Abstract
Non-enveloped viruses that are endocytosed employ numerous mechanisms to disrupt endosomal membranes for escape into the cellular cytoplasm. These include the use of amphipathic helices or sheets, hydrophobic loops, myristoylated peptides, and proteins with phospholipase activity. Some mechanisms result in immediate deterioration of the endosome, while others form pores in the membrane causing osmolysis to disrupt the endosome and allow viral escape. We describe an additional mechanism by a non-enveloped virus to disrupt endosomal membranes. Porcine Circovirus 2 (PCV2), possesses a forty-one amino acid arginine rich motif (ARM) at the N-terminus of its capsid protein that appears to be in the interior of the virus like particle (VLP). Using in-vitro membrane disruption assays we demonstrate that PCV2 VLP, unassembled capsid, and ARM peptide possess the ability to disrupt endosomal like membranes, whereas VLP lacking the ARM sequence do not possess this capability. Membrane disruption by VLP is insensitive to pH, but unassembled capsid protein and ARM peptide exhibit diminished activity at low pH. Our liposome disruption assays, circular dichroism and intrinsic tryptophan fluorescence assays allow us to propose a model for PCV2-endosomal membrane interaction wherein the ARM peptide externalizes from the capsid, its C-terminus (amino acids 28–40) anchors into the membrane, and the arginine rich N-terminus (amino acids 1–27) drives membrane disruption. To our knowledge, this is the first example of a non-enveloped virus using the arginines of an ARM to disrupt membranes. Also, this is the first example of such study for the Circoviridae family of viruses.
Keywords: Endosomal escape, Porcine circovirus, Arginine rich motif, liposome disruption, capsid protein
Introduction
Porcine circovirus 2 (PCV2), a prototypical representative of the Circoviridae family of viruses, is a small, non-enveloped, circular single stranded DNA (ssDNA) virus [1]. It is a major porcine pathogen that is ubiquitous and persistent in affecting the swine industries all over the world. PCV2 infection is associated with porcine circovirus associated disease (PCVAD) that causes enlarged lymph nodes, weight loss, jaundice, diarrhea, dermatitis, congenital tremors, congenital hypomyelination, nephropathy, respiratory and reproductive problems, and immunosuppression. PCVAD is responsible for a high mortality rate in swine [2]. The PCV2 genome (1766–1768 nucleotides) experiences mutation rates in the order of 1.2×10−3 substitutions per site per year, which is highest among DNA viruses [3]. The genome encodes for three larger open reading frames (ORF1 – ORF3) and multiple smaller reading frames [4, 5]. The larger ORFs encode for the replicase (ORF1) [6], the capsid protein (ORF2) [7], and a protein that might be responsible for the pathogenic nature of PCV2 by inducing apoptosis of infected cells (ORF3) [7]. PCV2 is able to replicate autonomously -without aid from other viruses. Consequently its small genome size designate PCV2 as the smallest pathogenic and autonomously replicating virus, and make PCV2 an excellent model system for understanding the minimum amount of genetic information necessary to produce an infectious and pathogenic virus [8–10]. Additional factors that make PCV2 an important subject of study are its ability to infect nearly every tissue within its host, its ability to infect and induce its pathogenic phenotype in multiple species of the animal kingdom, and its ability to rapidly mutate [11].
Cellular studies have shown that PCV2 attaches to cells via the heparan sulfate and chondroitin sulfate B (dermatan sulfate) glycosaminoglycans [12], and gains entry into porcine monocytic [13] and dendritic cells [14] via clathrin mediated endocytosis. PCV2 gains entry into epithelial cell lines PK15, SK and ST cells via caveolae-, clathrin and dynamin independent small GTPase regulated pathways [15]. Endocytosed macromolecules are either recycled to the surface of the cell or degraded by the cell as endosomes acidify to lysosome in what is referred to as the endosome-lysosome system [16]. Thus, PCV2 must escape from endosomes and enter the cells for infection to proceed.
Non-enveloped viruses utilize a variety of mechanisms for membrane disruption. Flock House Virus (FHV) and Nudaurelia capensis ω virus (NωV) have been shown to possess amphipathic helices at the C-termini of their capsid proteins that possess the ability to disrupt membranes. The C-termini are located inside the capsid, but autocatalytic cleavage at the C-termini generates amphipathic peptides (referred to as γ peptide) that can become exposed to the exterior via a process referred to as viral breathing [17]. The FHV γ peptide requires acidification to become active, whereas the NωV γ peptide is active at neutral pH [18, 19]. Binding of poliovirus to its receptor causes the capsid to undergo a conformational change to externalize the myristoylated amino acid of the VP4 capsid protein [20, 21] and the amphipathic helix at N-terminus of the VP1 capsid protein [22]. Both peptides insert into and disrupt the membrane [22]. The N-terminus of the minute virus of mice and B19 parvovirus capsid proteins possess phospholipase activity, and the low pH of endosomes potentiates this activity to disrupt the endosomal membrane [23, 24]. The N-terminus of the Reovirus μ1 capsid protein is hydrophobic and myristoylated [25]. Autocatalytic cleavage at the N-terminus of μ1 generates μ1N and μ1C fragments. The μ1N fragment is situated beneath the “protector” protein σ3. In the endosomes of infected cells, the cellular cathepsins digest σ3 and release μ1N which then proceeds to disrupt endosomal membrane [25–27]. Rotavirus gains cellular entry using hydrophobic loops in the VP5* fragment of the trimeric VP4 spike protein. Cellular trypsin digestion of VP4 generates VP8* and VP5*. Within the endosome, the hydrophobic loops of VP5* become exposed to disrupt the endosomal membrane for cellular entry [28, 29].
PCV2 does not possess any of the characteristics described above and little to nothing is known about how PCV2 may escape from the endosome. Sequence analysis of the PCV2 capsid protein does not identify amphipathic helices, or a myristoylated N-terminus; moreover, hydrophobic loops are not observed in the PCV2 virus like particle (VLP) structures [9, 30]. However, PCV2 possesses an arginine rich motif (ARM) at the N-terminus of the capsid protein that parallels the sequence properties of arginine-rich cell penetrating peptides (CPP) [31]. Arginine-rich CPP are positively charged peptides with few anionic or hydrophobic residues. These short peptides possess the ability to traverse plasma and endosomal membranes [31]. Indeed, such CPPs have successfully been used to transport 120 kDa proteins into the cellular cytoplasm [32, 33].
The 2.3 Å crystal structure of the PCV2 VLP visualized the capsid protein to adopt the canonical viral jelly roll, and identified the location of the ARM N-terminus (starting from amino acid 42) to the interior of the capsid (Fig. 1) [9, 30]. However, antigenic studies by multiple independent groups identified naturally occurring neutralizing antibodies that bind to the ARM region (amino acid 25–39) [34, 35]. Khayat et al. interpreted this observation to suggest that the ARM externalizes via viral breathing to sample the exterior of the capsid, and can thus be recognized by antibodies for viral neutralization [9]. Given the possibility that the ARM externalizes from the interior of the VLP, its sequence similarity to the well-documented arginine-rich CPP, and the requirement of PCV2 to escape from endosomes for delivery of the ssDNA genome into the nucleus for viral replication, we asked if PCV2 VLP possessed the ability to interact with endosomal-like membranes and if this interaction was driven by the ARM using an in vitro liposome disruption assay. The size of liposome and composition of lipids were chosen to closely mimic the composition of endosomal membranes. Liposome disruption assays involve loading liposomes with fluorophore at self-quenching concentrations. Such fluorophore exhibits minimal florescence when unperturbed, however fluorescence increases as the fluorophore is released from the liposome into the solution due to membrane disruption or leakage. Here we report that the PCV2 VLP possess the ability to rapidly release fluorophore from liposomes at pH 7 (early endosome) and pH 5 (endosome-lysosome) [16]. We further demonstrate that the unassembled capsid protein and the N-terminus ARM peptide also possess the ability to release fluorophore from liposomes at pH 7 and 5. The ARM peptide alone is poor in releasing fluorophore at pH 5, suggesting that the assembled capsid structure potentiates the ARM’s ability to disrupt membrane at pH 5. We also demonstrate that VLP lacking the ARM are incompetent in disrupting liposomes to release fluorophore. Taken together, our data suggests that the N-terminal ARM appears to be responsible for membrane disruption and fluorophore release. Further, our circular dichroism (CD) and intrinsic fluorescence studies point to the fact that the ARM region is intrinsically disordered with very little secondary structure and that the C-terminus (amino acids 28–40) of the peptide becomes buried inside the membrane whereas the N-terminus (amino acids 1–27) causes membrane disruption. While this manuscript was under preparation, a publication by Wu et al., further identifies the first 16 amino acids of the ARM peptide to exhibit cell penetrating properties (CPP) and corroborates with our results from liposome disruption assays [36]. To our knowledge this is the first example of a non-enveloped virus capable of disrupting membranes via the arginines of an ARM.
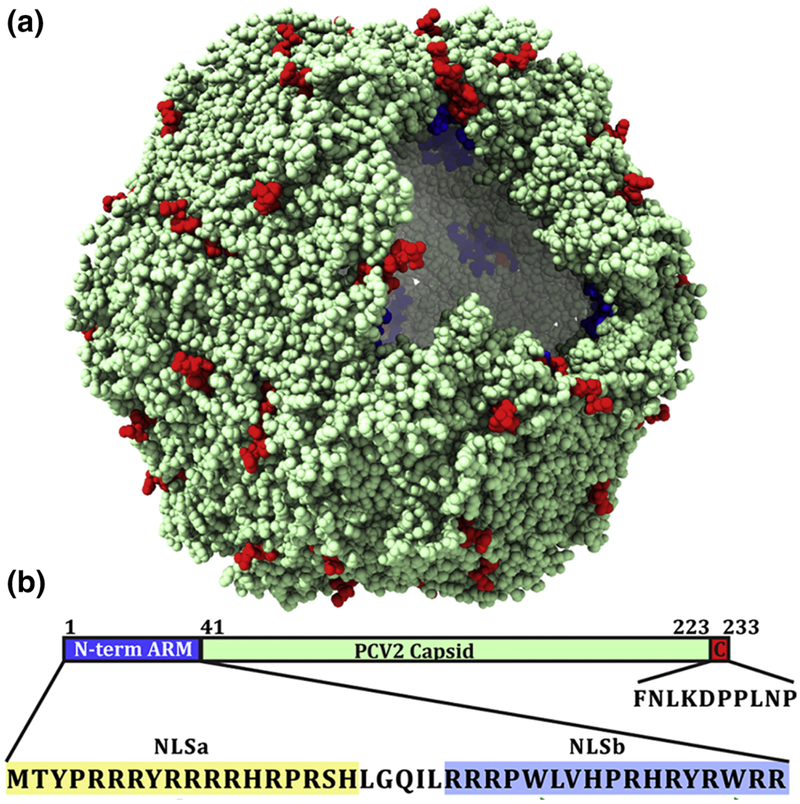
Figure 1.
a): Space filling representation of the PCV2 VLP crystal structure. A portion of the capsid has been computationally removed to visualize the interior. Blue spheres represent the first three observable amino acids at the N-terminus of PDB entry:3R0R, and the red spheres represent the last eight observable amino acid at the C-terminus. b): Schematic representation of the capsid protein sequence highlighting N-terminus arginine rich motif (ARM), and C-terminus. The sequence of the ARM and C-terminus peptide used in this study is shown below. The ARM sequence has been further classified on the basis of nuclear localization signal. The experimentally predicted secondary structure of ARM region is also represented. The brown color coil shows intrinsically disordered region and green arrows represent beta-sheet formation (see material and method section for the details).
Results
The PCV2 N-terminus ARM disrupts membranes
The ability of PCV2 VLP to disrupt membranes was examined using the liposome disruption assay at pH 7 and 5 to mimic the endosome-lysosome system [16]. We used a DOPC:DOPG ratio of 3:1 to generate 200 nm liposomes to mimic endosome. Such lipid content and ratio are routinely used to mimic the endosomal membrane [44]. The size of liposomes was confirmed with dynamic light scattering (DLS) measurements, wherein, the liposomes were reported to be homogenous (180–200 nm in diameter) (Fig. S1a, S1b). The size of the liposomes (200 nm) is similar to the size of endosomes observed via cellular cryo-electron tomography [45]. In all the liposome disruption assays, the ratio of lipid to protein (VLP, CPfl, ARM peptide, C-terminal peptide and CPΔARM) was estimated to be 1:408 for 0.5 μM; 1:272 for 0.75 μM and 1:204 for 1 μM concentrations (Fig. S2, Table 1).
Table 1:
Calculated phospholipid concentrations used in liposome disruption assays and their ratio with respect to the concentration of protein (VLP / CPfl / ARM peptide).
| Conc. of protein (μM) | Conc. of liposome (μM) | Ration: lipid/protein |
|---|---|---|
| 0.50 | 204.00 | 1:408 |
| 0.75 | 204.00 | 1:272 |
| 1.00 | 204.00 | 1:204 |
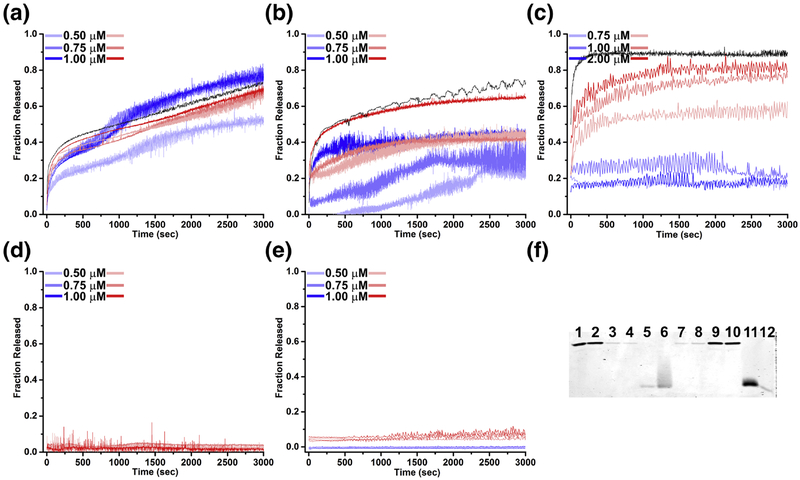
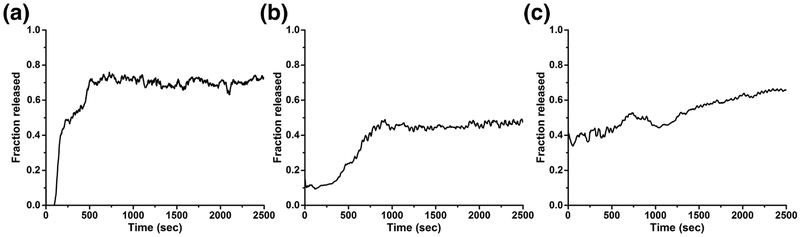
Our liposome disruption assays exhibit that increasing concentrations of VLP at pH 7 and 5 releases increasing amount of fluorophore (Fig. 2a). Fluorophore release is biphasic, with a rapid release within the first two minutes of the reaction at both pH values. This rapid release is more pronounced at pH 7 than at pH 5. Following the initial rapid release, the fluorophore release is approximately linear with respect to time for the remaining 48 minutes at pH 7. At pH 5, an additional increase of fluorophore release can be seen between 15 and 25 minutes followed by a release that is also linear with respect to time for the remainder of 48 minutes. We are uncertain to the cause of this rapid release. At 1 μM VLP concentration, the total amount of fluorophore release at pH 5 is comparable to that at pH 7 (Fig. 2a). The VLPs are also found to bind to the liposomal membranes as evidenced by the liposome-VLP pelleting assays (Fig. 2f; lanes 1, 2, 7, 8). VLPs bind to the liposome at both pH 7 and 5, though the binding is weaker when compared to unassembled full-length capsid protein or ARM peptide -see below.
Figure 2.
Liposome disruption and fluorophore release by PCV2 VLP and components of the capsid protein. The fraction of fluorophore release as compared to the liposome disruption by addition of 1% triton x-100 is represented. Red and blue shades represent fluorescence traces of fluorophore release from 200 nm DOPC:DOPG (75:25) liposomes at pH 7 and 5 respectively. a) VLP, b) capsid protein (CPfl), c) ARM peptide, d) VLP ARM, e) C-terminus peptide. Black curve in panel b and c represent fluorescence traces of fluorophore release from DOPC:DOPG:DOPG-PEG2000 (75:20:5) by capsid protein (CPfl) and ARM peptide, respectively. f) SDS-PAGE of the liposome pelleting assay to assess the association of ARM peptide, Lane 1 and 2: VLP supernatant at pH 7 and 5, Lane 3 and 4: CPfl supernatant at pH 7 and 5, Lane 5 and 6: ARM peptide supernatant at pH 7 and 5, Lane 7 and 8: VLP with liposome at pH 7 and 5, Lane 9 and 10: CPfl with liposome at pH 7 and 5, Lane 11 and 12: ARM peptide with liposome at pH 7 and 5.
To determine if components of the VLP also possessed membrane-disrupting capability, we examined the ability of full-length capsid protein (CPfl, possessing both ARM and C-terminus) to release fluorophore from liposomes. Liposome disruption assays with CPfl indicate that the unassembled capsid protein also possess the ability to disrupt membranes and release fluorophore (Fig. 2b). Similar to that of the VLP, a rapid release of fluorophore occurs within the first two minutes at pH 7 but appears to approach an asymptote for the remainder of 48 minutes. CPfl releases fluorophore at pH 5 with a much lower efficiency (Fig. 2b). The asymptote suggests that the CPfl may remain bound to the liposomes, such that it interacts with and releases fluorophore from a single liposome. To test this possibility, we performed liposome pelleting assays on CPfl at pH 7 and 5. Nearly all CPfl pellets with liposomes at pH 7 and 5 (Fig. 2f, lanes 3, 4, 9, 10).
We used a 41 amino acid peptide of the ARM sequence to test the ability of ARM for membrane disruption. Increasing concentrations of the peptide at pH 7 releases a greater fraction of the fluorophore (Fig. 2c). A rapid release of fluorophore can be observed within the first 2 minutes followed by an asymptote reached within the first 10 minutes of the reaction (Fig. 2c). Remarkably, the fluorophore release kinetic profile of CPfl and ARM peptide at pH 7 are very similar. The ARM peptide pellets with liposomes at pH 7 (Fig. 2f, lane 7, 11). The ARM peptide poorly releases fluorophore at pH 5.0 (Fig. 2c) and exhibits a diminished ability to bind liposomes at the lower pH (Fig. 2f, lane 5, 6, 11, 12). We note that the staining used for this assay may not be sensitive enough to detect liposome-ARM interaction -approximately 5 ng of sample can be detected by the stain according its manufacturer (Expedeon).
To test the ability of the N-terminal ARM in membrane disruption by VLP, we assembled capsid proteins lacking ARM (CPΔARM) in vitro into VLPΔARM (Fig. S3a, S3b) and tested its ability to release fluorophore from liposomes. VLPΔARM does not demonstrate any ability to release fluorophore at pH 7 or 5 (Fig. 2d). This loss of function provides further evidence that the ARM is indeed responsible for membrane disruption.
Further, we wanted to test the ability of the C-terminus to disrupt the membranes. The crystal structure and cryo-EM image reconstructions of PCV2 VLP visualize the C-terminus to radially protrude from the capsid while having limited interaction with the remaining portion of the capsid [4, 9, 46]. We thought that the position of the C-terminus and its limited interaction with the remaining capsid may allow it to possibly interact with and disrupt membranes. Our liposome disruption assays indicate that the C-terminus does not possess the ability to release fluorophore from liposomes at pH 7 or pH 5 (Fig. 2e).
PCV2 VLP, CPfl, and ARM peptide do not fuse membranes
Cell penetrating peptides like HIV-Tat, penetratin, and oligoarginines are believed to use a membrane fusion mechanism when releasing fluorophore from liposomes. In this mechanism the peptides drive membrane fusion, and their presence causes pore formation in the stressed membrane at the hemifusion stalk. The pores allow for the fluorophore to escape [44]. Membrane fusion can be prevented by incorporating poly(ethylene glycol)-lipid conjugates into the liposomes to prevent fusion by keeping two liposomes spatially separated [47].
To test if membrane fusion occurred when VLP, CPfl and ARM peptide were incubated with liposomes, we generated liposomes composed by of DOPC:DOPG:DOPG-PEG2000 in ratio of 75:20:5. The results from these assays also demonstrate robust fluorophore release, and exhibit similar kinetic profiles to those of the DOPC:DOPG liposomes. (Fig. 2a, b and c). This suggests that fluorophore release for VLP, CPfl and ARM peptide do not follow the membrane fusion mechanism described for HIV-Tat, penetratin, and oligoarginines [44]. Indeed, peptides that fuse membranes release less than 3% of the fluorophore, whereas our assays demonstrate nearly complete release of fluorophore (Fig. 2a–c). We are uncertain why additional fluorophore is released from the DOPC:DOPG:DOPG-PEG2000 liposomes when compared to the DOPC:DOPG liposomes.
The ARM peptide undergoes pH induced conformational change
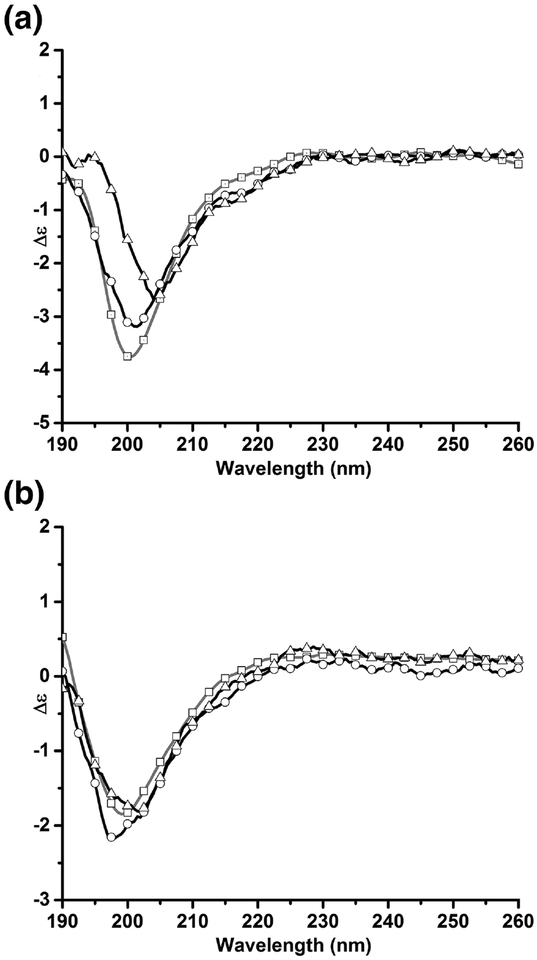
To assess if the interaction between the ARM peptide and liposome membrane affect the secondary structure composition of the peptide, we determined the CD spectra of the ARM and ARM-liposome mixture 10 minutes after the reactants were mixed (i.e. after the asymptote has been reached in our liposome disruption assays) (Fig. 3a). Analysis of the spectra with the BestSel server suggests that the antiparallel β-sheet content of 2 μM ARM peptide at pH 7 decreases while the α-helical content increases (Table 2). These differences are small and unlikely to be significant. To determine if a structural change in the ARM peptide was responsible for the lack of membrane disruption activity seen at pH 5, we collected CD spectra of ARM in the absence and presence of liposomes at pH 5. The addition of liposomes does not alter the secondary structure composition of the ARM peptide, as the β-sheet content of the peptide is 42% and 43% in the absence and presence of liposomes (Fig. 3b, Table 2). However, the greater β-sheet composition of the ARM peptide at lower pH indicates that the peptide may become more ordered at lower pH.
Figure 3: Circular dichroism spectra of ARM peptide changes in the presence of liposomes, and under acidic pH.
The circular dichroism (CD) spectra of ARM peptide were determined at pH 7 and 5. a) CD spectra at pH 7 (□) 1 μM peptide, (○) 2 μM peptide, (Δ) 2 μM peptide and liposomes. b) CD spectra at pH 5 (□) 1 μM peptide, (○) 2 μM peptide, (Δ) 2 μM peptide and liposomes.
Table 2: Secondary structure composition of ARM peptide in the presence of liposomes, and under acidic conditions.
The secondary structure was calculated using the BestSel server.
| Concentration (MM) | pH | Liposome | α-helix | Antiparallel β-sheet (%) | Parallel β-sheet (%) |
Turn (%) | Other (%) |
|---|---|---|---|---|---|---|---|
| 1.0 | 7 | NO | 0.0 | 32.0 | 0.0 | 11.2 | 56.8 |
| 2.0 | 7 | NO | 0.0 | 35.0 | 0.0 | 14.5 | 50.5 |
| 2.0 | 7 | YES | 3.2 | 29.5 | 0.0 | 14.9 | 52.4 |
| 1.0 | 5 | NO | 0.0 | 43.4 | 0.0 | 13.2 | 43.4 |
| 2.0 | 5 | NO | 0.0 | 42.0 | 0.0 | 13.5 | 44.5 |
| 2.0 | 5 | YES | 0.0 | 43.4 | 00 | 14.0 | 43.4 |
The Trp of the ARM inserts into the hydrophobic region of liposomal membrane
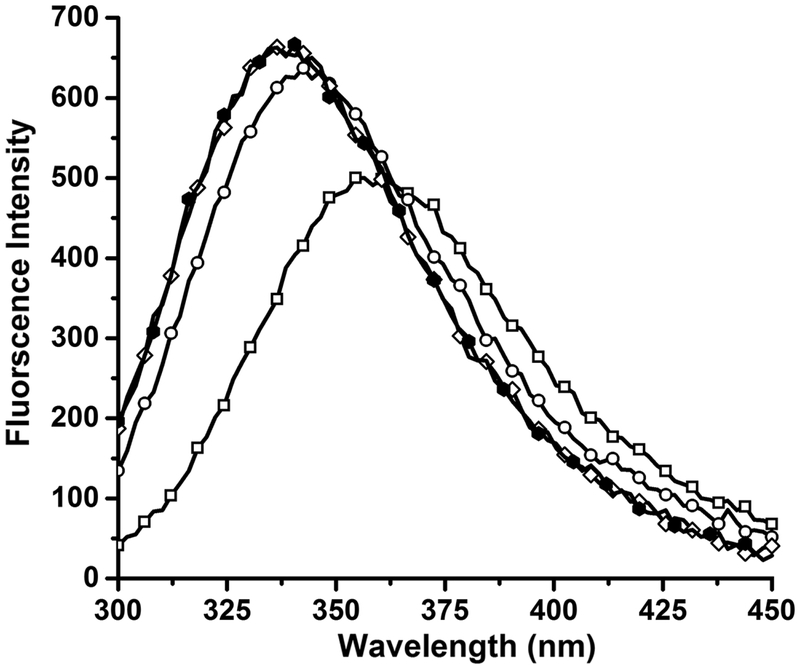
To further localize the amino acids involved in the binding of ARM peptide to the liposome, we performed tryptophan fluorescence spectroscopy to check the possibility of Trp binding to the liposomal membranes. There are 2 Trp amino acids in the ARM peptide, at position 28 and 38. A change in the intrinsic fluorescence of these amino acids in the presence of liposomes is indicative of a change in their environment. Trp fluorescence of the ARM peptide at 2 μM concentration was monitored prior to and after addition of increasing DOPC:DOPG (3:1) liposome concentrations. As shown in Fig. 4, λmax shifted to smaller wavelengths with an increase in fluorescence intensity upon the addition of liposomes. In the absence of liposomes, peptide at 2 μM concentration exhibited λmax at 355 nm that is observed for Trp in polar environment, whereas a blue shift in the λmax occurs with the addition of liposomes. In the presence of 200 μM liposome-lipid concentration, the λmax is at 335 nm. This change in emission is indicative of a change in the environment of Trp from hydrophilic to hydrophobic [48], and could be due to burying or insertion of Trp amino acids into the liposomal membranes.
Figure 4: Liposomes blue shift the intrinsic tryptophan fluorescence of ARM.
Tryptophan fluorescence excitation at λex = 5280 nm and emission at λem = 300–450 nm for 2 μM peptide in the absence of liposome ( ); presence of 75 μM liposome-lipid; (
); presence of 75 μM liposome-lipid; ( ); presence of 100 μM liposome-lipid (◇); and 200 μM liposome-lipid (
); presence of 100 μM liposome-lipid (◇); and 200 μM liposome-lipid ( ). Liposome-lipid refers to the concentration of lipid present in the liposomes.
). Liposome-lipid refers to the concentration of lipid present in the liposomes.
PCV2 disrupts membranes possessing cholesterol
The ability of PCV2 VLP to disrupt membranes at neutral pH suggests that the infectious virion may be able to disrupt plasma membranes or early endosomes. To test this possibility, we generated CH:DOPC:DOPG (50:44:6) membranes to partially mimic plasma membranes. Plasma membranes are composed of CH:DOPC:DOPE:SM:DOPS (50:20:11:13:6), where CH, DOPC, DOPE and SM carry a net charge of zero and DOPS carries a negative net charge [16, 49, 50]. Our results show that the PCV2 VLPs, CPfl and ARM peptide are capable of disrupting the liposomal membranes to release the fluorescent dye at pH 7 (Fig. 5 a, b and c). Thus, implying that VLP might also be active at early endosome or at the plasma membrane which are at pH 7.
Figure 5.
Liposome disruption assay with 200 nm CH:DOPC:DOPG (50:44:6) liposomes. The fraction of fluorophore release as compared to the liposome disruption by addition of 1% triton x-100 is represented for a) PCV2 virus like particles; b) CPfl and c) ARM peptide is depicted below.
Discussion
Members of the Circoviridae family (genre Circovirus and Cycloviridae) are small (~20 nm) icosahedral (T=1) non-enveloped viruses that infect or have been isolated from vertebrate and invertebrate members of the animal kingdom [51, 52]. The capsid proteins of these viruses share limited sequence conservation; however, all members of this family possess an N-terminus highly rich in arginines. For the case of PCV2, 18 of the 41 amino acids in N-terminus are arginines. The N-terminus is consequently referred to as an arginine rich motif (ARM). Sequence analysis of the PCV2 capsid protein with the cNLS mapper predicts the presence of bipartite nuclear localization signals (NLS) in the ARM -there are two stretches of NLS (NLSa and NLSb) parted by a short stretch of six amino acids [53] (Fig. 1). Both NLS are required by PCV1 (an apathogenic strain of PCV) for proper nuclear localization [54], and this is anticipated to be true for PCV2. Structural and neutralization studies of PCV2 VLP indicate that while the ARM is located to the interior of the capsid, it may externalize via a “breathing” mechanism [9]. Some ARM peptides are also cell penetrating peptides (CPP) and have been utilized to deliver cargo into the cell, and act as antimicrobial agents by penetrating membranes [55, 56]. Therefore, we asked if in addition to the nuclear localization signal, does the ARM of PCV2 also play a role in disrupting membranes?
Non-enveloped viruses have developed a number of mechanisms to disrupt membranes for completing their life-cycles [18]. These include the insertion of amphipathic α-helices or β-sheets into the membrane, use of phospholipase activity to hydrolyze the ester bonds of phospholipids, insertion of myristoylated peptides into membrane, and insertion of hydrophobic loops into the membranes [18]. PCV2 has been shown to be endocytosed into monocytic and dendritic cells [13, 14]. The mechanism by which PCV2 escapes from endosomes to acquire cellular entry remains to be described. To the best of our knowledge, we are providing the first report describing the mechanism by which members of the Circoviridae can disrupt membranes for endosomal escape. Our biochemical and biophysical studies of virus like particles (VLP) and their components demonstrate that the arginines of the PCV2 ARM are responsible for membrane disruption. These studies provide an additional mechanism that non-enveloped viruses may utilize for endosomal escape.
Our liposome disruption assays demonstrate that the VLP, unassembled PCV2 capsid protein (CPfl) and ARM peptide are capable of disrupting membranes (Fig. 2). VLP demonstrate higher efficiency in fluorophore release than the CPfl or ARM peptide. The VLP are found to be efficient in fluorophore release at both pH 7 and 5 (Fig. 2a). The fluorophore release profile of the VLP is reminiscent of burst kinetics observed for enzymes [57]. The initial rapid release of fluorophore release can result from free VLP interacting with liposome to rapidly release fluorophore (similar to pre-steady state kinetics where a free enzyme interacts with its substrate), this is followed by a slower and steady release of fluorophore that may be dominated by the slow release of the VLP (koff) from the liposomes (similar to steady state kinetics where the product is slowly released from the enzyme). The ability of the VLP to associate with the liposomes is demonstrated by the pelleting assays -we note that this association is much weaker than that of CPfl and ARM peptide (Fig. 2f, lanes 1–12). In contrast, the CPfl and ARM peptide only demonstrate the rapid fluorophore release. The absence of the second phase of fluorophore release (as observed for the VLP) suggests that both CPfl and ARM peptide may be very slow to dissociate from, or become embedded into, the liposome. Indeed, the liposome pelleting assays demonstrate a greater amount of CPfl and ARM peptide to pellet with liposomes (Fig. 2f, lanes 1–12). It is also possible that the differences in profiles is due to the high local concentration of ARMs present in a VLP, working together to disrupt the membrane.
The efficiency to release fluorophore is dramatically different between the VLP, CPfl and ARM peptide at pH 5 (Fig. 2 a, b and c). The ability of VLP to release fluorophore at the lower pH is comparable to that at neutral pH; however, both CPfl and ARM peptide exhibit a diminished efficiency to release fluorophore at the lower pH. An explanation for this could be the protonation of the four His amino acids in the ARM (Fig. 1). Some or all the His amino acids may insert themselves into the membrane when they are not protonated at pH 7, but unable to do so when protonated at pH 5. Indeed, it has been demonstrated that unprotonated His amino acids of CPP can insert into membranes, whereas the protonated His amino acids are unable to insert into membranes [59–61]. The local environment of the ARM is likely to be different in the context of the VLP, CPfl or peptide, and this environment is likely to affect its physicochemical properties. The ability of VLP to release fluorophore at both pH values suggest that the change in pH is not significantly affecting the physicochemical properties of the ARM; however, the diminished ability of CPfl and peptide to release fluorophore at pH 5 is indicative that the properties of the ARM are different at the two pH values. The observation that CPfl but not peptide pellets with liposomes at pH 5 (Fig. 2f) could be explained by the larger size of the CPfl ARM. The additional amino acids (42–233) present in the CPfl may provide additional sites for binding liposomes. This interaction locates the CPfl ARM in proximity of the liposomal membrane and thus increases the probability of the ARM to interact with and disrupt the membrane (Fig. 2b, c); however, the observed lack of association between the peptide and liposome suggests that peptide is less likely to appropriately interact with and disrupt the membrane at pH 5 (Fig. 2f).
In a recent published paper, Yu et al. were able to demonstrate CPP capability for a peptide consisting of the first 17 amino acids of the PCV2 capsid protein ARM [36]. The authors identified the NLSa region of the peptide to be responsible for this capability [36]. Secondary structure predictions with JPred, and de novo tertiary structure prediction with PEP-FOLD 3 and QUARK suggest that the ARM peptide is composed of two-to-three α-helices [62–64]. However, our results from CD spectroscopy do not identify the peptide to possess any α-helical secondary structures (Fig. 3a). Rather, the spectra demonstrate that only 35% of the peptide (around 13–14 amino acids) have the propensity to form secondary structure (β-sheets) (Table 2). This is in agreement with predictions made by Rosario et al. using intrinsically disordered protein (IDP) servers. These studies suggest the first 18 amino acids to be an intrinsically disordered region (IDR) [51]. IDR are low complexity sequences high in arginine and lysine and possess the ability to evolve faster than structured regions [65–67]. By combining our CD spectroscopy results with the IDR predictions generated by Rosario et al., we interpret the arginine rich NLSa region to be intrinsically disordered and the NLSb region possessing large aromatic amino acids (2 Trp and 1 Tyr) to have greater propensity to contribute towards the observed β-sheet content (Fig. 1, Fig. 3, Table 2). The blue shift of the Trp fluorescence (present in NLSb) is indicative of Trp becoming buried into a hydrophobic environment (Fig. 4). Thus, we conclude that the Trp amino acid(s) in NLSb become buried in the membrane while the arginines of NLSa are responsible for membrane disruption. The guanidinium group of arginine can hydrogen-bond (H-bond) to multiple lipid head groups concurrently. This capability allows the arginine to penetrate the membrane bilayer as it drags the head groups along with it and forces local thinning of the bilayer. As the arginine penetrates deeper into the bilayer, the guanidium group begins to form H-bonds with lipid groups in the second layer of the bilayer. The thinning of the membrane coupled to the H-bonding with head groups from both bilayers results in transmembrane pore formation [44].
The above studied characteristics of the ARM peptide can be extended to the membrane disruption capability of the VLP via multiple independent experiments that have demonstrated the NLSb to possess epitopes that interact with neutralizing antibodies [34, 35]. These experiments demonstrate that despite the structural evidences identifying the location of the ARM to be inside of the capsid, the ARM must externalize from amino acid 1 to 36 to interact with the antibodies. Combining the results from our study with the studies done by others, we propose the following sequence of events to occur following internalization of PCV2 into the endosome: amino acids 1–38 of the N-terminus ARM externalize from inside the capsid via viral breathing, the externalized NLSb region of the ARM interacts with the endosomal membrane by burying its Trp amino acid(s) into the membrane and anchors the VLP to the membrane, then the arginine rich NLSa with its cell penetrating or liposome disruption property disrupts the endosomal membrane to release the viral capsid into the cytoplasm. Our results that exhibit VLP to possess membrane disrupting capability at pH 7 in the presence of cholesterol (Fig. 5) suggest that PCV2 may also be capable of interacting with the plasma membrane or escaping from early-endosomes.
Materials and methods
Reagents
Mini extruder, lipids DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DOPG (1,2-dioleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (sodium salt) were purchased from Avanti Polar lipids in the powdered form. Sulforhodamine B dye was purchased from Santa Cruz Biotechnology. Cholesterol was purchased from Sigma Aldrich. Triton-x 100 was purchased from Fischer Scientific. Chloroform was acquired from Electron Microscopy Sciences. Formvar coated grids, stabilized with evaporated carbon film were purchased from Ted Pella (Prod No. 281217). Peptides N-term ARM PCV2 (residues 1–41) and C-term PCV2 (residues 223–233) were custom synthesized by Peptide2.0 Inc. ESF 921 media was purchased from Expression Systems. General laboratory reagents were purchased from VWR Life Science Amresco.
PCV2 VLP expression and purification
All PCV2 capsid protein constructs used in this manuscript are based on GenBank ACA51584.1. The services of GenScript (https://www.genscript.com) were utilized to generate the Baculovirus for expression of the PCV2 capsid protein in Trichoplusia ni (Ti ni) insect cells. Insect cells were grown in ESF921 medium at 27 °C with shaking at 120rpm till the cell count reached 2×106 cells/ml. Cell growth was monitored by staining with Trypan blue, digitized using a camera connected to a light microscope, and counted manually using a hemocytometer and Fiji software [37]. Cells were then infected with Baculovirus at a multiplicity of infection (MOI) of 0.1. Cells were grown at 27 °C with shaking at 120 rpm till their viability dropped to 50% (usually takes 5 days). Infected cells were then harvested by centrifuging at 3,000g for 20 minutes. The cells were suspended in the lysis buffer composed of 20 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) pH 7.5, 250 mM NaCl (sodium chloride), 2 mM MgCl2 (magnesium chloride), 1 mM TCEP (tris(2-carboxyethyl)phosphine hydrochloride) and 1% (v/v) NP-40 (nonylphenyl polyethylene glycol). Suspended cells were incubated on a revolver at 4 °C for 30 minutes and centrifuged for 720g at 4 °C to separate the cytoplasmic content from nuclear content. The nuclei pellet was suspended in nuclear lysis buffer, 20 mM HEPES pH 7.5, 500 mM NaCl, 2 mM MgCl2, 0.2 mM TCEP, 2 mM deoxycholate in the presence of salt activated nuclease (Sigma-Aldrich, SRE0015–5KU) and lysed using a sonicator. The nuclei lysate was centrifuged at 28,000g for 30 minutes at 4 °C. The supernatant was separated and loaded on a 30% sucrose cushion and centrifuged at 184,000g (Beckman Coulter rotor 50.2Ti), at 11°C for 2.5 hours. The pellet obtained above was suspended in 2ml of nuclear lysis buffer and loaded onto a discontinuous gradient of 20%, 30%, 40%, 50%, 60% and 70% sucrose and centrifuged at 160,000g (Beckman Coulter rotor SW 40Ti), at 4 °C for 18 hours. The fraction containing the VLP was visually identified using light scattering and SDS-PAGE. The fraction containing VLP was dialyzed overnight in 20 mM HEPES pH 7.0, 250 mM NaCl, 0.2 mM TCEP and 2 mM EDTA (ethylenediaminetetraacetate). Dialyzed sample was concentrated using a 100kDa MWCO ultrafiltration device (Pall Corporation) and stored at 4 °C. Sample were processed with SDS-PAGE, stained with InstantBlue (expedeon) and analyzed for concentration against a standard of similar molecular weight protein with known concentration.
SUMO-PCV2 expression and purification
The services of Biomatik (https://www.biomatik.com/) was utilized to codon optimize, synthesize and clone a fusion of His6-SUMO (GenBank: ABN58899.1), a SUMO-protease specific cleavage site and full-length PCV2 capsid protein gene (CPfl) into the pET 28a plasmid. The fusion protein SUMO-CPfl was expressed in One Shot™ BL21 Star™ (DE3) Escherichia coli cells in terrific broth (TB) media, induced by 100 μM IPTG and the cells were harvested after 16 hours of expression at 20°C. The cell pellet was suspended in lysis buffer constituting 20 mM CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) pH 10.5; 5 0mM NaCl, 0.2 mM PMSF (phenylmethane sulfonyl fluoride), 2 mM β-ME (2-mercaptoethanol) in the presence of DNAse and RNAseH. Cells were lysed using an ultra-sonicator and centrifuged at 16,000g for 30 min at 4 °C. Supernatant was injected and allowed to pass through the anion exchange column (HiTrap Q FF) followed by cation exchange column (HiTrap SP HP by GE Healthcare) and flow-through was collected. HiTrap QFF column was removed and SUMO-CPfl was eluted from the HiTrap SP HP column using a linear gradient from 0 to 1M NaCl. Fractions were collected and processed with SDS-PAGE, stained with Commassie blue and assessed for purity. Fractions with SUMO-CPfl were pooled and concentrated with a 30 kDa MWCO Pall ultrafiltration device. The concentration of the protein was determined using the method of Gill and von Hippel [38]. Protein samples were aliquot into 50 μl, flash frozen in N2(l), and stored in −80 °C.
SUMO-specific protease expression and purification
The SUMO-specific protease (dtUD1) construct was a kind gift from the laboratory of Dr. Patrick J. Loll [39]. The protein was expressed in E. coli and purified using the protocol provided in the publication [39].
PCV2 for liposome disruption assay
Prior to liposome disruption assay, SUMO-CPfl was digested to remove SUMO tag and release CPfl. For this, SUMO-CPfl was diluted accordingly and digested with SUMO-specific protease in the presence of DTT (dithiothreitol). The reaction mixture was kept at room temperature for 30 minutes. Sample was run on the SDS-PAGE to analyze the complete digestion.
SUMO-ΔARM PCV2 expression and purification
The services of GenScript (https://www.genscript.com/) was utilized to codon optimize, synthesize and clone a fusion of His6-SUMO (GenBank: ABN58899.1), a SUMO-protease specific cleavage site and PCV2 capsid protein with first 41 amino acids deleted (CPΔARM) into pET 28a plasmid. The fusion protein was expressed in One Shot™ BL21 Star™ (DE3) E. coli cells in terrific broth (TB) media, induced by 100 μM IPTG and grown for 16 hours at 20 °C before the cells were harvested. The cell pellet was suspended in lysis buffer consisting of 20 mM CAPS pH 10.5; 5 0mM NaCl; 0.2 mM PMSF; 0.5 mM TCEP in the presence of DNAse and RNAseH. Cells were lysed using an ultra-sonicator, followed by centrifugation at 17,000g for 60 minutes at 4 °C. The supernatant was collected and passed through the pre-equilibrated Ni-NTA resin packed into a gravity flow column. Column was washed with 10CV (column volume) of 20 mM CAPS pH 10.5, 1 M NaCl, 0.2 mM TCEP, 50 mM Imidazole pH 11. Protein was eluted and fractionated with 5CV of 20 mM CAPS pH 10.5, 50 mM NaCl, 0.2 mM TCEP and 1 M imidazole pH 11. Fractions were processed with SDS-PAGE, stained with Coomassie blue, and assessed for purity. Samples with SUMO-ΔARM PCV2 were pooled and concentrated. To the concentrated protein, arginine and NaCl were added to a final concentration of 0.25 M and 0.2 M respectively.
Assembly of CPΔARM into VLPΔARM
A final concentration of 1mg/ml of CPΔARM protein was used for VLPΔARM assembly. Protein was incubated with SUMO protease to release the free ΔARM PCV2 (CPΔARM). The digestion mixture was kept overnight and run on SDS-PAGE to analyze for complete digestion. The digested mixture was mixed with VLP assembly buffer in 1:1 ratio (vol/vol). The VLP assembly buffer is composed of 12% PEG 3350; 0.6 M Ammonium Citrate pH 5.0; 5.0% iso-propanol. The reaction mixture was kept at 4°C for 72 hours. Following the incubation, 0.3 M HEPPS buffer (3-[4-(2-Hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid) pH 9.0, 0.6 M NaCl, 0.25 M guanidine hydrochloride, 5 mM β-ME were added to the solution to adjust the volume to 500 μl. Reaction was allowed to sit at 4 °C for 24 hours followed by centrifuging the sample at 10,000g for 10 minutes at 4°C. The supernatant was loaded onto size exclusion column (Sephacryl S-500 from GE healthcare) pre-equilibrated with 50 mM HEPPS pH 9.0, 0.6 M NaCl, 7 mM β-ME. Fractions with assembled VLPs were concentrated and visualized using negative stained electron microscopy with a JEOL JEM-1230 electron microscope operating at 80kV. Images were recorded on a Gatan US-4000 CCD detector 4k × 4k using the Leginon software (Fig. S1) [40].
Liposome preparation
Lipids DOPC and DOPG were suspended separately in chloroform at a concentration of 25 mg/ml. DOPC and DOPG lipid solutions were then mixed in the ratio of 3:1 with total lipid concentration of 8mg/ml. The mixture was gently dried in a glass tube to a thin layer using a gentle stream of N2(g). The dried lipid mixture was kept under vacuum for ~3 hours to evaporate traces of residual chloroform. The 8 mg of lipid mixture was rehydrated in 600 μl of buffer 50 mM HEPES pH 7.0 and 50 mM NaCl with 17 mg of dissolved Sulforhodamine B dye for around 1 hour. The hydrated lipid dispersion was given five freeze-thaw cycles using N2(l) followed by 21 times extrusion through 0.2 μm polycarbonate membrane. The resultant liposomes were desalted and fractionated in 1 ml aliquots at 4 °C using a HiPrep G25 desalting column connected to an Äkta Pure (GE Healthcare Life Sciences) using 50 mM HEPES pH 7.0 and 50 mM NaCl. All the assays were done within 24 hours of liposome preparation. Similar protocol was followed for preparation of liposomes with cholesterol (CH) in the ratio of CH:DOPC:DOPG (50:44:6).
Liposome Lipid concentration measurement
The lipid concentration was determined to attain a ratio of liposome to CPfl, ARM peptide and VLP. The lipid content of the liposomes obtained after desalting the extruded liposomes was measured using Stewart assay [41]. This is a colorimetric method based on the formation of a complex between ammonium ferrothiocyanate and phospholipids. The method is sensitive over the range of 0.01–0.1 mg of phospholipids. The lipid sample used to prepare the test liposomes (8 mg of DOPC:DOPG in 3:1) was used for preparing the standard curve. The sample was diluted in chloroform to prepare aliquots of concentration in the range of 0.01–0.1 mg. For each aliquot, a spectrum scan was run from 200 to 800 nm to find the wavelength that gives maximum absorbance peak (Fig. S2a). For our samples, the maximum absorbance was obtained at 474 nm. A standard curve was plotted at this wavelength and used to quantify the concentration of the test sample (Fig. S2b). Because this method is unable to quantify phosphatidylglycerol, the amount of DOPG present in the liposomes was extrapolated according to the concentration of DOPC and the knowledge that the DOPC:DOPG ratio is 3:1.
Dynamic light scattering
The size of the prepared liposomes was analyzed using dynamic light scattering with the DynaPro® Platereader II (Wyatt Technology Corporation). Prepared liposomes were diluted to the same concentration as used in disruption assays and readings were taken in 96-well black clear flat bottom microplate from Corning. Measurements were taken at constant temperature of 25 °C, 10 acquisitions of 5 sec each were taken for each sample. Dynamics software v7 (supplied by the manufacturer) was used to analyze the data. Each experiment was repeated three times.
Liposome disruption assays
Liposome disruption assays were carried out in black 96-well, flat bottom microplates from Greiner Bio-one, at 25 °C using a SpectraMax M5 multi-mode microplate reader (Molecular Devices). The pH-based assays were performed at pH 7 (buffer 50 mM HEPES pH 7 and 50 mM NaCl), and at pH 5 (buffer 50 mM Ammonium citrate pH 5 and 50 mM NaCl). The assays were carried out in kinetic mode of the spectrophotometer at wavelength excitation (λex) and emission (λem) maxima of 565 nm and 586 nm respectively. The assays were initiated by adding 10μl of liposomes to 90μl of buffer containing 0.5 μM, 0.75 μM or 1 μM final concentrations of VLP, CPfl, VLP lacking the ARM VLPΔARM, and the C-term peptide. Thus, each well contained a total volume of 100 μl. The liposome disruption for the ARM peptide contained 0.75 μM, 1 μM or 2 μM of the peptide. The plate was shaken for three seconds before the run and readings were taken every second for 50 minutes. With each experiment, a control well was set with 10 μl liposome (or 204 μM as calculated by lipid concentration measurement) and 90 μl buffer. At the end of the experiment, 1 μl of 0.5% of triton-x 100 was added to all the wells and end-point readings were taken.
For every sample, percentage of dye release was calculated with respect to the control as:
where, I is the emission intensity of the sample; I0 is the emission intensity of the control and Imax is the emission intensity at the end of the experiment after adding Triton X-100. All the experiments were carried out at pH 7 and 5 and done in triplicates. Fraction of total dye release was plotted against time (in seconds) using ORIGIN software (OriginLab Corporation).
Liposome pelleting assay
Reactions identical to the disruption assays were performed at pH 7 and 5 for 1 μM VLP, CPfl, and 2 μM ARM peptide. After one hour of incubation, the reactions were centrifuged at 15,000 × g for 15 minutes at room temperature using a temperature-controlled centrifuge. The pellets were washed in 20 mM HEPES pH 7.0 and 50 mM NaCl for two times. Washing involved removal of supernatant and suspending the pellet in the fresh buffer followed by centrifugation. The pellets were suspended in the same volume as the initial supernatant. Samples (20 μl) were then run on polyacrylamide gel made with a 16% acrylamide and 6 M urea tricine, visualized by three step staining protocol involving first treating the gel with fixing solution (50% methanol, 10% acetic acid and 100 mM ammonium acetate), then staining the gel (10% acetic acid, 0.025% Coomassie blue) followed by de-staining of the gel with 10% acetic acid [42]. The gel was then quantified with a Li-Cor Odyssey CLx. The assay was repeated three times using freshly prepared liposomes each time.
Intrinsic Tryptophan fluorescence spectroscopy
Fluorescence emission spectra were recorded using the SpectraMax M5 multi-mode microplate reader (Molecular Devices). For the assays, a 100 μl reaction was set up with 2 μM peptide and increasing concentrations of liposomes (75, 100 and 200 μM: as calculated by lipid concentration measurement) in buffer (50 mM HEPES and 50 mM NaCl, and at pH 7). Fluorescence spectra for Trp emission were recorded at excitation wavelength of 280 nm and emission wavelength from 300 nm to 450 nm with step size of 2nm bandwidth. Background correction for each spectrum was performed by subtracting the liposome profile from the peptide/liposome fluorescence profile. The fluorescence intensity was plotted against wavelength using ORIGIN software (OriginLab Corporation).
Circular Dichroism
Circular dichroism (CD) experiments were carried out using Jasco J-1500 Circular Dichroism Spectrophotometer with a 10mm path-length cuvette at 20 °C. The ARM peptide and liposomes were independently dialyzed extensively against 10 mM phosphate buffer at pH 7 and 5. The presence of two tryptophan amino acids in the peptide allowed us to estimate the concentration of dialyzed peptide using UV absorbance at 280 nm. A final concentration of 1 μM and 2 μM peptide were used and far-UV CD spectra were recorded in the range of 190–260 nm. Readings were also taken in the presence of 200 μM liposomes to see the effect of liposomes on the secondary structure of peptide at pH 7 and 5. Baseline corrections were performed for the peptide by using the dialysis buffer, and the dialyzed liposome. The analysis of measured CD spectra were done by BestSel server [43].
Supplementary Material
Highlights.
Non-enveloped viruses employ several mechanisms to disrupt membranes to infect cells
Porcine circovirus 2 uses an Arginine rich motif (ARM) to accomplish this task
The ARM peptide is predominantly disordered
The Arginines of the ARM are responsible for membrane disruption
This is the first example of a non-enveloped virus using the Arginines of an ARM to disrupt membranes
Acknowledgement
Funds responsible for supporting these studies were provided by NIH National Institute of General medical Sciences and National institute of Allergy and Infectious Diseases (5SC1AI114843) and by Grant Number 5G12MD007603–30 from the National Institute on Minority Health and Health Disparities. Images for the assembly of CPΔARM into VLPΔARM were collected at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310). Images for the VLP were collected at the Imaging Facility of City University of New York (CUNY) Advanced Science Research Center (ASRC). We would like to acknowledge the scientific and technical assistance from the NYSBC and CUNY ASRC.
Abbreviation:
- PCV2
Porcine circovirus 2
- VLP
Virus like protein
- ARM
Arginine rich motif
- CPP
cell penetrating peptide
- CPfl
full length capsid protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tischer I, Gelderblom H, Vettermann W, Koch M (1982). A very small porcine virus with circular single-stranded DNA. Nature.295:64. [DOI] [PubMed] [Google Scholar]
- [2].Nayar G, Hamel A, Lin L (1997). Detection and characterization of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. Can Vet J.38:385. [PMC free article] [PubMed] [Google Scholar]
- [3].Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC (2009). Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J Virol.83:12813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM (2010). Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J.7:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, et al. (1998). Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol.79:2171–9. [DOI] [PubMed] [Google Scholar]
- [6].Mankertz A, Mankertz J, Wolf K, Buhk H-J (1998). Identification of a protein essential for replication of porcine circovirus. J Gen Virol.79:381–4. [DOI] [PubMed] [Google Scholar]
- [7].Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS (2000). Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol.81:2281–7. [DOI] [PubMed] [Google Scholar]
- [8].Liu Z, Guo F, Wang F, Li T-C, Jiang W (2016). 2.9 Å resolution cryo-EM 3D reconstruction of close-packed virus particles. Structure.24:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, et al. (2011). The 2.3 Å Structure of Porcine Circovirus 2. J Virol:JVI. 00737–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Crowther R, Berriman J, Curran W, Allan G, Todd D (2003). Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J Virol.77:13036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Franzo G, Tucciarone CM, Cecchinato M, Drigo M (2016). Porcine circovirus type 2 (PCV2) evolution before and after the vaccination introduction: a large scale epidemiological study. Sci Rep.6:39458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Misinzo G, Delputte PL, Meerts P, Lefebvre DJ, Nauwynck HJ (2006). Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J Virol.80:3487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Misinzo G, Meerts P, Bublot M, Mast J, Weingartl H, Nauwynck H (2005). Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J Gen Virol.86:2057–68. [DOI] [PubMed] [Google Scholar]
- [14].Vincent IE, Carrasco CP, Guzylack-Piriou L, Herrmann B, McNeilly F, Allan GM, et al. (2005). Subset-dependent modulation of dendritic cell activity by circovirus type 2. Immunology.115:388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nauwynck H, Sanchez R, Meerts P, Lefebvre D, Saha D, Huang L, et al. (2012). Cell tropism and entry of porcine circovirus 2. Virus Res.164:43–5. [DOI] [PubMed] [Google Scholar]
- [16].Huotari J, Helenius A (2011). Endosome maturation. EMBO J.30:3481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bothner B, Schneemann A, Marshall D, Reddy V, Johnson JE, Siuzdak G (1999). Crystallographically identical virus capsids display different properties in solution. Nat Struct Mol Biol.6:114. [DOI] [PubMed] [Google Scholar]
- [18].Johnson JE, Banerjee M (2008). Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr Protein Pept Sci.9:16–27. [DOI] [PubMed] [Google Scholar]
- [19].Domitrovic T, Matsui T, Johnson JE (2012). Dissecting quasi-equivalence in non-enveloped viruses: membrane disruption is promoted by lytic peptides released from subunit pentamers, not hexamers. J Virol:JVI. 01089–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].de Sena J, Mandel B (1977). Studies on the in vitro uncoating of poliovirus II. Characteristics of the membrane-modified particle. Virology.78:554–66. [DOI] [PubMed] [Google Scholar]
- [21].Chow M, Newman J, Filman D, Hogle J, Rowlands D, Brown F (1987). Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature.327:482. [DOI] [PubMed] [Google Scholar]
- [22].Fricks CE, Hogle JM (1990). Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol.64:1934–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dorsch S, Liebisch G, Kaufmann B, von Landenberg P, Hoffmann JH, Drobnik W, et al. (2002). The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J Virol.76:2014–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mani B, Baltzer C, Valle N, Almendral JM, Kempf C, Ros C (2006). Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J Virol.80:1015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC (2002). Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell.108:283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sturzenbecker L, Nibert M, Furlong D, Fields B (1987). Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol.61:2351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang L, Agosto MA, Ivanovic T, King DS, Nibert ML, Harrison SC (2009). Requirements for the formation of membrane pores by the reovirus myristoylated μ1N peptide. J Virol.83:7004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gutiérrez M, Isa P, Sánchez-San Martin C, Pérez-Vargas J, Espinosa R, Arias CF, et al. (2010). Different rotavirus strains enter MA104 cells through different endocytic pathways: the role of clathrin-mediated endocytosis. J Virol.84:9161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim IS, Trask SD, Babyonyshev M, Dormitzer PR, Harrison SC (2010). Effect of mutations in VP5* hydrophobic loops on rotavirus cell entry. J Virol.84:6200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Harrison S, Olson A, Schutt C, Winkler F, Bricogne G (1978). Tomato bushy stunt virus at 2.9 Å resolution. Nature.276:368. [DOI] [PubMed] [Google Scholar]
- [31].Bechara C, Sagan S (2013). Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett.587:1693–702. [DOI] [PubMed] [Google Scholar]
- [32].Olson ES, Aguilera TA, Jiang T, Ellies LG, Nguyen QT, Wong EH, et al. (2009). In vivo characterization of activatable cell penetrating peptides for targeting protease activity in cancer. Integr Biol (Camb).1:382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Orange J, May M (2008). Cell penetrating peptide inhibitors of nuclear factor-kappa B. Cell Mol Life Sci 65:3564–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mahe D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, et al. (2000). Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol.81:1815–24. [DOI] [PubMed] [Google Scholar]
- [35].Guo L, Lu Y, Huang L, Wei Y, Liu C (2011). Identification of a new antigen epitope in the nuclear localization signal region of porcine circovirus type 2 capsid protein. Intervirology.54:156–63. [DOI] [PubMed] [Google Scholar]
- [36].Yu W, Zhan Y, Xue B, Dong Y, Wang Y, Jiang P, et al. (2018). Highly efficient cellular uptake of a cell-penetrating peptide (CPP) derived from the capsid protein of porcine circovirus type 2. J Biol Chem.293:15221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods.9:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gill SC, Von Hippel PH (1989). Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem.182:319–26. [DOI] [PubMed] [Google Scholar]
- [39].Weeks SD, Drinker M, Loll PJ (2007). Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif.53:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, et al. (2009). Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol.167:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stewart JCM (1980). Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem.104:10–4. [DOI] [PubMed] [Google Scholar]
- [42].Schägger H (2006). Tricine–sds-page. Nat Protoc.1:16. [DOI] [PubMed] [Google Scholar]
- [43].Micsonai A, Wien F, Kernya L, Lee Y-H, Goto Y, Réfrégiers M, et al. (2015). Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci U S A:201500851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thorén PE, Persson D, Lincoln P, Nordén B (2005). Membrane destabilizing properties of cell-penetrating peptides. Biophys Chem.114:169–79. [DOI] [PubMed] [Google Scholar]
- [45].Adell MAY, Migliano SM, Upadhyayula S, Bykov YS, Sprenger S, Pakdel M, et al. (2017). Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. Elife.6:e31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dhindwal S, Avila B, Feng S, Khayat R (2019). Porcine circovirus 2 uses a multitude of weak binding sites to interact with heparan sulfate, and the interactions do not follow the symmetry of the capsid. J Virol. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Holland JW, Hui C, Cullis PR, Madden TD (1996). Poly (ethylene glycol)− lipid conjugates regulate the calcium-induced fusion of liposomes composed of phosphatidylethanolamine and phosphatidylserine. Biochemistry.35:2618–24. [DOI] [PubMed] [Google Scholar]
- [48].Zhao H, Lappalainen P (2012). A simple guide to biochemical approaches for analyzing protein–lipid interactions. Mol Biol Cell.23:2823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kobayashi T, Beuchat M-H, Chevallier J, Makino A, Mayran N, Lebrand C, et al. (2002). Separation and characterization of late endosomal membrane domains. J Biol Chem.277:32157–64. [DOI] [PubMed] [Google Scholar]
- [50].Darwich Z, Klymchenko AS, Dujardin D, Mély Y (2014). Imaging lipid order changes in endosome membranes of live cells by using a Nile Red-based membrane probe. RSC Adv.4:8481–8. [Google Scholar]
- [51].Rosario K, Schenck RO, Harbeitner RC, Lawler SN, Breitbart M (2015). Novel circular single-stranded DNA viruses identified in marine invertebrates reveal high sequence diversity and consistent predicted intrinsic disorder patterns within putative structural proteins. Front Microbiol.6:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rosario K, Mettel KA, Benner BE, Johnson R, Scott C, Yusseff-Vanegas SZ, et al. (2018). Virus discovery in all three major lineages of terrestrial arthropods highlights the diversity of single-stranded DNA viruses associated with invertebrates. PeerJ.6:e5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, et al. (2009). Six classes of nuclear localization signals specific to different binding grooves of importin α. J Biol Chem.284:478–85. [DOI] [PubMed] [Google Scholar]
- [54].Shuai J, Wei W, Jiang L, Li X. l., Chen N, Fang W (2008). Mapping of the nuclear localization signals in open reading frame 2 protein from porcine circovirus type 1. Acta Biochim Biophys Sin (Shanghai).40:71–7. [DOI] [PubMed] [Google Scholar]
- [55].Chan DI, Prenner EJ, Vogel HJ (2006). Tryptophan-and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta Biomembr.1758:1184–202. [DOI] [PubMed] [Google Scholar]
- [56].Yang C-H, Chen Y-C, Peng S-Y, Tsai AP-Y, Lee TJ-F, Yen J-H, et al. (2018). An engineered arginine-rich α-helical antimicrobial peptide exhibits broad-spectrum bactericidal activity against pathogenic bacteria and reduces bacterial infections in mice. Sci Rep.8:14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Johnson KA Transient-State Kinetic Analysis of Enzyme Reaction Pathways. The Enzymes 1992. p. 1–61. [Google Scholar]
- [58].Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res.31:3784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Georgescu J, Munhoz VH, Bechinger B (2010). NMR structures of the histidine-rich peptide LAH4 in micellar environments: membrane insertion, pH-dependent mode of antimicrobial action, and DNA transfection. Biophys J.99:2507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ladokhin AS, White SH (2004). Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry.43:5782–91. [DOI] [PubMed] [Google Scholar]
- [61].Martfeld AN, Greathouse DV, Koeppe RE (2016). Ionization properties of histidine residues in the lipid-bilayer membrane environment. J Biol Chem:jbc. M116. 738583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Drozdetskiy A, Cole C, Procter J, Barton GJ (2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Res.43:W389–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P (2016). PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res.44:W449–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xu D, Zhang Y (2012). Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins.80:1715–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Xue B, Dunker AK, Uversky VN (2012). Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn.30:137–49. [DOI] [PubMed] [Google Scholar]
- [66].Xue B, Blocquel D, Habchi J, Uversky AV, Kurgan L, Uversky VN, et al. (2014). Structural disorder in viral proteins. Chem Rev.114:6880–911. [DOI] [PubMed] [Google Scholar]
- [67].Pushker R, Mooney C, Davey NE, Jacqué J-M, Shields DC (2013). Marked variability in the extent of protein disorder within and between viral families. PloS one.8:e60724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.