Abstract
To personalize treatment for renal cell carcinoma (RCC), it would be ideal to confirm the activity of druggable protein pathways within individual tumors. We have developed a high-resolution nanoimmunoassay (NIA) to measure protein activity with high precision in scant specimens (eg, fine needle aspirates [FNAs]). Here, we used NIA to determine whether protein activation varied in different regions of RCC tumors. Since most RCC therapies target angiogenesis by inhibiting the vascular endothelial growth factor (VEGF) receptor, we quantified phosphorylation of extracellular signal-regulated kinase (ERK), a downstream effector of the VEGF signaling pathway. In 90 ex vivo FNA biopsies sampled from multiple regions of 38 primary clear cell RCC tumors, ERK phosphorylation differed among patients. In contrast, within individual patients, we found limited intratumoral heterogeneity of ERK phosphorylation. Our results suggest that measuring ERK in a single FNA may be representative of ERK activity in different regions of the same tumor. As diagnostic and therapeutic protein biomarkers are being sought, NIA measurements of protein signaling may increase the clinical utility of renal mass biopsy and allow for the application of precision oncology for patients with localized and advanced RCC.
Keywords: Kidney cancer, Renal cell carcinoma, Fine needle aspirate, Extracellular signal-regulated kinase, Protein signaling, Intratumor heterogeneity, Intertumor heterogeneity, Phosphorylation
Patient summary:
In this report, we applied a new approach to measure the activity of extracellular signal-regulated kinase (ERK), a key cancer signaling protein, in different areas within kidney cancers. We found that ERK activity varied between patients, but that different regions within individual kidney tumors showed similar ERK activity. This suggests that a single biopsy of renal cell carcinoma may be sufficient to measure protein signaling activity to aid in precision oncology approaches.
Tissue sampling approaches have been proposed to address the fundamental need for diagnostic and predictive biomarkers in renal cell carcinoma (RCC). However, few patients with a renal mass receive a biopsy to confirm the diagnosis of RCC due to concerns of nondiagnostic procedures, sampling errors, and the associated risk of complications [1]. In cases where a renal mass biopsy is performed, the scant amount of biopsy material poses a challenge for quantitative protein analysis. Moreover, genomic multiregion analyses have demonstrated significant spatial intratumoral heterogeneity (ITH) in primary and metastatic RCC tumors, and raise concerns that the area sampled may not be representative of the entire tumor [2]. Nevertheless, some of these genomic alterations suggest convergence on a limited set of protein signaling pathways required for RCC progression. Many alterations that constitute genomic heterogeneity may represent passenger mutations that do not change the cancer phenotype. Whether genomic ITH translates into heterogeneity of protein expression and activity in signaling pathways that are relevant to the diagnosis of RCC and to the efficacy of targeted therapies is not well characterized. Here, to determine how protein signals can vary between patients and within each tumor, we measure the activity of the extracellular signal-regulated kinase (ERK) protein in fine needle aspirates (FNAs) from different regions of primary clear cell RCC tumors.
ERK is a downstream effector in the vascular endothelial growth factor (VEGF) signaling pathway (Supplementary Fig. 1). ERK is activated by phosphorylation of two amino acids: a threonine and a tyrosine residue. ERK phosphorylation is associated with tumor invasiveness, growth, and angiogenesis in RCC [3,4]. Treatment with VEGF receptor inhibitors reduces ERK phosphorylation in RCC cells [5–8]. We hypothesized that within a kidney tumor, there may be limited variation in the activation of key signaling proteins such as ERK. As standard techniques to measure protein phosphorylation require relatively large amounts of tissue and often cannot distinguish between single- and multiphosphorylated protein isoforms, we developed a highly sensitive assay, nanoimmunoassay (NIA), to resolve and quantify the phosphorylation status of proteins in FNA specimens [9]. Here, we apply NIA to evaluate the ITH of ERK activity in RCC tumors.
We performed ex vivo FNAs in up to three regions of 38 primary clear cell RCC tumors, resulting in 90 total biopsies. NIA was used to quantify ERK activation in FNA lysates, with up to three technical replicate measurements per sample. NIA analysis detected three isoforms of ERK1: unphosphorylated ERK1, monophosphorylated ERK1 (pERK1), and dual-phosphorylated ERK1 (ppERK1). NIA detected four isoforms of ERK2: unphosphorylated ERK2 (ERK2a); a second, more acidic isoform of unphosphorylated ERK2 (ERK2b) [10]; monophosphorylated ERK2 (pERK2); and dual-phosphorylated ERK2 (ppERK2; Fig. 1). Activity was assessed by calculating the relative abundance of each isoform, expressed as a percentage of total ERK1 or ERK2. ITH was quantified as the standard deviation (SD) of the abundance of each isoform across the regional FNAs from each tumor. For patient demographics, tumor characteristics, and statistical methods, see Supplementary Table 1 and the Supplementary materials (Methods).
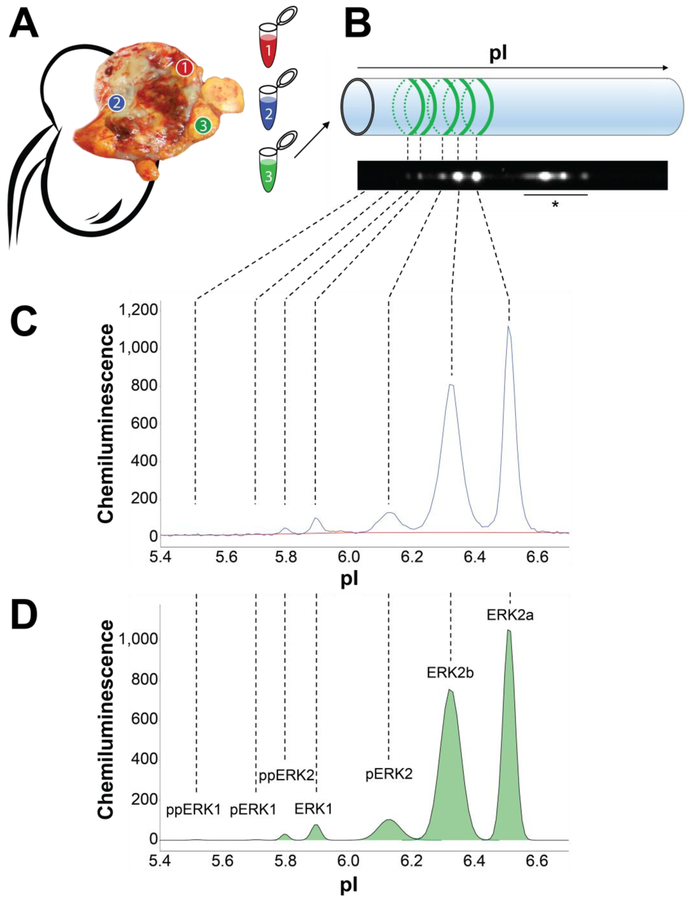
Fig. 1 –
Multiregion FNA sampling and nanoimmunoassay analysis. (A) Immediately after surgical extirpation, the sample was bivalved and FNA biopsies were obtained in grossly representative regions. (B) FNAs were lysed and analyzed using charge separation by isoelectric focusing. ERK isoforms were detected by chemiluminescence using a pan-ERK antibody. (C) NIA chemiluminescence intensity trace (blue curve) of one representative RCC FNA. Background intensity was subtracted by using a baseline fit (red curve). (D) Area under the curve (green) was calculated for each peak using a fitted curve (black), and peaks were assigned to specific ERK isoforms according to their isoelectric point (pI). Percent phosphorylation was calculated by dividing the area under the curve of each isoform by the sum of the area of all isoforms of ERK1 or ERK2, respectively. ERK = extracellular signal-regulated kinase; FNA = fine needle aspirate; RCC = renal cell carcinoma. * Nonspecific antibody binding.
ERK1 was infrequently activated, with no detectable phosphorylation in 58% of samples (Supplementary Fig. 2A). In contrast, ERK2 was phosphorylated in all but one sample. Interestingly, monophosphorylated ERK2 (pERK2, abundance ranging from 0% to 58%) was more abundant than dual-phosphorylated ERK2 (ppERK2, 0–26%). For each tumor, ITH of ERK2 activation was low (estimated SD: 3 percentage points for pERK2, 2 percentage points for ppERK2) compared with the overall variability in ERK2 activation across all 38 tumors (SD: 11 percentage points for pERK2, 4 percentage points for ppERK2; Fig. 2). Activation of ERK1 and ERK2 in adjacent normal kidney tissue was not markedly different from tumor tissue: in some cases, ERK activation was higher in RCC, in other cases, it was higher in adjacent normal kidney (Supplementary Fig. 2B). ERK activation in RCC tumors or its difference to adjacent normal kidney was not associated with tumor size, stage, or grade (Supplementary Fig. 3).
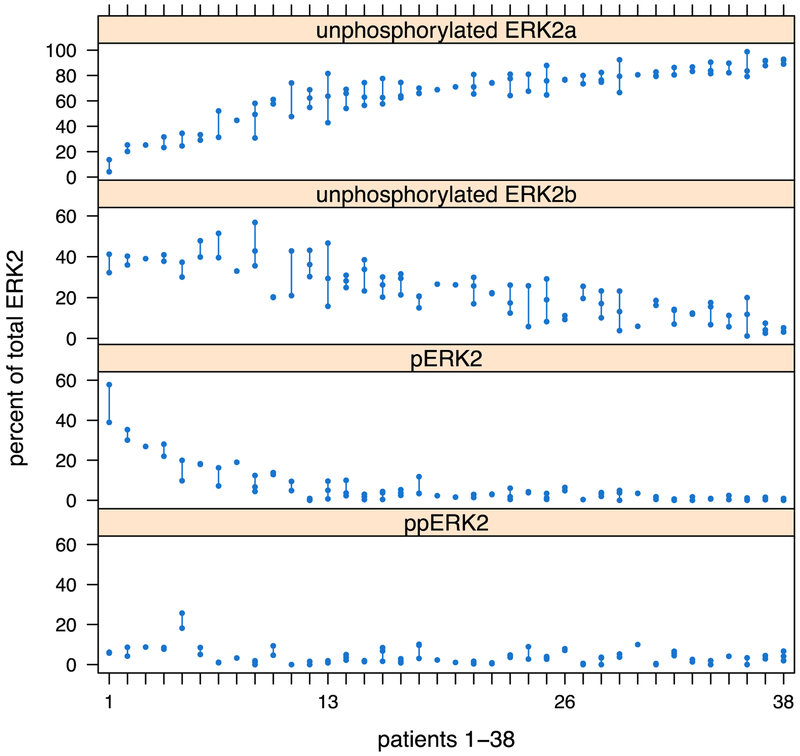
Fig. 2 –
Variation in ERK2 phosphorylation across 90 fine needle aspirates from 38 clear cell RCC tumors. Each column represents an individual tumor, while each dot representing a distinct region sampled. Differences between regions of the same tumor are illustrated by the line connecting each dot. Overall, variability between tumors was much higher than that seen across regions of an individual tumor. ERK = extracellular signal-regulated kinase; RCC = renal cell carcinoma.
To our knowledge, this is the first report to quantify protein activation in one of the largest cohorts to investigate multiregion samples of RCC tumors. We note several important findings. First, NIA provided the required sensitivity and retained high levels of precision across technical replicates in scant FNA specimens (overall estimated SD: 0.9 percentage points, range 0.4–1.4 percentage points for individual isoforms; Supplementary Fig. 4). Using this approach, we found that ERK1 was frequently inactive, whereas ERK2 had a broad range of activation states between RCC tumors. Moreover, we determined that differences in ERK2 activity were mostly due to differences in percentage of the monophosphorylated isoform (pERK2). Initially, it seems surprising that there is a predominance of monophosphorylated ERK2 and that this finding could be clinically significant. However, we note that prior antibody-based studies of ERK activation in RCC could not distinguish between mono- and dual-phosphorylated isoforms, and our prior work in leukemia patients showed that changes in the abundance of monophosphorylated ERK2, in the absence of dual-phosphorylated ERK2, could predict response to targeted therapy [9]. Second, we found little ITH in ERK activation among different regions of individual tumors. This suggests that a single biopsy may be sufficient to assess ERK protein signaling activity within the entire tumor. We acknowledge that this initial study reports ITH data from a single protein (ERK). Future work will be required to validate these data in larger cohorts, including metastatic tumors and protein markers for RCC subtype, tumor aggressiveness, and activity of additional signaling proteins in pathways that are targeted by drug therapies for RCC. There are additional caveats to our findings. We analyzed ex vivo FNAs, and assay performance may change with in vivo percutaneous biopsies. NIA of FNAs provides a signal average of all the cells in the specimen. To determine which cell types show specific activation states of signaling pathways, complementary protein analysis methods in intact tumor tissue or single cell approaches will be needed. Based on our simulations (see the Supplementary material), it is unlikely that our findings are due to inadequate spatial sampling. Some previous studies reported a correlation between ERK activity and clinicopathologic parameters (eg, tumor size or stage). Our study does not identify such correlations, possibly because previous studies used qualitative scoring of immunohistochemistry staining intensity in fixed tissue sections, whereas our study precisely quantifies ERK activity in fresh-frozen FNAs. Last, future studies comparing ITH on the protein and genomic levels will help identify the subset of genomic alterations that are relevant for the tumor phenotype and may lead to complementary genetic biomarkers for the diagnosis and treatment of RCC. In summary, we found little ITH in ERK activation using a sensitive NIA to compare multiple regions of clear cell RCCs.
The extent of genomic ITH in RCC and other solid tumors has raised concerns that a single biopsy may not be representative of the tumor phenotype. Our proof-of-concept study of ERK suggests that ITH is limited on the level of protein activity. Sensitive and precise quantification of protein expression and signaling activity in tumor FNAs by NIA may eventually facilitate future precision medicine approaches for RCC patients. To personalize treatment of localized renal masses, the ability to interrogate multiple protein markers in a single FNA may expand the clinical utility of renal mass biopsy and could aid in diagnosis and clinical decision making by identifying RCC tumors requiring surgery versus benign tumors amenable to “watchful waiting.” For metastatic RCC, it would be ideal to confirm the activity of protein pathways within individual tumors to select the drug that precisely targets the aberrant signaling pathway(s) in the tumor. NIA enables measuring such protein activity in scant specimens obtained by minimally invasive sampling.
Supplementary Material
Acknowledgments:
This work is in the memory of Professor Kuang Huan Fan. We wish to thank Dean W. Felsher for his support of this work. We wish to thank Rachael Curtis, David Praharaj, and the Translational Applications Service Center at Stanford University for their contributions.
Funding/Support and role of the sponsor: This work was supported by the US National Institute of Health/NCI R21169964 (J.L., A.C.F.), NIH/NCI U54 CCNE 19907502 (A.C.F., C.R.H., T.J.M.), NIH/NCI K23 140722 (A.C.F.), NIH/NCI R21 CA185804 (CS), and the Stanford Clinical and Translational Science Award (CTSA) to Spectrum UL1 TR001085 (L.S.). The CTSA program is led by the National Center for Advancing Translational Sciences (NCATS) at the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional support was provided by the Stanford Cancer Institute Translational Developmental Cancer Research Award (J.L., A.C.F.), the Stanford Translational Research and Applied Medicine (TRAM) Program Pilot Grant (J.L., A.C.F.), the Department of Defense Peer Reviewed Cancer Research Program 2015 11981051 (J.L.), and an ASCO Career Development Award (A.C.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: John T. Leppert certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Alice Fan: founder of Molecular Decisions Inc., with Ownership Stock; patent filed: U.S. App. No. 62/480,044.
References
- [1].Leppert JT, Hanley J, Wagner TH, et al. Utilization of renal mass biopsy in patients with renal cell carcinoma. Urology 2014;83:774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Campbell L, Nuttall R, Griffiths D, Gumbleton M. Activated extracellular signal-regulated kinase is an independent prognostic factor in clinically confined renal cell carcinoma. Cancer 2009;115: 3457–67. [DOI] [PubMed] [Google Scholar]
- [4].Huang D, Ding Y, Luo W-M, et al. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Res 2008;68:81–8. [DOI] [PubMed] [Google Scholar]
- [5].Sakai I, Miyake H, Fujisawa M. Acquired resistance to sunitinib in human renal cell carcinoma cells is mediated by constitutive activation of signal transduction pathways associated with tumour cell proliferation. BJU Int 2013;112:E211–20. [DOI] [PubMed] [Google Scholar]
- [6].Diaz-Montero CM, Mao FJ, Barnard J, et al. MEK inhibition abrogates sunitinib resistance in a renal cell carcinoma patient-derived xenograft model. Br J Cancer 2016;115:920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhou L, Liu X-D, Sun M, et al. Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma. Oncogene 2016;35:2687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao H, Nolley R, Chan AMW, Rankin EB, Peehl DM. Cabozantinib inhibits tumor growth and metastasis of a patient-derived xenograft model of papillary renal cell carcinoma with MET mutation. Cancer Biol Ther 2017;18:863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fan AC, Deb-Basu D, Orban MW, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med 2009;15:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].O’Neill RA, Bhamidipati A, Bi X, et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc Natl Acad Sci U S A 2006;103:16153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.