Abstract
The maternal-fetal interface represents a unique immune privileged site that maintains the ability to defend against pathogens while orchestrating the necessary tissue remodeling required for placentation. The recent discovery of novel cellular families (innate lymphoid cells, tissue-resident NK cells) suggests that our understanding of the decidual immunome is incomplete. To understand this complex milieu, new technological developments allow reproductive immunologists to collect increasingly complex data at a cellular resolution. Polychromatic flow cytometry allows for greater resolution in the identification of novel cell types by surface and intracellular protein. Single-cell RNAseq coupled with microfluidics allows for efficient cellular trascriptomics. The extreme dimensionality and size of datasets generated, however, requires the application of novel computational approaches for unbiased analysis. There are now multiple dimensionality reduction (t-SNE, SPADE) and visualization tools (SPICE) that allow researchers to efficiently analyze flow cytometry data. Development of computational tools has also been extended to RNAseq data (including scRNAseq), which requires specific analytical tools. Here, we provide an overview and a brief primer for the reproductive immunology community on data acquisition and computational tools for the analysis of complex flow cytometry and RNAseq data.
Keywords: flow cytometry, RNA-seq, dimensionality reduction, data visualization
Introduction
Pregnancy is a time of maternal adaptation to the semiallogeneic fetus, requiring a unique immunome to support the developing placenta1,2. The maternal-fetal interface maintains a specific milieu of immune cells, consisting mostly of decidual natural killer cells (dNKs)1–4, with smaller populations of antigen presenting cells, such as dendritic cells (DCs) and macrophages5,6, and T cells7,8, including unconventional subsets9–11, and B cells12. Further, there is evidence of cellular interactions between dNKs and DCs in early pregnancy13–15, suggesting an important role in proper development of the placenta. However, the nature of these interactions remains unclear. Chemokine silencing is thought to limit T cell traffic into the decidua16, an important tolerance-inducing mechanism. Despite limited traffic, both CD4+ and CD8+ T cells have been identified at maternal-fetal interface17,18 and implicated in preterm labor18. Recently, multiple groups have identified non-dNK members of the innate lymphoid cell (ILC) family at the maternal-fetal interface19,20 with a specific tissue-resident NK cell (trNK) in the decidua21,22. This adds a layer of complexity to our understanding of the decidual immunome. However, our ability to assign unique and unambiguous cellular identities depends on improved methods of assigning cellular phenotypes.
Technological advances allows researchers to collect increasing amount of data per cell23. Advances in flow cytometry enable researchers to assess the expression of multiple proteins simultaneously from single cells. Similarly, advances in RNA-seq enable researchers to analyze the transcriptome of sorted cell groups or even individual cells. The tsunami of data, however, presents entirely novel challenges in visualization and require computational tools for thorough analysis.
This review aims to highlight some of the technological advances in (1) flow cytometry, including its sister platform mass cytometry (CyTOF™), (2) RNA-seq, and (3) data analysis workflows to handle complex data. We open with a discussion of data acquisition, outline best practices in flow cytometry and RNA-seq, and discuss some of the benefits and limitations of analysis tools for cytometry and sequencing data. We highlight some recent advances like expansion of fluorochrome detection in flow cytometry and single-cell RNA-seq (scRNA-seq). We then transition to data analysis and discuss dimensionality reduction tools that are available for the analysis of flow cytometry data, outline a workflow for RNA-seq data analysis, and discuss user-friendly packages available for the analysis of RNA-seq data. This review aims to provide reproductive immunologists with a go-to guide for single-cell data analysis to aid in the advancement of the field.
Data Acquisition
Flow cytometry
Flow cytometry is a method that allows rapid assessment of multiple parameters simultaneously for a single cell23–28. The earliest version of flow cytometry was developed as a fast method for individual cell counting and separation using hydrodynamic flow focusing29,30. Subsequently, cell sorting using a conducting medium was introduced25. The introduction of fluorescent proteins to flow cytometry allowed for the development of fluorescence activated cell sorting (FACS)31, combining both parameter acquisition (i.e. surface marker expression) and live cell collection, giving rise to modern flow cytometry. Early applications of flow cytometry used one or a small number of fluorochromes, each with a specific absorption and emission spectrum, for the identification of cell populations. Development of additional fluorochromes and hardware (e.g. lasers, detectors, etc.) now allows detection of up to 18 parameters23 by commercially available flow cytometers, with experimental cytometers able to detect more than 30 parameters32. In addition to traditional hydrodynamic focusing technology that employs laminar flow to focus cell suspensions through the flow cell33,34, acoustic focusing cytometers have recently entered the market, with the advantage of faster analysis of large sample inputs32,35. Flow cytometry is a powerful tool in immunology, including reproductive immunology, and has been broadly adopted, increasing our understanding of the maternal-fetal interface immunome19,20,36–41.
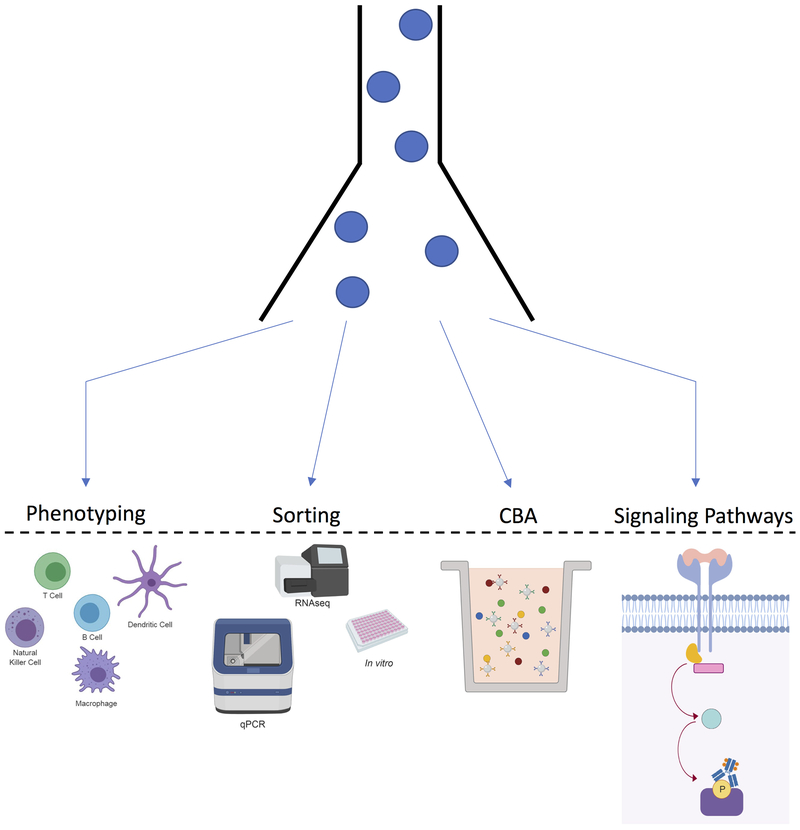
Flow cytometry has applications beyond cell classification and assessment of activation status and interfaces with many other single cell-based modern applications (Figure 1). FACS, for example, is used to obtain pure cell populations for downstream applications such as gene expression analysis (RNA-seq, qPCR)23,32 and recovery of live cells to conduct in vitro experiments. Moreover, the versatility of flow cytometry has been leveraged to create flow bead-based variants of protein detection assays (cytometric bead analysis) using fluorescent sandwich-antibody capture beads32, analogous in principle to ELISA. Signaling pathway analysis using phospho-protein detection allows researchers to investigate signaling events in individual cells42,43. Phospho-protein labeling can also be applied as a barcoding technique44, allowing researchers to barcode different samples and pool them for staining and acquisition, thus minimizing reagent use and increasing reliability of comparative measures.
Figure 1. Flow cytometry applications.
Flow cytometry has many applications including: phenotyping, allowing for the identification of various cells in a specimen; sorting of specific cellular populations for downstream applications; cytometric bead arrays (CBA), which allows for the quantification of soluble proteins; analysis of signaling pathways, detecting phosphorylation events.
There are some limitations when conducting flow cytometry experiments. Spectral overlap between available fluorochromes limits the number and fidelity of channels available23. Spectral overlap, or ‘spillover’, occurs when the same fluorochrome can be detected in more than one detector, due to wide emission spectra of the natural and especially tandem fluorochromes45,46. Spectral overlap can be corrected for mathematically by applying a spillover coefficient within the analysis software with appropriate controls45,46. Despite correction, designing and optimizing multispectral flow cytometry panels to achieve high levels of sensitivity remains a challenge as each additional color correction increases the coefficient of variance47. To overcome this, a number of resources are available, including well-characterized Optimized Multicolor Immunofluorescence Panels, or OMIPs, that demonstrate optimized and validated panels47. In addition, online tools, such as FluoroFinder (www.FluoroFinder.com), provide a panel building platform that allows researchers to visualize spectral overlap, match marker density to fluorochrome intensity, and display antibody availability with vendors, thus facilitating panel design.
Reproducibility constraints are a key and often underappreciated consideration in flow cytometry. To improve the integrity and quality of flow cytometry data the MIFlowCyt (The Minimum Information About a Flow Cytometry Experiment)48 standards have been developed to address necessary reporting. Adhering to the MIFlowCyt guidelines allows for greater collaboration and improves reproducibility. In addition, the use of a standardizing method for flow cytometer set up/calibration should be considered, specifically when analyzing fluorescence intensities, as these values depend on laser intensity and detector settings that can vary between experimental runs.
Similar to flow cytometry, mass cytometry allows researchers to collect single cell data by combining both flow cytometry and mass spectrometry23,32. Mass cytometry, or Cytometry by Time-Of-Flight (CyTOF™), uses heavy metal isotopes as tags attached to antibodies49,50, thus allowing for assessment of up to 40 markers, achieving higher resolution compared to flow cytometry32,50. However, the sensitivity and throughput of mass cytometry remains low23,50. Furthermore, use of mass cytometry leads to loss of cellular material during data acquisition, with up to 70% of cells being lost23, a particularly important limitation for reproductive immunologists who sample from very limited tissue sources. An additional obstacle are longer acquisition times required for mass cytometry compared to flow cytometry23,32. Mass cytometry, however, has been successfully used to immunophenotype maternal and fetal peripheral blood at term pregnancy51, thus illustrating that mass cytometry can be a useful technique in long-term studies of immune changes across pregnancy.
RNA-seq
Cellular transcriptome has become an important tool to study the biology of cell populations. Recently, RNA-seq has emerged as a valuable tool that allows us to understand the transcriptome of tissue52 or cells53 of interest. In the 1990s, microarrays were the standard analysis platform for gene expression analysis in large studies52,54,55. However, microarrays have high degrees of background hybridization and relative gene expression analysis of different transcripts within the same array is difficult to interpret. Furthermore, scanning of transcripts is only possible for those whose probes are present in the predesigned microarray52, thus restricting discovery of novel observations. RNA-seq addresses some of the limitations of microarrays and allows researchers to sequence the entire transcriptome without limiting the scope of transcripts analyzed.
Experimental design is imperative in obtaining good quality data from RNA-seq. Traditionally, RNA-seq has been performed on bulk tissue52; however, analysis of bulk tissue meant that cell heterogeneity was underappreciated. Depending on the research question or limitations in obtaining samples, RNA-seq on bulk tissue might be a viable option, although most studies now rely on purified cells. Whether performing RNA-seq on bulk tissue or purified cells, it is important that researchers follow the guidelines established by the ENCODE (Encyclopedia of DNA Elements) consortium56–58. High-quality RNA is essential to the production of good poly(A) libraries58; therefore, researches need to follow an RNA isolation protocol that minimizes introduction of additional biases59, including the removal of ribosomal RNA58,59 while maintaining RNA integrity.
Designing RNA-seq experiments requires sequencing depth and number of biological replicates to be determined beforehand, as these two variables affect the power of statistical analysis. Increasing sequencing depth, or the numbers of reads for a given sample, allows for detection and quantification of a greater number of transcripts, especially less abundant transcripts59,60. There is a trade-off between sequencing depth and number of replicates58, with a minimum of three replicates needed for gene expression comparisons59. Tools that help calculate the best experimental design based on researcher’s budget59,61 are also available, allowing for the best balance between cost and statistical confidence.
Single-cell RNA-seq (scRNA-seq) has been developed to address cellular heterogeneity within tissues. The simplest workflow for scRNA-seq includes sorting of single cells, RNA isolation, library preparation, and sequencing. A major limitation is the difficulty in isolation of high-quality RNA, given that cells are subjected to staining and sorting pressures, consequently leading to some amount of cell death resulting in low RNA quality.
Microfluidic technologies, specifically droplet microfluidics, have been instrumental in the development of high-throughput protocols for scRNA-seq that requires fewer reagents and less time while increasing the amount of data acquired62,63. Numerous droplet encapsulation protocols have been developed and applied to various biological systems64–67. They all involve using beads to encapsulate cells in a high-throughput fashion and differ only in the encapsulation method68,69. Droplet encapsulation methods for scRNA-seq are commercially available (reviewed in70) thus making this technology accessible to many labs and centers. Encapsulation of doublets is a problem of droplet microfluidics. However, commercially available systems have been optimized to minimize doublet encapsulation, thereby allowing for true single-cell analysis, with the 10X Genomics platform providing the lowest doublet frequency69,70.
Newer technologies can couple surface expression data with transcriptomics, thus allowing researchers to match protein expression with transcript levels71,72. Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq; available commercially as TotalSeq™ through BioLegend®), allows simultaneous acquisition of protein and mRNA expression71. CITE-seq technology uses antibodies tagged with an oligo containing a PolyA tail, thus mimicking an mRNA molecule and providing a unique ‘barcode’ for the antibody. The labeled single cells are then encapsulated with beads, using either the 10X Genomics and Drop-seq platforms, followed by reverse transcription of all mRNAs, including the antibody oligo tag, and sequencing of resulting full length and tagged cDNAs. Relative abundance of cDNA tags is used to approximate protein expression, thus providing both protein expression and transcriptome information for single cells. A similar approach is REAP-seq, which differs only in how the antibody is conjugated to the DNA barcode72,73. Overall, these technologies will further our understanding of the maternal-fetal interface immunome.
Data Analysis
The pivotal challenge arising from acquisition of high complexity single cell data is data analysis. Advances in the data acquisition methods also require high computational resources. To address this, many computational algorithms have been developed to properly visualize, filter, and interpret high-dimensional data. Outlined here are some of the algorithms used to analyze such high-dimensional data sets.
Flow Cytometry
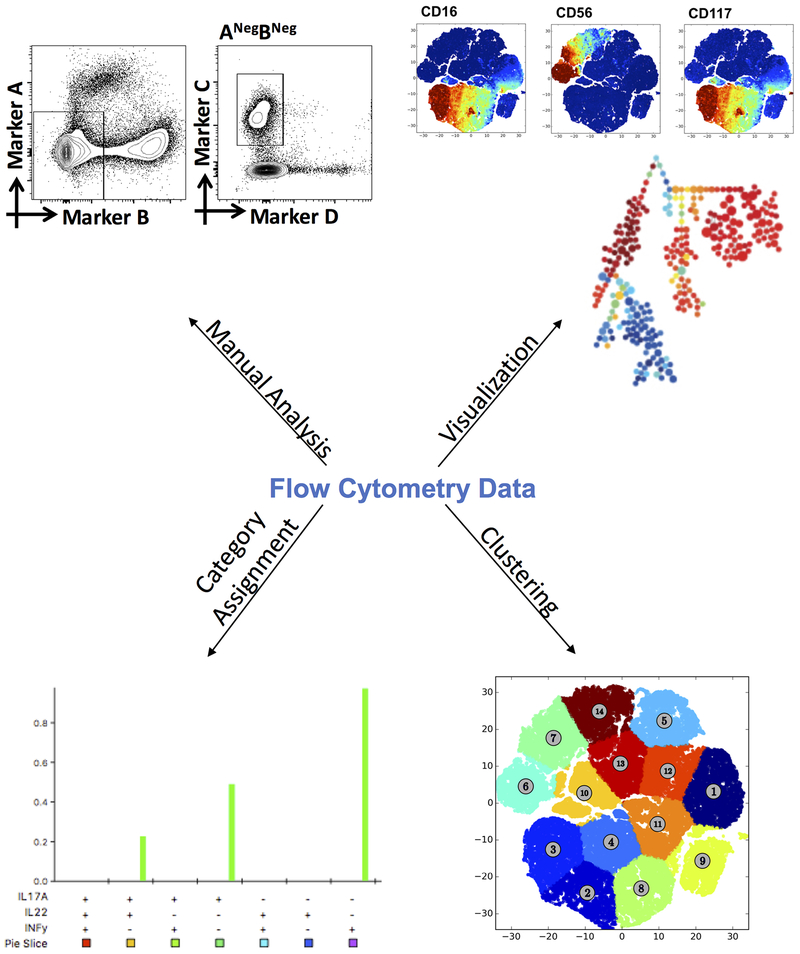
Flow cytometry has emerged as an important tool in single cell analysis. Advances in fluorochrome availability and detection capabilities by flow cytometers has led to an expansion of computational tools that allow researchers to analyze polychromatic flow cytometry data. Flow cytometry data is recorded in Flow Cytometry Standard (FCS) files which can be analyzed by multiple methods. Below, we discuss a series of computational tools that are available for visualization (t-SNE, SPADE, Cytosplore+HSHE), clustering (DensVM, k-means), and category assignment (SPICE) (Figure 2).
Figure 2. Options for flow cytometry data analysis.
Flow cytometry data can be analyzed by various methods. For high-dimensional flow cytometry, the use of dimensionality reduction algorithms for visualization is recommended. Coupled with clustering algorithms, dimensionality reduction can aid in the identification of novel or rare cellular populations.
Manual analysis
Manual analysis involves the construction of one- or two-dimensional plots for a subset of markers and selecting cellular populations of interest (i.e. gating) in a sequential manner27,28. Manual analysis remains the main method of analysis in many labs, however it has limitations which include user bias and operator variability28. Multiple studies have shown a high degree of variability between different users, even when analyzing identical data sets28,74–76. Having a pre-established gating scheme, based on well established phenotypes77,78, as part of the analytical pipeline in the laboratory can help reduce variability between users. This can, however, inadvertently reduce exploration of the entire dataset, thus decreasing the possibility of identifying novel cellular subsets79. Alternatively, laboratories and centers can assign one individual to analyze all data generated from one study74–76, although this might no be feasible for small laboratories or longitudinal studies were personnel turnover maybe encountered. Despite its limitations, manual analysis remains an important component of data analysis and is required for data filtering and preprocessing in preparation for computational analysis.
Computational Approaches
As data acquisition technology has advanced in recent years, computational analysis tools have become readily available to researchers. In flow cytometry, computational algorithms have allowed researchers to address the shortcomings of manual analysis28,74,75. Additionally, use of unbiased computational algorithms give researchers the ability to discover novel cell types that they might have otherwise overlooked using manual analysis79. More importantly, computational approaches are capable of recapitulating manual analysis results75, thus reinforcing their applicability in our understanding of the immune system. Despite the advantages in implementing computational analysis, one needs to have the computational skills and resources required for program implementation. Fortunately, there are easy-to-implement computer programs that require minimal computer coding knowledge, thus making them accessible to novice users. Below, we outline programs that are useful to reproductive immunologists to further our understanding of the immunome at the maternal-fetal interface.
t-SNE
A dimensionality reduction method that allows for visualization of data points on a two-dimensional map is t-distributed stochastic neighbor embedding (t-SNE)80. Unlike principal component analysis (PCA), which is well suited for linear data, t-SNE is better suited for the analysis of flow cytometry data, which is logarithmically distributed. Coupled with clustering methods, t-SNE allows for the partitioning and grouping of cells by similarity in an unbiased, data driven manner. Implemented in the immunophenotyping of human term decidua41, amniotic fluid36, and the tracking of immunome changes across murine gestation38, t-SNE visualization has proven to be a powerful tool in the understanding of the maternal-fetal interface immunome.
To implement t-SNE, researchers can use Cytofkit81, available as an R package through BioConductor82,83. The user-friendly graphical user interface (GUI) allows easy implementation of Cytofkit in R, thus minimizing the need for extensive R language. Originally developed for the analysis of mass cytometry data, Cytofkit has been successfully used to adequately phenotype the mouse myeloid compartment84 and human helper T cells85. Cytofkit combines t-SNE with the machine-learning algorithm DensVM (density-based clustering aided by support Vector Machine) for the unbiased partitioning of cells, and it includes additional visualization algorithms such as phenograph and ClusterX81.
Further, t-SNE can be implemented using the Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) software86. ACCENSE, like Cytofkit, is an easy-to-use tool that does not require understanding of programming language. Because ACCENSE uses k-means clustering to assign cells to a particular group, some cells remain unclassified81 thus limiting the power of ACCENSE. A fast, distributed version of t-SNE is viSNE87. Once available as a stand-alone MatLab program called cyt, viSNE is now part of the software suite available through Cytobank©, which requires a paid subscription. Additionally, t-SNE is available as a plug-in feature in FlowJo® and requires only installation of required R packages.
Although t-SNE is widely available to researchers and is useful for the identification of rare cell types, it has considerable limitations. Experimental variability represents a major challenge in t-SNE analysis, and results in significant batching of experimental runs41,86,88. This variability arises at multiple experimental stages, including minor deviations in staining procedures and set-up of voltages on the flow cytometer’s detectors. These can affect the fluorescence intensity measures of the markers assessed since these values depend on both antibody concentration and voltage levels. Laboratories can establish standard operating procedures for staining to minimize variability between experimental runs74. For flow cytometer set-up, users can employ standardizing beads41 which allow detector voltages to be set at the same level across experiments. Because t-SNE is computationally expensive, minimizing data input by downsampling is necessary, thus limiting the power of t-SNE, specifically in identifying rare populations89. Despite these limitations, t-SNE still represents a viable option for researchers in reproductive immunology.
SPADE
Spanning-tree progression analysis of density-normalized events (SPADE) allows multiple cell types to be visualized in a branched tree90,91. Tree nodes represent cellular clusters connected in a minimum spanning tree based on phenotypic similarity92. The resulting SPADE tree allows for visualization of cluster density, with node size indicating the number of cells within clusters92 and color scale indicating marker expression levels93. SPADE has been successfully implemented to understand changes in the immune system during cancer treatment94, characterize the diversity of extracellular vesicles95, and asses immune drug responses96. SPADE is part of Cytobank’s suite of computational tools. It is also available as a standalone package that can be installed in both Macs and PCs and as a Matlab program (http://pengqiu.gatech.edu/software/SPADE/).
Although SPADE has proven useful in visualizing flow cytometry data, there are some limitations to its use. First, unlike t-SNE/DensVM analysis which determines the number of clusters in a user-independent fashion, SPADE users have to specify the initial number of clusters believed to be in the data93,97, potentially leading to underestimation of cellular clusters. If the number of clusters is overestimated, clusters can be expertly merged97, by merging two clusters that seem phenotypically similar; however, leading to the introduction of user bias. Second, SPADE down-sampling occurs in a stochastic manner, thus leading to potential exclusion of rare cellular populations. Users interested in identifying rare populations should indicate a low down-sampling percentage, to increase the likelihood that low density regions are assigned their own cluster23,90,97. Third, SPADE does not allow for group-level statistical comparisons when analyzing multiple FCS files simultaneously98. The most recent version of SPADE includes several improvements, including a deterministic density-dependent down-sampling method and the implementation of a deterministic k-means clustering algorithm, reducing the randomness of down-sampling and clustering in the original implementation of SPADE91. This newest version also includes a tree-partitioning algorithm that automatically suggests a tree partition based on the largest phenotypic difference in high-dimensional space. The user can either accept or reject the suggested partition, thus allowing for a semiautomated interpretation of the SPADE tree91. Similar to t-SNE, SPADE uses fluorescence intensity as data input, therefore experimental standardization should be adopted to maintain consistency across experiments and minimize any batch effects.
Despite its limitations, SPADE is a useful analytical tool in reproductive immunology. Because SPADE allows the user to predetermine the number of clusters (i.e. categories), allowing additional datasets to be added and fitted into the existing clusters. This can prove useful in longitudinal studies involving the immune profiling of women across pregnancy and into the postpartum period, thereby allowing researchers to map peripheral immune changes across pregnancy progression.
Cytosplore+HSNE
Cytosplore+HSNE implements Hierarchical Stochastic Neighbor Embedding (HSNE)99 in an integrated fashion, thus allowing for the interactive exploration of high-dimensional single-cell datasets and identification of distinct populations in a data-driven fashion89,100. Cytosplore+HSNE has been applied to the analysis of innate lymphoid cells in fetal human intestine101. One of the limitations of t-SNE is the computational time required to analyze a dataset, which often requires down-sampling. To address this, Cytosplore+HSNE uses A-tSNE, a modified version of t-SNE that aims at minimizing the precomputation times required to analyze high-dimensional datasets102. Cytosplore+HSNE also uses SPADE clustering which allows for high-level partitioning of the data. Coupling SPADE with A-tSNE reduces the input size of each embedding and makes analysis feasible. To address some of the shortcomings that come with the use of ACCENSE, Cytosplore+HSNE uses Gaussian Mean Shift (GMS), which can create arbitrarily shaped clusters and leads to all the available data being clustered.
One of the main benefits of Cytosplore+HSNE is that it can be applied to many different types of data, such as that from flow and mass cytometry and scRNA-seq. Because of the computational benefits designed into the program, downsampling is not required, allowing the analysis of the entire data set. This added benefit allows one to identify rare populations that would have been missed using traditional t-SNE89.
SPICE
SPICE, or Simplified Presentation of Incredibly Complex Evaluations, allows users to evaluate multiple parameters, such as age or treatment, using an easy-to-use graphical interface103. Unlike the computational tools described above, SPICE is not a dimensionality reduction tool. Rather, SPICE allows grouping of samples based on any number of categories to be displayed graphically, either in pie charts or bar graphs. Pestle© is an accompanying program that allows users to reformat FlowJo® data tables for SPICE analysis104. Although Pestle© is not necessary for the current version of SPICE (v.6), it is useful to subtract background noise, thus allowing for cleaner visualization103. SPICE includes multiple statistical tests such as permutation tests to compare pie charts and Student’s t tests to compare different groups. SPICE has been applied to understand the functional properties of mucosal associated invariant T (MAIT) cells in the female genital mucosa105 as well as analyzing the immune modulating effects of progesterone during pregnancy106.
SPICE is freely distributed by the NIH and is presented in a user-friendly interface that is easy to implement. However, a drawback is that it is only supported in Mac with no plans to support a PC version. Formatting data for SPICE can also be a challenge but can be overcome with a working knowledge of table formatting in FlowJo. Additionally, SPICE depends on Boolean gates of manually gated populations, which can lead to biases107, as manual gating can be subjective as discussed above. To address this, it is possible to integrate dimensionality reduction techniques and draw Boolean gates on identified clusters, thus making SPICE analysis data-driven and user independent.
RNA-seq
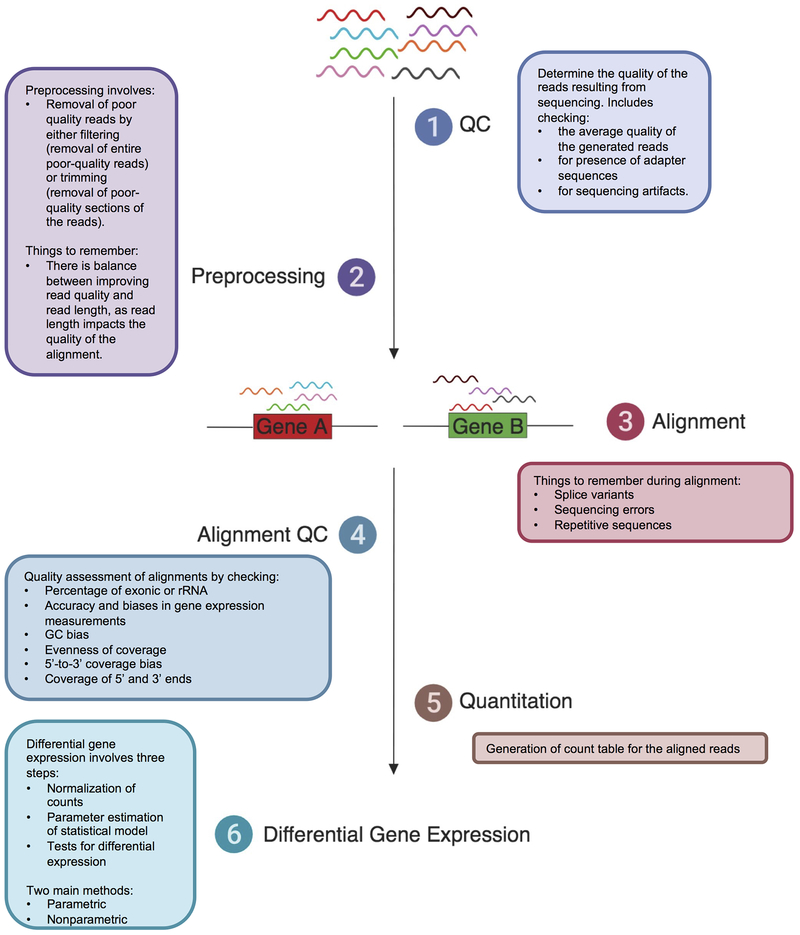
Data analysis for RNA-seq experiments can be broken down into the following steps: quality control, preprocessing, alignment, alignment quality control, and differential gene expression (DGE) (Figure 3; Table 1). Below we discuss the analysis of scRNA-seq which follows a similar approach. We then provide an overview of additional analysis options, including pathway analysis and deconvolution approaches. We end discussing user interface packages that might prove useful and accessible.
Figure 3. Steps for RNA-seq data analysis.
Proper RNAseq data analysis requires multiple steps. Outlined in this figure are the six basic steps: (1) Quality control check of raw reds; (2) Preprocessing of reads, including the removal of adaptor sequences, poor quality reads, and trimming of reads; (3) Alignment of reads to the reference genome; (4) Quality control check of the resulting read alignment; (5) Quantification of the read alignment and count table generation; and (6) Differential gene expression. Table 1 describes programs of interest for each of the steps outlined above.
Table 1.
Programs used to complete steps in RNA-seq data analysis outline in Figure 3.
| Step | Tool | Description | Ease of Use | Ref. |
|---|---|---|---|---|
| 1 | FastQC | Provides a report of raw read quality. Implemented in JAVA and accepts BAM, SAM, and FastQ file formats. Available at (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). | GUI* | 152 |
| FastX | Part of the FastX-Tool kit. Implemented either through Galaxy or through command-line. Pre-compiled binaries are available in Linux and MacOS X platforms. | Command line | 153 | |
| PRINSeq | Used to check quality of RNA-seq data. Can also filter, reformat, and trim reads. Provides summary statistics of the reads in both graphical and tabular format. | GUI | 154 | |
| 2 | Trimmomatic | Is a flexible trimmer that can handle paired-end data. It is implemented through Java and is available at www.usadellab.org/cms/index.php?page=trimmomatic. Only works with Illumina generate data. Does not automatically detect the PHRED score automatically. | Command line | 155 |
| AdapterRemoval | Trimming tool that can remove adaptor sequences. Implemented in C++. Useful in processing large data sets, with longer reads, on a desktop machine. | Command line | 156 | |
| TagCleaner | Automatically detects an adaptor sequence. It is available at (http://edwards.sdsu.edu/tagcleaner) and is implemented using Perl 5.8, through a user web-interface. | GUI | 157 | |
| 3 | BWA | Burrows-Wheeler Aligner’s (BWA) is able to align both short and long reads. It allows for mismatches and gaps. Performance is faster compared to other aligners such as MAQ. Available at http://bio-bwa.sourceforge.net | Command line | 158,159 |
| Bowtie | Aligns shorts reads and requires less memory allowing implementation in a desktop computer. It is faster than comparable programs. It is available at http://bowtie.cbcb.umd.edu | Command line | 160 | |
| STAR | Aligns non-contiguous sequences directly to the reference genome. Is able to detect splice junctions, multiple mismatches, and indels. Benefits include its ability to accurately align long reads, having the lowest false-positive rate while maintaining high sensitivity, and being fast. Implemented in C++. | Command line | 161 | |
| 4 | RNA-SeQC | Provides important measures of alignment quality including: yield, alignment and duplicate rates, GC bias, rRNA content, regions of alignment, continuity of coverage, 3’/5’ bias, and count of detectable transcripts. Implemented through Java or through the GenePattern web interface (www.GenePattern.org). | GUI | 162 |
| RSeQC | Can evaluate sequence quality, GC bias, PCR bias, nucleotide composition bias, sequencing depth, strand specificity, coverage uniformity, and read distribution over the genome structure. It is the most comprehensive and efficient program. | Command line | 163 | |
| Qualimap 2 | Can compare multiple sequencing data sets and includes a novel mode that aids in the discovery of biases and problems specific to RNA-seq technology. It is available in a user-friendly interface at http://qualimap.bioinfo.cipf.es | GUI | 164 | |
| 5 | Flux Capacitor | Quantifies the abundance of annotated alternatively spliced transcripts by distributing the reads mapping to a given splice junction among the transcripts including the exon. Written in Java; requires a Java Virtual Machine; platform independent. | Command line | 165 |
| Cufflink | Allows for the probabilistic deconvolution of RNA-seq fragment densities and accounts for cases in which genome alignments of fragments do not uniquely correspond to source transcripts. It is an open-source C++ program and can be implemented in Linux and Mac OS X. | Command line | 166 | |
| HTSeq | Using the Htseg-count function, it counts the overlap between reads and genes, and counts only reads that map unambiguously to a single gene. Implemented in Python. | Command line | 167 | |
| 6 | EBSeq | Uses an empirical Bayes hierarchical model approach to identify differentially expressed isoforms. It can compare two or more biological conditions. It is a robust method for identifying differentially expressed genes. Implemented in R and can be implemented through a user-friendly interface available at https://www.biostat.wisc.edu/ñingleng/EBSeq_Package/EBSeq_Interface/ | GUI | 168 |
| DESeq2 | Uses shrinkage estimators for dispersion and fold change which improves its stability and reproducibility. Ideal for analysis of small studies with few replicates. Allows for a more quantitative analysis focused on the strength rather than the mere presence of differential expression. Implemented in R. | Command line | 169 | |
| Limma+Voom | Transforms the normalized counts to logarithmic base 2 and adds a precision weight for each observation. Can model the data in normal Gaussian distribution, thus allowing the data to be tested statistically. It is computationally fast and can be used with small sample sizes, with a minimum of two replicates per group. Implemented in R. | Command line | 170 |
GUI = Graphical user interface.
Analysis of scRNA-seq
Single-cell RNA-seq has emerged as a powerful tool that provides the ability to analyze the transcriptome at the single-cell level108, thus allowing researchers to identify rare cell populations, infer cell lineage trajectories, and assess cellular differentiation109. The workflow for scRNA-seq data analysis follows the same steps outlined above. However, there are special considerations for scRNA-seq data analysis. For instance, scRNA-seq data is sparse, compared to bulk RNA-seq, either because of the nature of transcription and temporal gene expression or event dropout resulting from inefficient reverse transcription reactions of low abundance genes70. As a result, assumptions about data distribution in bulk RNA-seq do not apply to scRNA-seq70.
Quality control, alignment, and quantification of scRNA-seq reads can be done with the same programs used for bulk RNA-seq110. Because of dropout in scRNA-seq data, it is important that additional data filtering is done. Gene filtering allows for the removal of low quality genes and samples110. The best way to filter out poor quality genes and samples is to use External RNA Controls Consortium (ERCC) spike-ins that are added at the start of the experiment, thus providing calibration of the relative amount of starting material111,112. OEFinder is a program that can be used for the removal of poor quality genes113. Certain confounders such as batch effects and cell-cycle induced variation also need to be removed from scRNA-seq data for adequate analysis110. For most studies, down-sampling can eliminate batch effects, however this reduces the complexity of the data110. There are packages that can be used for the removal of batch effects, such as COMBAT114, and for cell-cycle induced variation, scLVM115 and ccRemover116.
Normalization of scRNA-seq data, as with bulk RNA-seq, is an important step in data analysis. For experiments that did not use ERCC spike-ins, DESeq2 and edgeR can be used for normalization, just as in bulk RNA-seq110. For scRNA-seq experiments that do contain ERCC spike-ins, programs such as GRM117, BASiCS118, and SAMstrt119 can use spike-ins as internal controls for normalization110.
Although DGE is not usually the main objective of scRNA-seq experiments, there are programs that are capable of doing this analysis110. There are specific programs that address dropout in scRNA-seq data, such as Single-Cell Differential Expression (SCDE)120, PAGODA121, and MAST122. Other programs available, such as Monocle123, address the bimodality of gene expression. Programs used for bulk RNA-seq, such as edgeR and DESeq2, can be applied to scRNA-seq data and have been shown to perform better than programs specific for scRNA-seq data110,124.
The power of scRNA-seq comes from the ability to identify rare subpopulations and to infer developmental trajectories of single cells110. For subpopulation identification, dimensionality reduction algorithms such as PCA and t-SNE can be applied110,125–127. There are specific dimensionality reduction algorithms for scRNA-seq such as Zero-inflated factor analysis (ZIFA)128 and PAGODA121 that can be implemented for the identification of subpopulations. Furthermore, there are multiple programs, such as Monocle123, Waterfall129, SLICER130, and SCUBA131 that can infer differentiation pathways from scRNA-seq data.
Gene Pathway and Deconvolution
Gene pathway analysis takes into account the relationship between expressed genes, thus allowing researchers to determine whether biological processes differ between groups132. Gene pathway analysis workflow involves a list of genes of interest from the RNA-seq data set and the application of statistical methods to test for gene enrichment132,133. There are multiple packages that can perform pathway analysis, such as GSEA134 and GSVA135. Recently, programs have been developed to account for the pathway topology132. Multiple programs, such as SPIA136 and TAPPA137, provide pathway topology information by taking into account gene interactions and weighing interacting genes more than non-interacting genes, while DEAP138 identifies the pathway that is most differentially expressed.
Deconvolution programs are capable of determining cellular composition of heterogeneous samples based on gene expression patterns across different cell subtypes139,140. In most instances, these programs require gene expression datasets as a reference to determine the composition of bulk RNA-seq samples140. There are multiple programs that are capable of determining the composition of bulk samples all using a specific mathematical approach139. CIBERSORT141, for example, is a partial deconvolution algorithm that uses nu support vector regression to estimate the cellular fractions in bulk samples139. However, the effectiveness of CIBERSORT depends heavily on the reference profiles used141. Multi-Subject Single Cell (MuSiC) is another deconvolution algorithm that uses scRNA-seq data obtained from a cross section of individuals as a reference to then analyze bulk RNA-seq data from different individuals142. MuSiC might prove of particular interest to reproductive immunologist as only a small set of samples would be required for scRNA-seq, thus limiting costs, with the remaining of the study samples requiring bulk RNA-seq.
Integrated Graphical User Interface Packages
Most of the programs/packages outlined here require some coding knowledge. Fortunately, there are some software packages that combine some of the tools listed above that make RNA-seq data feasible to users who might not have extensive coding skills. Here we cover Chipster and Galaxy.
Chipster143 is a collection of up-do-date analysis and visualization tools and is available as a graphical Java desktop application. Chipster easily integrates tools regardless of how the tool is implemented143. Chipster allows users to track their analysis workflow and save any series of steps144. In addition to RNA-seq data, Chipster is able to analyze other types of data, including microarrays, miRNA-seq, ChIP-seq, and whole genome sequencing143,145. Chipster is freely available and can be used with a free short-term evaluation account through Chipster’s server143. Additional benefits of Chipster include the ability for users to save their workflows and share them with other users, thus allowing for collaboration and reproducibility143. Users are able to view the original tool code and integrate additional code143. Chipster, however, is not designed to be integrated to a laboratory information management system (LIMS), limiting Chipster’s use to laboratories that do not have a large amount of NGS data145.
Galaxy146–148 is a popular, web-based framework148 that provides computational tools with an intuitive user interface146,149. Most importantly, Galaxy does not require substantial programming skills147. Galaxy includes a unique history system that allows the user to organize and save workflows, and maintain quality control of the analysis performed149, thus allowing for reproducibility146. Although written in Python, Galaxy does not require the user to have Python programming knowledge. Galaxy hides the computational details from the user, thus eliminating the need for extensive bioinformatics expertise146,149. Despite this, Galaxy is limited because pipeline output is not standardized and depends on the user input145. Because of the high level of flexibility within Galaxy, its usage might be limited towards those that have a good knowledge of the tools being applied, compared to Chipster145.
Verifying biological veracity of single cell data sets
Single cell data presents are wealth of information. Despite its advantages there are some considerations to keep in mind, both as users and readers. Below, we outline advice on how to minimize common problems and how to improve data quality.
To address batch effects and document the extent of experimental variation in flow cytometry, users should create additional parameters in the FCS files that document user/operator and experiment number. Downstream analysis would reveal whether there is grouping based on experimental runs and/or operator, allowing researchers to properly interpret their data.
We recommend that researchers take advantage of user-friendly programs such as Chipster to check the quality of their RNA-seq data. At a minimum, researchers would be able to check read quality and determine the presence of outliers. This would allow the researchers to make experimental adjustments if needed. Having knowledge of outliers or batching effects would also allow researchers to have an informed conversation with bioinformaticians that could then guide them on how to handle downstream analysis. In addition, researchers should perform some validation of their RNA-seq data, either by performing qPCR or protein level detection (flow cytometry, imaging). As readers, we should also expect some level of validation on reported RNA-seq results.
When working with bioinformaticians, we recommend that researchers request all downstream data files (count tables, DE analysis tables). This is particularly important when looking at gene pathway analysis, were the bioinformatician might be unfamiliar with the biology and might consider some results irrelevant.
Finally, we recommend that researchers become familiar with basic bioinformatics language (Table 2) and basic data analysis terms (Figure 3). This will facilitate conversations with bioinformaticians and will make undertaking RNA-seq experiments manageable.
Table 2.
Common useful terms
| Term | Definition |
|---|---|
| Alignment | The process of matching reads to a particular region of the genome. |
| Counts | Number of reads that map to a particular gene. |
| Coverage | The extent to which a genomic region of interest was sequenced. |
| Dimensionality reduction | The processed by which the most important features of a high-dimensional data set are extracted thus resulting in a data set with reduced dimensions. |
| Dropout events | Missing data values due to low transcript expression and the stochastic nature of gene expression. |
| Embedding | Mapping of high-dimensional features onto a low dimensional space. |
| Feature extraction | Transformed data set built with the most predictive features of a high-dimensional data set. |
| Feature selection | Selection of most predictive features in a high-dimensional data set. |
| Features | Variables of a particular data set (i.e. fluorescence intensity or gene counts). |
| Machine learning | Process of building mathematical models that allow computers to be trained to be predictive. Requires training with a “training dataset”. |
| Partitioning | Division of data. |
| Quantitation | Generation of count table that integrates that number of reads per gene that were aligned successfully. |
| Reads | Nucleotide sequences as a result of sequencing. |
| Sequencing depth | The number of reads per genomic region. |
Conclusion
Expanding our knowledge of the reproductive immunome presents a great opportunity to improve fetal and maternal health. Many immunological questions remain obscure at the maternal-fetal interface, including (1) how decidual immune cells interact with non-immune cells (trophoblasts, stromal cells, endothelial cells) and other decidual immune cells, (2) if and how decidual immune cell dysfunction lead to pregnancy pathologies, and (3) how leveraging single-cell data could lead to reliable diagnostic tools. Already, tools discussed here, such as the 10X Genomics platform and scRNA-seq, have been leveraged to map cellular interactions in first trimester decidua150,151, providing insight into these important cellular relationships. This review, although not exhaustive, highlights practical tools that will help the reproductive immunology community decipher the mysteries of immune cellular networks governing gestational outcomes.
Acknowledgements
We thank C. Zhou and D. Boeldt for review of manuscript and suggestions. J.V. was supported by NIH Ruth L Kirschstein National Research Award (T32-HD041921), UW SciMed GRS Fellowship. I.M.O was supported by University of Wisconsin Carbone Cancer Center Support Grant (P30 CA014520), Wisconsin Partnership Program, NIH NIAID U19AI104317, NIH NCI R01CA204320, and NIH NCI R01CA219154–01. AKS was supported by grant K12HD000849–28 awarded to the Reproductive Scientist Development Program by the Eunice Kennedy Shriver National Institute of Child Health & Human Development and March of Dimes Basil O’Connor Award (5-FY18–541). Additional support (to A.K.S.) was provided by Burroughs Wellcome Fund, March of Dimes, and American Society for Reproductive Medicine, as part of the Reproductive Scientist Development Program.
REFERENCES
- 1.Hanna J, Goldman-Wohl D, Hamani Y, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12(9):1065–1074. [DOI] [PubMed] [Google Scholar]
- 2.Monk JM, Leonard S, McBey BA, Croy BA. Induction of murine spiral artery modification by recombinant human interferon-gamma. Placenta. 2005;26(10):835–838. [DOI] [PubMed] [Google Scholar]
- 3.Croy BA, Chantakru S, Esadeg S, Ashkar AA, Wei Q. Decidual natural killer cells: key regulators of placental development (a review). J Reprod Immunol. 2002;57(1):151–168. [DOI] [PubMed] [Google Scholar]
- 4.Ashkar AA, Croy BA. Functions of uterine natural killer cells are mediated by interferon gamma production during murine pregnancy. Semin Immunol. 2001;13(4):235–241. [DOI] [PubMed] [Google Scholar]
- 5.Gardner L Dendritic Cells in the Human Decidua. Biol Reprod. 2003;69(4):1438–1446. [DOI] [PubMed] [Google Scholar]
- 6.Hunt JS, King CR, Wood GW. Evaluation of human chorionic trophoblast cells and placental macrophages as stimulators of maternal lymphocyte proliferation in vitro. J Reprod Immunol. 6:377–391. [DOI] [PubMed] [Google Scholar]
- 7.Sindram-Trujillo A, Scherjon S, Kanhai H, Roelen D, Claas F. Increased T-Cell Activation in Decidua Parietalis Compared to Decidua Basalis in Uncomplicated Human Term Pregnancy. Am J Reprod Immunol. 2003;49(5):261–268. [DOI] [PubMed] [Google Scholar]
- 8.Sindram-Trujillo AP, Scherjon SA, Miert PP van H, Kanhai HHH, Roelen DL, Claas FHJ. Comparison of decidual leukocytes following spontaneous vaginal delivery and elective cesarean section in uncomplicated human term pregnancy. J Reprod Immunol. 2004;62(1–2):125–137. [DOI] [PubMed] [Google Scholar]
- 9.Bonney EA, Pudney J, Anderson DJ, Hill JA. Gamma-delta T cells in midgestation human placental villi. Gynecol Obstet Invest. 2000;50:153–157. [DOI] [PubMed] [Google Scholar]
- 10.Solders M, Gorchs L, Erkers T, et al. MAIT cells accumulate in placental intervillous space and display a highly cytotoxic phenotype upon bacterial stimulation. Sci Rep. 2017;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilburgs T, van der Mast BJ, Nagtzaam NMA, Roelen DL, Scherjon SA, Claas FHJ. Expression of NK cell receptors on decidual T cells in human pregnancy. J Reprod Immunol. 2009;80(1–2):22–32. [DOI] [PubMed] [Google Scholar]
- 12.Leng Y, Romero R, Xu Y, et al. Are B Cells Altered in the Decidua of Women with Preterm or Term Labor? Am J Reprod Immunol. February 2019:e13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kämmerer U, Eggert AO, Kapp M, et al. Unique appearance of proliferating antigen-presenting cells expressing DC-SIGN (CD209) in the decidua of early human pregnancy. Am J Pathol. 2003;162(3):887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leno-Durán E, Muñoz-Fernández R, Olivares EG, Tirado-González I. Liaison between natural killer cells and dendritic cells in human gestation. Cell Mol Immunol. 2014;11(5):449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tirado-González I, Muñoz-Fernández R, Prados A, et al. Apoptotic DC-SIGN+ cells in normal human decidua. Placenta. 2012;33(4):257–263. [DOI] [PubMed] [Google Scholar]
- 16.Nancy Patrice, Tagliani Elisa, Tay Chin-Siean, Asp Patrik, S. P. Strom, Adrian Erlebacher. Chemokine Gene Silencing in Decidual Stromal Cells Limits T Cell Access to the Maternal-Fetal Interface. Science. 2012;336(6086):1317–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tilburgs T, Strominger JL. CD8+ Effector T Cells at the Fetal-Maternal Interface, Balancing Fetal Tolerance and Antiviral Immunity. Am J Reprod Immunol. 2013;69(4):395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arenas-Hernandez M, Romero R, Xu Y, et al. Effector and Activated T Cells Induce Preterm Labor and Birth That Is Prevented by Treatment with Progesterone. J Immunol. March 2019:ji1801350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacca P, Montaldo E, Croxatto D, et al. Identification of diverse innate lymphoid cells in human decidua. Mucosal Immunol. 2015;8(2):254–264. [DOI] [PubMed] [Google Scholar]
- 20.Doisne J-M, Balmas E, Boulenouar S, et al. Composition, Development, and Function of Uterine Innate Lymphoid Cells. J Immunol. 2015;195(8):3937–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montaldo E, Vacca P, Chiossone L, et al. Unique Eomes+ NK Cell Subsets Are Present in Uterus and Decidua During Early Pregnancy. Front Immunol. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sojka DK, Plougastel-Douglas B, Yang L, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33(7):323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards BS, Kuckuck F, Sklar LA. Plug flow cytometry: An automated coupling device for rapid sequential flow cytometric sample analysis. Cytometry. 1999;37(2):156–159. [PubMed] [Google Scholar]
- 25.Fulwyler MJ. Electronic Separation of Biological Cells by Volume. Science. 1965;150(3698):910–911. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro Howard M. Practical Flow Cytometry. 4th ed. Wiley-Liss; 2003. [Google Scholar]
- 27.Robinson JP, Rajwa B, Patsekin V, Davisson VJ. Computational analysis of high-throughput flow cytometry data. Expert Opin Drug Discov. 2012;7(8):679–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16(7):449–462. [DOI] [PubMed] [Google Scholar]
- 29.Crosland-Taylor PJ. A device for counting small particles suspended in a fluid through a tube. Nature. 1953;171(4340):37–38. [DOI] [PubMed] [Google Scholar]
- 30.Moldavan A PHOTO-ELECTRIC TECHNIQUE FOR THE COUNTING OF MICROSCOPICAL CELLS. Science. 1934;80(2069):188–189. [DOI] [PubMed] [Google Scholar]
- 31.Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Fluorescence Activated Cell Sorting. Rev Sci Instrum. 1972;43(3):404–409. [DOI] [PubMed] [Google Scholar]
- 32.McKinnon KM. Flow Cytometry: An Overview: Flow Cytometry: An Overview In: Coligan JE, Bierer BE, Margulies DH, Shevach EM, Strober W, eds. Current Protocols in Immunology. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2018:5.1.1–5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golden JP, Justin GA, Nasir M, Ligler FS. Hydrodynamic focusing—a versatile tool. Anal Bioanal Chem. 2012;402(1):325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sa Y, Feng Y, Jacobs KM, et al. Study of low speed flow cytometry for diffraction imaging with different chamber and nozzle designs: Study of Low Speed Flow Cytometry. Cytometry A. 2013;83(11):1027–1033. [DOI] [PubMed] [Google Scholar]
- 35.Piyasena ME, Austin Suthanthiraraj PP, Applegate RW, et al. Multinode Acoustic Focusing for Parallel Flow Cytometry. Anal Chem. 2012;84(4):1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Lopez N, Romero R, Xu Y, et al. The immunophenotype of amniotic fluid leukocytes in normal and complicated pregnancies. Am J Reprod Immunol. 2018;79(4):e12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis EL, Sierra L-J, Barila GO, Brown AG, Porrett PM, Elovitz MA. Placental immune state shifts with gestational age. Am J Reprod Immunol. 2018;79(6):e12848. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Lopez GE, Vazquez J, et al. Decidual-Placental Immune Landscape During Syngeneic Murine Pregnancy. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller D, Motomura K, Garcia-Flores V, Romero R, Gomez-Lopez N. Innate Lymphoid Cells in the Maternal and Fetal Compartments. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sojka DK, Yang L, Yokoyama WM. Uterine natural killer cells: To protect and to nurture. Birth Defects Res. November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vazquez J, Chavarria M, Li Y, Lopez GE, Stanic AK. Computational flow cytometry analysis reveals a unique immune signature of the human maternal-fetal interface. Am J Reprod Immunol. 2018;79(1):e12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: Monitoring single cell signaling events. Cytometry. 2003;55A(2):61–70. [DOI] [PubMed] [Google Scholar]
- 43.Krutzik PO, Irish JM, Nolan GP, Perez OD. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol. 2004;110(3):206–221. [DOI] [PubMed] [Google Scholar]
- 44.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3(5):361–368. [DOI] [PubMed] [Google Scholar]
- 45.Roederer M Spectral compensation for flow cytometry: Visualization artifacts, limitations, and caveats. Cytometry. 2001;45(3):194–205. [DOI] [PubMed] [Google Scholar]
- 46.Szalóki G, Goda K. Compensation in multicolor flow cytometry: Compensation in Multicolor Flow Cytometry. Cytometry A. 2015;87(11):982–985. [DOI] [PubMed] [Google Scholar]
- 47.Mahnke Y, Chattopadhyay P, Roederer M. Publication of optimized multicolor immunofluorescence panels. Cytometry A. 2010;77A(9):814–818. [DOI] [PubMed] [Google Scholar]
- 48.Lee JA, Spidlen J, Boyce K, et al. MIFlowCyt: the minimum information about a Flow Cytometry Experiment. Cytometry A. 2008;73(10):926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bandura DR, Baranov VI, Ornatsky OI, et al. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal Chem. 2009;81(16):6813–6822. [DOI] [PubMed] [Google Scholar]
- 50.Simoni Y, Chng MHY, Li S, Fehlings M, Newell EW. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr Opin Immunol. 2018;51:187–196. [DOI] [PubMed] [Google Scholar]
- 51.Fragiadakis GK, Baca QJ, Gherardini PF, et al. Mapping the Fetomaternal Peripheral Immune System at Term Pregnancy. J Immunol. 2016;197(11):4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16(3):133–145. [DOI] [PubMed] [Google Scholar]
- 54.Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and Microarray in Transcriptome Profiling of Activated T Cells. Zhang S-D, ed. PLoS ONE. 2014;9(1):e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Yu Y, Hertwig F, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. [DOI] [PubMed] [Google Scholar]
- 57.The ENCODE Project Consortium. A User’s Guide to the Encyclopedia of DNA Elements (ENCODE). Becker PB, ed. PLoS Biol. 2011;9(4):e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15(2):121–132. [DOI] [PubMed] [Google Scholar]
- 59.Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. [DOI] [PubMed] [Google Scholar]
- 61.Busby MA, Stewart C, Miller CA, Grzeda KR, Marth GT. Scotty: a web tool for designing RNA-Seq experiments to measure differential gene expression. Bioinformatics. 2013;29(5):656–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joensson HN, Andersson Svahn H. Droplet Microfluidics-A Tool for Single-Cell Analysis. Angew Chem Int Ed. 2012;51(49):12176–12192. [DOI] [PubMed] [Google Scholar]
- 63.Price AK, Paegel BM. Discovery in Droplets. Anal Chem. 2016;88(1):339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zilionis R, Nainys J, Veres A, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc. 2016;12(1):44–73. [DOI] [PubMed] [Google Scholar]
- 65.Rosenberg AB, Roco CM, Muscat RA, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360(6385):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daniel E Wagner Caleb Weinreb, Collins Zach M, Briggs James A, Sean G Megason, Allon M Klein. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 2018;360:981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon H-S, Je K, Min J-W, et al. Inertial-ordering-assisted droplet microfluidics for high-throughput single-cell RNA-sequencing. Lab Chip. 2018;18(5):775–784. [DOI] [PubMed] [Google Scholar]
- 69.Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen A, Khoo WH, Moran I, Croucher PI, Phan TG. Single Cell RNA Sequencing of Rare Immune Cell Populations. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterson VM, Zhang KX, Kumar N, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35(10):936–939. [DOI] [PubMed] [Google Scholar]
- 73.Todorovic V Gene expression: Single-cell RNA-seq—now with protein. Nat Methods. 2017;14(11):1028–1029. [Google Scholar]
- 74.Nomura L, Maino VC, Maecker HT. Standardization and optimization of multiparameter intracellular cytokine staining: Standardization and Optimization of Multiparameter ICS. Cytometry A. 2008;73A(11):984–991. [DOI] [PubMed] [Google Scholar]
- 75.Gouttefangeas C, Chan C, Attig S, et al. Data analysis as a source of variability of the HLA-peptide multimer assay: from manual gating to automated recognition of cell clusters. Cancer Immunol Immunother. 2015;64(5):585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pachón G, Caragol I, Petriz J. Subjectivity and flow cytometric variability. Nat Rev Immunol. 2012;12(5):396–396. [DOI] [PubMed] [Google Scholar]
- 77.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.on behalf of the EuroFlow Consortium (EU-FP6, LSHB-CT-2006–018708), van Dongen JJM, Lhermitte L, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26(9):1908–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Irish JM. Beyond the age of cellular discovery. Nat Immunol. 2014;15(12):1095–1097. [DOI] [PubMed] [Google Scholar]
- 80.Laurens van der Maaten, Geoffrey Hinton. Visualizing Data using t-SNE. J Mach Learn Res. 2008;1:1–48. [Google Scholar]
- 81.Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. Schneidman D, ed. PLOS Comput Biol. 2016;12(9):e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Becher B, Schlitzer A, Chen J, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15(12):1181–1189. [DOI] [PubMed] [Google Scholar]
- 85.Wong MT, Chen J, Narayanan S, et al. Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis. Cell Rep. 2015;11(11):1822–1833. [DOI] [PubMed] [Google Scholar]
- 86.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE). Proc Natl Acad Sci. 2014;111(1):202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amir ED, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hahne F, Khodabakhshi AH, Bashashati A, et al. Per-channel basis normalization methods for flow cytometry data. Cytometry A. 2009;9999A:NA–NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Unen V, Höllt T, Pezzotti N, et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun. 2017;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qiu P Toward deterministic and semiautomated SPADE analysis: Deterministic SPADE. Cytometry A. 2017;91(3):281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chester C, Maecker HT. Algorithmic Tools for Mining High-Dimensional Cytometry Data. J Immunol. 2015;195(3):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mair F, Hartmann FJ, Mrdjen D, Tosevski V, Krieg C, Becher B. The end of gating? An introduction to automated analysis of high dimensional cytometry data: Highlights. Eur J Immunol. 2016;46(1):34–43. [DOI] [PubMed] [Google Scholar]
- 94.Lohmann L, Janoschka C, Schulte-Mecklenbeck A, et al. Immune Cell Profiling During Switching from Natalizumab to Fingolimod Reveals Differential Effects on Systemic Immune-Regulatory Networks and on Trafficking of Non-T Cell Populations into the Cerebrospinal Fluid—Results from the ToFingo Successor Study. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marcoux G, Duchez A-C, Cloutier N, Provost P, Nigrovic PA, Boilard E. Revealing the diversity of extracellular vesicles using high-dimensional flow cytometry analyses. Sci Rep. 2016;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diggins KE, Ferrell PB, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anchang B, Hart TDP, Bendall SC, et al. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protoc. 2016;11(7):1264–1279. [DOI] [PubMed] [Google Scholar]
- 99.Pezzotti N, Höllt T, Lelieveldt B, Eisemann E, Vilanova A. Hierarchical Stochastic Neighbor Embedding. Comput Graph Forum. 2016;35(3):21–30. [Google Scholar]
- 100.Höllt T, Pezzotti N, van Unen V, et al. Cytosplore: Interactive Immune Cell Phenotyping for Large Single-Cell Datasets. Comput Graph Forum. 2016;35(3):171–180. [Google Scholar]
- 101.Li N, van Unen V, Höllt T, et al. Mass cytometry reveals innate lymphoid cell differentiation pathways in the human fetal intestine. J Exp Med. 2018;215(5):1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pezzotti N, Lelieveldt BPF, L van der Maaten, Hollt T, Eisemann E, Vilanova A Approximated and User Steerable tSNE for Progressive Visual Analytics. IEEE Trans Vis Comput Graph. 2017;23(7):1739–1752. [DOI] [PubMed] [Google Scholar]
- 103.Roederer M, Nozzi JL, Nason MC. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79A(2):167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roederer M PestleDocumentation. October 2011. [Google Scholar]
- 105.Gibbs A, Leeansyah E, Introini A, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017;10(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shah NM, Imami N, Johnson MR. Progesterone Modulation of Pregnancy-Related Immune Responses. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lugli E, Roederer M, Cossarizza A. Data analysis in flow cytometry: The future just started. Cytometry A. 2010;77A(7):705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perraudeau F, Risso D, Street K, Purdom E, Dudoit S. Bioconductor workflow for single-cell RNA sequencing: Normalization, dimensionality reduction, clustering, and lineage inference. F1000Research. 2017;6:1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.AlJanahi AA, Danielsen M, Dunbar CE. An Introduction to the Analysis of Single-Cell RNA-Sequencing Data. Mol Ther - Methods Clin Dev. 2018;10:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Poirion OB, Zhu X, Ching T, Garmire L. Single-Cell Transcriptomics Bioinformatics and Computational Challenges. Front Genet. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brennecke P, Anders S, Kim JK, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10(11):1093–1095. [DOI] [PubMed] [Google Scholar]
- 112.Treutlein B, Brownfield DG, Wu AR, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leng N, Choi J, Chu L-F, Thomson JA, Kendziorski C, Stewart R. OEFinder: a user interface to identify and visualize ordering effects in single-cell RNA-seq data. Bioinformatics. 2016;32(9):1408–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. [DOI] [PubMed] [Google Scholar]
- 115.Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–160. [DOI] [PubMed] [Google Scholar]
- 116.Barron M, Li J. Identifying and removing the cell-cycle effect from single-cell RNA-Sequencing data. Sci Rep. 2016;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ding B, Zheng L, Zhu Y, et al. Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics. 2015;31(13):2225–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vallejos CA, Marioni JC, Richardson S. BASiCS: Bayesian Analysis of Single-Cell Sequencing Data. Morris Q, ed. PLOS Comput Biol. 2015;11(6):e1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Katayama S, Töhönen V, Linnarsson S, Kere J. SAMstrt: statistical test for differential expression in single-cell transcriptome with spike-in normalization. Bioinformatics. 2013;29(22):2943–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11(7):740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fan J, Salathia N, Liu R, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods. 2016;13(3):241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schurch NJ, Schofield P, Gierliński M, et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA. 2016;22(6):839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bacher R, Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haque A, Engel J, Teichmann SA, Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zappia L, Phipson B, Oshlack A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. Schneidman D, ed. PLOS Comput Biol. 2018;14(6):e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pierson E, Yau C. ZIFA: Dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 2015;16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shin J, Berg DA, Zhu Y, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17(3):360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Welch JD, Hartemink AJ, Prins JF. SLICER: inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome Biol. 2016;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marco E, Karp RL, Guo G, et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci. 2014;111(52):E5643–E5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Han Y, Gao S, Muegge K, Zhang W, Zhou B. Advanced Applications of RNA Sequencing and Challenges. Bioinforma Biol Insights. 2015;9s1:BBI.S28991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 134.Mootha VK, Lindgren CM, Eriksson K-F, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. [DOI] [PubMed] [Google Scholar]
- 135.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao S, Wang X. TAPPA: topological analysis of pathway phenotype association. Bioinformatics. 2007;23(22):3100–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haynes WA, Higdon R, Stanberry L, Collins D, Kolker E. Differential Expression Analysis for Pathways. Bonneau R, ed. PLoS Comput Biol. 2013;9(3):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67(7):1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ren X, Kang B, Zhang Z. Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol. 2018;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X, Park J, Susztak K, Zhang NR, Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kallio MA, Tuimala JT, Hupponen T, et al. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Spjuth O, Bongcam-Rudloff E, Hernández GC, et al. Experiences with workflows for automating data-intensive bioinformatics. Biol Direct. 2015;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bianchi V, Ceol A, Ogier AGE, et al. Integrated Systems for NGS Data Management and Analysis: Open Issues and Available Solutions. Front Genet. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blankenberg D, Kuster GV, Coraor N, et al. Galaxy: A Web-Based Genome Analysis Tool for Experimentalists In: Ausubel FM, Brent R, Kingston RE, et al. , eds. Current Protocols in Molecular Biology. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Giardine B Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Goecks J, Nekrutenko A, Taylor J, Galaxy Team T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using Galaxy to Perform Large-Scale Interactive Data Analyses In: Baxevanis AD, Davison DB, Page RDM, Petsko GA, Stein LD, Stormo GD, eds. Current Protocols in Bioinformatics. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vento-Tormo R, Efremova M, Botting RA, et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563(7731):347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Suryawanshi H, Morozov P, Straus A, et al. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv. 2018;4(10):eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Andrews S FastQC. 2010.
- 153.Gregory J Hannon. FASTX-Toolkit: FASTQ/A short-reads pre-processing tools.
- 154.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An Extensive Evaluation of Read Trimming Effects on Illumina NGS Data Analysis. Seo J-S, ed. PLoS ONE. 2013;8(12):e85024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lindgreen S AdapterRemoval: Easy Cleaning of Next Generation Sequencing Reads. BMC Res Notes. 2012;5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schmieder R, Lim YW, Rohwer F, Edwards R. TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics. 2010;11:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. [DOI] [PubMed] [Google Scholar]
- 164.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. October 2015:btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464(7289):773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Leng N, Dawson JA, Thomson JA, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]