Abstract
This study investigated the prevalence of Orientia tsutsugamushi, Anaplasma phagocytophilum, and Leptospira interrogans in wild rodents through molecular detection using organ samples and through serological assay using blood samples of mice collected from two distinct sites in Gwangju Metropolitan City, Republic of Korea (ROK). A total of 47 wild rodents, identified as Apodemus agrarius (A. agrarius), were captured from June to August 2016. The seroprevalence of antibodies against bacterial pathogens in A. agrarius sera was analyzed; 17.4% (8/46) were identified as O. tsutsugamushi through indirect immunofluorescence assay and 2.2% (1/46) were identified as Leptospira species through passive hemagglutination assay. Using polymerase chain reaction, the spleen, kidney and blood samples were investigated for the presence of O. tsutsugamushi, A. phagocytophilum, and L. interrogans. Out of the 47 A. agrarius, 19.1% (9/47) were positive for A. phagocytophilum and 6.4% (3/47) were positive for L. interrogans, while none were positive for O. tsutsugamushi. Four out of 46 (8.7%) blood samples, six out of 45 (13.3%) spleen samples, and one out of 47 (2.1%) kidney samples were positive for A. phagocytophilum. Three out of 47 (6.4%) kidney samples were positive for L. interrogans. The sequencing results of PCR positive samples demonstrated > 99% similarity with A. phagocytophilum and L. interrogans sequences. A. phagocytophilum was mostly detected in the spleen, whereas L. interrogans was mostly detected in the kidneys. Notably, A. phagocytophilum and L. interrogans were detected in A. agrarius living in close proximity to humans in the metropolitan suburban areas. The results of this study indicate that rodent-borne bacteria may be present in wild rodents in the metropolitan suburban areas of ROK.
Introduction
Rodents are known carriers of zoonotic pathogenic agents that are usually transmitted to humans through direct or indirect contact [1].
Scrub typhus, caused by Orientia tsutsugamushi, is an infectious disease and has become one of the most prevalent human diseases in the Asia-Pacific region, including in the Republic of Korea (ROK) [2]. Since 2004, the disease has spread widely in the southwestern provinces of the country and is endemic in nature [3]. In 1995, 274 cases of scrub typhus were reported in ROK, whereas 10,365 cases were reported in 2013, indicating a 38.1-fold increase in the incidence of this disease [4].
Anaplasma phagocytophilum causes human granulocytic anaplasmosis (HGA). A. phagocytophilum has been detected in Hemaphysalis longicornis (H. longicornis), Ixodes nipponensis (I. nipponensis), and Ixodes persulcatus (I. persulcatus) ticks in ROK [5]. HGA was first identified in the United States in 1994 [6]. The incidence of anaplasmosis has also increased from 1.4 cases per million in 2000 to 6.1 cases per million in 2010 [7, 8]. The first HGA case in ROK was reported in 2013 [9].
Leptospirosis has been one of the most important endemic diseases in ROK since 1984. Wild rodents, particularly A. agrarius, are a common source of infection in ROK, especially during the harvest season in the rural areas. The prevalence of Leptospira infection in field rodents has been previously investigated via detection of leptospiral DNA in rodent kidneys [10, 11].
The presence of scrub typhus, anaplasmosis, and leptospirosis in ROK indicates that O. tsutsugamushi, A. phagocytophilum, and Leptospira interrogans have infected both rodents and humans. Therefore, it is necessary to monitor the prevalence of pathogens in animal hosts, such as wild rodents, which harbor such pathogens and vectors like ticks and mites involved in disease transmission. A limited number of studies have evaluated the prevalence of pathogens in wild rodents in the suburban areas of metropolitan cities in ROK. Especially, studies on the issue of organ tropism (i.e., prevalence rates of O. tsutsugamushi, A. phagocytophilum, and L. interrogans in different tissues) are very limited.
This study investigated the prevalence of O. tsutsugamushi, A. phagocytophilum, and L. interrogans in wild rodents through molecular detection of the pathogens in the organs of rodents collected from two distinct sites in Gwangju City, ROK. This study may broaden the understanding of health risks associated with wild rodents living in close proximity to humans.
Materials and methods
Collection of wild rodent samples
The rodents were captured using Sherman traps (3” × 3.5” × 9’, USA) at two sylvatic habitats located in the suburban areas of Buk-gu (35° 10′ 12″ N, 126° 54′ 36″ E) and Gwangsan-gu (35° 8′ 5″ N, 126° 47′ 40″ E) in the Gwangju metropolitan city of ROK from June to August 2016. The captured rodents were numbered sequentially. All the animals were euthanized by inhalation of 5% isoflurane in accordance with an approved animal use protocol. Blood, spleen, and kidney tissues were obtained and stored at– 80 °C for use in future experiments. All the samples were tested for the presence of O. tsutsugamushi, A. phagocytophilum, and L. interrogans.
Ethics statement
This study was approved by the institutional review board (IRB) of Chosun University. Approval for the capturing and experiments of the animals were obtained from Chosun University Institutional Animal Care and Use Committee (CIACUC) (approval no. CIACUC2016-A0003).
Indirect immunofluorescence assay (IFA)
The blood samples were centrifuged at 3000 rpm for 20 min, to separate the serum which was stored at 4 °C. Indirect immunofluorescence assay (IFA) and passive hemagglutination assay (PHA) were performed after 24–36 h of euthanizing the mice.
For IFA, 10 μL of sera from each A. agrairus was used in two-fold serial dilutions of 1:16 to 1:2048 in phosphate-buffered saline (PBS, pH 7.2; Welgene Inc., Korea). The diluted sera were incubated on antigen slides at 37 °C for 30 min in a humidified chamber, washed twice with sterile PBS, and again washed twice with distilled water. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Sigma, St. Louis, Missouri, USA) (25 μL) was added to each slide, incubated at 37 °C for 30 min in a humidified chamber, washed twice with sterile PBS and distilled water, and air-dried. A mounting medium (Sigma) was added to each slide which was covered with a cover slip. The slides were examined for specific spots under a fluorescence microscope (Carl Zeiss, Oberkochen, Germany). A cutoff titer of 1:16 was used for interpreting infection in the wild rodents. In-house antigen slides for O. tsutsugamushi were provided by the Korea Center for Disease Control (KCDC). O. tsutsugamushi-infected human serum and normal human serum provided by KCDC were used as positive and negative controls. The in-house O. tsutsugamushi-injected mouse serum and normal mouse serum were used as positive and negative controls.
Passive hemagglutination assay (PHA)
For PHA, Genedia Lepto PHA (Green Cross, Seoul, Korea) kit reagents were used. Serum (25 μL) from each rodent was added to a 96-well plate, and diluted to 1:80 in a dissolved solution of Genedia Lepto kit. Sheep blood (75 μL) containing red blood cells sensitized to Leptospira species was placed in diluted serum for the agglutination assay. The Ag-Ab agglutination reaction at 1:80 dilution was found to be positive for leptospirosis.
DNA extraction
A small piece of tissue (around ~50 mg) in 200 μL of ATL buffer of QIAamp Tissue & Blood Mini Kit (QIAGEN, Germany) was homogenized with a bead beater (BioSpec 3110BX Mini-Beadbeater-1 High-Energy Cell Disrupter). The tissue lysates were incubated at 56 °C overnight, and the genomic DNA was extracted using QIAamp Tissue & Blood Mini Kit following the manufacturer’s instructions.
Polymerase chain reaction (PCR)
To detect the presence of O. tsutsugamushi DNA, the 16S ribosomal RNA gene (rrs) and 56 kDa gene (56 kDa) of O. tsutsugamushi were targeted. PCR was also performed using the INNOPLEX TSUTSU detection kit (cat. no. IPC10040; Intron Biotechnology, Korea). The kit contained primer sets designed to detect the 475-bp fragment of a gene encoding the 56 kDa antigen of O. tsutsugamushi. PCR was performed with 2 μL of DNA extract and 18 μL of distilled water treated with diethyl pyrocarbonate (DEPC, Gendepot, Barker, Texas, USA) in PCR premix tubes following the manufacturer’s instructions.
The heat shock protein gene (groEL) and ankyrin-related protein gene (ankA) were targeted to detect A. phagocytophilum. To detect the presence of L. interrogans DNA, RNA polymerase subunit beta (rpoB), outer membrane lipoprotein (LipL32), and DNA gyrase subunit B (gyrB) were targeted. All the primers used for the specific target genes, PCR conditions, and product sizes are given in Table 1. Nested PCR (nPCR) was performed in 20 μL reaction volumes using the AccuPowerR PCR PreMix (Bioneer Corp., Korea). Each PCR mixture consisted of 16 μL of distilled water, 1 μL of each primer (10 pmol/μL), and 2 μL of DNA template.
Table 1. Nucleotide sequences and PCR conditions for the detection of rodent-borne bacteria in the rodent tissue.
| Bacteria | Target gene | Primer name | Nucleotide sequence (5′-3′) | Product size (bp) | PCR profile (°C/s) | Reference | |||
|---|---|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | Cycles | ||||||
| Orientia tsutsugamushi | rrs | ot-16sRF1 | AGGGATGATAATGACAGTACCTACAG | 199 | 94/60 | 57/30 | 72/45 | 36 | In this study |
| ot-16sRR1 | CCTCTACCATACTCTAGCCTAACAG | ||||||||
| 56 kDa | 56BO-144F | YGYAGAATCTRCTCGCTTGG | 1250 | 94/60 | 60/60 | 72/60 | 35 | In this study | |
| 56BO-1395R | AGCTAMCCCTRCACCAABAC | ||||||||
| 56BO-406F | CCWCCTCARCCTACTATRATGC | 680 | 94/30 | 61/30 | 72/45 | 30 | |||
| 56BO-1088R | GCWGCTGCTRCTGCTTCTTG | ||||||||
|
Anaplasma phagocytophilum |
groEL | GRO607F | GAAGATGCWGTWGGWTGTACKGC | 688 | 95/30 | 54/30 | 72/60 | 30 | [12] |
| GRO1294R | AGMGCTTCWCCTTCWACRTCYTC | ||||||||
| GRO677F | ATTACTCAGAGTGCTTCTCARTG | 445 | 95/30 | 57/30 | 72/60 | 30 | |||
| GRO1121R | TGCATACCRTCAGTYTTTTCAAC | ||||||||
| ankA | ANK-F1 | GAAGAAATTACAACTCCTGAAG | 705 | 95/30 | 53/30 | 72/60 | 35 | [13] | |
| ANK-R1 | CAGCCAGATGCAGTAACGTG | ||||||||
| ANK-F2 | TTGACCGCTGAAGCACTAAC | 664 | 95/30 | 52/30 | 72/60 | 5 | |||
| ANK-R2 | ACCATTTGCTTCTTGAGGAG | 95/30 | 54/30 | 72/60 | 25 | ||||
|
Leptospira interrogans |
rpoB | rpoB-1889F | GTTCCAACATGCAACGYCAR | 1649 | 94/60 | 52/60 | 72/60 | 35 | In this study |
| rpoB-3537R | GTTGAAGGATTCRGGRATAC | ||||||||
| rpoB-2438F | TYATGCCKTGGGAAGGWTAC | 1023 | 94/30 | 56/30 | 72/45 | 30 | |||
| rpoB-3460R | GCATRTCRTCKGACTTGATG | ||||||||
| LipL32 | hap1-435F | GGGAATACGTAGAMGTTCG | 1435 | 94/60 | 56/60 | 72/60 | 35 | In this study | |
| hap1-1870R | GTTTATAGTAGGTTGAAGCTTG | ||||||||
| L-hap1-217F | CCGTGATTTTCCTAACTAAGG | 848 | 94/30 | 58/20 | 72/45 | 30 | [14] | ||
| L-hap1-218R | CAGATTACTTAGTCGCGTCAGA | ||||||||
| gyrB | LeptoB2F | TGAGCCAAGAAGAAACAAGCTACA | 500 | 94/30 | 59/30 | 72/45 | 35 | [15] | |
| LeptoB504R | MATGGTTCCRCTTTCCGAAGA | ||||||||
All amplifications were performed in an AB thermal cycler (Applied Biosystem, Foster City, CA, USA). The amplified products were separated by electrophoresis on a 1.2% agarose gel and stained with ethidium bromide for visualization.
Sequencing and phylogenetic analysis
Sequencing of PCR amplicons from rodent-borne pathogens was conducted by Solgent Inc. (Daejeon, Korea). The obtained sequences were compared for similarity with the sequences deposited in the GenBank using BLAST. The gene sequences excluding the primer regions were aligned using the multisequence alignment program of Lasergene version 8 (DNASTAR, USA), and phylogenetic analysis was performed using the MEGA 6 software.
Phylogenetic trees were constructed using ClustalW of the MegAlign Program (DNASTAR, USA) based on the alignments of positive gene sequences using the neighbour-joining method. Bootstrap analysis (1,000 replicates) was performed according to the Kimura 2-parameter method. Pairwise alignments were performed with an open-gap penalty of 10 and a gap extension penalty of 0.5.
Results
Wild rodent collection
A total of 47 wild rodents were captured from June to August 2016 at the two trapping sites in the vicinity of Gwangju city in ROK. The wild rodents collected were: 16 mice (captured in June), 18 mice (captured in July), and 13 mice (captured in August). All the wild rodents were identified to be A. agrarius which were captured in fallow ground, around water, a boundary area between forest and field, and around a tomb.
Seropositivity for O. tsutsugamushi and L. interrogans in rodent serum
The serum samples collected from 46 A. agrarius were subjected to IFA. Eight samples out of 46, (17.4%) were seropositive for O. tsutsugamushi with a cutoff titer of ≥ 1:16 for IgG. In addition, PHA was performed for the detection of antibodies against Leptospira species and one sample out of 46 (2.2%) serum was positive with a cutoff titer of 1:160 (Table 2).
Table 2. Rate of positive bacterial infections in 46 Apodemus agrarius rodents, as indicated by serological assays.
| Name of positive sera | Seropositive | |
|---|---|---|
| IFA titer for O. tsutsugamushi a (% positive rate) | PHA for Leptospira species b (% positive rate) |
|
| 6–6 | 1:128 | – c |
| 6–7 | 1:64 | – |
| 7–4 | 1:16 | – |
| 7–13 | 1:16 | – |
| 7–17 | 1:16 | 1:160 |
| 7–18 | 1:32 | – |
| 8–7 | 1:256 | – |
| 8–9 | 1:64 | – |
| No. of positive sera | 8 (17.4) | 1 (2.2) |
a Cutoff titer of immunofluorescence assay, immunoglobulin G ≥ 1:16,
b Cutoff for passive hemagglutination assay ≥ 1:80,
c–: negative.
Molecular detection of bacterial pathogens in rodent tissue
The tissue samples were obtained from blood, spleen, and kidneys of 47 A. agrarius. Among the total mice samples, four blood samples out of 46 (8.7%), six spleen samples out of 45 (13.3%), and one kidney sample out of 47 (2.1%) was positive for A. phagocytophilum. Three kidney samples out of 47 (6.4%) were positive for L. interrogans, whereas and all the spleen and blood samples were negative. In contrary, all the A. agrarius tissues were negative for O. tsutsugamushi (Table 3). No co-infection was found in our study (S1 Table).
Table 3. Number of positive cases for Orientia tsutsugamushi, Anaplasma phagocytophilum, and Leptospira interrogans, among the 47 Apodemus agrarius rodents obtained by PCR targeting different genes.
| Specimen | No. of samples | O. tsutsugamushi | A. phagocytophilum | L. interrogans | |||||
|---|---|---|---|---|---|---|---|---|---|
| rrsa | TSUTSU Kitb | 56 kDac | groELd | ankAe | rpoBf | LipL32g | gyrBh | ||
| Blood | 46 | 0 | 0 | 0 | 4 | 4 | 0 | 0 | NAi |
| Spleen | 45 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | NA |
| Kidney | 47 | 0 | 0 | 0 | 0 | 1 | 3 | 2 | 2 |
| No. of positive rodents | 0 | 9 (19.1)j | 3 (6.4) | ||||||
a 16S ribosomal RNA,
b INNOPLEX TSUTSU detection kit for O. tsutsugamushi,
c 56 kDa gene,
d Heat shock protein chaperone,
e Ankyrin-related protein gene,
f RNase polymerase subunit beta,
g Outer membrane lipoprotein,
h DNA gyrase subunit B,
i NA: not available,
j % Positive rate.
Sequencing and phylogenetic analysis
The genes of rodent-borne bacteria were sequenced and aligned with the sequences obtained from the GenBank database using ClustalW. The neighbor-joining tree was constructed using the Kimura 2-parameter model (1,000 bootstrap replicates) in the MEGA 6 software.
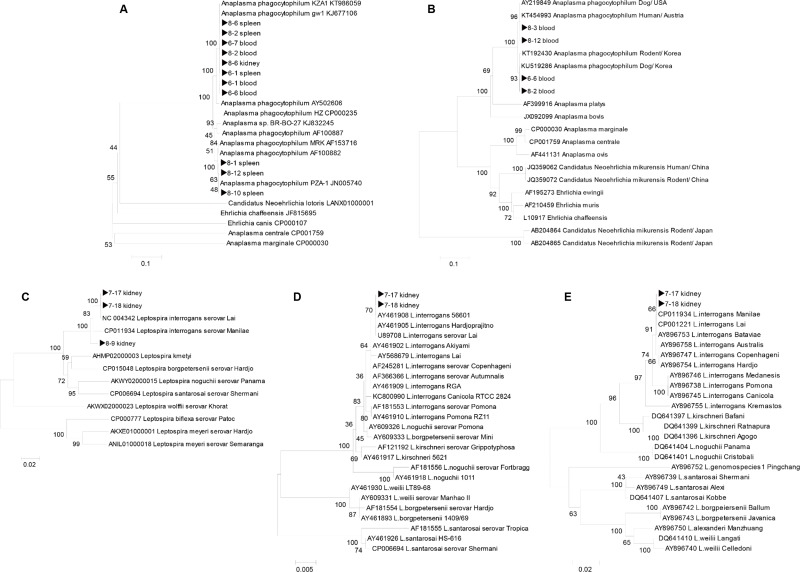
The ankA gene sequences collected from the rodent tissues demonstrated 99% similarity with that the of A. phagocytophilum strain isolated from humans in ROK (accession no. KJ677106 and KT986059, 100% bootstrap support, Fig 1A). The groEL gene sequences demonstrated 99% similarity with the A. phagocytophilum strain isolated from the rodents and dogs in ROK (accession no. KT192430 and KU519286, 93% bootstrap support, Fig 1B), and with A. phagocytophilum strain isolated from humans in Austria (accession no. KT454993, 96% bootstrap support, Fig 1B).
Fig 1. Phylogenetic trees generated based on nucleotide sequences of A. phagocytophilum and L. interrogans in tissues obtained from the Apodemus agrarius rodents captured in Gwangju.
(A) 560 bp of ankA. (B) 330 bp of groEL gene sequences for A. phagocytophilum. (C) 890 bp of rpoB gene. (D) 780 bp of LipL32 gene. (E) 400 bp of gyrB gene sequences for L. interrogans.
The rpoB gene sequences collected from the rodent tissues showed 99–100% similarities with L. interrogans serovar lai isolated from China (accession no. NC004342, 100% bootstrap support, Fig 1C), while the other samples exhibited 99–100% similarities with L. interrogans serovar manilae isolated from the mouse kidneys in Japan (accession no. CP011934, 100% bootstrap support, Fig 1C). The LipL32 gene sequences demonstrated 100% similarities with L. interrogans serovar lai isolated from sewage in China (accession no. AY461908) and with L. interrogans serovar hardjoprajitno isolated from humans in Indonesia (accession no. AY461905, 70% bootstrap support, Fig 1D). The gyrB gene sequences demonstrated 100% similarities with L. interrogans serovar lai isolated from sewage in China (accession no. CP001221) and with L. interrogans serovar manilae isolated from mouse kidneys in Japan (accession no. CP011934, 66% bootstrap support, Fig 1E).
Discussion
Scrub typhus, which is caused by O. tsutsugamushi, is transmitted to humans by the bites of chigger mites (Trombiculidae) [16]. The incidence of scrub typhus is influenced by multiple factors, such as the behavior and population density of Trombiculidae and wild rodents. Human activity may also influence the infection rates [17, 18]. A. agrarius is a dominant wild rodent, which harbors chigger mites carrying O. tsutsugamushi [19]. In this study, O. tsutsugamushi was not detected in 47 A. agrarius through PCR. Eight out of 46 (17.4%) sera were found to be seropositive through IFA with a cutoff titer of 1:16 for IgG. The gold standard for serological detection of scrub typhus is IFA [20] however, it lacks standardization and the use of variable cutoff titers has shown lab-wise variability in the results [21]. The usage varied significantly across different geographical areas where scrub typhus is endemic and the cutoff ranged from 1:10 to 1:400 [21]. Our IFA and PCR results did not corroborate because the time of wild rodents being infected was unclear. Kim, et al. [22] observed that IgG antibody titer increases abruptly over the first 2 weeks and reaches its peak at about 4 weeks in scrub typhus patients. The molecular techniques, such as PCR provide the highest sensitivity and specificity for the detection of scrub typhus, especially in the early period of infection, due to the specificity of primers and low detection limits [23–27]. Thus, although the antibody reaction was positive, molecular detection was not achieved in the positive mice. It may be preferable to perform both molecular and serological assays for precisely detecting the pathogens.
A. phagocytophilum has long been recognized as an animal and human pathogen and is one of the major concern to public health [28]. In previous studies, serological and molecular evidence indicated that A. phagocytophilum has spread to humans, rodents, and ticks in many Asian countries, including Korea, China, and Japan [29–32]. Chae et al. [5], have confirmed that ticks, rodents, and shrews near the Demilitarized Zone (DMZ) in Korea were positive for A. phagocytophilum, and 20 out of 403 rodent spleens (around ~5%) were tested positive for A. phagocytophilum in species-specific PCR assays. In the present study, nine out of 47 A. agrarius (19.1%) were positive for A. phagocytophilum. Moreover, six out of 45 spleen samples, four out of 46 blood samples, and one out of 47 kidney samples were positive for A. phagocytophilum. These results displayed 99% similarities with that of the isolates obtained from humans and dogs in ROK (Fig 1A and 1B). This suggests the possibility that A. agrarius may be the reservoirs of A. phagocytophilum in ROK.
L. interrogans causes leptospirosis in both humans and animals. The rodents asymptomatically carry the bacteria in their kidneys and excrete them in urine, thus contaminating the environment [33, 34]. L. interrogans was detected in only 6.4% of the kidney samples in this study. In addition, PHA results indicate that only one out of 46 (2.2%) blood samples and only one of the 3 PCR-positive kidney samples were serologically positive for Leptospira species. This suggests that the kidneys of A. agrarius may be the main reservoirs of L. interrogans. From the results of phylogenetic tree analysis, the positive samples demonstrated 99–100% similarity with serovar lai isolated from sewage in China and serovar manilae isolated from mouse kidney in Japan (Fig 1C–1E). Since 1984, L. interrogans isolates identified from A. agrarius field rodents were classified under serogroup Icterohaemorrhagiea as serovar lai and hongchon in ROK [35].
Our results demonstrated the rate of rodent-borne bacterial infections in different tissues of 47 A. agrarius collected from June to August 2016 in Gwangju City of ROK. A total of 12 (25.5%) mice were positive for A. phagocytophilum and L. interrogans as determined through PCR, and eight (17.4%) sera samples were seropositive as determined by IFA and PHA. A. phagocytophilum and L. interrogans were detected in A. agrarius living in close proximity to humans in the suburban areas, such as Gwangju City. Considering the tissue detection levels of A. phagocytophilum, the spleen was found to be a more sensitive organ to detect the presence of the pathogen. In contrast, the kidneys were more sensitive to detect the presence of L. interrogans. All the blood samples were found to be negative for the pathogens by PCR. This signifies that the overall rates of PCR detection can be lower if only blood samples were tested.
The serological assay of A. phagocytophilum was not performed in this study due to the lack of an antigen slide for A. phagocytophilum. The data were limited as only a few samples were obtained from only two sites. Future studies, using larger samples collected over a longer period of time, may lead to a better understanding of the nature and spread of these infectious pathogens. Further research is required to monitor bacterial prevalence rates in wild rodents, which serve as reservoir hosts for many infectious pathogens.
In conclusion, the present study indicates that rodent-borne bacterial infections circulating in wild rodent populations may be prevalent in the metropolitan suburban areas of ROK. In particular, the spleen was found to be more sensitive to detect the presence of A. phagocytophilum, whereas the kidneys were more sensitive for L. interrogans.
Supporting information
(DOCX)
(ZIP)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by a grant of the Korean Health Technology Research & Development Project, Ministry of Health & Welfare, Republic of Korea (HI16C2118) awarded to DMK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kurucz K, Madai M, Bali D, Hederics D, Horváth G, Kemenesi G, et al. Parallel Survey of Two Widespread Renal Syndrome-Causing Zoonoses: Leptospira spp. and Hantavirus in Urban Environment, Hungary. Vector-Borne and Zoonotic Diseases. 2018;18(4):200–5. 10.1089/vbz.2017.2204 [DOI] [PubMed] [Google Scholar]
- 2.Kelly DJ, Fuerst PA, Ching W-M, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clinical Infectious Diseases. 2009;48(Supplement_3):S203–S30. [DOI] [PubMed] [Google Scholar]
- 3.Lee HW, Cho PY, Moon SU, Na BK, Kang YJ, Sohn Y, et al. Current situation of scrub typhus in South Korea from 2001–2013. Parasit Vectors. 2015;8:238 Epub 2015/05/01. 10.1186/s13071-015-0858-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roh JY, Song BG, Park WI, Shin EH, Park C, Park MY, et al. Coincidence between geographical distribution of Leptotrombidium scutellare and scrub typhus incidence in South Korea. PLoS One. 2014;9(12):e113193 Epub 2014/12/17. 10.1371/journal.pone.0113193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chae J-S, Yu D-H, Shringi S, Klein TA, Kim H-C, Chong S-T, et al. Microbial pathogens in ticks, rodents and a shrew in northern Gyeonggi-do near the DMZ, Korea. Journal of veterinary science. 2008;9(3):285–93. 10.4142/jvs.2008.9.3.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S-M, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. Journal of clinical microbiology. 1994;32(3):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demma LJ, Holman RC, McQuiston JH, Krebs JW, Swerdlow DL. Epidemiology of human ehrlichiosis and anaplasmosis in the United States, 2001–2002. The American journal of tropical medicine and hygiene. 2005;73(2):400–9. [PubMed] [Google Scholar]
- 8.You M-J, Kim W-I, Cho H-S, Shin G-W, Hwang J-H, Lee C-S. Human Anaplasmosis in Acute Febrile Patients during Scrub Typhus Season in Korea. Infection & chemotherapy. 2015;47(3):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K-H, Yi J, Oh WS, Kim N-H, Choi SJ, Choe PG, et al. Human granulocytic anaplasmosis, South Korea, 2013. Emerging infectious diseases. 2014;20(10):1708 10.3201/eid2010.131680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yim EK, Kim YW, Lee JS, Chang I, Baek LJ, Song JW, et al. Detection of leptospiral DNA from field rodents by PCR. Journal of Bacteriology and Virology. 2003;33(3):177–81. [Google Scholar]
- 11.Chang WH, Kim YW, Oh HB, Cho MK, Kee SH, Kim HJ. Serovar identification and genetic characterization of Leptospira isolates by arbitrarily primed PCR and ribotyping. Journal of the Korean Society for Microbiology. 1999;34(4):409–21. [Google Scholar]
- 12.Pritt BS, Sloan LM, Johnson DKH, Munderloh UG, Paskewitz SM, McElroy KM, et al. Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. New England Journal of Medicine. 2011;365(5):422–9. 10.1056/NEJMoa1010493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massung RF, Levin ML, Munderloh UG, Silverman DJ, Lynch MJ, Gaywee JK, et al. Isolation and propagation of the Ap-Variant 1 strain of Anaplasma phagocytophilum in a tick cell line. Journal of clinical microbiology. 2007;45(7):2138–43. 10.1128/JCM.00478-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branger C, Blanchard B, Fillonneau C, Suard I, Aviat F, Chevallier B, et al. Polymerase chain reaction assay specific for pathogenic Leptospira based on the gene hap1 encoding the hemolysis-associated protein-1. FEMS microbiology letters. 2005;243(2):437–45. 10.1016/j.femsle.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 15.Slack AT, Symonds ML, Dohnt MF, Smythe LD. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC microbiology. 2006;6(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traub R, Wisseman CL Jr, Jones MR, O'Keefe JJ. The acquisition of Rickettsia tsutsugamushi by chiggers (trombiculid mites) during the feeding process. Annals of the New York Academy of Sciences. 1975;266(1):91–114. [DOI] [PubMed] [Google Scholar]
- 17.Ree H-I, Lee I-Y, Cho M-K. Determination of the vector species of tsutsugamushi disease in Korea. Korean J Parasitol. 1991;29(1):87–92. [DOI] [PubMed] [Google Scholar]
- 18.Lee IY, Kim HC, Lee Y-S, Seo JH, Lim JW, Yong TS, et al. Geographical distribution and relative abundance of vectors of scrub typhus in the Republic of Korea. The Korean journal of parasitology. 2009;47(4):381 10.3347/kjp.2009.47.4.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Łopucki R, Mróz I, Berliński Ł, Burzych M. Effects of urbanization on small-mammal communities and the population structure of synurbic species: an example of a medium-sized city. Canadian journal of zoology. 2013;91(8):554–61. [Google Scholar]
- 20.La Scola B, Raoult D. Laboratory diagnosis of rickettsioses: current approaches to diagnosis of old and new rickettsial diseases. Journal of clinical microbiology. 1997;35(11):2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blacksell SD, Bryant NJ, Paris DH, Doust JA, Sakoda Y, Day NP. Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: a lack of consensus leads to a lot of confusion. Clinical infectious diseases. 2007;44(3):391–401. 10.1086/510585 [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Lee YM, Back JH, Yang T, Lee J, Song HJ, et al. A serosurvey of Orientia tsutsugamushi from patients with scrub typhus. Clinical Microbiology and Infection. 2010;16(5):447–51. 10.1111/j.1469-0691.2009.02865.x [DOI] [PubMed] [Google Scholar]
- 23.Sugita Y, Nagatani T, Okuda K, Yoshida Y, Nakajima H. Diagnosis of typhus infection with Rickettsia tsutsugamushi by polymerase chain reaction. Journal of medical microbiology. 1992;37(5):357–60. 10.1099/00222615-37-5-357 [DOI] [PubMed] [Google Scholar]
- 24.Furuya Y, Yoshida Y, Katayama T, Yamamoto S, Kawamura A. Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. Journal of clinical microbiology. 1993;31(6):1637–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levett PN, Branch SL, Whittington CU, Edwards CN, Paxton H. Two methods for rapid serological diagnosis of acute leptospirosis. Clinical and diagnostic laboratory immunology. 2001;8(2):349–51. 10.1128/CDLI.8.2.349-351.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyokawa T, Ohnishi M, Koizumi N. Diagnosis of acute leptospirosis. Expert review of anti-infective therapy. 2011;9(1):111–21. 10.1586/eri.10.151 [DOI] [PubMed] [Google Scholar]
- 27.Luce-Fedrow A, Mullins K, Kostik AP, St John HK, Jiang J, Richards AL. Strategies for detecting rickettsiae and diagnosing rickettsial diseases. Future microbiology. 2015;10(4):537–64. 10.2217/fmb.14.141 [DOI] [PubMed] [Google Scholar]
- 28.Dumler JS, Madigan JE, Pusterla N, Bakken JS. Ehrlichioses in humans: epidemiology, clinical presentation, diagnosis, and treatment. Clinical infectious diseases. 2007;45(Supplement_1):S45–S51. [DOI] [PubMed] [Google Scholar]
- 29.Cao W-c, Zhan L, He J, Foley JE, De Vlas SJ, Wu X-m, et al. Natural Anaplasma phagocytophilum infection of ticks and rodents from a forest area of Jilin Province, China. The American journal of tropical medicine and hygiene. 2006;75(4):664–8. [PubMed] [Google Scholar]
- 30.CHAE JS, KIM CM, KIM EH, HUR EJ, Klein TA, KANG TK, et al. Molecular epidemiological study for tick‐borne disease (Ehrlichia and Anaplasma spp.) surveillance at selected US military training sites/installations in Korea. Annals of the New York Academy of Sciences. 2003;990(1):118–25. [DOI] [PubMed] [Google Scholar]
- 31.Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, et al. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Applied and environmental microbiology. 2006;72(2):1102–9. 10.1128/AEM.72.2.1102-1109.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohashi N, Inayoshi M, Kitamura K, Kawamori F, Kawaguchi D, Nishimura Y, et al. Anaplasma phagocytophilum–infected ticks, Japan. Emerging infectious diseases. 2005;11(11):1780 10.3201/eid1111.050407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Faria MT, Athanazio DA, Ramos EG, Silva EF, Reis MGd, Ko AI. Morphological alterations in the kidney of rats with natural and experimental Leptospira infection. Journal of comparative pathology. 2007;137(4):231–8. 10.1016/j.jcpa.2007.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Athanazio DA, Silva EF, Santos CS, Rocha GM, Vannier-Santos MA, McBride AJ, et al. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta tropica. 2008;105(2):176–80. 10.1016/j.actatropica.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 35.Cho M, Kee S, Song H, Kim K, Song K, Baek L, et al. Infection rate of Leptospira interrogans in the field rodent, Apodemus agrarius, in Korea. Epidemiology & Infection. 1998;121(3):685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(ZIP)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.