Abstract
Previous studies from others and us have demonstrated that CRISPR genome editing could offer a promising therapeutic strategy to restore dystrophin expression and function in the skeletal muscle and heart of Duchenne muscular dystrophy (DMD) mouse models. However, the long-term efficacy and safety of CRISPR genome-editing therapy for DMD has not been well established. We packaged both SaCas9 and guide RNA (gRNA) together into one AAVrh.74 vector, injected two such vectors (targeting intron 20 and intron 23, respectively) into mdx pups at day 3 and evaluated the mice at 19 months. We found that AAVrh.74-mediated life-long CRISPR genome editing in mdx mice restored dystrophin expression and improved cardiac function without inducing serious adverse effects. PCR analysis and targeted deep sequencing showed that the DSBs were mainly repaired by the precise ligation of the two cut sites. Serological and histological examination of major vital organs did not reveal any signs of tumor development or other deleterious defects arising from CRISPR genome editing. These results support that in vivo CRISPR genome editing could be developed as a safe therapeutic treatment for DMD and potentially other diseases.
Keywords: cardiomyopathy, Cas9, CRISPR, Duchenne muscular dystrophy, dystrophin, genome editing, heart
CRIPSR has been harnessed as a promising therapy for Duchenne muscular dystrophy, but its long-term efficacy and safety remain elusive. In this issue of Molecular Therapy, Han and colleagues (2019) demonstrate that systemic delivery of AAV-CRISPR-SaCas9 into dystrophic mouse pups improves cardiomyopathy without serious lesions at 19 months of age.
Introduction
CRISPR-mediated genome editing has been harnessed as an exciting therapeutic platform for a number of human diseases. Duchenne muscular dystrophy (DMD) is a progressive muscle-wasting disease affecting both skeletal and cardiac muscles in approximately 250–300 thousand young males worldwide.1 DMD is caused by mutations in the dystrophin gene.2 We and others have recently shown that it could be used to correct the genetic base of DMD in short-term animal models using intramuscular or systemic delivery of recombinant adeno-associated virus (rAAV).3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14
Although highly encouraging, these short-term studies did not reveal whether a one-time AAV-CRISPR treatment can lead to a life-long benefit as for the restoration of dystrophin expression and disease amelioration. As DMD is a chronic disease, it is important for a therapy to result in sustained dystrophin expression. Moreover, the long-term safety of AAV-CRISPR-mediated genome-editing therapy has not been well established in preclinical animal models. Recent studies showed that repair of double-strand break (DSB) induced by CRISPR leads to large deletions and complex genomic rearrangements at the target site,15, 16 potentially causing pathogenic consequences. Owing to the intrinsic infidelity of RNA-guided Cas9 targeting,17, 18, 19, 20, 21 continued expression of the active CRISPR components may result in accumulation of unwanted mutations at off-target sites. Moreover, two recent studies alarmed that CRISPR-edited cells may seed for tumor due to p53 deficiency.22, 23 Therefore, the long-term safety of CRISPR genome-editing therapy has been in question.
To address these critical questions, we injected AAVrh.74-SaCas9/guide RNA (gRNA) into 3-day-old mdx pups and examined dystrophin expression and disease pathology at 19 months of age. The mdx mouse carries a nonsense mutation in exon 23 of the dystrophin gene and this mutation can be removed by CRISPR targeting both intron 20 and 23 as we previously showed.
Results
Long-Term Rescue of Dystrophin Expression following Systemic AAV-CRISPR Therapy in mdx Mice
We administered rAAVrh.74-CRISPR,3, which was designed to remove exons 21–23 of the Dmd gene (Figure 1A), into mdx neonates at day 3 by intraperitoneal injection. The mice were maintained and studied along with untreated mdx littermates and wild-type (WT) controls for 19 months (close to the reported lifespan of mdx mice, ∼22 months24). Two of the five untreated mdx mice developed visible sarcoma in their hind limbs at 19 months of age, while none of the five treated mdx mice developed any visible signs of tumor throughout the experimental period (Figure S1). Previous studies also showed that aging mdx mice are prone to the development of spontaneous rhabdomyosarcoma.24
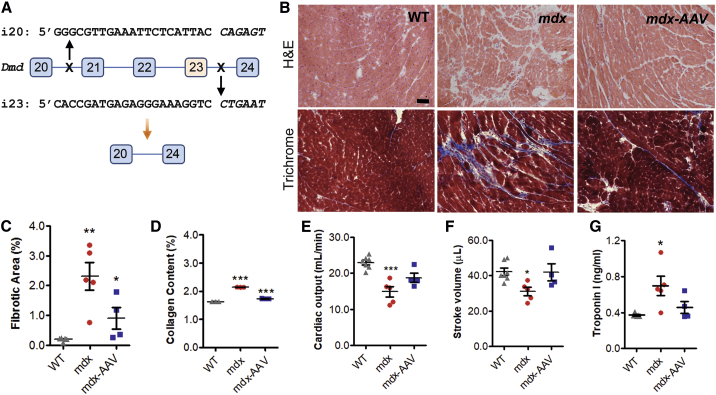
Figure 1.
Life-Long AAV-Mediated CRISPR Genome Editing Improved Cardiomyopathy in mdx Mice
(A) Scheme illustrating the CRISPR/SaCas9-mediated genome-editing strategy to delete the exons 21–23 in order to restore the dystrophin reading frame. The sequences for the two gRNAs were shown with the PAM sequences italicized. (B) Representative H&E staining and Masson’s trichrome staining images of heart sections from 19-month-old WT and mdx mice treated with or without rAAV-CRISPR (1 × 1012 vg, intraperitoneal [i.p.] at day 3). Scale bar, 50 μm. (C) Quantification of cardiac fibrotic area in trichrome stained heart sections. (D) Quantification of collagen content in Sirius red/fast green-stained heart sections. (E and F) Cardiac output (E) and stroke volume (F) were measured in 19-month-old WT and mdx mice by echocardiography. (G) Measurements of cardiac troponin I in the serum samples of mice at 19 months of age. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars indicate mean ± SEM.
At 19 months of age, the mice were sacrificed for genome-editing outcome and cardiac-histopathology analysis. H&E and Masson’s trichrome staining showed that hearts from the mdx mice had increased fibrosis as compared to those from WT mice (Figure 1B). Both the fibrotic area (Figure 1C) and collagen content (Figure 1D) in mdx hearts were significantly decreased by AAV-CRISPR genome editing. The cardiac function was assessed by echocardiography. The cardiac output and stroke volume in mdx mice were significantly improved following AAV-CRISPR treatment (Figures 1E and 1F). Moreover, reduced levels of cardiac troponin I in serum samples derived from the AAV-CRISPR-treated mice were measured in comparison to the untreated mdx mice (Figure 1G).
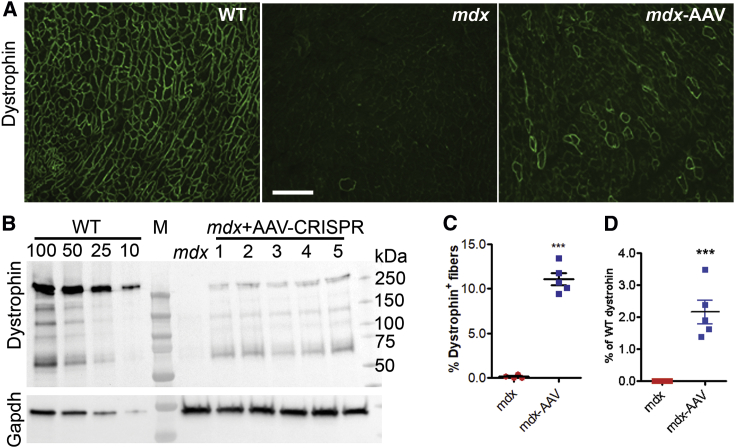
Immunofluorescence staining revealed that dystrophin expression was restored in 11.1% ± 0.7% of cardiomyocytes of the mdx mice at 19 months after AAV-CRISPR treatment (Figures 2A and 2C), whereas the untreated mdx mice showed little to no dystrophin-positive fibers (0.1% ± 0.1%, p < 0.0001). Western blot analysis showed that the total dystrophin expression was restored to 2.16% ± 0.37% of that in WT heart (Figures 2B and 2D). Consistent with the long-term restoration of dystrophin in the cardiac muscle at 19 months of age, quantification of the AAV vector genome showed that the highest AAV vector genome titer was present in the heart (Figure S2). Utrophin is a homolog of dystrophin, and increased expression of utrophin can compensate for the loss of dystrophin. To examine if the AAV-CRISPR altered the expression of utrophin and thus contributed to the functional improvement in the dystrophic heart, we performed western blot analysis and found that utrophin expression was not statistically affected by AAV-CRISPR treatment (Figure S3).
Figure 2.
Life-Long AAV-Mediated CRISPR Genome Editing Restored Dystrophin Expression in mdx Heart
(A) Representative immunofluorescence images of heart sections stained with the antibodies against dystrophin (green). Scale bar, 500 μm. (B) Western blot analysis of dystrophin expression in heart lysates from 19-month-old WT and mdx mice treated with or without rAAV-CRISPR (1 × 1012 vg, i.p. at day 3). Gapdh was used as a gel loading control. (C) Quantification of dystrophin-positive cardiomyocytes on immunofluorescence stained sections. (D) Densitometry quantification of relative expression of total dystrophin on western blot. Error bars indicate mean ± SEM.
Precision of the DSB Repair following AAV-CRISPR-Mediated Two DNA Cuts
One major concern with in vivo CRISPR genome editing is the cancerous potential of unexpected DSB repair events. First of all, we attempted to detect if any large deletions occurred by PCR analysis with primer pairs to amplify a genomic region of ∼2 kb at the target sites (Figure 3A). However, no smaller products were detected except the amplicon of expected size (Figure 3B). This suggests that either the rate of the large deletion events was below the detection limit or the cells with such large deletions had not survived during the course of 19 months of life.
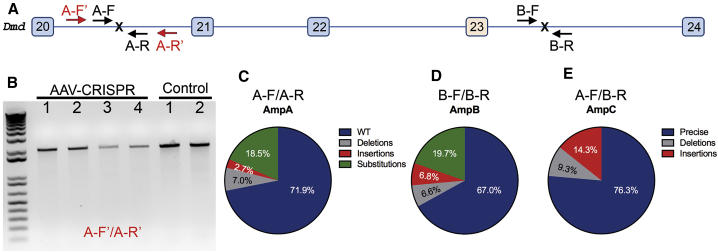
Figure 3.
CRISPR-Mediated DSBs at Dystrophin Locus in Dystrophic Mouse Heart Were Repaired Mainly by Precise Ligation of the Two Cut Ends
(A) Scheme illustrating the primers for PCR to detect larger deletions at the intron 20 target site (A-F′ and A-R′, 2 kbp), and for targeted deep-sequencing analysis (amplicon A, A-F/A-R; amplicon B, B-F/B-R; and amplicon C, A-F/B-R, 150 bp each). Since the unedited locus from A-F to B-R is over 23 kbp, PCR with A-F/B-R would not amplify the unedited DNA, while the correctly edited DNA would result in amplification of an ∼150-bp product. (B) Genomic DNA PCR showed only the expected 2 kbp product without smaller bands, indicating that no larger deletions at the target site were detected. (C–E) Targeted deep sequencing analyses of the amplicon A (C), B (D), and C (E) from pooled heart samples of five mdx mice treated with rAAV-CRISPR.
We then assessed the precision of the DSB repair at the junction of two edited sites by targeted next-generation deep sequencing (NGS). Three 150-bp amplicons (A, B, and C) with primers covering each of the target sites or the ligation products following deletion of exons 21–23 (Figure 3A) were subjected to NGS. For amplicons A and B, 28.1% and 33.0% of the reads carried small insertions or deletions (indels) or substitutions, respectively (Figures 3C and 3D). Surprisingly, for amplicon C, 76.3% of the total reads had the precise ligation product (e.g., without any indels at the ligation junction), while 14.3% had small insertions and 9.3% had small deletions (Figure 3E). These data suggest that the DSBs induced by CRISPR were repaired mainly by direct ligation of the two cut sites.
Undetectable Adverse Impact of Long-Term AAV-CRISPR Therapy in mdx Mice
To determine if long-term AAV-CRISPR treatment caused any kidney toxicity, we measured the blood urea nitrogen (BUN) and found that both mdx and AAV-CRISPR treated mdx mice showed similar levels of BUN as compared to WT mice (Figure 4A). Although mdx mice at 19 months of age displayed elevation in alanine transaminase (ALT) and aspartate transaminase (AST) as compared to WT mice, their levels were either decreased or unaltered following AAV-CRISPR treatment (Figures 4B and 4C), indicating that AAV-CRISPR genome editing did not result in increased liver toxicity in mdx mice. Previous studies showed that the elevation of serum ALT and AST in mdx mice are likely from injured muscle.25
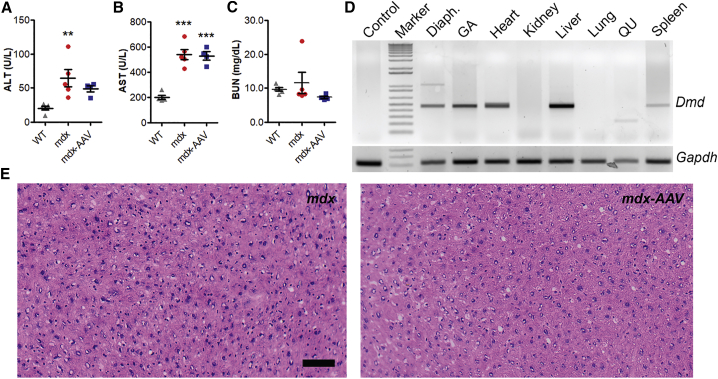
Figure 4.
Life-Long AAV-Mediated CRISPR Genome Editing Did Not Induce Detectable Toxicity or Tumor Development in mdx Mice
(A–C) Measurements of serum BUN (A), ALT (B), and AST (C) in WT, mdx, and mdx-AAV mice. *p < 0.05, **p < 0.01, ***p < 0.001. (D) Genomic DNA PCR analysis of dystrophin gene editing of various tissues (Diaph., diaphragm; GA, gastrocnemius; QU, quadriceps) from an AAV-treated mdx mouse. (E) Representative H&E-stained images of the liver from mdx and AAV-treated mdx mice. Scale bar, 50 μm. Error bars indicate mean ± SEM.
Since the heart is generally resistant to development of tumor, we examined if the life-long CRISPR genome editing induced any lesions in other tissues such as liver. We first surveyed the on-target genome-editing events in different tissues of all five rAAV-injected mice by performing genomic DNA PCR. The heart and liver from all five treated mice consistently showed positive genome editing as evident by the presence of 500-bp editing PCR products (Figure 4D). Liver tissues were assessed histologically by board-certified pathologist and no signs of pathological changes including hyperplasia were identified (Figure 4E). Also, analysis of the other major vital organs did not reveal any signs of tumor development either (Figure S4).
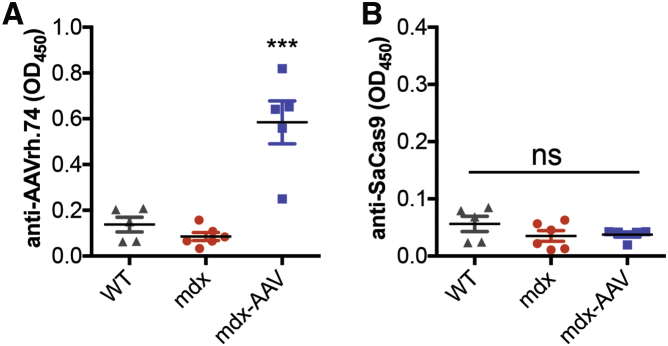
Another important consideration to long-term therapeutic benefit of in vivo CRISPR genome editing is the host response to bacteria-derived Cas9 proteins. In our study, all injected mdx mice developed substantial humoral immune response against AAV viral particles (Figure 5A); however, no humoral immune response against the SaCas9 protein was detected in any of the mice injected as neonates (Figure 5B). Consistent with a recent report by Nelson et al.,11 our data suggests that a significant host response is avoided if AAV-CRISPR is administered at the neonatal stage.
Figure 5.
Host Response to AAV-CRISPR in mdx Mice
Antibodies against AAVrh.74 (A) but not SaCas9 (B) are detected in mice treated with AAV-CRISPR as neonates after 19 months of age. Error bars indicate mean ± SEM. ***p < 0.001; ns, not significant.
In conclusion, our study demonstrated that life-long AAV-CRISPR-mediated genome editing led to sustained restoration of dystrophin expression and improved cardiac pathology without causing deleterious consequences in mdx mice. These findings highlight the potential promise of CRISPR genome editing as a safe therapy for DMD and other diseases.
Discussion
Previous studies, including our own, have demonstrated the promise of CRISPR-mediated genome editing in treating DMD by permanently restoring the reading frame of the mutated DMD gene using cultured muscle cells and short-term studies in animal models.3, 4, 5, 6, 7, 8 In this study, we further demonstrated that long-life AAV-mediated CRISPR genome editing restored dystrophin expression in over 10% cardiomyocytes of mdx mice with no detectable adverse effects. Cardiac function and pathology were also observed to show improvement in animals treated with AAV-CRISPR. This is consistent with two recent studies by Hakim et al.10 and Nelson et al.11 that also found AAV-CRISPR genome editing improved cardiac and muscle function of aged mdx mice when treated at their early life.
There are several potential safety concerns regarding AAV-CRISPR genome-editing therapy for DMD. First, the CRISPR/Cas9 system has been well known for its off-target activity.17, 18, 19, 20 Second, repair of DSBs induced by CRISPR/Cas9 could lead to large deletions and/or complex rearrangements.15, 16 Third, CRISPR/Cas9 genome editing may select p53 deficiency,22, 23 thus increasing the risk of tumor development. Fourth, pre-existing anti-Cas926, 27 and anti-AAV immunity may prevent effective genome editing. In addition, the body may generate immunity against the newly produced dystrophin protein.28 All these possibilities could potentially pose significant challenges in translating the CRISPR genome-editing therapy into clinics for treating DMD.
Despite the well-documented mismatch tolerance within the protospacer for CRISPR/Cas9 targeting,17, 18, 19, 20, 21 several recent studies found that careful selection of the guide RNA can effectively minimize the chance of off-target events for Cas9 in mice.29, 30, 31, 32 The guide RNAs chosen in this study have previously been validated and have no off-target sites with less than three mismatches in the mouse genome.3 Kosicki et al.15 and others recently reported that repair of DSB induced by CRISPR/Cas9 at on-target sites leads to large deletions and complex rearrangements in cultured cells.16 In our in vivo study with the mdx mice, we performed PCR analysis and failed to detect any large deletion events in any of the AAV-CRISPR-treated heart samples. We further performed deep-sequencing studies and found that the majority of the DNA repair products for the two DSBs induced by CRISPR were direct ligations of the two blunted ends at the cutting sites, consistent with recent reports that repair of DSBs induced by CRISPR/Cas9 is highly predictable.13, 14, 33, 34, 35 However, these data do not exclude the possibility that a negligible number of cells may carry large deletions associated with CRISPR genome editing that is beyond the sensitivity of the genomic DNA sequencing and PCR analysis at the tissue level. If this were the case, these cells had not acquired growth advantage over other cells, as otherwise they should have outnumbered over a 19-month period.
Additional concern regarding the safety of in vivo AAV-CRISPR genome editing comes from several recent studies that found selection of tumor suppressor p53 deficiency is associated with positive CRISPR genome editing.22, 23 Our study did not reveal any evidence for tumor development in major vital organs or tissues (heart, lung, diaphragm, spleen, kidney, liver, quadriceps, and gastrocnemius) as examined by H&E staining. It is noted that old mdx mice are susceptible to spontaneous rhabdomyosarcoma.24 Indeed, we found two of the five untreated mice developed spontaneous rhabdomyosarcoma, while none of the long-term AAV-CRISPR-treated mdx mice had similar tumors observed. Clearly, our study showed that after 19 months of treatment, the edited cells in liver and heart did not present remarkable pathological lesions.
In this study, the AAV-CRISPR was administered into young mdx pups, for which the immune system is not fully developed.36, 37 In this scenario, we found no humoral response against SaCas9, although anti-AAV immunity was detected. Thus, it is possible to avoid anti-Cas9 immunity by delivering AAV-CRISPR in newborns. However, it is not yet clear to what extent this approach can be translated into newborn humans. For patients whose immune system is fully developed, alternative approaches, such as transient depletion of B cells,38 induction of immune tolerance,39 and removal of T cell epitopes,40 would be required to avoid anti-Cas9 immune responses. Moreover, recent studies found that pre-existing anti-Cas9 immunity is widely present in human population.26, 27 Whether the pre-existing anti-Cas9, anti-AAV, and anti-dystrophin immunity affect AAV-CRISPR genome-editing therapy in patients would need to be addressed in future animal studies and clinical investigations.
Overall, these studies highlight the safety and efficacy of the long-term AAV-CRISPR genome-editing therapy in mdx mice. These findings are highly encouraging for developing a safe CRISPR-based genome-editing therapy in future clinical trials with DMD patients.
Materials and Methods
Mice and Experimental Protocol
Mice (C57BL/10ScSn and C57BL/10ScSn-Dmdmdx/J) were purchased from the Jackson Laboratory and maintained at The Ohio State University Laboratory Animal Resources in accordance with animal use guidelines. All the experimental procedures were approved by the Animal Care, Use, and Review Committee of the Ohio State University.
The mdx pups (day 3) were injected systemically with 1 × 1012 viral particles via intraperitoneal approach as previously described.3 After the final echocardiography assessment, mice from WT, mdx, and mdx/AAV were sacrificed by CO2 inhalation at 19 months of age. The major vital organs or tissues (heart, lung, diaphragm, spleen, kidney, liver, quadriceps, and gastrocnemius) were embedded in optimal cutting temperature (OCT; Sakura Finetek, the Netherlands) compound and snap-frozen in cold isopentane for cryosectioning. The tissues were stored at −80°C and processed for biochemical analysis and histology assessment.
Generation of AAV Particles
AAV vectors were produced at the viral vector core of the Nationwide Children’s Hospital as previously described.3 The SaCas9 and gRNA (i20, 5′-GGGCGTTGAAATTCTCATTAC CAGAGT-3′; and i23, 5′-CACCGATGAGAGGGAAAGGTC CTGAAT-3′; note, the underlined PAM sequences were not included in the gRNA) expression cassettes were packaged into AAV serotype rh74 capsid using the standard triple transfection protocol.41 A qPCR-based titration method42 was used to determine an encapsulated vg titer utilizing a Prism 7500 Fast Taqman detector system (PE Applied Biosystems Grand Island, NY, USA). The following primers were used: 5′-GGATTTCCAAGTCTCCACCC-3′ and 5′-TCCCACCGTACACGCCTAC-3′. Titers are expressed as DNase-resistant particles per mL (DRP/mL) and rAAV titers used for injection in mice were 9.1 × 1012 DRP/mL.
AAV Titer Detected by Real-Time PCR
Genomic DNA was extracted from different tissues by using DNeasy blood and tissue kit (QIAGEN) according to the manufacturer’s instructions. 50 ng of DNA from each sample was used for PCR. Regular PCR to detect edited mouse dystrophin gene was performed using GoTaq green master (Promega, Madison, WI, USA) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used as a reference for PCR analysis. The primers used for mouse dystrophin genomic DNA were as follows: 5′-GGCCAAAGCAAACTCTGGTA-3′ and 5′-TTTAATCCCACGTCATGCAA-3′. For real-time-PCR, all the samples were run on Roche480. The shuttle plasmid for the rAAV production was prepared with different dilutions as copy number controls. All samples and controls were run in triplicate. The primers used for real time PCR were as follows: AAV-ITR-F, 5′-CTCACATGTCCTGCAGGCAGCTGCGCGCT-3′; and AAV-ITR-R, 5′-TGGGCATGCAGGGACGCGTAGGAACCCCTAGTGATGGA-3′.
Illumina NGS
PCR products were purified using a commercial purification kit (Promega, Madison, WI, USA), and electrophoresed on an agarose gel, showing a single sharp peak. Their quality and quantity were assayed using an Agilent Bioanalyzer 2100 (Genomics Shared Resource, Ohio State University Comprehensive Cancer Center). The purified PCR products were polyadenylated, and a generic Illumina sequencing adaptor was ligated using T4 DNA ligase and amplified by PCR for 2–3 cycles to enrich for ligated adaptors using appropriate primers. Ligation products were cleaned up using Agencourt ampure beads. Samples were amplified again by PCR for 4–5 cycles to incorporate flanking Agilent SureSelect indices A01, B01, and C04 and again cleaned up using AMpure beads. Quality again was checked using a Bioanalyzer and Qubit and then sent for sequence clustering using the Illumina cBot. Sequence data were generated for the pooled samples using a MiSeq nano-scale flow cell (Molecular and Cellular Imaging Center, OSU Wooster campus).
Ion Torrent NGS
PCR products were checked for quality. Following instructions for the Ion Torrent Fragment DNA library preparation kit, they were sheared using the Covaris S2 following manufacturer’s recommendation to result in 200-nt products. Ends were repaired using T4 DNA polymerase Klenow fragment. Adaptors for Ion Torrent P1 and index were ligated and cleaned up, quality was checked, and products were amplified by PCR for a few cycles, and were cleaned up and sequenced. Beads were templated on the One Touch, then loaded for 316 chip. Outputs were processed using the Ion Torrent server software at default settings to remove adaptor sequences, resulting in FASTQ files.
Informatics Analysis of Targeted Sequencing Data
Adaptor and primer sequences were removed using standard Illumina MiSeq BCL2FASTQ software. For amplicons A and B, we confirmed where the amplicons started and stopped by aligning the data to the reference genome (hg19). For amplicon C, we generated an artificial template sequence comprised of amplicons A and B, joined by 50-nt buffer sequences to the right and to the left of the amplicons, respectively. We provided these reference template amplicon sequences using CRISPResso with default parameters.43 For Ion Torrent sequence data, to confirm the specificity of the target amplicons A, B, and C, we aligned each set first to the reference human genome (hg19) using GSNAP.44 We extracted the ∼350-nt target region for each amplicon. Then, we used GSNAP to locally realign the sequences to these short reference templates. Variants were visualized using the integrative genomics viewer (IGV; Broad Institute).
Echocardiography
To assess cardiac function, mice were anesthetized with isoflurane as described previously.45 The echocardiographic measurements were performed blinded to the animal groups using a 30-MHz transducer with the Vevo 2100 imaging system (Visual Sonic, Toronto, Canada). One day prior to experiment, the hair was removed from the anterior chest with depilatory cream. Mice were anesthetized with 1%–2% isoflurane in oxygen at flow rate 1 L/min, and the animals were placed on imaging stage at the Vevo Imaging Station to monitor the animal’s blood pressure, electrocardiogram, body temperature, and heart rate. Ultrasound gel was applied to the chest wall to obtain a two-dimensional and Doppler images using ultrasound transducer attached to a 3D motor system. A complete two-dimensional short-axis view was acquired at the mid-papillary muscle level and M-mode images of the left ventricle were recorded. Left ventricular ejection fraction (EF) and fractional shortening (FS) were determined and analyzed from the averaged of three consecutive beats with the Vevo LAB desktop Software (v1.7.1, VisualSonics, Toronto, ON, Canada).
Serological Analysis
The blood samples from all groups were collected before they were sacrificed. The blood was allowed to clot for 15 min to 30 min and centrifuged at 5,000 rpm for 10 min in room temperature. The supernatant was collected as serum and stored at −80°C for the biochemical assay. Measurement of ALT (BioVision Incorporated), AST (BioVision Incorporated), BUN (Arbor Assays, Michigan, USA), and troponin (Life Diagnostics) were performed according to the manufacturer’s protocols.
Antibody ELISA
Antibodies against AAVrh.74 and SaCas9 were detected by adapting previously published protocols.11, 46 In brief, recombinant AAVrh.74-CRISPR (2 × 109 vg/well) and SaCas9 protein (0.2 μg/well) were diluted in 1× coating buffer A (BioLegend) and used to coat a 96-well Nunc MaxiSorp plate. Proteins were incubated overnight at 4°C to adsorb to the plate. Plates were washed four times for 5 min each with PBS plus 0.05% Tween-20 and then blocked with 1× assay diluent A (BioLegend) for 1 h at room temperature. The anti-SaCas9 antibody (Diagenode C15200230) was used as positive control for detection of anti-SaCas9 antibodies. For AAVrh.74, we found that the anti-AAV2 antibody (cat. #03-65155, American Research Products) did not recognize AAVrh.74, and we do not realize there is any validated commercial anti-AAVrh.74 antibody. Serum samples were added in 1:50 dilution and plates were incubated for 2 h at room temperature with shaking. Plates were washed four times for 5 min each, and 100 μL of blocking solution containing goat anti-mouse immunoglobulin G (IgG) (Sigma 1:3,000) was added to each well and incubated for 1 h at room temperature. Plates were washed four times for 5 min each and 100 μL of freshly mixed TMB substrate solution (BioLegend) was added to each well and incubated in the dark for 20 min. The reaction was stopped by adding 100 μL 2N H2SO4 stop solution. Optical density at 450 nm was measured with a plate reader.
Histopathology Assessment of Tissues
For histological examinations, frozen cryosections of 8 μm were prepared from each group and were fixed in 4% paraformaldehyde for 15 min at room temperature. The slides were then processed according to the standard protocol of H&E and Masson-trichrome staining (American MasterTech Scientific Incorporated, Lodi, CA, USA) for histological and pathological examination. All images were taken under a Zeiss Axiovert 200 phase-contrast microscope, magnification, X200 (Zeiss, Oberkochen, Germany). For quantification of myocardial collagen content in heart tissues, whole images were scanned with Olympus cellSens, version 2.1 (Olympus Corporation, Center Valley, PA, USA). The images were then quantified from Masson-trichrome staining (blue stained) using NIS-Elements AR version 4.30 (Nikon, Melville, NY, USA). Data were expressed as total area of fibrosis/total tissue area (%).
Immunofluorescence Staining
Frozen sections of hearts were fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS, the slides were blocked with 5% BSA for 1 h. The slides were incubated with primary antibodies against dystrophin (ab15277, 1:200, Abcam), caveolin 3 (sc-5310, 1:500, Santa Cruz Biotechnology, Dallas, TX, USA), CD3 (ab5690, 1:500, Abcam), and laminin-α2 (ALX-804-190, 1:100, Alexis, San Diego, CA, USA) at 4°C overnight. After that, the slides were washed extensively with PBS and incubated with secondary antibodies (Alexa Fluor 488 goat anti-rat IgG, Invitrogen, Carlsbad, CA, USA, or Alexa Fluor 594 goat anti-mouse IgG, Invitrogen) for 2 h at room temperature. The slides were sealed with VECTASHIELD Antifade Mounting Medium with DAPI (vector laboratory, Burlingame, CA, USA). All images were taken under a Nikon Ti-E fluorescence microscope, magnification ×200 (Nikon, Melville, NY, USA). Caveolin-3-positive cardiomyocytes and dystrophin-positive cardiomyocytes were counted using NIS-Elements AR version 4.30 (Nikon, Melville, NY, USA). The amount of dystrophin-positive cardiomyocytes is represented as a percentage of total caveolin-3-positive cardiomyocytes.
Statistical Analysis
The data were expressed as mean ± SEM and analyzed with GraphPad Prism 5.02 software (San Diego, CA, USA). Statistical significance were determined using one-way ANOVA followed by Bonferroni post-hoc tests. A p value of less than 0.05 is regarded as significant.
Author Contributions
R.H. conceived the study and wrote the manuscript. Y.S.L. and L.X. performed the serological studies, immunofluorescence, and western blot experiments and analyzed the data. Y.G. performed the echocardiography recording and ELISA experiments, maintained the mouse colony, assisted the rAAV injections, and collected mouse tissues. H.L. assisted L.X. for western blot experiments.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We thank the Viral Vector Core at the Nationwide Children’s hospital for producing the AAV. R.H. is supported by US NIH grants (R01 HL116546 and R01 AR064241).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.05.001.
Supplemental Information
References
- 1.Emery A.E. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul. Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E.P., Brown R.H., Kunkel L.M. Dystrophin: the protein product of the Duchene muscular dystrophy locus. 1987. Biotechnology. 1992;24:457–466. [PubMed] [Google Scholar]
- 3.El Refaey M., Xu L., Gao Y., Canan B.D., Adesanya T.M.A., Warner S.C., Akagi K., Symer D.E., Mohler P.J., Ma J. In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circ. Res. 2017;121:923–929. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2015;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Long C., Li H., McAnally J.R., Baskin K.K., Shelton J.M., Bassel-Duby R., Olson E.N. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 2017;3:e1602814. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakim C.H., Wasala N.B., Nelson C.E., Wasala L.P., Yue Y., Louderman J.A., Lessa T.B., Dai A., Zhang K., Jenkins G.J. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight. 2018;3:124297. doi: 10.1172/jci.insight.124297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson C.E., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D., Robinson-Hamm J.N., Bulaklak K., Castellanos Rivera R.M., Collier J.H. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amoasii L., Hildyard J.C.W., Li H., Sanchez-Ortiz E., Mireault A., Caballero D., Harron R., Stathopoulou T.R., Massey C., Shelton J.M. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyombe-Engembe J.P., Ouellet D.L., Barbeau X., Rousseau J., Chapdelaine P., Lagüe P., Tremblay J.P. Efficient Restoration of the Dystrophin Gene Reading Frame and Protein Structure in DMD Myoblasts Using the CinDel Method. Mol. Ther. Nucleic Acids. 2016;5:e283. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duchene B.L., Cherif K., Iyombe-Engembe J.P., Guyon A., Rousseau J., Ouellet D.L., Barbeau X., Lague P., Tremblay J.P. CRISPR-Induced Deletion with SaCas9 Restores Dystrophin Expression in Dystrophic Models In Vitro and In Vivo. Mol. Ther. 2018;26:2604–2616. doi: 10.1016/j.ymthe.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adikusuma F., Piltz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J., Beard M.R., Hughes J., Pomerantz R.T., Thomas P.Q. Large deletions induced by Cas9 cleavage. Nature. 2018;560:E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 17.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 18.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cradick T.J., Fine E.J., Antico C.J., Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 23.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 24.Chamberlain J.S., Metzger J., Reyes M., Townsend D., Faulkner J.A. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 25.McMillan H.J., Gregas M., Darras B.T., Kang P.B. Serum transaminase levels in boys with Duchenne and Becker muscular dystrophy. Pediatrics. 2011;127:e132–e136. doi: 10.1542/peds.2010-0929. [DOI] [PubMed] [Google Scholar]
- 26.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simhadri V.L., McGill J., McMahon S., Wang J., Jiang H., Sauna Z.E. Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol. Ther. Methods Clin. Dev. 2018;10:105–112. doi: 10.1016/j.omtm.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flanigan K.M., Campbell K., Viollet L., Wang W., Gomez A.M., Walker C.M., Mendell J.R. Anti-dystrophin T cell responses in Duchenne muscular dystrophy: prevalence and a glucocorticoid treatment effect. Hum. Gene Ther. 2013;24:797–806. doi: 10.1089/hum.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer V., Shen B., Zhang W., Hodgkins A., Keane T., Huang X., Skarnes W.C. Off-target mutations are rare in Cas9-modified mice. Nat. Methods. 2015;12:479. doi: 10.1038/nmeth.3408. [DOI] [PubMed] [Google Scholar]
- 30.Iyer V., Boroviak K., Thomas M., Doe B., Riva L., Ryder E., Adams D.J. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 2018;14:e1007503. doi: 10.1371/journal.pgen.1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akcakaya P., Bobbin M.L., Guo J.A., Malagon-Lopez J., Clement K., Garcia S.P., Fellows M.D., Porritt M.J., Firth M.A., Carreras A. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–419. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Páleníkova P., Khodak A., Kiselev V., Kosicki M. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4317. Published online November 27, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarti A.M., Henser-Brownhill T., Monserrat J., Poetsch A.R., Luscombe N.M., Scaffidi P. Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol. Cell. 2019;73:699–713.e6. doi: 10.1016/j.molcel.2018.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu C., Lipshutz G.S. AAV-based neonatal gene therapy for hemophilia A: long-term correction and avoidance of immune responses in mice. Gene Ther. 2012;19:1166–1176. doi: 10.1038/gt.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee E.K., Hu C., Bhargava R., Rozengurt N., Stout D., Grody W.W., Cederbaum S.D., Lipshutz G.S. Long-term survival of the juvenile lethal arginase-deficient mouse with AAV gene therapy. Mol. Ther. 2012;20:1844–1851. doi: 10.1038/mt.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu S., Dunn R., Kehry M.R., Braley-Mullen H. B cell depletion inhibits spontaneous autoimmune thyroiditis in NOD.H-2h4 mice. J. Immunol. 2008;180:7706–7713. doi: 10.4049/jimmunol.180.11.7706. [DOI] [PubMed] [Google Scholar]
- 39.Zhang P., Sun B., Osada T., Rodriguiz R., Yang X.Y., Luo X., Kemper A.R., Clay T.M., Koeberl D.D. Immunodominant liver-specific expression suppresses transgene-directed immune responses in murine pompe disease. Hum. Gene Ther. 2012;23:460–472. doi: 10.1089/hum.2011.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferdosi S.R., Ewaisha R., Moghadam F., Krishna S., Park J.G., Ebrahimkhani M.R., Kiani S., Anderson K.S. Multifunctional CRISPR/Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019;10:1842. doi: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grose W.E., Clark K.R., Griffin D., Malik V., Shontz K.M., Montgomery C.L., Lewis S., Brown R.H., Jr., Janssen P.M., Mendell J.R., Rodino-Klapac L.R. Homologous recombination mediates functional recovery of dysferlin deficiency following AAV5 gene transfer. PLoS ONE. 2012;7:e39233. doi: 10.1371/journal.pone.0039233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnepp B.C., Jensen R.L., Chen C.L., Johnson P.R., Clark K.R. Characterization of adeno-associated virus genomes isolated from human tissues. J. Virol. 2005;79:14793–14803. doi: 10.1128/JVI.79.23.14793-14803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J., El Refaey M., Xu L., Zhao L., Gao Y., Floyd K., Karaze T., Janssen P.M., Han R. Genetic disruption of Ano5 in mice does not recapitulate human ANO5-deficient muscular dystrophy. Skelet. Muscle. 2015;5:43. doi: 10.1186/s13395-015-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y., Ng A.H., Zhu K., Wagers A.J., Church G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.