Abstract
Cardiolipin is an anionic lipid found in the mitochondrial membranes of eukaryotes ranging from unicellular microorganisms to metazoans. This unique lipid contributes to various mitochondrial functions, including metabolism, mitochondrial membrane fusion and/or fission dynamics, and apoptosis. However, differences in cardiolipin content between the two mitochondrial membranes, as well as dynamic fluctuations in cardiolipin content in response to stimuli and cellular signaling events, raise questions about how cardiolipin concentration affects mitochondrial membrane structure and dynamics. Although cardiolipin’s structural and dynamic roles have been extensively studied in binary mixtures with other phospholipids, the biophysical properties of cardiolipin in higher number lipid mixtures are still not well resolved. Here, we used molecular dynamics simulations to investigate the cardiolipin-dependent properties of ternary lipid bilayer systems that mimic the major components of mitochondrial membranes. We found that changes to cardiolipin concentration only resulted in minor changes to bilayer structural features but that the lipid diffusion was significantly affected by those alterations. We also found that cardiolipin position along the bilayer surfaces correlated to negative curvature deflections, consistent with the induction of negative curvature stress in the membrane monolayers. This work contributes to a foundational understanding of the role of cardiolipin in altering the properties in ternary lipid mixtures composed of the major mitochondrial phospholipids, providing much-needed insights to help understand how cardiolipin concentration modulates the biophysical properties of mitochondrial membranes.
Significance
Cardiolipin is an intrinsic component of the lipid milieu that makes up the membranes of eukaryotic mitochondria, and the distinctive lipid plays roles in both mitochondrial structure and function. Despite the many studies that establish that cardiolipin has unique properties in membranes, there are still questions about how much cardiolipin directly contributes to structural differences between the inner and outer mitochondrial membranes, as well as questions about how natural fluctuations in cardiolipin concentration alter the structure and dynamics of mitochondrial membranes. This study provides quantitative predictions for cardiolipin-dependent properties of lipid bilayer systems that mimic the major components of mitochondrial membranes. As such, it provides further insights into cardiolipin’s role in modulating the biophysical properties of mitochondrial membranes.
Introduction
Cardiolipin (CL) is an anionic lipid found in the mitochondrial membranes of eukaryotes (1), including unicellular microorganisms as well as metazoans. This structurally unique phospholipid is composed of two phosphatidylglycerol moieties connected by a single bridging glycerol group (2), thereby conferring it with a small polar headgroup with limited flexibility and mobility. It has been shown in previous work that the small size of the polar headgroup imparts CL with a propensity to form inverted hexagonal lipid phases (3, 4), whereas the restrictions placed on the polar headgroup flexibility and mobility impart CL with unique membrane properties (5). CL contributes to various mitochondrial functions, such as metabolism (6), mitochondrial membrane fusion and/or fission dynamics (7), and apoptosis (8, 9) in higher organisms. Furthermore, dysregulation of mitochondrial CL synthesis and remodeling has been implicated in several human diseases, including diabetes (10, 11), heart disease (11, 12), and Barth syndrome (13, 14).
Within the mitochondrion, CL is primarily localized in the inner mitochondrial membrane (IMM) (15, 16, 17, 18, 19, 20), where it accounts for ∼10 mol% of IMM lipid content. However, CL is also present at much lower concentrations (∼1–5 mol% of lipids) in the outer mitochondrial membrane (OMM) (15, 16, 17, 18, 19, 20). Because the two membranes are structurally dissimilar, the disparity in CL concentration between them raises questions about the role of CL in shaping mitochondrial membrane structure. For example, there is evidence that CL plays a role in maintaining IMM superstructure (21, 22, 23, 24, 25), affecting the shape and stability of cristae. Yet, it is unclear to what extent CL plays a role in directly modulating the in vivo structure of the IMM because the IMM also has a high content of membrane proteins, to which CL molecules bind promiscuously (1, 26), that also likely play a role in stabilizing the IMM’s highly curved interfaces (27). Moreover, the mitochondrion is a dynamic organelle, and both the total amount and intramitochondrial distribution of CL can change in response to stimuli and cellular signaling events (28, 29, 30, 31, 32), thus raising further questions about how CL concentration modulates mitochondrial membrane properties.
The main purpose of this work is to investigate the CL-dependent properties of lipid bilayer systems that mimic the major components of mitochondrial membranes. The two major phospholipid species in mitochondrial membranes are phosphatidylcholine (PC) and phosphatidylethanolamine (PE) lipid species. A variety of studies, including monolayer experiments (33, 34, 35, 36), vesicle and bilayer experiments (33, 37, 38, 39), and simulations (40, 41, 42), have probed the biophysical properties of binary mixtures of CL with PC or PE lipids. A limited number of experimental studies have also probed the biophysical properties of more complex lipid mixtures (more than three lipid species) using OMM-like supported lipid bilayers (39) and IMM-like monolayers (36). However, the studies that investigate the properties of CL in mixed PC-PE lipid membranes are limited, so there is still a shortage of basic biophysical data to describe CL in ternary lipid membrane systems. We have therefore run a series of atomistic molecular dynamics (MD) simulations of model mitochondrial membranes composed of PC and PE lipids with CL concentrations consistent with the natural ranges of CL content found in the OMM and IMM, as well as their contact sites in the OMM, which have been reported to be enriched in CL (up to ∼25 mass%) (20). From these simulations, we have estimated a variety of structural and dynamic bilayer properties and show their dependence on the concentration of CL. We also compare our results to those of previous studies of the thermodynamic, structural, and dynamic properties of similar ternary PC, PE, and CL lipid membranes. This work contributes toward a foundational understanding of the role of CL in altering the properties of ternary lipid mixtures composed of PC, PE, and CL molecules. This work also helps to illuminate the concentration-dependent role of CL in mitochondrial membrane structure and lipid dynamics, contributing to the biophysical data needed to help understand CL’s role in shaping the structural differences between the inner and outer membranes, as well as how mitochondrial membrane properties are altered when CL content changes in response to stimuli and cellular signaling events.
Methods
Model systems
We have constructed five atomistic models of ternary lipid bilayer systems mimicking mitochondrial membranes. The membrane bilayers were square patches with sizes of ∼(14–15 nm)2 and were composed of PC, PE, and varying proportions of CL molecules consistent with those found in the outer (∼1–5 mol%, ∼15 mol% at contact sites) and inner (∼5–10 mol%) mitochondrial membranes. The classes of lipids were modeled using the following lipids:
-
1)
PC: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC).
-
2)
PE: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE).
-
3)
CL: 1,3-Bis-[1,2-di-(9,12-octadecadienoyl)-sn-glycero-3-phospho]-sn-glycerol (tetralinoleoyl CL, (18:2)4-CL).
We chose POPC and DOPE as the model PC and PE lipids, respectively, because of the relatively high prevalence of 16:0 and 18:1 fatty acid chains among mitochondrial lipids (43). We chose polyunsaturated tetralinoleoyl CL ((18:2)4-CL) because of its high relative abundance among CL species in human mitochondria, particularly in heart and skeletal muscle tissues (44, 45, 46). Tetralinoleoyl CL is also the primary CL species affected in Barth syndrome (13, 45).
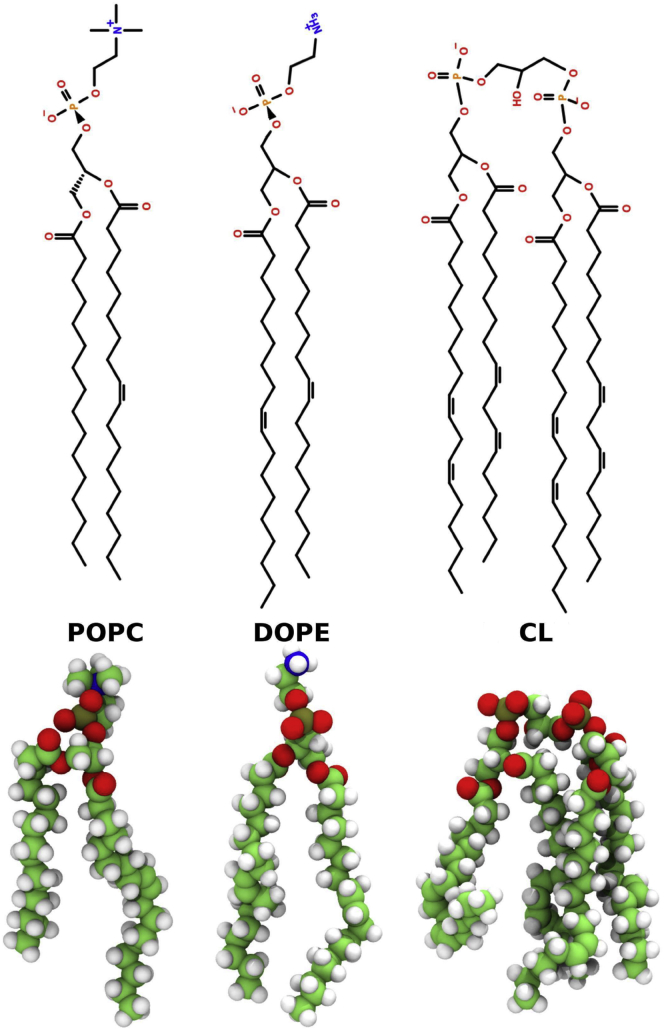
Because they have two phosphate groups, CL molecules can carry up to two negative charges. However, the ionization state of CL at neutral pH (7.0) and physiological pHs (around pH 7.4) is a contentious issue, with some studies (47, 48, 49) suggesting that a mixture of CL ionization states predominated by the singly deprotonated (−1e charge) species would likely form, whereas other studies (50, 51, 52) suggest that CLs would be fully deprotonated (−2e charge) at that pH range. Because even in the former case, the fully ionized CL species may be expected to be present in reasonable proportion, we modeled CL lipids as fully ionized. The structures of the lipids used in our computational models are shown in Fig. 1.
Figure 1.
The line (top) and three-dimensional structures (bottom) of the lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and the −2e charge version of the lipid tetralinoleoyl cardiolipin (CL) used in the model mitochondrial outer membranes. The line structure drawings were generated using Lipid MAPS (125) and Open Babel tools (126) tools. The three-dimensional structure images were generated using the Tachyon renderer (127) in VMD (128). To see this figure in color, go online.
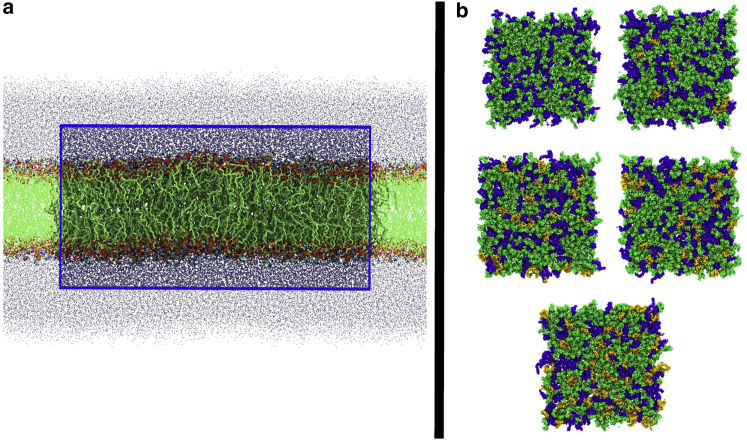
All-atom lipid bilayers were constructed using the Membrane Builder Input Generator of the CHARMM-GUI web toolkit (53, 54, 55, 56, 57, 58). Each MOM-like bilayer system consisted of 600 lipids (300 per leaflet) solvated with 22.5 Å of explicit water molecules on either side of the bilayer (37–43 water molecules per lipid). We generated membranes with CL content 0, 2, 7, 10, and 15 mol% (0, 3.8, 12.6, 17.6, and 25.3 mass%, respectively); the total number of lipids (600) was held constant. The numbers of POPC and DOPE lipids were adjusted such that an approximate 2:1 ratio (POPC/DOPE) was maintained. Sodium chloride ions were added to neutralize excess lipid charges and set the salt concentration to ∼0.2 M in the 0, 7, 10, 15 mol% systems. The 2 mol% system was only charge neutralized with Na+ ions; although this gives the 2 mol% system slightly different ionic conditions than the other model systems, based on previous studies (59, 60), we expect this difference in ion conditions to have a negligible effect on the properties reported in this study. Interactions were modeled using the CHARMM36 force field (61) with the TIP3P water model (62). Representations of the model systems are depicted in Fig. 2.
Figure 2.
Snapshots of lipid bilayers used to model mitochondrial outer membranes. (a) A representative side view of one of the model systems is shown. For clarity, the hydrogen atoms were removed. Water oxygen atoms are shown in ice blue. The simulation cell is outlined by the blue rectangular box, and the material surrounding the simulation cell shows the periodic images. (b) A top view of each model bilayer after initial system minimization and relaxation is shown. The CL concentrations are in increasing order, going from left to right and top to bottom. POPC lipids are shown in green, DOPE lipids in blue, and CL lipids in gold. The images of simulation snapshots were generated using the Tachyon renderer (127) in VMD (128). To see this figure in color, go online.
MD simulations
Simulations were run employing the GROMACS software (version 5.1.4) (63, 64). The preproduction minimization and relaxation of each system was carried out using the GROMACS protocol generated by the CHARMM-GUI Input Generator, which consists of
-
1)
Energy minimization for 5000 steps.
-
2)
Constant volume and 303.15 K (NVT) using the Berendsen thermostat for 50,000 steps with a 1 fs time step.
-
3)
1.0 bar and 303.15 K (NPT) using the Berendsen thermostat and barostat for 25,000 steps with a 1 fs time step.
-
4)
1.0 bar and 303.15 K (NPT) using the Berendsen thermostat and barostat for 150,000 steps with a 2 fs time step.
For the production runs, each system was simulated with a reference pressure of 1.0 bar and a temperature of 303.15 K (NPT) using the Parrinello-Rahman barostat and the Nosé-Hoover thermostat, respectively. The simulation time step was set to 2 fs. A single MD simulation was launched for each system; the 2, 7, and 10% CL systems were simulated for a total of 1 μs, whereas the 0 and 15% CL systems were simulated for a total of 0.9 μs.
All simulations were carried out with covalent bonds to hydrogen atoms constrained via the LINCS algorithm (65). A 12 Å spherical cutoff was used for short-range nonbonded interactions, with a force-switching function from 10 Å for the van der Waals term and shifted electrostatics. Long-range electrostatic interactions were computed using the particle-mesh Ewald method (66) with a grid spacing on the order of 1 Å or less. The equations of motion were integrated using the default GROMACS “md” integrator. Periodic boundary conditions were applied to all simulations, and all NPT ensemble simulations were run under zero tension and semi-isotropic pressure coupling.
Analysis
Analysis was performed using tools from a Python-based lipid bilayer analysis toolkit, PyBILT (https://github.com/LoLab-VU/PyBILT), that is currently being developed in our lab. PyBILT-based analysis included the use of MDAnalysis (67, 68), NumPy (69, 70), and SciPy (71) software; data plots were generated using Matplotlib (72) and seaborn (73) software. Descriptions of individual methods for analysis are provided in subsequent sections. In lipid membrane systems, the dynamics of some properties, such as ion binding and unbinding to the membrane-water interface (59) and lipid diffusion (74), can have characteristic times on the order of 100 ns or greater. Therefore, the first 400 ns of each production run were discarded as a long-timescale equilibration time and not included in the analysis. Unless otherwise noted, thermodynamic averages and their standard errors were estimated by using the block averages taken over nonoverlapping 100 ns segments of the remaining trajectory; key quantities were examined using blocked standard error analysis with varying block sizes (Figs. S1 and S2) as described by Grossfield and Zuckerman (75), and it was determined from this analysis that block sizes of at least 50 ns resulted in uncorrelated blocks for block averaging. The reported error for our data was computed as 1.96 times the standard error estimate (from block averaging with 100 ns blocks) and thus represent estimates of the 95% confidence intervals. The resulting observable estimates as reported here were then rounded to include only one uncertain digit.
Average area per lipid molecule
The average area per lipid molecule was computed using the area projected onto the xy-plane, which was taken as the xy simulation box area, divided by the number of lipid molecules per bilayer leaflet:
| (1) |
where Axy is the instantaneous xy-area of the simulation box (used to estimate the lateral surface area of the bilayer) at each simulation snapshot and Nt/2 is the number of lipids per bilayer leaflet; Nt = 600 was the total number of lipids in the bilayer and was constant for all simulated systems.
Average area per lipid phosphate headgroup
The average area per lipid phosphate headgroup (abbreviated hereafter as average area per phosphate) was estimated by computing the average area per lipid phosphate headgroup projected onto the xy-plane, calculated as the xy simulation box area divided by the number of lipid phosphate headgroups per leaflet:
| (2) |
where Axy is the instantaneous xy-area of the simulation box (used to estimate the lateral surface area of the bilayer) at each simulation snapshot and (Nt + NCL)/2 is the number of lipid phosphate headgroups per bilayer leaflet; NCL is the total number of CL lipid molecules in the bilayer. Note that this quantity is directly related to the average area per lipid through the conversion factor Nt/(Nt + NCL); i.e., .
Partial area per lipid phosphate headgroup
The partial area per lipid phosphate headgroup was computed using a grid-based tessellation scheme adapted from Allen et al. (76). Briefly, at each snapshot of the simulation trajectory, the lipids are mapped to a two-dimensional (2-d) grid based on Euclidean distance in the lateral plane of the bilayer (xy-plane). The positions of the phosphate phosphorous atoms are projected into the xy-plane as the reference points for distance calculations and subsequent mapping of the lipid headgroups to the 2-d grids. For each grid element, the nearest lipid is assigned to that grid. We performed this analysis with 100 × 100 grids, resulting in grid elements with an area <3 Å2. Gapsys et al. (77) reported that an area per grid element of <5 Å2 is sufficient for convergence of this type of grid-based area analysis. A grid map is generated for each leaflet of the bilayer, resulting in two grid maps per analyzed snapshot. The partial area per lipid phosphate headgroup is computed for lipid type l from the grids via the following equation,
| (3) |
where Ng,l is the total number of grid elements occupied by lipid type l (including both leaflets), axy = Axy/1002 is the area per grid element, and Np,l is the number of phosphate headgroups that lipid type l has in the bilayer. Note that as described, this analysis yields the composite partial areas over both bilayer leaflets and assumes molecules of each lipid type are equally distributed across those leaflets.
Average area per lipid acyl chain
The average area per lipid acyl chain was computed in the same manner as the average area per lipid molecule (Average Area per Lipid Molecule) or the average area per phosphate (Average Area per Lipid Phosphate Headgroup), except in this case, the xy simulation box area (Axy) was divided by the number of lipid acyl chains per leaflet. Note that because each lipid has two acyl per chains per phosphate headgroup, the average area per lipid acyl chain is equivalent to the average area per phosphate multiplied by a factor of 1/2.
Partial area per lipid acyl chain
The partial areas per lipid acyl chain were computed in the same manner as the partial areas per phosphate (Partial Area per Lipid Phosphate Headgroup), except in this case, the positions of the terminal acyl chain carbon atoms projected into the xy-plane were used as the reference points for distance calculations and subsequent mapping of the lipids to the 2-d grids.
Relative fractions of lipid-lipid interactions
The relative fractions of lipid-lipid interactions were computed using a nearest-neighbor analysis as in de Vries et al. (78). For each lipid-lipid interaction of type X-Y, the fraction fX-Y was computed:
| (4) |
with NY the number of lipids of type Y among the Nnn nearest neighbors of lipid type X. We used Nnn = 5 for our calculations, and the positions of the lipids’ centers of mass projected into the xy-plane were used as the reference points for distance calculations and subsequent construction of nearest-neighbor lists.
Dimensionless packing shape parameter
The dimensionless packing shape parameter (79), P = v/(lcao), of each lipid type can be used to quantify lipid shape (80); in that expression, v is the volume occupied by the lipid, lc is the length of its hydrophobic region, and ao is the area occupied by its polar headgroup. For our analysis, we assumed each lipid within the bilayer was a truncated cone, cylinder, or inverted truncated cone shape (using shapes as defined in Fig. 1 of Dutt et al. (81)). In this case, another parameter for the tail-end area occupied by the lipid acyl chains, ah, can be introduced (80), and the equation for the packing shape parameter can be rewritten as
| (5) |
this parameter is equal to 1 for cylinder shaped lipid molecules, <1 for truncated cone-shaped lipid molecules, and >1 for inverted truncated-cone-shaped lipid molecules. For each lipid type, we used ao equal to the partial area per phosphate times the number of phosphate headgroups per lipid of that type (i.e., times 1 for POPC/DOPE and times 2 for CL) and ah equal to the partial area per lipid acyl chain of times the total number of acyl chains per lipid of that type (i.e., times 2 for POPC/DOPE and times 4 for CL). This yields an estimate of the average value of P, quantifying the effective shape of the lipids within the bilayer.
Hydrophobic thickness
The hydrophobic thickness, hC, of each bilayer was approximated by estimating the average distance between the C2 carbon atoms of the lipid acyl chains (82, 83), computed using a 2-d grid mapping tessellation procedure (see thickness calculations from Allen et al. (76) or Gapsys et al. (77)). This procedure was the same as that used in estimating the partial area per phosphate (Average Area per Lipid Phosphate Headgroup), except the positions of the C2 carbon atoms of the lipid acyl chains projected into the xy-plane were used as the reference points for distance calculations and subsequent mapping of lipids to the grid elements, and in addition to mapping lipids to grid elements, the z-positions of the C2 atoms were also mapped to the grid elements. The hydrophobic distance for a snapshot was then computed as the average z-distance between corresponding grid elements of the opposing bilayer leaflets.
P-P thickness
The phosphorus-to-phosphorus (P-P) thickness, hPP, of each bilayer was computed in the same manner as the hydrophobic thickness (Hydrophobic Thickness), except that the positions of the phosphorus atoms of the lipid headgroup phosphates projected into the xy-plane were used as the reference points for distance calculations and subsequent mapping of lipids and z-positions to the grid elements.
Lipid lengths
The length of each individual lipid was estimated by computing the distance between the position of the lipid’s headgroup phosphorus atom and the center of mass of its terminal acyl chain carbons. The average lipid length of lipid type l, Ll, was then estimated by taking the ensemble average of the lengths over lipids of type l.
Bilayer surface fluctuations
We examined the bilayer surface fluctuations using the rectangular grid-based tessellation procedure with grids generated in the same manner as described in P-P Thickness for the P-P thickness, but additionally, the area per grid element was fixed to 1 Å2, and the z-positions mapped to the grid elements were filtered through a Gaussian filter with σ = 5.0 Å; the filter reduced noise and smoothed the surface representation generated by the grid assignments. For simplicity, we only analyzed the upper leaflet of the bilayer. At each snapshot, a new grid was generated, and the surface fluctuations or roughness was quantified by computing the SD of the z-values for grid points that compose the surface, σz. After analyzing all the snapshots from a trajectory, the time average, maximal, and minimal values of the σz-values were computed.
Correlation between local bilayer surface deflections and lipid molecule localization
The correlation between local bilayer surface deflections and lipid molecule localization was estimated using the procedure described in Koldsø et al. (84) (under the section “Correlation between bilayer surface curvature and the clustering of lipid molecules”). At each snapshot of the analysis, the lateral area (xy box dimensions) of the bilayer was divided into blocks; each leaflet is considered individually. Then, the cross correlation between the deflection of lipids along the bilayer normal (in our case, the z-dimension) relative to the mean position within the blocks and the local lipid composition within that block are estimated, yielding a normalized correlation for each lipid species:
| (6) |
where Ln is the number of lipids of a given species in a grid box n, is the average of Ln over all grid boxes, is the average z coordinate zn of the lipid reference atom(s) (excluding the current lipid species being calculated) within grid block n, and is the average of across all grid boxes. 3 × 3 grids (∼48 × 48 Å2 grid elements) were used for the computation. The correlations for each leaflet at each snapshot of the analysis were time-averaged to yield estimates of the correlation between bilayer surface curvature and the clustering of lipid molecules at each leaflet. The correlation for the upper and lower leaflet values of each lipid type were then averaged according to
| (7) |
to yield a composite estimate combining the upper and lower leaflet values.
Diffusion coefficients
The diffusion coefficients were estimated by applying the mean-square displacement relation of the Einstein model of Brownian motion,
| (8) |
where the left-hand side is the mean-squared displacement as a function of time t and on the right-hand side, d is the dimensionality and D the diffusion coefficient. The mean-squared displacement of each lipid type (averaged over all lipids of that type) was computed for the lateral motion of the center of mass of lipids in each leaflet of the bilayer; the lipid coordinates were adjusted at each step to remove the center-of-mass motion of the bilayer before estimating the mean-squared displacements. The mean-squared displacement curves were then fitted with a linear equation, the slope of which was used to estimate the diffusion coefficient according to Eq. 8; note that d = 2 for 2-d lateral motion.
The mean-squared displacement curves were estimated by segmenting the analyzed portion of the production trajectories into nonoverlapping 50 ns portions. For each portion, a new mean-squared displacement curve was estimated. Then, the resulting set of mean-squared displacement curves were averaged to generate a composite estimate of the curve with low noise (Fig. S3, solid curves). The composite mean-squared displacement curves were then fitted in the time range 10–50 ns to a linear equation via the least-squares optimization method (Fig. S3, dashed lines). The slope of the curve was used to estimate the diffusion coefficient in accordance with Eq. 8. The standard error from the least-squares fitting procedure was used to estimate error in the diffusion coefficient, which was then multiplied by 1.96 to make an estimate of the 95% confidence intervals.
Results
CL increases the average area per lipid molecule in ternary mixtures with PC and PE lipids
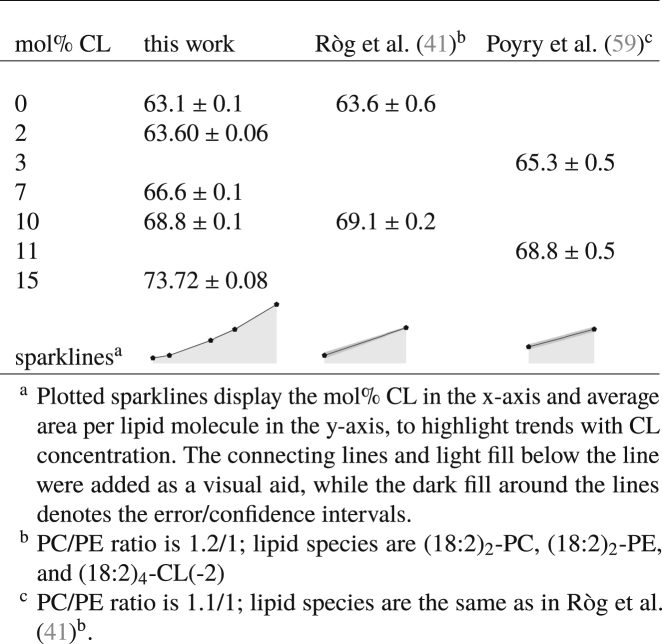
Many of the membrane structural parameters are directly affected by the lateral packing density of the membrane lipid molecules. To characterize the lateral packing density of the lipid molecules in our model mitochondrial membranes, we estimated the average area per lipid molecule as described in Average Area per Lipid Molecule. The values obtained from our simulations are reported in Table 1, along with other previously reported simulation estimates for the average area per lipid molecule in similar planar ternary lipid bilayers from the literature. The data show that increasing the mole percentage of CL increases the average area per lipid molecule. Our simulation data further indicate that the increase is nonlinear relative to the increase in CL content within the simulated concentration range. The increase of area per lipid molecule with CL content is consistent with trends reported for monolayer experiments (33, 34, 35, 36, 85) (at given surface pressure) and bilayer simulations (40, 41, 42) of CL in binary mixtures with PC or PE lipids. Furthermore, because the total number of lipids was fixed in our simulations, the increase in the average area per lipid indicates that the average lateral area of the bilayer is expanded by the replacement of POPC and DOPE lipid molecules with CL lipid molecules. This observation intuitively aligns with the fact that CL molecules are larger than their two-acyl-chain counterparts and display average area per lipid molecule values that exceed 100 Å2 within pure CL bilayers in the liquid phase (86, 87).
Table 1.
Average Area per Lipid Molecule at Various Concentrations of CL in Mixtures of PC and PE Lipids
Ternary mixtures of CL and PC and PE lipids have slightly higher lateral headgroup and acyl chain packing densities
The average area per lipid calculations presented above did not take into account the unique diglycerophospholipid structure of CL, which is comparable to that of two glycerophospholipids. Therefore, we further characterized the lateral lipid packing density by calculating both the average area per lipid phosphate group and the average area per lipid acyl chain, as well as their corresponding partial molar quantities.
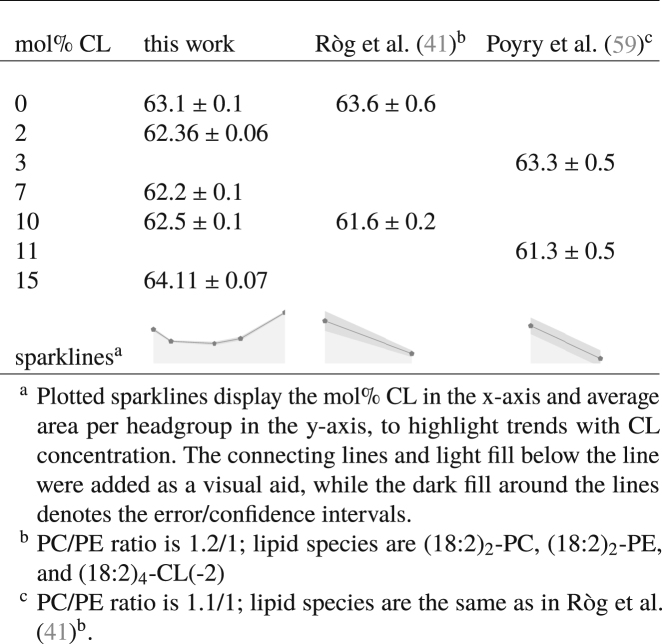
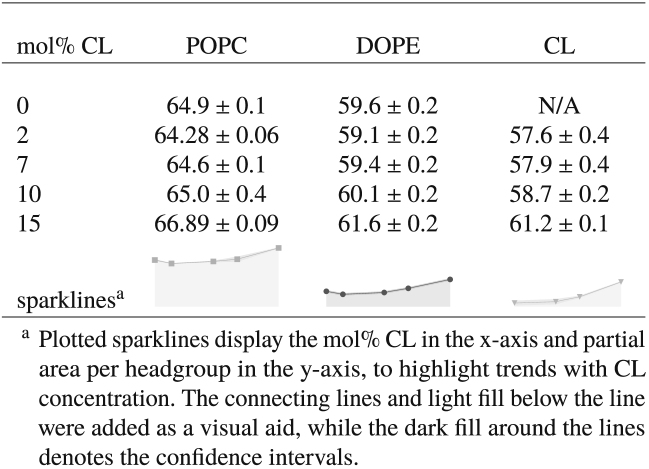
The average area per lipid phosphate headgroup (abbreviated hereafter as “average area per phosphate”) was computed in the manner described in Average Area per Lipid Phosphate Headgroup. The values obtained from our simulations are reported in Table 2, together with corresponding literature results for the average area per phosphate. It can be seen from our simulation data that the addition of up to 10 mol% CL into a mixture of PC and PE lipids results in a relative decrease in the average area per phosphate as compared to the CL-free bilayer, with essentially no difference between the results for the 2, 7, and 10 mol% systems. The data from Róg et al. (41) indicate that the addition of 10 mol% CL decreases the average area per phosphate as compared to the CL-free bilayer, whereas the data from Pöyry et al. (59) indicate that increasing the CL content from 3 to 11 mol% also results in a relative decrease in the average area per phosphate. Altogether, these results indicate that at low concentrations, CL tends to decrease the average area per phosphate in ternary mixtures with PC and PE lipids; the relative differences in the levels of area per phosphate as shown by our simulation data and those of Róg et al. (41) and Pöyry et al. (59) may be attributed, at least in part, to differences in length and saturation of the PC and PE lipid acyl chains between our studies, as well as differences in the relative proportion of PC-PE lipids. In contrast to the relative decreases in area per phosphate at lower concentrations, our simulation data indicated a relative increase in area per phosphate at 15 mol% CL as compared to both the other CL-containing bilayers and the CL-free bilayer that we modeled. The average area per phosphate of our model mitochondrial membranes therefore exhibited a nonmonotonic dependence on CL content.
Table 2.
Average Area per Lipid Phosphate Headgroup; Recall that Each CL Molecule Has Two Phosphate Headgroups
To better understand the effects that CL has on the headgroup packing of the different lipid types, we estimated the partial area per phosphate of each lipid type using the approach described in Partial Area per Lipid Phosphate Headgroup. The results of this analysis are reported in Table 3. Our simulation data indicated that the partial area per phosphate of POPC and DOPE lipid molecules only underwent a small decrease when 2 mol% CL was added to the bilayer. Further increasing the mol% of CL resulted in small increases in their partial areas per phosphate. Interestingly, the partial areas per phosphate of POPC and DOPE lipids at 10 mol% either match or exceed their values in the CL-free bilayer, suggesting that the relative decrease in the average area per phosphate maintained at 10 mol% CL (see Table 2) is primarily driven by the lower partial area per phosphate of the CL molecules and that the headgroup area of the PC and PE lipids are no longer compressed by the inclusion of CL. The partial area per phosphate of CL molecules increased monotonically but nonlinearly as their concentration increased. In all cases, the 15 mol% CL system exhibited the largest differences from the other systems. Finally, as shown by our data, the area occupied by the headgroups of each lipid type followed the general trend POPC > DOPE > CL.
Table 3.
Partial Areas per Phosphate for Each Lipid Type
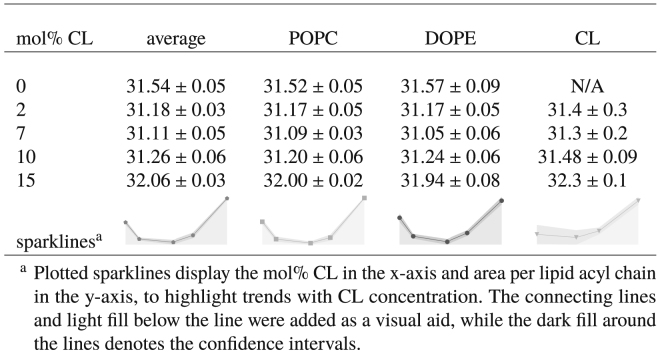
In addition to examining the packing densities of the lipid headgroups, we also computed the average and partial areas per lipid acyl chain. The average area per lipid acyl chain was estimated in the manner described in Average Area per Lipid Acyl Chain, and the partial areas per lipid acyl chain of each lipid type were computed using the method outlined in Partial Area per Lipid Acyl Chain. The results of these analyses are reported in Table 4. As can be seen from our data, the trends in acyl chain areas are similar to those seen for the headgroups. The average and partial areas for POPC and DOPE were nonmonotonic with CL concentration, showing a slight compression of the areas for 2–10 mol% CL relative to the CL-free bilayer along with a relative expansion at 15 mol% CL. Because the average area per lipid acyl chain is related to the average area per phosphate by a constant multiplicative factor, it has the same trends as that quantity. However, the partial areas per lipid acyl chain for POPC and DOPE were not perfectly correlated with their corresponding partial areas per phosphate, remaining compressed at 10 mol% CL relative to the CL-free system; their trends are much more closely aligned with the average area per lipid acyl chain. Additionally, the partial areas per lipid acyl chain are much more consistent across the different lipid types than the corresponding headgroup areas. With the headgroup areas, there was a clear trend in relative sizes. However, with the acyl chain areas, the values for POPC and DOPE are indistinguishable, whereas CL has only a slight tendency to larger areas. Presumably, the higher density of the lipid acyl chains within the bilayer forces a more uniform packing of the lipid tails.
Table 4.
Average and Partial Areas per Lipid Acyl Chain
CL molecules repel one another, interacting preferentially with POPC molecules
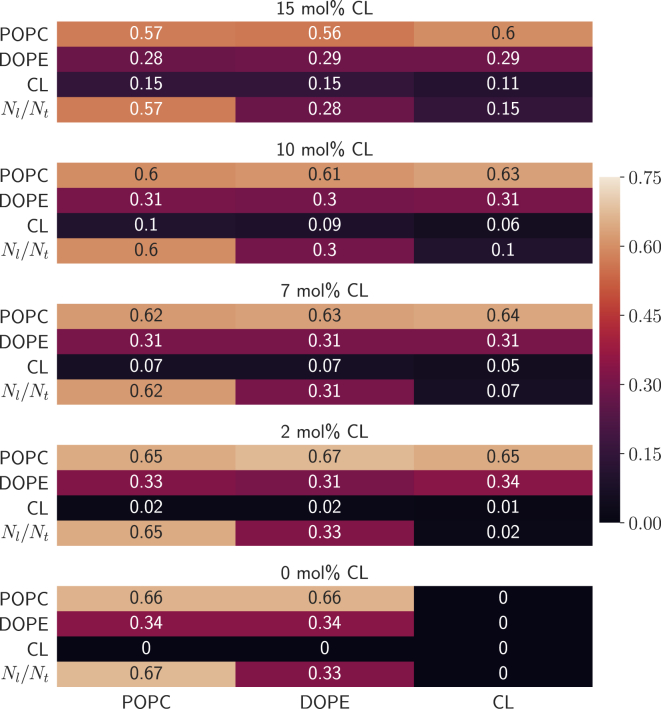
Although CL lipid molecules are miscible in binary mixtures with PC lipid molecules (33, 34, 88), formation of laterally segregated domains has been reported for binary mixtures of CL and PE lipids (4, 34, 88) and in a ternary mixture of PC, PE, and CL (89). Additionally, CL-enriched subdomains within the OMM have been suggested to serve as an activating platform for mitochondrial apoptosis (9). To explore lipid aggregation, we characterized the lateral organization and mixing behavior of our model mitochondrial membranes. To do this, we characterized the lipid-lipid interactions and their lateral organization within the mitochondrial bilayers using a nearest-neighbor analysis and estimated the relative fractions of lipids interacting with their nearest neighbors as described in Relative Fractions of Lipid-Lipid Interactions. The results of this analysis are displayed in Fig. 3.
Figure 3.
Lipid-lipid fractional interactions computed using the nearest-neighbor analysis described in Relative Fractions of Lipid-Lipid Interactions. Lipid-lipid interactions are read first up along the column and then across the row as interaction lipidcolumn-lipidrow. The additional rows labeled Nl/Nt provide the lipid mole fractions for reference. Errors are less than 0.02. To see this figure in color, go online.
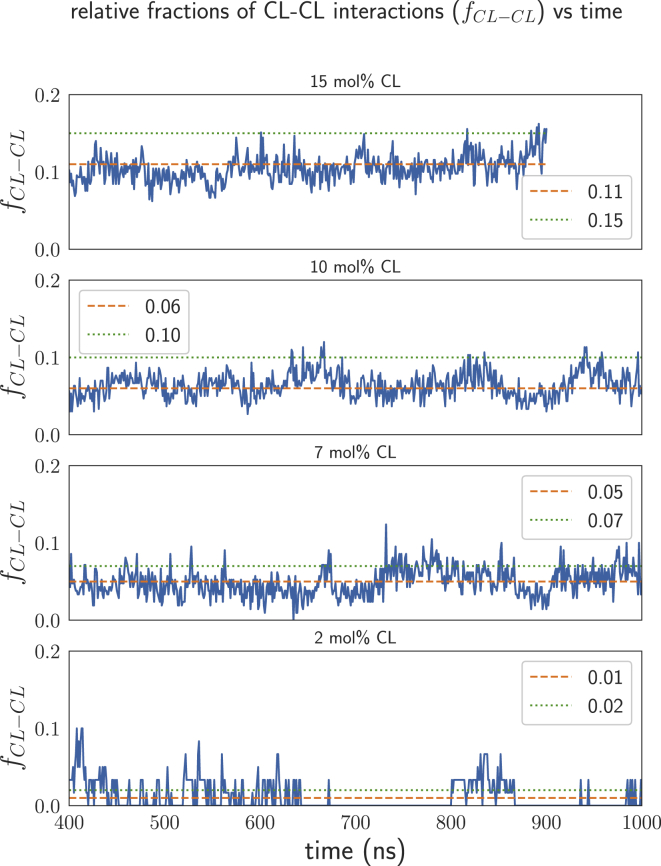
In general, the CL-CL interaction fractions are consistently lower than the CL’s mole fraction in each system, as can be seen from the data in Fig. 3; the effect is most notable in the 10 and 15 mol% CL systems. This suggests that the CL-CL interactions are less favorable, presumably because of repulsion between the negatively charged headgroups. Additionally, CL-CL radial distribution functions in the 7, 10, and 15 mol% CL systems (Fig. S4) also suggest that CL molecules have little interaction within a distance of 40 Å. However, it is possible that these results could be an artifact of CL lipid molecules’ low proportion and slow diffusion within the bilayers. Therefore, we also analyzed the time course of the CL-CL interaction fractions for each CL-containing system. The results of this analysis are displayed in Fig. 4. As can be seen in Fig. 4, although the fluctuations present in the 2 and 7 mol% CL systems obscure the small difference between the average value of fCL-CL and CL’s mole fraction, the time-course data for the interactions in the 10 and 15 mol% CL clearly show that despite oscillations, the CL-CL fractional interactions tend to be lower than the mole fraction of CL in those systems. These results are consistent with those displayed in Fig. 3, and they further confirm that at least in the timescale of these simulations, the CL-CL interactions appear to be repulsive.
Figure 4.
CL-CL fractional interactions (f<SUB>CL-CL</SUB>) from nearest-neighbor analysis plotted versus time. To see this figure in color, go online.
Additionally, the CL-POPC interaction fractions exceeded POPC’s mole fraction in the 7, 10, and 15 mol% CL systems, suggesting that the CL molecules interact preferentially with POPC molecules. The radial distribution functions (Fig. S4) also show that CL-POPC interactions tend to have slightly higher peak densities than the CL-DOPE interactions, which also suggests that CL lipids interact preferentially with POPC lipids. Indeed, CL molecules have been reported to have a stronger condensing effect in binary mixtures with PC lipids than in binary mixtures with PE lipids (41, 42), suggesting that CL molecules tend to have stronger attractive interactions with PC lipids than with PE lipids. Róg et al. (41) also reported that PC molecules preferentially formed charge pairs with CL molecules within a bilayer composed of a ternary mixture of PC, PE, and CL lipid molecules.
CL-dependent changes in lipid packing do not induce significant changes in bilayer thickness
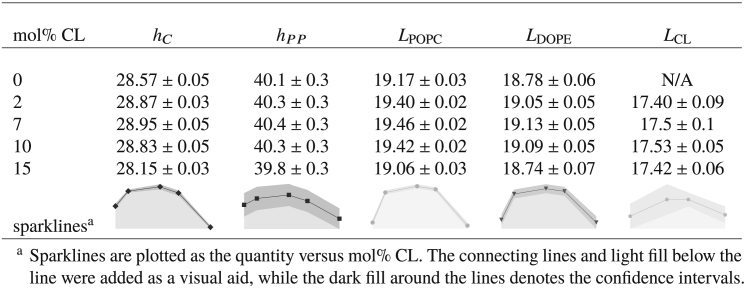
Lipid monolayers and bilayers exhibit many of the properties of an elastic sheet (90, 91). Under this model, alterations in the bilayer area result in commensurate alterations in the bilayer thickness. Because the changes in CL content resulted in minor changes to the lipid packing areas, we estimated some of the bilayer thickness properties to see if they also changed with CL content. The results of these analyses are reported in Table 5. First, we estimated the hydrophobic thickness, hC, of each bilayer using the method described in Hydrophobic Thickness. The hydrophobic thickness quantifies the thickness of a bilayer’s hydrophobic core (hydrocarbon region) and is particularly important for protein-membrane interactions, driving integral protein-membrane interaction energetics through hydrophobic mismatch (92, 93). As the data show, the variation of hC with CL content was minimal (<1 Å), but the changes followed the trends (negatively correlated) in the areas per phosphate and acyl chain (see Ternary Mixtures of CL and PC and PE Lipids Have Slightly Higher Lateral Headgroup and Acyl Chain Packing Densities). The hC values, 28–29 Å, agree with the average length (28.6 ± 1.4 Å) of the hydrophobic transmembrane domains of IMM proteins reported by Pogozheva et al. (94).
Table 5.
Hydrophobic Thickness, h<SUB>C</SUB>, P-P Thickness, h<SUB>PP</SUB>, and Lipid Lengths, Ll
In general, membrane thickness plays an important role in membrane-protein membrane interactions (95, 96), affecting the structure (93, 94, 97, 98), dynamics (99, 100), and distribution (101) of integral and associated proteins, as well as directly affecting membrane permeability (102). To gauge the overall thickness of the bilayer membranes, we computed the average phosphorus-to-phosphorus (P-P) thickness, hPP, of the bilayers using the approach described in P-P Thickness. As the data in Table 5 show, the hPP values appear to follow a similar trend with CL content as the hC values, but the small variations in hPP are obscured by statistical error. Regardless, values of hPP ≈ 40 Å compare favorably with the average P-P thickness (42 Å) reported for the bilayer with a ternary mixture of PC, PE, and CL simulated by Róg et al. (41), as well as the average distance between phosphate groups (41 ± 1 Å) reported by Dahlberg and Maliniak (40) for coarse-grained simulations of a binary mixture of teraoleoyl CL (9.2%) in POPC.
To better understand the molecular contributions of the different lipid types to the bilayer thickness, we estimated the average length of lipids, Ll, using the approach described in Lipid Lengths. As can be seen from the data in Table 5, similar to the membrane thickness metrics, the variation of the Ll values with CL content were minimal for POPC and DOPE (<0.5 Å) but followed the same overall trends; the changes in Ll with CL content correlated reasonably well to trends those observed for the partial areas per lipid acyl chain (see Ternary Mixtures of CL and PC and PE Lipids Have Slightly Higher Lateral Headgroup and Acyl Chain Packing Densities). Within the simulated concentration range, the average length of CL molecules was virtually unaffected by their concentration. Additionally, the differences between lipid types corresponded well to acyl chain unsaturation, following the trend LPOPC > LDOPE > LCL.
CL concentration in the membrane has a significant impact on lateral lipid diffusion
Fluidity (i.e., viscosity) is another physical property of membranes that is important for lipid distribution and protein-membrane interactions (101, 103, 104). Membrane fluidity plays a key role in controlling the diffusion (100, 105) and conformational changes of membrane-bound proteins (93, 98), making the property particularly important in signal transduction cascades that depend on membrane-bound diffusion-limited protein-protein interactions and protein conformational changes, such as in mitochondrial apoptosis regulation.
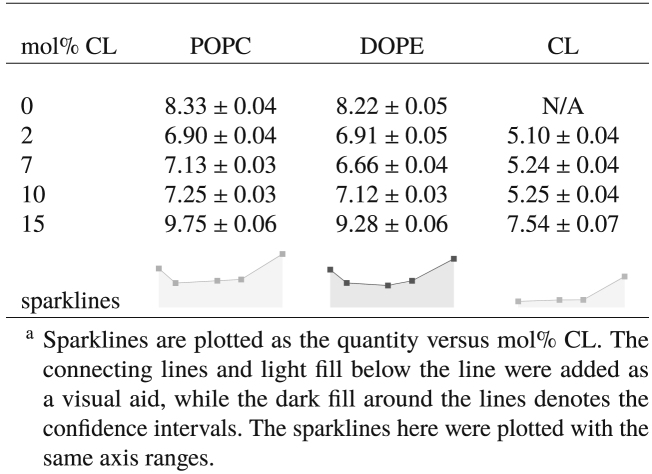
Membrane fluidity (or viscosity) is intrinsically linked to the lateral diffusion coefficients of its lipid constituents. Therefore, we estimated the lateral diffusion coefficients, Dl, of each lipid species using the method described in Diffusion Coefficients. Our simulation results are reported in Table 6.
Table 6.
Lateral Lipid Diffusion Coefficients, Dl
In general, the diffusion coefficients for POPC and DOPE match reasonably well with those expected from phospholipids in the liquid state (∼3–11 × 10−8 cm2/s) (106). The values for CL are comparable to previously published diffusion coefficients for tetramyristol CL (∼5 × 10−8 cm2/s) (107) and tetraoleoyl CL (∼4 × 10−8 cm2/s) (108) in liquid-phase unary CL bilayers. The trends in Dl values with CL content are correlated with changes in the partial areas per lipid acyl chain, consistent with the correlation between the lateral lipid packing density and lipid diffusion reported by Javanainen et al. (74). The resultant decrease in POPC and DOPE diffusion at at 2, 7, and 10% CL suggests a relative decrease in membrane fluidity at those concentrations, which is reversed at 15% CL. Interestingly, even though the partial areas per lipid acyl chain were only condensed by ∼3% at 2, 7, and 10% CL, Dl values for POPC and DOPE lipids were decreased by ∼20% at those concentrations. Thus, even though the changes to the membrane structural properties are subtle, corresponding changes in the lipid lateral diffusion are significant.
The effective cylindrical packing shape of lipid molecules has a small bias toward truncated conical shapes
Under the conditions in which we simulated CL (−2 charge), lipids would be expected to be stable in the lamellar phase, but under certain pH and salt conditions, lamellar CL membranes transition to a nonlamellar, inverted hexagonal phase (3, 4). The transition is driven by an effective decrease in the head/tail size ratio as the CL headgroups are protonated or sufficiently screened. Similarly, PE lipids are able to form inverted hexagonal phases (109), and inverted hexagonal phases have also been observed for mixtures of CL and PE lipids (4, 34, 88). The ability to form inverted hexagonal phases signifies an inherent bias toward an inverted conical packing shape. Therefore, we quantified the effective shape of lipid molecules within our model mitochondrial bilayers by estimating the average values of the dimensionless packing shape parameter (79) based on their lateral packing areas using the approach described in Dimensionless Packing Shape Parameter; this parameter is equal to 1 for cylinder shaped lipid molecules, <1 for truncated cone-shaped lipid molecules, and >1 for inverted truncated cone-shaped lipid molecules. The results of this analysis are reported in Table 7.
Table 7.
Effective Critical Packing Parameters of Each Lipid Type in the Model Mitochondrial Membranes
| Mol% CL | PPOPC | PDOPE | PCL |
|---|---|---|---|
| 0 | 0.99 | 1.03 | N/A |
| 2 | 0.98 | 1.03 | 1.03 |
| 7 | 0.98 | 1.03 | 1.03 |
| 10 | 0.98 | 1.02 | 1.03 |
| 15 | 0.98 | 1.02 | 1.02 |
Errors are 0.01 or less.
As can be seen from the data in Table 7, all the lipids have packing shape parameters ∼1, which indicates that the lipids are packed with effectively cylindrical shapes. Note that values near 1 were to be expected because the lipid systems we modeled are planar bilayers (lamellar phase), and lamellar bilayer lipids have cylindrical shapes with packing parameters ∼1 (79, 80). However, CL and DOPE have a small but consistent bias to values of 1, in accordance with a slight preference to take on truncated cone shapes. In contrast, POPC has a slight bias to values <1, suggesting a slight preference to inverted truncated cone shapes. And, because averaging the packing parameters across lipid types results in an average value of 1.00, the slight differences in packing shape between POPC and CL/DOPE appear to be complementary.
CL and DOPE lipid molecules correlate to negative deviations in the bilayer surface curvature
Both CL and DOPE lipids have a propensity to form inverted hexagonal phases and are thus expected to have a preference toward inverted conical shapes and negative curvatures. However, as our estimates of the effective packing shape parameter (The Effective Cylindrical Packing Shape of Lipid Molecules Has a Small Bias Toward Truncated Conical Shapes) indicated, all the lipids were constrained to approximately cylindrical shapes within the bilayer. Restriction of the lipids to less favorable packing shapes can result in curvature stress within the membrane monolayers (110) and a tendency toward spontaneous local curvature fluctuations (42). In the case of the interactions that regulate mitochondrial apoptosis, a curvature-dependent mechanism has been implicated in tBid-interactions (111, 112), whereas permeabilization via Bax-type molecules was reported to be sensitive to intrinsic monolayer curvature (113). Therefore, we evaluated the relationship between lipid localization and local curvature fluctuations within the bilayer surface. To quantify this relationship, we computed the cross correlation between local bilayer surface deflections and lipid molecule localization, , as described in Correlation between Local Bilayer Surface Deflections and Lipid Molecule Localization. The results of this analysis are reported in Table 8.
Table 8.
Cross Correlations between Local Bilayer Surface Deflections and Lipid Molecule Localization,
| Mol% CL | POPC | DOPE | CL |
|---|---|---|---|
| 0 | 0.0 ± 0.1 | −0.3 ± 0.1 | N/A |
| 2 | 0.04 ± 0.06 | −0.18 ± 0.06 | −0.05 ± 0.09 |
| 7 | 0.02 ± 0.08 | −0.2 ± 0.1 | −0.2 ± 0.1 |
| 10 | 0.04 ± 0.09 | −0.11± 0.07 | −0.09 ± 0.08 |
| 15 | 0.1 ± 0.1 | −0.3 ± 0.1 | −0.1 ± 0.1 |
As the data show, DOPE and CL localization tended toward negative correlation coefficients, indicating a bias toward negative curvature deflections in the bilayer surface. Consistent with this result, Dahlberg and Maliniak (42) reported that CL induced negative spontaneous curvature in coarse-grained lipid bilayer simulations. Additionally, Dahlberg and Maliniak (42) also reported that at low concentrations of a fully ionized CL, DOPE/CL bilayers had more negative spontaneous curvature than another system containing partially ionized CL in POPC.
Our results show that the correlation coefficient for POPC lipids was zero in the 2–10 mol% systems. This is not too surprising because POPC is the highest mole fraction component in each of the bilayers and therefore has the greatest weight in determining their average surface properties. Interestingly, POPC had a small positive correlation coefficient in the 15 mol% system, consistent with a its mild bias toward a truncated cone packing shape (The Effective Cylindrical Packing Shape of Lipid Molecules Has a Small Bias Toward Truncated Conical Shapes).
The magnitude of bilayer surface fluctuations is not significantly affected by CL content
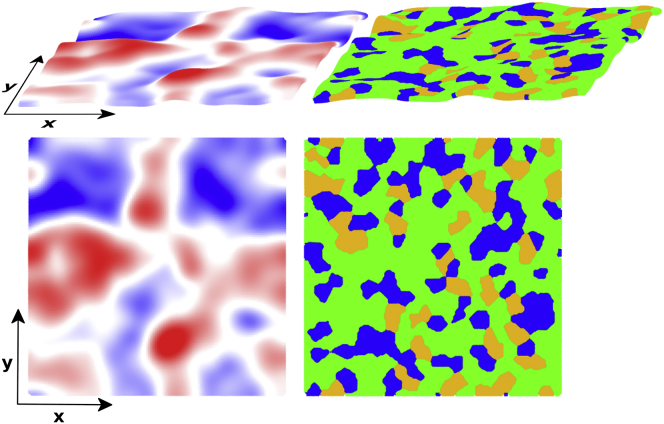
For a planar bilayer under zero tension, the surface is expected to be relatively flat. However, as can be seen in Fig. 5, there were small deformations in the surface of the bilayers, contributing to roughness and small local changes in surface curvature. To determine whether CL concentration affected the overall surface roughness and the magnitude of surface fluctuations, we computed the SD of the z-positions associated with the bilayer surface, including its time average, minimal, and maximal values from each simulation using the analysis methods described in Bilayer Surface Fluctuations. Our simulation results from this analysis are reported in Table 9. As the data indicate, increasing the mol% of CL did not have a significant effect on the overall surface roughness or the magnitude of surface fluctuations. We did not examine the low-frequency bilayer undulations; however, previous coarse-grained simulations (42) of binary PC/CL and PE/CL bilayers reported that increased CL content, particularly CL with a single −1 charge, resulted in greater deformability and larger undulations. Additionally, the results from another set of coarse-grained simulations (114) suggested that CL increased a bilayer’s ability to deform when under strain.
Figure 5.
Sample snapshot taken from the 10 mol% CL system of the surface of the upper bilayer leaflet, demonstrating surface roughness and curvature fluctuations. Left: heatmapped according to the Δz value at each point of the surface; the heatmap is color-coded blue → red: −4 Å → 4 Å. Right: color-coded at point according to the lipid type; lipid colors are POPC = green, DOPE = blue, and CL = gold. The images were generated using the Tachyon renderer (127) in VMD (128). To see this figure in color, go online.
Table 9.
SD of the Surface z-Coordinates from the Upper Leaflet of the Bilayers
| Mol% CL | Minimum | Time Average | Maximum |
|---|---|---|---|
| 0 | 1.3 | 2.2 | 3.6 |
| 2 | 1.2 | 2.2 | 4.2 |
| 7 | 1.3 | 2.2 | 3.6 |
| 10 | 1.2 | 2.2 | 3.6 |
| 15 | 1.5 | 2.4 | 3.9 |
Units are in Å.
Discussion
In this article, we have constructed five atomistic models of ternary lipid bilayer systems mimicking mitochondrial membranes. The membrane bilayers were square patches with sizes of ∼(14–15 nm)2 and were composed of POPC, DOPE, and varying proportions of CL molecules consistent with those in the outer (∼1–5 mol%, ∼15 mol% at contact sites) and inner (∼5–10 mol%) mitochondrial membranes. We characterized the properties of the membranes by microsecond MD simulations. To understand how the CL molecules influenced the properties of these model mitochondrial membranes, we estimated a variety of structural and dynamic bilayer properties from those simulations.
From our simulations, we found that CL content up to 15 mol% had only small effects on model membrane structural properties (see CL Increases the Average Area per Lipid Molecule in Ternary Mixtures with PC and PE Lipids–CL-Dependent Changes in Lipid Packing Do Not Induce Significant Changes in Bilayer Thickness). The addition of 2 mol% CL into the mixed POPC-DOPE matrix resulted in a slight bilayer condensation (Ternary Mixtures of CL and PC and PE Lipids Have Slightly Higher Lateral Headgroup and Acyl Chain Packing Densities) that was accompanied by a significant decrease in membrane fluidity (CL Concentration in the Membrane Has a Significant Impact on Lateral Lipid Diffusion). Further structural and fluidity changes with CL content were negligible between systems with 2–10 mol% CL. Interestingly, the condensing effect was reversed for the 15 mol% system, which was slightly expanded relative to all other systems and exhibited greater fluidity; we suspect the transition could be driven by electrostatic repulsion between the CL molecules. The results from the simulations by Róg et al. (41) indicated that the ternary mixture of PC, PE, and 10 mol% CL they modeled was only mildly condensed as compared to the corresponding CL-free binary PC-PE bilayer (see Table 2). Similarly, the results from the simulations by Pöyry et al. (59) indicated a small condensation in a ternary lipid bilayer when going from 3 to 11 mol% CL (also Table 2). Additionally, fluorescence experiments performed by Khalifat et al. (23) suggested that although the addition of 10% bovine cardiac CL to PC bilayers (at pH 7.4) leads to condensation, there was no discernible effect on mixed PC-PE bilayers. Taken together, these studies, along with our data, suggested that CL induces a mild condensing or ordering effect in mixed PC-PE lipid bilayers when the concentration of CL is <15 mol%. Furthermore, our data indicated that the system with 15 mol% CL had greater membrane fluidity. To the best of our knowledge, there are no other studies of similar ternary mixtures that examine the lipid packing properties at a CL concentration of ∼15 mol%. Therefore, we are unable to make a direct comparison for our results of the 15 mol% system.
Although the formation of laterally segregated domains has been reported for binary mixtures of CL and PE lipids (4, 34, 88) and for a ternary mixture of PC, PE, and 20 mol% CL (89), we did not find any evidence of domain formation within our model systems. Indeed, our analysis of the lateral lipid-lipid interactions and mixing suggested that lipids were for the most part well mixed (see CL Molecules Repel One Another, Interacting Preferentially with POPC Molecules). The one major exception were the CL-CL interactions, which we found to be consistently lower than dictated by the CL mole fraction, suggesting that CL-CL interactions were at least somewhat repulsive on average. We suspect this effect is driven by electrostatic repulsion between CL molecules’ negatively charged headgroups. Regardless, it should be noted that the initial configurations of our systems were approximately well mixed, which may have biased the systems against domain formation within the timescales that we simulated. And, although considerably larger than similar bilayer systems previously simulated, it is also possible the size of the bilayer patches used in our simulations was still too small to accurately gauge the formation of laterally segregated domains. Additionally, it should be noted that the ionic conditions play an important role in CL lamellar-to-nonlamellar phase transitions (3, 115, 116), and, in particular, divalent ions (e.g., Ca2+) are more effective at inducing this phase transition than monovalent ions (e.g., Na+). However, the experimental studies that reported lateral segregation all used NaCl as the salt subphase (4, 34, 88, 89), so divalent ions are not required for lateral segregation in CL-containing membranes. Overall, our result should be taken as suggesting an underlying repulsion between the negatively charged CL molecules but not definitive evidence against domain formation in these ternary PC, PE, and CL lipid systems.
Our analysis of the effective packing shape of the lipids revealed that the lipids took on near cylindrical shapes within the bilayers (see The Effective Cylindrical Packing Shape of Lipid Molecules Has a Small Bias Toward Truncated Conical Shapes). And, although analysis of the surface roughness indicated that CL content did not have a significant effect on the magnitude of surface roughness fluctuations (see The Magnitude of Bilayer Surface Fluctuations Is Not Significantly Affected by CL Content), analysis of the correlation between CL localization and surface deflection revealed that CL tended to cause local deflections in the bilayer surface with negative curvature (see CL And DOPE Lipid Molecules Correlate to Negative Deviations in the Bilayer Surface Curvature). However, it is unclear from these analyses whether negative curvature deflections in the bilayer surface associated with CL (or DOPE) are due primarily to the lipid’s shorter average length (Table 5) or coupling with undulatory modes. Additionally, it should be noted that the patch size of the bilayers likely affects the magnitude of curvature fluctuations and the correlation estimates. A recent simulation study performed by Boyd et al. (114) reported that CL became locally enriched in the negative curvature regions of buckled bilayers, suggesting that lateral reorganization helped stabilize buckling deformations. Together with our results, these data suggest that CL is constrained to a nonfavorable packing shape within bilayers that results in curvature frustration and a tendency to negative curvature fluctuations. As such, when the bilayer is perturbed by external forces into a highly curved state, as in the buckling simulations, CL responds by reorganizing into the negatively curved regions, alleviating its own inherent curvature strain and stabilizing the negatively curved regions of the bilayer.
Conclusions
We used atomistic MD simulations to investigate the CL-dependent properties in ternary lipid bilayer systems composed of PC, PE, and CL; the CL content encompassed the natural range found in mitochondrial membranes, including the inner and outer mitochondrial membrane and their contact sites. From these simulations, we estimated a variety of membrane properties known to influence the energy and kinetics of protein-membrane and protein-protein interactions at membranes (93, 117) and consequently affect membrane-protein properties such as conformation (98, 104, 118, 119, 120), activity (121, 122, 123, 124), and distribution (101, 103). Our analysis suggested that CL plays a minor role in altering membrane structural features in ternary lipid bilayers with a background matrix of PC and PE lipids but significantly alters lipid diffusion and thus bilayer fluidity. Furthermore, our data indicated that these CL-dependent effects are nonmonotonic, indicating a delicate balance between attractive and repulsive interactions within the bilayers. Finally, examination of the lipid packing shapes and curvature fluctuations suggested that CL correlates to negative curvature deflections of the bilayer surface, which likely induces negative curvature strain within the membrane monolayers.
This work contributes to a foundational understanding of the role of CL in altering the properties ternary lipid mixtures composed of PC, PE, and CL molecules. And, because these are the primary components of mitochondrial membranes, this work also serves to illuminate the concentration-dependent role of CL in the mitochondrial membranes. However, our models did not capture the full complexity of the lipid milieu of mitochondrial membranes, including the inclusion of various minor components (e.g., phosphatidylserine, phosphatidylinositol, and cholesterol) and the distributions of acyl chain length and saturation. Furthermore, in our model membranes, CL lipids were distributed equally among bilayer leaflets, not accounting for the possibility of asymmetric lipid distribution. Additional experiment and simulation of more complex lipid mixtures will be required to determine what role these differences have on bilayer structure and dynamics and ultimately, mitochondrial function.
Author Contributions
B.A.W., A.R., and C.F.L. designed the research. B.A.W. performed the research and analyzed the data. B.A.W., A.R., and C.F.L. wrote the manuscript.
Acknowledgments
This work was supported by a grant from the National Science Foundation (MCB1411482 to C.F.L.), the National Institutes of Health (U01CA215845 to C.F.L.), The Incyte-Vanderbilt Research Alliance (M. Savona PI, C.F.L. co-I), and the Oak Ridge Leadership Computing Facility at the Oak Ridge National Laboratory, through the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725. A.R. is now at Argonne National Laboratory.
Editor: Alemayehu Gorfe.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.06.023.
Supporting Material
References
- 1.Hoch F.L. Cardiolipins and biomembrane function. Biochim. Biophys. Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 2.Lecocq J., Ballou C.E. On the structure of cardiolipin. Biochemistry. 1964;3:976–980. doi: 10.1021/bi00895a023. [DOI] [PubMed] [Google Scholar]
- 3.Seddon J.M., Kaye R., Marsh D. Induction of the lamellar-inverted hexagonal phase transition in cardiolipin by protons and monovalent cations. Biochim. Biophys. Acta. 1983;734:347–352. [Google Scholar]
- 4.Domènech O., Morros A., Hernández-Borrell J. Thermal response of domains in cardiolipin content bilayers. Ultramicroscopy. 2007;107:943–947. doi: 10.1016/j.ultramic.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Lewis R.N., McElhaney R.N. The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim. Biophys. Acta. 2009;1788:2069–2079. doi: 10.1016/j.bbamem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Ren M., Phoon C.K., Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog. Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Kameoka S., Adachi Y., Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018;28:67–76. doi: 10.1016/j.tcb.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalvez F., Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 9.Sorice M., Manganelli V., Garofalo T. Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett. 2009;583:2447–2450. doi: 10.1016/j.febslet.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Han X., Yang J., Gross R.W. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: a shotgun lipidomics study. Biochemistry. 2007;46:6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y. Emerging roles of cardiolipin remodeling in mitochondrial dysfunction associated with diabetes, obesity, and cardiovascular diseases. J. Biomed. Res. 2010;24:6–15. doi: 10.1016/S1674-8301(10)60003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sparagna G.C., Chicco A.J., Moore R.L. Loss of cardiac tetralinoleoyl cardiolipin in human and experimental heart failure. J. Lipid Res. 2007;48:1559–1570. doi: 10.1194/jlr.M600551-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Schlame M., Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalvez F., D’Aurelio M., Petit P.X. Barth syndrome: cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochim. Biophys. Acta. 2013;1832:1194–1206. doi: 10.1016/j.bbadis.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Colbeau A., Nachbaur J., Vignais P.M. Enzymac characterization and lipid composition of rat liver subcellular membranes. Biochim. Biophys. Acta. 1971;249:462–492. doi: 10.1016/0005-2736(71)90123-4. [DOI] [PubMed] [Google Scholar]
- 16.Comte J., Maïsterrena B., Gautheron D.C. Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes. Biochim. Biophys. Acta. 1976;419:271–284. doi: 10.1016/0005-2736(76)90353-9. [DOI] [PubMed] [Google Scholar]
- 17.Chang M.C., Roots B.I. The lipid composition of mitochondrial outer and inner membranes from the brains of goldfish acclimated at 5 and 30°C. J. Therm. Biol. 1989;14:191–194. [Google Scholar]
- 18.Hovius R., Thijssen J., de Kruijff B. Phospholipid asymmetry of the outer membrane of rat liver mitochondria. Evidence for the presence of cardiolipin on the outside of the outer membrane. FEBS Lett. 1993;330:71–76. doi: 10.1016/0014-5793(93)80922-h. [DOI] [PubMed] [Google Scholar]
- 19.de Kroon A.I., Dolis D., de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim. Biophys. Acta. 1997;1325:108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- 20.Daum G., Vance J.E. Import of lipids into mitochondria. Prog. Lipid Res. 1997;36:103–130. doi: 10.1016/s0163-7827(97)00006-4. [DOI] [PubMed] [Google Scholar]
- 21.Acehan D., Xu Y., Schlame M. Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography. Lab. Invest. 2007;87:40–48. doi: 10.1038/labinvest.3700480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalifat N., Puff N., Angelova M.I. Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys. J. 2008;95:4924–4933. doi: 10.1529/biophysj.108.136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalifat N., Fournier J.B., Puff N. Lipid packing variations induced by pH in cardiolipin-containing bilayers: the driving force for the cristae-like shape instability. Biochim. Biophys. Acta. 2011;1808:2724–2733. doi: 10.1016/j.bbamem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 24.van der Laan M., Horvath S.E., Pfanner N. Mitochondrial contact site and cristae organizing system. Curr. Opin. Cell Biol. 2016;41:33–42. doi: 10.1016/j.ceb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Ikon N., Ryan R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017;1859:1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlame M., Ren M. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta. 2009;1788:2080–2083. doi: 10.1016/j.bbamem.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 28.Yamauchi T., Ohki K., Nozawa Y. Thermal adaptation of Tetrahymena membranes with special reference to mitochondria. Role of cardiolipin in fluidity of mitochondrial membranes. Biochim. Biophys. Acta. 1981;649:385–392. doi: 10.1016/0005-2736(81)90428-4. [DOI] [PubMed] [Google Scholar]
- 29.Gallet P.F., Petit J.M., Julien R. Asymmetrical distribution of cardiolipin in yeast inner mitochondrial membrane triggered by carbon catabolite repression. Biochem. J. 1997;324:627–634. doi: 10.1042/bj3240627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia Fernandez M., Troiano L., Cossarizza A. Early changes in intramitochondrial cardiolipin distribution during apoptosis. Cell Growth Differ. 2002;13:449–455. [PubMed] [Google Scholar]
- 31.Crimi M., Esposti M.D. Apoptosis-induced changes in mitochondrial lipids. Biochim. Biophys. Acta. 2011;1813:551–557. doi: 10.1016/j.bbamcr.2010.09.014. Published online October 1, 2010. [DOI] [PubMed] [Google Scholar]
- 32.Chu C.T., Ji J., Kagan V.E. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols-Smith S., Teh S.Y., Kuhl T.L. Thermodynamic and mechanical properties of model mitochondrial membranes. Biochim. Biophys. Acta. 2004;1663:82–88. doi: 10.1016/j.bbamem.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Domènech O., Sanz F., Hernández-Borrell J. Thermodynamic and structural study of the main phospholipid components comprising the mitochondrial inner membrane. Biochim. Biophys. Acta. 1758;1758:213–221. doi: 10.1016/j.bbamem.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Phan M.D., Shin K. Effects of cardiolipin on membrane morphology: a Langmuir monolayer study. Biophys. J. 2015;108:1977–1986. doi: 10.1016/j.bpj.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pennington E.R., Fix A., Shaikh S.R. Distinct membrane properties are differentially influenced by cardiolipin content and acyl chain composition in biomimetic membranes. Biochim. Biophys. Acta Biomembr. 2017;1859:257–267. doi: 10.1016/j.bbamem.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata A., Ikawa K., Terada H. Significant stabilization of the phosphatidylcholine bilayer structure by incorporation of small amounts of cardiolipin. Biochim. Biophys. Acta. 1994;1192:71–78. doi: 10.1016/0005-2736(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 38.Chen Q.P., Li Q.T. Effect of cardiolipin on proton permeability of phospholipid liposomes: the role of hydration at the lipid-water interface. Arch. Biochem. Biophys. 2001;389:201–206. doi: 10.1006/abbi.2001.2319. [DOI] [PubMed] [Google Scholar]
- 39.Unsay J.D., Cosentino K., García-Sáez A.J. Cardiolipin effects on membrane structure and dynamics. Langmuir. 2013;29:15878–15887. doi: 10.1021/la402669z. [DOI] [PubMed] [Google Scholar]
- 40.Dahlberg M., Maliniak A. Molecular dynamics simulations of cardiolipin bilayers. J. Phys. Chem. B. 2008;112:11655–11663. doi: 10.1021/jp803414g. [DOI] [PubMed] [Google Scholar]
- 41.Róg T., Martinez-Seara H., Vattulainen I. Role of cardiolipins in the inner mitochondrial membrane: insight gained through atom-scale simulations. J. Phys. Chem. B. 2009;113:3413–3422. doi: 10.1021/jp8077369. [DOI] [PubMed] [Google Scholar]
- 42.Dahlberg M., Maliniak A. Mechanical properties of coarse-grained bilayers formed by cardiolipin and zwitterionic lipids. J. Chem. Theory Comput. 2010;6:1638–1649. doi: 10.1021/ct900654e. [DOI] [PubMed] [Google Scholar]
- 43.Daum G. Lipids of mitochondria. Biochim. Biophys. Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 44.Schlame M., Shanske S., Blanck T.J. Microanalysis of cardiolipin in small biopsies including skeletal muscle from patients with mitochondrial disease. J. Lipid Res. 1999;40:1585–1592. [PubMed] [Google Scholar]
- 45.Schlame M., Towbin J.A., Blanck T.J. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann. Neurol. 2002;51:634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- 46.Schlame M., Kelley R.I., Blanck T.J. Phospholipid abnormalities in children with Barth syndrome. J. Am. Coll. Cardiol. 2003;42:1994–1999. doi: 10.1016/j.jacc.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Kates M., Syz J.Y., Haines T.H. pH-dissociation characteristics of cardiolipin and its 2′-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 48.Hielscher R., Wenz T., Hellwig P. Monitoring the redox and protonation dependent contributions of cardiolipin in electrochemically induced FTIR difference spectra of the cytochrome bc(1) complex from yeast. Biochim. Biophys. Acta. 2009;1787:617–625. doi: 10.1016/j.bbabio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Sidiq S., Verma I., Pal S.K. pH-driven ordering transitions in liquid crystal induced by conformational changes of cardiolipin. Langmuir. 2015;31:4741–4751. doi: 10.1021/acs.langmuir.5b00798. [DOI] [PubMed] [Google Scholar]
- 50.Olofsson G., Sparr E. Ionization constants pKa of cardiolipin. PLoS One. 2013;8:e73040. doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malyshka D., Pandiscia L.A., Schweitzer-Stenner R. Cardiolipin containing liposomes are fully ionized at physiological pH. An FT-IR study of phosphate group ionization. Vib. Spectrosc. 2014;75:86–92. [Google Scholar]
- 52.Sathappa M., Alder N.N. The ionization properties of cardiolipin and its variants in model bilayers. Biochim. Biophys. Acta. 2016;1858:1362–1372. doi: 10.1016/j.bbamem.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu E.L., Cheng X., Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jo S., Lim J.B., Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks B.R., Brooks C.L., III, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J., Cheng X., Im W. CHARMM-GUI Input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 59.Pöyry S., Róg T., Vattulainen I. Mitochondrial membranes with mono- and divalent salt: changes induced by salt ions on structure and dynamics. J. Phys. Chem. B. 2009;113:15513–15521. doi: 10.1021/jp905915m. [DOI] [PubMed] [Google Scholar]
- 60.Miettinen M.S., Gurtovenko A.A., Karttunen M. Ion dynamics in cationic lipid bilayer systems in saline solutions. J. Phys. Chem. B. 2009;113:9226–9234. doi: 10.1021/jp810233q. [DOI] [PubMed] [Google Scholar]
- 61.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 63.Berendsen H., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 64.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 65.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 66.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 67.Michaud-Agrawal N., Denning E.J., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gowers R.J., Linke M., Beckstein O. MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations. In: Benthall S., Rostrup S., editors. Proceedings of the 15th Python in Science Conference. 2016. [Google Scholar]
- 69.Oliphant T.E. Python for scientific Computing. Computing in Science & Engineering. 2006;9:10–20. [Google Scholar]
- 70.Van Der Walt S., Colbert S.C., Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011;13:22–30. [Google Scholar]
- 71.Jones E., Oliphant T., Peterson P. SciPy: open source scientific tools for Python. 2001. http://www.scipy.org/
- 72.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 73.Waskom M., Botvinnik O., Qalieh A. mwaskom/seaborn: v0.9.0 (July 2018) 2018. [DOI]
- 74.Javanainen M., Hammaren H., Vattulainen I. Anomalous and normal diffusion of proteins and lipids in crowded lipid membranes. Faraday Discuss. 2013;161:397–417. doi: 10.1039/c2fd20085f. discussion 419–459. [DOI] [PubMed] [Google Scholar]
- 75.Grossfield A., Zuckerman D.M. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allen W.J., Lemkul J.A., Bevan D.R. GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 2009;30:1952–1958. doi: 10.1002/jcc.21172. [DOI] [PubMed] [Google Scholar]
- 77.Gapsys V., de Groot B.L., Briones R. Computational analysis of local membrane properties. J. Comput. Aided Mol. Des. 2013;27:845–858. doi: 10.1007/s10822-013-9684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Vries A.H., Mark A.E., Marrink S.J. The binary mixing behavior of phospholipids in a bilayer: a molecular dynamics study. J. Phys. Chem. B. 2004;108:2454–2463. [Google Scholar]
- 79.Israelachvili J.N., Mitchell D.J., Ninham B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. 1976;72:1525–1568. [Google Scholar]
- 80.Cullis P.R., Hope M.J., Tilcock C.P. Lipid polymorphism and the roles of lipids in membranes. Chem. Phys. Lipids. 1986;40:127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- 81.Dutt S., Siril P.F., Remita S. Swollen liquid crystals (SLCs): a versatile template for the synthesis of nano structured materials. RSC Advances. 2017;7:5733–5750. [Google Scholar]
- 82.de Planque M.R., Killian J.A. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 83.Kim T., Lee K.I., Im W. Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers. Biophys. J. 2012;102:1551–1560. doi: 10.1016/j.bpj.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koldsø H., Shorthouse D., Sansom M.S. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput. Biol. 2014;10:e1003911. doi: 10.1371/journal.pcbi.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nichols-Smith S., Kuhl T. Electrostatic interactions between model mitochondrial membranes. Colloids Surf. B Biointerfaces. 2005;41:121–127. doi: 10.1016/j.colsurfb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 86.Aguayo D., González-Nilo F.D., Chipot C. Insight into the properties of cardiolipin containing bilayers from molecular dynamics simulations, using a hybrid all-atom/united-atom force field. J. Chem. Theory Comput. 2012;8:1765–1773. doi: 10.1021/ct200849k. [DOI] [PubMed] [Google Scholar]
- 87.Pan J., Cheng X., Katsaras J. Structural and mechanical properties of cardiolipin lipid bilayers determined using neutron spin echo, small angle neutron and X-ray scattering, and molecular dynamics simulations. Soft Matter. 2015;11:130–138. doi: 10.1039/c4sm02227k. [DOI] [PubMed] [Google Scholar]
- 88.Sennato S., Bordi F., Rufini S. Evidence of domain formation in cardiolipin-glycerophospholipid mixed monolayers. A thermodynamic and AFM study. J. Phys. Chem. B. 2005;109:15950–15957. doi: 10.1021/jp051893q. [DOI] [PubMed] [Google Scholar]
- 89.Domènech O., Redondo L., Hernández-Borrell J. Atomic force microscopy characterization of supported planar bilayers that mimic the mitochondrial inner membrane. J. Mol. Recognit. 2007;20:546–553. doi: 10.1002/jmr.849. [DOI] [PubMed] [Google Scholar]
- 90.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 91.Campelo F., Arnarez C., Kozlov M.M. Helfrich model of membrane bending: from Gibbs theory of liquid interfaces to membranes as thick anisotropic elastic layers. Adv. Colloid Interface Sci. 2014;208:25–33. doi: 10.1016/j.cis.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 92.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andersen O.S., Koeppe R.E., II Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 94.Pogozheva I.D., Tristram-Nagle S., Lomize A.L. Structural adaptations of proteins to different biological membranes. Biochim. Biophys. Acta. 2013;1828:2592–2608. doi: 10.1016/j.bbamem.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li L.B., Vorobyov I., Allen T.W. The role of membrane thickness in charged protein-lipid interactions. Biochim. Biophys. Acta. 2012;1818:135–145. doi: 10.1016/j.bbamem.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 96.Karabadzhak A.G., Weerakkody D., Engelman D.M. Bilayer thickness and curvature influence binding and insertion of a pHLIP peptide. Biophys. J. 2018;114:2107–2115. doi: 10.1016/j.bpj.2018.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kleinschmidt J.H., Tamm L.K. Secondary and tertiary structure formation of the β-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J. Mol. Biol. 2002;324:319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- 98.Burgess N.K., Dao T.P., Fleming K.G. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu Q., Kim M., Cafiso D.S. Membrane hydrocarbon thickness modulates the dynamics of a membrane transport protein. Biophys. J. 2008;95:2849–2858. doi: 10.1529/biophysj.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weiß K., Neef A., Enderlein J. Quantifying the diffusion of membrane proteins and peptides in black lipid membranes with 2-focus fluorescence correlation spectroscopy. Biophys. J. 2013;105:455–462. doi: 10.1016/j.bpj.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schäfer L.V., de Jong D.H., Marrink S.J. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. USA. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paula S., Volkov A.G., Deamer D.W. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 1996;70:339–348. doi: 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sánchez S.A., Tricerri M.A., Gratton E. Lipid packing determines protein-membrane interactions: challenges for apolipoprotein A-I and high density lipoproteins. Biochim. Biophys. Acta. 2010;1798:1399–1408. doi: 10.1016/j.bbamem.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barrera F.N., Fendos J., Engelman D.M. Membrane physical properties influence transmembrane helix formation. Proc. Natl. Acad. Sci. USA. 2012;109:14422–14427. doi: 10.1073/pnas.1212665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Venable R.M., Ingólfsson H.I., Pastor R.W. Lipid and peptide diffusion in bilayers: the saffman-delbrück model and periodic boundary conditions. J. Phys. Chem. B. 2017;121:3443–3457. doi: 10.1021/acs.jpcb.6b09111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Almeida P.F., Vaz W.L., Thompson T.E. Lipid diffusion, free area, and molecular dynamics simulations. Biophys. J. 2005;88:4434–4438. doi: 10.1529/biophysj.105.059766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lemmin T., Bovigny C., Dal Peraro M. Cardiolipin models for molecular simulations of bacterial and mitochondrial membranes. J. Chem. Theory Comput. 2013;9:670–678. doi: 10.1021/ct300590v. [DOI] [PubMed] [Google Scholar]
- 108.Boyd K.J., Alder N.N., May E.R. Molecular dynamics analysis of cardiolipin and monolysocardiolipin on bilayer properties. Biophys. J. 2018;114:2116–2127. doi: 10.1016/j.bpj.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Siegel D.P., Epand R.M. The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: implications for membrane fusion mechanisms. Biophys. J. 1997;73:3089–3111. doi: 10.1016/S0006-3495(97)78336-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Frolov V.A., Shnyrova A.V., Zimmerberg J. Lipid polymorphisms and membrane shape. Cold Spring Harb. Perspect. Biol. 2011;3:a004747. doi: 10.1101/cshperspect.a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Terrones O., Antonsson B., Basañez G. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J. Biol. Chem. 2004;279:30081–30091. doi: 10.1074/jbc.M313420200. [DOI] [PubMed] [Google Scholar]
- 112.Yethon J.A., Epand R.F., Andrews D.W. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J. Biol. Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- 113.Basañez G., Sharpe J.C., Zimmerberg J. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J. Biol. Chem. 2002;277:49360–49365. doi: 10.1074/jbc.M206069200. [DOI] [PubMed] [Google Scholar]
- 114.Boyd K.J., Alder N.N., May E.R. Buckling under pressure: curvature-based lipid segregation and stability modulation in cardiolipin-containing bilayers. Langmuir. 2017;33:6937–6946. doi: 10.1021/acs.langmuir.7b01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rand R., Sengupta S. Cardiolipin forms hexagonal structures with divalent cations. Biochim. Biophys. Acta. 1972;255:484–492. doi: 10.1016/0005-2736(72)90152-6. [DOI] [PubMed] [Google Scholar]
- 116.Cullis P.R., Verkleij A.J., Ververgaert P.H. Polymorphic phase behaviour of cardiolipin as detected by 31P NMR and freeze-fracture techniques. Effects of calcium, dibucaine and chlorpromazine. Biochim. Biophys. Acta. 1978;513:11–20. doi: 10.1016/0005-2736(78)90107-4. [DOI] [PubMed] [Google Scholar]
- 117.Cho W., Stahelin R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 118.White S.H., Ladokhin A.S., Hristova K. How membranes shape protein structure. J. Biol. Chem. 2001;276:32395–32398. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- 119.Hunte C., Richers S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 120.Gupta K., Donlan J.A.C., Robinson C.V. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smith A.D., Stubbs C.D. Modulation of membrane protein function by bilayer lipids. In: Stam H., van der Vusse G.J., editors. Lipid metabolism in the normoxic and ischaemic heart. Steinkopff; 1987. pp. 93–97. [Google Scholar]
- 122.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 123.Teague W.E., Jr., Soubias O., Gawrisch K. Elastic properties of polyunsaturated phosphatidylethanolamines influence rhodopsin function. Faraday Discuss. 2013;161:383–395. doi: 10.1039/c2fd20095c. discussion 419–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Malhotra K., Modak A., Alder N.N. Cardiolipin mediates membrane and channel interactions of the mitochondrial TIM23 protein import complex receptor Tim50. Sci. Adv. 2017;3:e1700532. doi: 10.1126/sciadv.1700532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fahy E., Sud M., Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O’Boyle N.M., Banck M., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stone J. University of Missouri-Rolla; 1998. An efficient library for parallel ray tracing and animation. Master’s thesis. [Google Scholar]
- 128.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. , 27–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.