at W2041 seems not to be caused by steric hindrance. Disruption of hydrophobic pocket by mutations is more likely the reason for disruption of rapamycin binding.
The tor2–2041 mutant is defective in both the shared and unique functions of Tor2, which is surprising given the fact that the point mutation is within the FRB domain of the Tor2 protein. This observation suggests that the FRB domain is also important for the function of Tor2 in TORC2. The defect in TORC1 function of the Tor2W2041R mutant protein appears to be caused by its impaired ability to interact with Kog1, which leads to dissociation of Kog1 from Tor2 (Fig. 3C). This finding is consistent with the notion that the FRB domain is involved in the association of the Tor proteins with Kog1 within TORC1 (Adami et al., 2007). However, the subsequent degradation of Kog1 is unexpected, since the protein itself is not affected by mutations. This finding indicates that Kog1 is an unstable protein that is consistently degraded when it is not in complex with the Tor proteins. Consistent with the notion, we found that the presence of wild type Tor1 in the tor2–2041 mutant cells is able to partially stabilize Kog1 (Fig. 3E), presumably, by binding with Kog1 and protecting it from degradation.
In an attempt to understand how the W2041R mutation affects Tor2 function, we identified ubiquitin as a dosage suppressor for the growth defect of tor2–2041. While the essential role of G76 in the suppressor activity of ubiquitin appears to suggest an involvement of covalently attached ubiquitination, we fail to find evidence indicating that any of the TORC1 components is modified by ubiquitination under various conditions, including starvation, high osmotic stress, and elevated temperature (data not shown). In addition, the findings that K48 and other lysine residues in ubiquitin are dispensable for its suppressive activity rule out the possibility of polyubiquitination. These observations suggest that a covalently attached modification of TORC1 components by ubiquitin is unlikely to underlie its suppressor activity. Instead, our finding that ubiquitin is able to non-covalently interact with Kog1 offers an explanation for its role in TORC1 function. A previous study has shown that ubiquitin is able to bind to a recombinant protein containing the WD repeats from the C-terminal region of Kog1 (Pashkova et al., 2010). It is thus likely that ubiquitin stabilizes the association of Kog1 with the mutant Tor2 and hence prevent its degradation by direct binding with Kog1. The fact that the binding between ubiquitin and Kog1 is induced under elevated temperatures further suggests that the binding may play a role in the stress response of TORC1. In a previous study, we show that heat stress induce a transient dissociation of TORC1 from membranes, which disrupts its function (Yan et al., 2012b, Yan et al., 2012a). It is possible that the binding of ubiquitin under the stress condition stabilizes TORC1 in the cytosol and facilitate its reassociation with membrane when the stressed cells adapted to the stress condition.
In summary, our study reveals a novel function of ubiquitin in TORC1 regulation. This regulation is not dependent on a covalent attachment of ubiquitin as in traditional ubiquitination. Our findings suggest that ubiquitin is a TORC1 associated factor that may control TORC1 function through binding with Kog1 and stabilizing its association with the Tor proteins.
Experimental Procedures
Strains, plasmids and media
Yeast strains used in this study are listed in Table I and plasmids in Table II. Standard YEPD and dropout synthetic complete (SC) media were used.
Table I.
Yeast strains used in this study
| Strains | Genotypea | Reference |
|---|---|---|
| Y031 | MATa tor2::LEU2 tor1::HIS3 trp1 ade2 ura3 leu2 his3 [TOR2S1975R-YCp50 (CEN, URA3)] | Lab Stock |
| Y110 | MATα tor2::HA3-TOR2 tor1::HIS3 ura3–52 leu2 his3–1 trp1 ade2–101 lys2 | Lab Stock |
| Y662 | MATα ura3–1 trp1–1 can1–100 ade2–1 his3–11,15 leu2–3, 112 | Lab Stock |
| Y812 | MATa tor2::LEU2 tor1::HIS3 trp1 ade2 ura3 leu2 his3 [tor2–2041-pRS314 (CEN, TRP1)] | This Study |
| Y817 | MATα tor2::HIS3 his3 ura3 leu2 trp1 ade2 [tor2–2041-pRS314 (CEN, TRP1)] | This Study |
| Y818 | MATα tor2::LEU2 tor1::HIS3 trp1 ade2 ura3 leu2 his3 [TOR2-pRS314 (CEN, TRP1)] | This Study |
| Y864 | MATα tor2::tor2–2041 tor1::HIS3 ura3–52 leu2 his3–1 trp1 ade2–101 lys2 | This Study |
| Y985 | MATα KOG1-HA3 ura3–1 trp1–1 can1–100 ade2–1 his3–11,15 leu2–3, 112 | This Study |
| Y989 | MATα kog1::KOG1-HA3:URA3 tor1::HIS3 ura3–52 leu2 his3–1 trp1 ade2–101 lys2 | Lab Stock |
| Y1035 | MATα tor2::HA3-tor2–2041 tor1::HIS3 ura3–52 leu2 his3–1 trp1 ade2–101 lys2 | This Study |
Plasmids are marked by square brackets. Yeast markers carried on each plasmid are listed in parentheses following the plasmid designation.
Table II.
Plasmids used in this study
| Plasmid | Characteristics | Reference |
|---|---|---|
| pKJ107 | RPL40A-pYEp24 (2μ, URA3) | This study |
| pJY136 | TOR2-pRS426 (2μ, URA3) | This study |
| pJY206 | HA3-TOR2-pRS314 (CEN, TRP1) | (Jiang & Broach, 1999) |
| pJY806 | tor2–2041-pRS314 (CEN, TRP1) | This study |
| pJY941 | HA3-tor2–2041-pRS406(CEN, URA3) | This study |
| pJY942 | HA3-tor2–2041-pRS314(CEN, TRP1) | This study |
| pJY948 | kog1-Δ5-HA3-pRS406 (URA3) | (Yan et al., 2006) |
| pJY949 | avo1-Δ5-HA3-pRS406 (URA3) | (Yan et al., 2006) |
| pJY966 | pCUP1-UbNo-K (2μ, TRP1) | This study |
| pJY973 | pCUP1-UbK63R (2μ, TRP1) | This study |
| pJY988 | pCUP1-UbK29R (2μ, TRP1) | This study |
| pJY991 | pCUP1-UbK29R, K48R, K63R | This study |
| pJY1066 | kog1-Δ5-myc13-pRS406 | (Yan et al., 2006) |
| pJY1573 | avo1-Δ5-myc13-pRS406 | This study |
| pΔUb | YEP96 deleted of Ub | This study |
| pUb202 | pCUP1-Ub UbG76A (2μ, TRP1) | (Ecker et al., 1987) |
| pUb203 | pCUP1-UbK48R (2μ, TRP1) | (Ecker et al., 1987) |
| YEP96 | pCUP1-Ub (2μ, TRP1) (a synthetic yeast Ub gene controlled by CUP1 promoter) | (Ecker et al., 1987) |
| LHP306 | pCUP1-UbNo-K (2μ, URA3) (substitutions all the seven lysines with arginine on Ub). | (Terrell et al., 1998) |
Plasmid YEP96, pUb202 and pUb203 are the kind gifts from Dr. Finley (Harvard University). YEP96 plasmid bears a synthetic yeast ubiquitin gene under the control of a copper-inducible (CUP1) promoter (Ecker et al., 1987). Expression from the promoter is induced by including copper sulfate (50 μM) in culture media. pUB202 and pUB203 are derivatives of YEP96 which contain mutant ubiquitin K48R and G76A, respectively. pJB966 was created by replacing the ubiquitin gene in YEP96 with ubiquitinNo-K from LHP306 (a gift from Dr. Hicke at Northwestern University) in which all the seven lysine residues of ubiquitin substituted with arginine (Terrell et al., 1998).
3’-end triple HA (pJY948) or 13 x myc (pJY1066) epitope tagged KOG1 and 3’-end triple HA tagged AVO1 (pJY949) constructs were created previously (Yan et al., 2006). A 3’-end 13x myc tagged AVO1 construct was created by replacing the HA tags in pJY949 with a DNA fragment containing 13x myc and ADH1 termination sequence. These constructs were used to replace the corresponding endogenous genes by two-step gene replacement. The 5’-end triple-HA tagged tor2–2041 gene (HA3-tor2–2041) was generated by site directed mutagenesis that replaced the tryptophan residue with arginine at position 2041 in HA3-TOR2-pRS314 plasmid (pJY206) that was created previously (Jiang & Broach, 1999). HA3-tor2–2041-pRS406 was created by cloning the triple HA tagged mutant tor2 gene into pRS406 (pJY941).
Construction of FRB mutant library
The FRB mutant library was created using the PCR directed mutagenesis approach (Muhlrad et al., 1992). The 2.2 kb region of TOR2 spanning nucleotides +4720 to 6920 of the gene was amplified with PCR in the presence of 1 mM MnCl2, which caused nucleotide misincorporation during PCR. The mutagenized PCR products were transformed into Y031 (tor1 tor2 pTOR2-Ycp50) together with plasmid TOR2-pRS314 that was digested with BamHI and SpeI. Transformants were selected on Trp dropout plates and TRP+ colonies were replicated onto Trp dropout plates containing FOA to select colonies that lost YCp50-TOR2 plasmid. The resulting colonies were further replicated onto YPD plates and analyzed for their growth at 23 and 37°C or in the presence or absence of 100 nM of rapamycin. Colonies display temperature sensitive growth and/or rapamycin resistant were isolated.
Screening for dosage suppressors of tor2–2041
The temperature sensitive tor1 tor2–2041 strain (Y864) was transformed with a yeast genomic library constructed with YEP24 (2μ, URA3). A total of 160 colonies that were able to grow at 37°C were obtained after screening ~100,000 Ura+ colonies. These positive clones were grouped into three categories based on their growth rates at the nonpremissive temperature: strong (58), moderate (58) and weak (44) growth. The plasmids were recovered from the colonies from the first two groups, and then re-introduced into Y864. Of those, 80 plasmids retained the suppression for the growth defect of Y864 at the restrictive temperature. Restriction mapping was then performed to eliminate the possible TOR2 clones, and to identify the redundant clones for the remaining suppressors. The selected suppressor clones were then sequenced to identify the suppressor genes.
Site-Directed Mutagenesis
Point mutation of ubiquitin was performed using QuikChange® Site-Directed Mutagenesis Kit (Stratagene). Substitutions of lysine to arginine were made using the synthetic WT ubiquitin on plasmid YEP96 as the template. All mutagenesis was confirmed by sequencing. An ubiquitin deletion plasmid pΔUb was also prepared by deleting the Bgl II and Xho I fragment from YEP96 and re-circularizing the plasmid after treating with Klenow.
Preparation of yeast protein extracts, fractionation of membrane protein and co-immunoprecipitation
Exponentially growing yeast cells were collected and lysed by vortexing with glass beads at 4 °C in lysis buffer containing 50 mM Tris (pH 7.4), 100 mM NaCl, 2 mM EDTA, 1mM PMSF, 2× protease inhibitor cocktail (Roche). The lysate was centrifuged at 500 × g for 5 min to remove unbroken cells and debris. Protein concentration was determined using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories). For fractionation of membrane and soluble proteins, the clear lysate obtained above was further centrifuged at 100, 000 × g for 1 h at 4 °C. The pellet after centrifugation represents the membrane-bound fraction (P) and the supernatant cytosolic soluble fraction (S). For co-immunoprecipitation, Triton-X-100 was added to cell lysate to a final concentration of 1%. After incubation at 4 °C for 15 min with agitation, insoluble cell debris were removed by centrifugation at 10,000 × g for 10 min. Clear lysate was then incubated with anti-HA antibody overnight followed by addition of Protein A agarose beads. After incubation for 1.5 hr with agitation, beads were washed four times with lysis buffer containing 1 % Triton-X-100, once with 20 mM Tris-Cl buffer and boiled for 3 min in the presence of 50 μl of SDS sample buffer. The precipitated sample was subsequently analyzed by western blotting.
Acknowledgements
We thank Dan Finley, Linda Hicke, and Michael Hall for generously providing plasmids and other laboratory members for comments and discussion during the course of the study. This study was supported by NIH grants (GM068832 and CA169186) to YJ. The authors have no conflict of interest to declare for publishing the present study.
References
- Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D & Llorca O, (2007) Structure of TOR and its complex with KOG1. Molecular cell 27: 509–516. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ & Schreiber SL, (1995) Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A 92: 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL & Clardy J, (1996) Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273: 239–242. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Khan MI, Marsh J, Butt TR & Crooke ST, (1987) Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem 262: 3524–3527. [PubMed] [Google Scholar]
- Finley D, Ozkaynak E & Varshavsky A, (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Freeman K & Livi GP, (1996) Missense mutations at the FKBP12-rapamycin-binding site of TOR1. Gene 172: 143–147. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hiestand PC & Hall MN, (1991) FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 88: 1948–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R & Hall MN, (1994) TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell 5: 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, (1983) Ubiquitin: roles in protein modification and breakdown. Cell 34: 11–12. [DOI] [PubMed] [Google Scholar]
- Hicke L & Dunn R, (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172. [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen K, Zhang J, Li Y, Wang H, Cui D, Tang J, Liu Y, Shi X, Li W, Liu D, Chen R, Sucgang RS & Pan X, (2013) A functional variomics tool for discovering drug-resistance genes and drug targets. Cell Rep 3: 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E & Hall MN, (2003) Tor signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol 4: 117–126. [DOI] [PubMed] [Google Scholar]
- Jiang Y & Broach JR, (1999) Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J 18: 2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK & Livi GP, (1991) Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol 11: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P & Hall MN, (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molecular cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Lorenz MC & Heitman J, (1995) TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem 270: 27531–27537. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R & Parker R, (1992) A rapid method for localized mutagenesis of yeast genes. Yeast 8: 79–82. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D & Riezman H, (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205. [DOI] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S & Piper RC, (2010) WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Molecular cell 40: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, (2001) Ubiquitin enters the new millennium. Molecular cell 8: 499–504. [DOI] [PubMed] [Google Scholar]
- Stan R, McLaughlin MM, Cafferkey R, Johnson RK, Rosenberg M & Livi GP, (1994) Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem 269: 32027–32030. [PubMed] [Google Scholar]
- Sun L & Chen ZJ, (2004) The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 16: 119–126. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Suzuki T & Chiba T, (1998) The ligation systems for ubiquitin and ubiquitin-like proteins. Mol Cells 8: 503–512. [PubMed] [Google Scholar]
- Terrell J, Shih S, Dunn R & Hicke L, (1998) A function for monoubiquitination in the internalization of a G protein-coupled receptor. Molecular cell 1: 193–202. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R & Hall MN, (2006) TOR signaling in growth and metabolism. Cell 124: 471–484. [DOI] [PubMed] [Google Scholar]
- Yan G, Lai Y & Jiang Y, (2012a) The TOR complex 1 is a direct target of Rho1 GTPase. Molecular cell 45: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Lai Y & Jiang Y, (2012b) TOR under stress: targeting TORC1 by Rho1 GTPase. Cell Cycle 11: 3384–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Shen X & Jiang Y, (2006) Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. Embo J 25: 3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XF, Florentino D, Chen J, Crabtree GR & Schreiber SL, (1995) TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82: 121–130. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A & Sabatini DM, (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]