Abstract
The growth plate provides a substantial source of mesenchymal cells in the endosteal marrow space during endochondral ossification. The current model postulates that a group of chondrocytes in the hypertrophic zone can escape from apoptosis and transform into cells that eventually become osteoblasts in an area beneath the growth plate. The growth plate is composed of cells with various morphologies; particularly, at the periphery of the growth plate immediately adjacent to the perichondrium are ‘borderline’ chondrocytes, which align perpendicularly to other chondrocytes. However, in vivo cell fates of these special chondrocytes have not been revealed. Here we show that borderline chondrocytes in growth plates behave as transient mesenchymal precursor cells for osteoblasts and marrow stromal cells. A single cell RNA-seq analysis revealed subpopulations of Col2a1-creER-marked neonatal chondrocytes and their cell-type specific markers. A tamoxifen pulse to Pthrp-creER mice in the neonatal stage (before the resting zone was formed) preferentially marked borderline chondrocytes. Following the chase, these cells marched into the nascent marrow space, expanded in the metaphyseal marrow and became Col(2.3kb)-GFP+ osteoblasts and Cxcl12-GFPhigh reticular stromal ‘CAR’ cells. Interestingly, these borderline chondrocyte-derived marrow cells were short-lived, as they were significantly reduced during adulthood. These findings demonstrate based on in vivo lineage-tracing experiments that borderline chondrocytes in the peripheral growth plate are a particularly important route for producing osteoblasts and marrow stromal cells in growing murine endochondral bones. A special microenvironment neighboring the osteogenic perichondrium might endow these chondrocytes with an enhanced potential to differentiate into marrow mesenchymal cells.
Keywords: Genetic animal models, Developmental modeling, Osteoblasts, Stromal/Stem Cells, Growth plate
Introduction
Growth of bones and their marrow space requires a constant supply of mesenchymal cells to build the new stromal compartment. The growth plate can provide a substantial source of mesenchymal cells in endochondral ossification, which has been supported by a series of murine lineage-tracing studies using cre/creER transgenes active in chondrocytes and their close relatives, such as Sox9-creER, type II collagen (Col2)-creER, aggrecan (Acan)-creER, type X collagen (Col10)-cre/creER(1–3). Our lineage-tracing study using a Pthrp-creER line demonstrated that PTHrP+ resting chondrocytes can establish columnar chondrocytes, some of which can further differentiate into osteoblasts and marrow stromal cells through chondrocyte-to-osteoblast transformation(4). However, these columnar chondrocytes of the central growth plate can contribute only to a minor fraction of marrow mesenchymal cells in adulthood; therefore, chondrocytes in other locations may contribute more significantly in earlier stages. ‘Borderline’ chondrocytes at the periphery of the growth plate in a special microenvironment immediately adjacent to the perichondrium(5) can produce osteoblast-like cells in cultured conditions. In this study, we set out to reveal cellular heterogeneity of neonatal chondrocytes by a single cell RNA-seq analysis, and in vivo cell fates of borderline chondrocytes by taking advantage of a Pthrp-creER line.
Materials and Methods
Mice.
Pthrp-creER(4), Cxcl12GFP/+(6) mice have been described elsewhere. Col1a1(2.3kb)-GFP (JAX013134), Col2a1-creER (JAX006774), Rosa26-CAG-loxP-stop-loxP-tdTomato (Ai14: R26R-tdTomato, JAX007914) mice were acquired from the Jackson laboratory. FVB/N mice were acquired from the Charles River Laboratories. All procedures were conducted in compliance with the Guidelines for the Care and Use of Laboratory Animals approved by the University of Michigan’s Institutional Animal Care and Use Committee (IACUC), protocol 7681 (Ono) and the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. All mice were housed in a specific pathogen-free condition, and analyzed in a mixed background. For neonatal experiments, tamoxifen (Sigma T5648) or 4-hydroxytamoxifen (4-OHT, Sigma H6278) was dissolved in sunflower seed oil (Sigma S5007) and injected intraperitoneally (0.125 – 0.25mg) shortly after birth. For prenatal experiments, tamoxifen and progesterone (Sigma P3972) were dissolved and injected intraperitoneally (4mg each) at an indicated embryonic day. Mice were used for analysis regardless of the sex. No statistical method was used to predetermine sample size. Sample size was determined on the basis of previous literature and our previous experience to give sufficient standard deviations of the mean so as not to miss a biologically important difference between groups. The experiments were not randomized. All of the available mice of the desired genotypes were used for experiments. The investigators were not blinded during experiments and outcome assessment. One femur from each mouse was arbitrarily chosen for histological analysis. Genotypes were not particularly highlighted during quantification.
Histology.
Frozen sections were prepared using a cryostat (Leica CM1850) and adhered on positively charged glass slides (Fisherbrand ColorFrost Plus). Sections were stained with anti-active Caspase-3 polyclonal (1:250; Promega PR-G7481), anti-Runx2 polyclonal (1:100; Novus Biologicals NBP1–89104) or anti-Fra1 polyclonal (1:100; Invitrogen PA5–40361) antibodies overnight at 4°C, followed by staining with appropriate Alexa Fluor 488/647-conjugated secondary antibodies for 3 hours at 4°C and DAPI (4’,6-diamidino-2-phenylindole, 5μg/ml, Invitrogen D1306). In situ hybridization was performed with RNAscope 2.5 HD Reagent kit Brown (Advanced Cell Diagnostics 322300) using following probes: Col2a1 (407221), Acan (439101) and Col10a1 (426181) according to the manufacturer’s protocol. Sections were analyzed using an automated inverted fluorescence microscope with a structured illumination system (Zeiss Axio Observer Z1 with ApoTome.2 system). Images were typically tile-scanned with a motorized stage, Z-stacked and reconstructed by a maximum intensity projection (MIP) function. Differential interference contrast (DIC) was used for objectives higher than 10x. Serial sections spanning over 1mm thickness (up to 20 sections) of distal femurs were used for cell quantification.
Single cell RNA-seq analysis.
Femur growth plate cells were dissociated enzymatically (Sigma/Roche, Liberase TM) and loaded onto a five laser BD FACSAriaIII high-speed cell sorter with a 100μm nozzle. Approximately 18,000 tdTomato+ cells were loaded onto 10X Genomics Chromium 3’ Single Cell Gene Expression Solution v3 (10X Genomics Inc., Pleasanton, CA) microfluidics chip. The 10X Cell Ranger output matrix file was analyzed by Seurat R package(7) with cell cycle regression. For alignment purposes, we used a custom genome fasta and index file by including the sequences of tdTomato-WPRE (woodchuck hepatitis virus post-transcriptional regulatory element, a 3’UTR of the tdTomato sequence) to the mouse genome (mm10). The single cell dataset discussed in this publication are accessible in the NCBI’s Gene Expression Omnibus under accession number GSE125464.
Results
Single cell RNA-seq characterization of neonatal growth plate chondrocytes
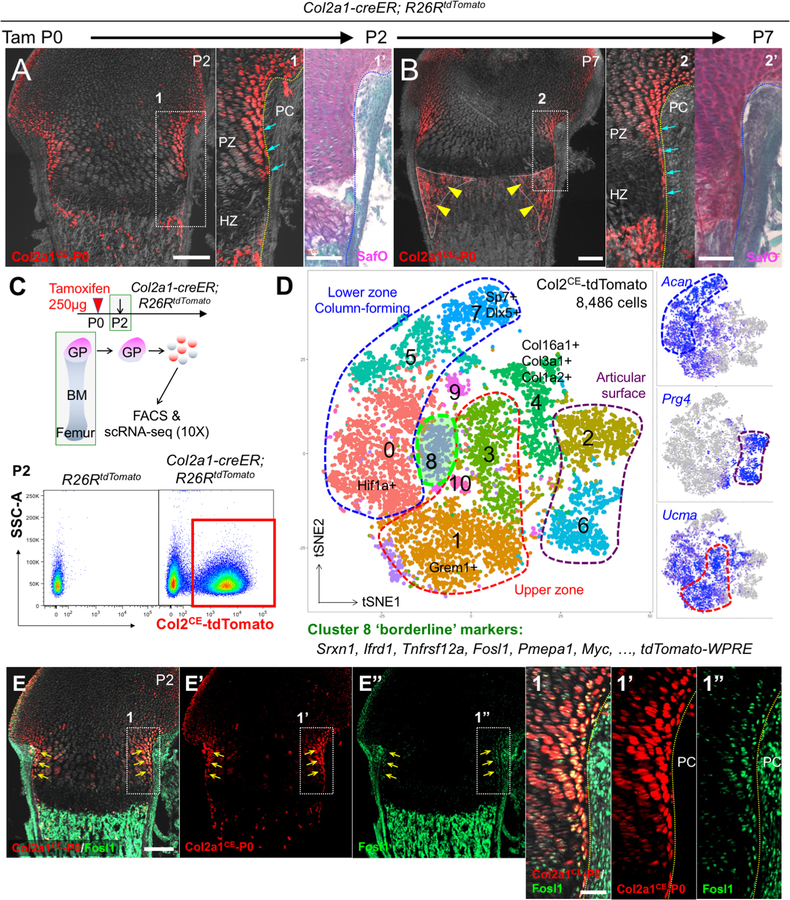
Col2a1-creER marks a group of early skeletal progenitors in growing bones during the perinatal stage(2). A tamoxifen pulse to Col2a1-creER; R26RtdTomato mice at P0 predominantly marked chondrocytes at the periphery of the growth plate, including those termed as borderline chondrocytes located immediately adjacent to the perichondrium, shortly after tamoxifen injection at P2 (Fig.1A). After a week of chase at P7, these chondrocytes continued to occupy the periphery of the growth plate in a borderline-enriched manner (Fig.1B, arrows), and their descendants contributed to well-defined reverse triangular areas adjacent to the metaphyseal endosteal surface (Fig.1B, arrowheads). Therefore, Col2a1-creER can mark a group of chondrocytes including precursors for borderline chondrocytes in the neonatal growth plate, which further contribute to mesenchymal cells in the marrow space.
Figure 1. Single cell RNA-seq characterization of neonatal growth plate chondrocytes.
(A,B) Cell-fate analysis of Col2a1-creER+ neonatal growth plate chondrocytes (P0-pulsed) at P2 (A) and P7 (B). PC: perichondrium, PZ: proliferating zone, HZ: hypertrophic zone. Right panels: SafraninO & FastGreen staining of identical sections. Arrows: tdTomato+ borderline chondrocytes, arrowheads: tdTomato+ cells in peripheral metaphyseal marrow space. Scale bars: 200μm (left panels), 100μm (right panels). n=3 mice for each experiment.
(C,D) Single cell RNA-seq analysis of Col2a1-creER+ neonatal growth plate chondrocytes. (D) t-SNE-based visualization of major classes of FACS-sorted Col2a1CE-tdTomato+ single cells at P2 (Cluster 0–10). Cluster 0,5,7: lower zone column-forming chondrocytes, Cluster1,3: upper zone chondrocytes, Cluster 2,6: articular surface chondrocytes, Cluster 4: collagen-rich articular cells, Cluster 8: borderline chondrocytes, Cluster 9,10: cells in cell cycle. Dots: individual cells, color: cell type. Green contour: borderline chondrocytes. Right panels: feature plots of Acan, Prg4 and Ucma. Blue: high expression, grey: no expression. n=8,486 cells.
(E) In situ validation of the identified marker, Fosl1. Col2a1-creER; R26RtdTomato distal femurs at P2 (P0-pulsed). (E’,E”): tdTomato, Fosl1-Alexa488 single channel, (1–1”): magnified views of the dotted area. Scale bars: 200μm (E-E”), 50μm (1–1”). n=3 mice.
We utilized a single cell RNA-seq analysis to define the characteristics of borderline chondrocytes. Flow cytometry analysis of cells dissociated from Col2a1-creER; R26RtdTomato femur growth plates at P2 (pulsed at P0) revealed that tdTomato+ cells accounted for approximately two-thirds of harvested cells (Fig.1C). We subsequently profiled 8,486 Col2a1CE-tdTomato+ single cells isolated by fluorescence-activated cell sorting (FACS) using the 10X Genomics Chromium platform. An initial t-distributed stochastic neighbor embedding (t-SNE) analysis revealed 11 clusters (Fig.1D). Of the 9 clusters of interest, we discovered three clusters of column-forming chondrocytes in the lower zone growth plate abundant in Acan (Cluster 0,5,7), two clusters of chondrocytes in the upper zone growth plate abundant in Ucma (Cluster 1,3)(8,9) and two cluster of cells in the articular surface abundant in Prg4 (Cluster 2,6)(10) (Fig.1D, Fig.S1A). Interestingly, chondrocytes in Cluster 8 were abundant in tdTomato-WPRE and exhibited a borderline state between the upper and lower zone growth plate; their cell-type specific markers included Srxn1, Ifrd1, Tnfrsf12a, Fosl1, Pmepa1 and Myc (Fig.S1B). These cells expressed genes encoding secreted factors Pthlh (Pthlh is also known as Pthrp) and Cxcl14, but not Ihh, Grem1 or Cxcl12 (Fig.S1A,C) Histological validation of an identified marker revealed that FOSL1 (also knowns as Fra1) was indeed expressed at a particularly high level by tdTomatobright chondrocytes at the periphery of the growth plate (Fig.1E). Moreover, these cells did not express hypertrophic markers Col10a1 or Sp7 (Fig.S1D). Therefore, these analyses reveal fundamental transcriptional heterogeneity and cellular diversity of neonatal growth plate chondrocytes.
Pthrp-creER specifically marks borderline chondrocytes in the fetal and neonatal stage
PTHrP is initially expressed in the periarticular region of the fetal and neonatal growth plate, and later in the resting zone of the postnatal growth plate(4).
First, we set out to determine cell fates of fetal PTHrP+ cells by in vivo lineage-tracing experiments. We injected tamoxifen to Pthrp-creER; R26RtdTomato mice carrying Cxcl12GFP/+ reporter at embryonic day 12.5 (E12.5) and analyzed these mice at postnatal day 9 (P9) (Fig.S2A). The descendant of PTHrP+ periarticular cells at E12.5 (PTHrPCE-E12.5 cells) became a small number of cells on the metaphyseal endosteal region (Fig.S2B). Second, we pulsed these mice at later fetal stages at E14.5 or E17.5. Both PTHrPCE-E14.5 and PTHrPCE-E17.5 cells became cells on the metaphyseal endosteal region, Cxcl12-GFPhigh stromal cells in the metaphyseal marrow space and cells on the articular surface (Fig.S2C,D). Therefore, Pthrp-creER marks precursors for metaphyseal marrow mesenchymal cells during the fetal stage, while also marking precursors for articular chondrocytes in later fetal stages. These precursors might reside in the perichondrium, therefore representing a discrete population from borderline chondrocytes.
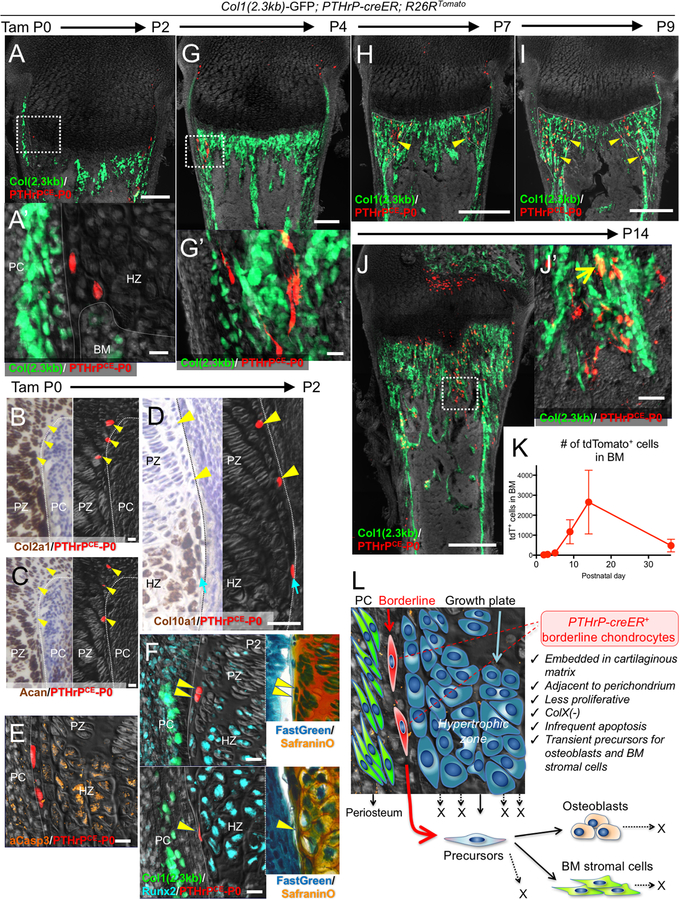
Second, we pulsed Pthrp-creER; R26RtdTomato mice carrying Col1(2.3kb)-GFP reporter at birth (postnatal day 0, P0) and analyzed these mice at 48 hours later at P2. A small number of chondrocytes at the periphery of the growth plate, including ‘borderline chondrocytes’ adjoining the perichondrium, was marked by tdTomato (PTHrPCE-P0 cells, 25.6±5.8 tdTomato+ cells per growth plate) (Fig.2A). These PTHrPCE-P0 cells were localized within the cartilage, expressing type II collagen (Col2a1) (18/18 tdTomato+ cells examined, 100%) and aggrecan (Acan) (21/23 tdTomato+ cells examined, 91%); some of them also expressed type X collagen (Col10a1) mRNAs (4/25 tdTomato+ cells examined, 16%) (Fig.2B–D). PTHrPCE-P0 cells underwent apoptosis less frequently; only a small fraction of these tdTomato+ cells were positive for activated Caspase 3 (4.4±8.8% of PTHrPCE-P0 cells, in contrast to 27.0±8.2% of hypertrophic chondrocytes) (Fig.2E, Fig.S3A). These tdTomato+ cells were embedded in the cartilaginous matrix, with a fraction of them expressing transcription factors RUNX2 (13.8±10.5%) (Fig.2F, Fig.S3B). Following the chase, PTHrPCE-P0 cells started to move into the metaphyseal marrow space (Fig.S3C, Fig.2G). To determine the cell cycle status of tdTomato+ peripheral chondrocytes, we performed an EdU label-exclusion assay. Pthrp-creER; R26R-tdTomato mice were first pulsed with tamoxifen at P0, then serially pulsed with EdU for three days and analyzed at P3. A great majority of tdTomato+ cells were resistant to EdU incorporation (Fig.S3D, EdU+; 4.6±4.4% of tdTomato+ cells, compared to 97.7±0.7% of cells in the adjacent perichondrium, n=3 mice), indicating that most of these peripheral chondrocytes were not dividing. Therefore, borderline chondrocytes can passively translocate to the marrow space without involving active cell proliferation.
Figure 2. Pthrp-creER+ borderline chondrocytes in the neonatal stage behave as transient mesenchymal precursor cells.
Cell-fate analysis of Pthrp-creER+ borderline chondrocytes (P0-pulsed) at P2 (A-F), P4 (G), P7 (H), P9 (I) and P14 (J). (A,F-J): Col1(2.3kb)-GFP; Pthrp-creER; R26RtdTomato distal femurs, (B-E): Pthrp-creER; R26RtdTomato distal femurs. (A’,G’,J’): magnified views of the dotted areas. In (B-D), left panels (bright field) and right panels (dark field) represent the identical sections. Right panels in (F): SafraninO & FastGreen staining of left panels. Arrowheads in (B-F): tdTomato+ borderline chondrocytes denoting identical cells. PC: perichondrium, PZ: proliferating zone, HZ: hypertrophic zone. Arrowheads in (H,I): tdTomato+ cells in peripheral metaphyseal marrow space, arrow in (J): GFP+tdTomato+ osteoblasts. Scale bars: 200μm (A,G), 500μm (H-J), 20μm (A-F), 50μm (J’). n=3 mice for each experiment. (K): Quantification of tdTomato+ marrow cells at each time point. n=6 (P2), n=4 (P4), n=3 (P5, P9, P14, P36) mice, data are presented as mean ± S.D. (L): Concluding diagram. Borderline chondrocytes in growth plates can translocate into marrow space and behave as transient precursors for osteoblasts and stromal cells by escaping hypertrophy and apoptosis.
Neonatal borderline chondrocytes behave as transient mesenchymal precursor cells
We interrogated fates of PTHrPCE-P0 cells at sequential time points after the pulse. After 5 days of chase, PTHrPCE-P0 cells further moved into the endosteal region, some of which became Cxcl12-GFPhigh stromal cells in the adjacent marrow space (Fig.S4A). After a week of chase at P7, PTHrPCE-P0 cells became Col1(2.3kb)-GFP+ osteoblasts on the metaphyseal trabecular bone surface and reticular stromal cells in the inter-trabecular marrow space (Fig.2H). After 9 days of chase at P9, PTHrPCE-P0 cells occupied a well-defined reverse triangular area of the spongiosa at the corner of the hypertrophic zone and the endosteal surface, demonstrating that borderline chondrocytes can particularly contribute to marrow mesenchymal cells of the peripheral areas (Fig.2I). After two weeks of chase at P14, PTHrPCE-P0 cells continued to become Col1(2.3kb)-GFP+ osteoblasts and Cxcl12-GFPhigh stromal cells in the metaphyseal marrow space (Fig.2J, Fig.S4B). Quantification of Col1(2.3kb)-GFP+tdTomato+ and Cxcl12-GFPhightdTomato+ cells after being pulsed at P0 or P6 demonstrated that PTHrPCE-P0 cells, but not PTHrPCE-P6 cells, became osteoblasts and marrow stromal cells at this stage, indicating that these tdTomato+ cells were derived from neonatal borderline chondrocytes (Fig.S4C). In addition, columnar chondrocytes derived from PTHrPCE-P0 cells resting chondrocytes had not yet reached the entire length of the growth plate at this stage. We further chased PTHrPCE-P0 cells for a longer period into adulthood. At 5 weeks of chase at P36, PTHrPCE-P0 cells sporadically remained as Col1(2.3kb)-GFP+ osteoblasts and Cxcl12-GFPhigh stromal cells in the metaphyseal marrow space, without any expansion of tdTomato+ areas (Fig.S4D,E), although we could no longer discriminate the origin of these cells at this stage because columnar chondrocytes originating from these resting chondrocytes had already reached the primary spongiosa. After 9.5 weeks of chase at P67, a vast majority of PTHrPCE-P0 cells disappeared from the marrow space, with only a small number of tdTomato+ cells remaining on trabecular bone surfaces (Fig.S4F). Quantification of tdTomato+ cells in the marrow space revealed that PTHrPCE-P0 cells transiently increased their number in the marrow space at P14, then drastically decreased their number toward P36 (Fig.2K). Taken together, these data suggest that PTHrP+ borderline chondrocytes behave as mesenchymal precursor cells for osteoblasts and marrow stromal cells that transiently expand in the marrow space during the early phase of postnatal bone growth.
Discussion
Our lineage-tracing study provides evidence that borderline chondrocytes in the fetal and neonatal growth plate can generate osteoblasts and marrow stromal cells in vivo in a mouse model. Further, our single cell RNA-seq analysis reveals cellular identity of these special chondrocytes. Chondrocytes at the periphery of the growth plate appear to represent an important pathway to generate marrow mesenchymal cells particularly in rapidly growing endochondral bones (see diagram, Fig.2L); these cells go through a unique state expressing Col10a1 without hypertrophy on their way to becoming mesenchymal cells in the marrow space. In combination with our previous finding that hypertrophic chondrocytes in the central growth plate can produce marrow mesenchymal cells predominantly in the adult stage(4), we speculate that different routes of chondrocytes are utilized to generate osteoblasts and marrow stromal cells in a stage-specific manner. Additionally, our data indicate that Pthrp-creER+ borderline chondrocytes represent a non-self-renewing subset of Col2a1-creER+ chondrocytes; we observed that borderline chondrocytes-derived marrow mesenchymal cells could only transiently persist within the marrow space. More immature Col2a1+PTHrPneg chondrocytes within the growth plate such as Grem1+ cells in the upper zone(11), rather than immature perichondrial cells, are likely to serve as precursors for PTHrP+ borderline chondrocytes that continue to feed into the nascent marrow space. Additionally, Pthrp-creER can mark perichondrial precursor cells during the fetal stage. Marked by the same Pthrp-creER transgene at different time points, the characteristics of PTHrP+ borderline chondrocytes appear to be fundamentally different from those of PTHrP+ resting chondrocytes that can self-renew long term within the resting zone. We speculate that differentiation of borderline chondrocytes occurs more effectively than transdifferentiation of hypertrophic chondrocytes in generating mesenchymal cells of the marrow space. How PTHrP+ chondrocytes at the periphery and the center are functionally different needs to be addressed through further studies.
Supplementary Material
Acknowledgements
We thank T. Nagasawa of Osaka University for providing Cxcl12-GFP mice. This research was supported by NIH R01DE026666 to N.O. and R03DE027421 to W.O. Authors’ roles: study design & conduct, data collection, analysis & interpretation: K.M., M.N., Y.M. and N.O. Drafting manuscript: K.M. and N.O., revising manuscript content: W.O. Approving final version of manuscript: K.M., M.N., Y.M., W.O. and N.O. N.O. takes responsibility for the integrity of the data analysis.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A. August 2014;111(33):12097–102. Epub 2014/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. December 2014;16(12):1157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. December 2014;10(12):e1004820 Epub 2014/12/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuhashi K, Ono W, Matsushita Y, Sakagami N, Takahashi A, Saunders TL, et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. November 2018;563(7730):254–8. Epub 2018/10/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianco P, Cancedda FD, Riminucci M, Cancedda R. Bone formation via cartilage models: the “borderline” chondrocyte. Matrix Biol. July 1998;17(3):185–92. [DOI] [PubMed] [Google Scholar]
- 6.Ara T, Itoi M, Kawabata K, Egawa T, Tokoyoda K, Sugiyama T, et al. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J Immunol. May 2003;170(9):4649–55. [DOI] [PubMed] [Google Scholar]
- 7.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. June 2018;36(5):411–20. Epub 2018/04/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tagariello A, Luther J, Streiter M, Didt-Koziel L, Wuelling M, Surmann-Schmitt C, et al. Ucma--A novel secreted factor represents a highly specific marker for distal chondrocytes. Matrix Biol. Jan 2008;27(1):3–11. Epub 2007/07/13. [DOI] [PubMed] [Google Scholar]
- 9.Surmann-Schmitt C, Dietz U, Kireva T, Adam N, Park J, Tagariello A, et al. Ucma, a novel secreted cartilage-specific protein with implications in osteogenesis. J Biol Chem. March 2008;283(11):7082–93. Epub 2007/12/21. [DOI] [PubMed] [Google Scholar]
- 10.Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, et al. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nat Genet. November 1999;23(3):319–22. [DOI] [PubMed] [Google Scholar]
- 11.Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. January 2015;160(1–2):269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.